Abstract
Structural studies of polytopic membrane proteins are often hampered by the vagaries of these proteins in membrane mimetic environments and by the difficulties in handling them with conventional techniques. Designing and creating water-soluble analogues with preserved native structures offer an attractive alternative. We report here solution NMR studies of WSK3, a water-soluble analogue of the potassium channel KcsA. The WSK3 NMR structure (PDB ID code 2K1E) resembles the KcsA crystal structures, validating the approach. By more stringent comparison criteria, however, the introduction of several charged residues aimed at improving water solubility seems to have led to the possible formations of a few salt bridges and hydrogen bonds not present in the native structure, resulting in slight differences in the structure of WSK3 relative to KcsA. NMR dynamics measurements show that WSK3 is highly flexible in the absence of a lipid environment. Reduced spectral density mapping and model-free analyses reveal dynamic characteristics consistent with an isotropically tumbling tetramer experiencing slow (nanosecond) motions with unusually low local ordering. An altered hydrogen-bond network near the selectivity filter and the pore helix, and the intrinsically dynamic nature of the selectivity filter, support the notion that this region is crucial for slow inactivation. Our results have implications not only for the design of water-soluble analogues of membrane proteins but also for our understanding of the basic determinants of intrinsic protein structure and dynamics.
Keywords: membrane protein, protein design, potassium channels, slow inactivation
Less than 1% of known protein structures belong to transmembrane (TM) proteins. The scarcity of structural data is due to the difficulties associated with handling membrane proteins for structure determination. With few exceptions, membrane proteins are often present at low levels in natural tissues, and the available cellular machinery for membrane insertion often limits the capacity of high-level expression systems. Outside their native environment, membrane proteins are unpredictable in behavior, with proper folding and stability depending on the choice of the membrane mimetic environment in addition to the common variables affecting soluble proteins. Although some membrane proteins can be correctly refolded after misfolding or aggregation during expression and purification, others are affected by these processes irreversibly. Currently, no reliable method is available to predict which membrane mimetic will stabilize a given protein in its native conformation without aggregation, and often an arduous process of trial and error using expensive reagents must be carried out to find appropriate conditions.
An intriguing alternative to inserting TM proteins into a membrane mimetic environment for structure determination is to alter the proteins such that a membrane is no longer required. Designing water-soluble analogues of membrane proteins challenges the basic understanding of what stabilizes protein structure in both membranous and aqueous environments as well as to what degree the membrane lipids are an integral part of TM protein structures. The current theory (1, 2) suggests that the side-to-side TM helix packing and the interaction with lipids can be distinguished. The lipid-facing surfaces of TM proteins act primarily to position the protein in the membrane. Thus, it seems reasonable to hypothesize that mutating hydrophobic residues to hydrophilic ones at the protein–lipid interface will not significantly change the folding process and the overall structure of membrane proteins but will prevent them from entering the membrane (1, 3, 4). Whether this hypothesis is valid is an open question that we address in this study. It is yet to be determined how other aspects of membrane protein structure, such as the stabilizing effect of specific lipids and the formative role of the membrane during synthesis, can be incorporated into the design of water-soluble analogues.
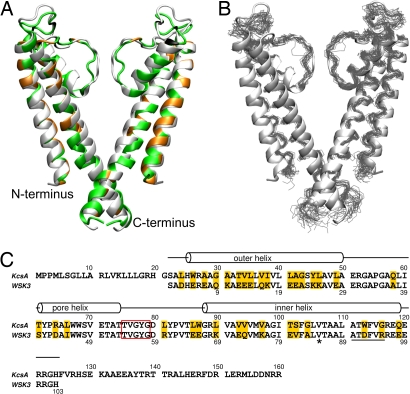
The potassium channel from the bacterium Streptomyces lividans, KcsA, is one of the first TM proteins modified in this fashion (4). KcsA was chosen because it represents a family of interesting ion channels. It forms a symmetric tetramer of 4 identical 160-residue subunits, each comprising an inner and an outer helix traversing the membrane, a pore helix dipping into the membrane, and an extended structure rising from the pore helix toward the external vestibule to form the selectivity filter. KcsA has the characteristic protein–protein, protein–lipid, and protein–water interfaces; its crystal structures under various conditions have been solved; and a wealth of data is available from biochemical, biophysical, and molecular-dynamics studies of the native protein and variants thereof (5, 6). These data provide the basis for evaluating the success in obtaining a water-soluble analogue of KcsA. The sequence and structure of KcsA are shown in Fig. 1, along with those of WSK3, the herein-studied water-soluble analogue. WSK3 was designed by using a statistical approach (7) to predict which amino acids were likely to reside at the 35 lipid-exposed positions to make the designed protein water-soluble while keeping the structure of KcsA (4). This resulted in 30 mutations plus 3 additional ones to confer agitoxin-2 binding as a functional measure of a proper fold (8). The mutations are highlighted in Fig. 1 A and C. The WSK3 sequence was renumbered with S22 in KcsA designated as S1 in WSK3 (Fig. 1C).
Fig. 1.
Comparison of WSK3 with KcsA. (A and B) The averaged NMR structure (green in A) and the 20 lowest energy structures (black in B) of WSK3 are superimposed on the low-K+ KcsA crystal structure (1K4D in light gray). In A, the mutations made to facilitate water solubility and agitoxin-2 binding are highlighted in orange and black, respectively. (C) Sequence alignment and relative numbering of KcsA and WSK3. The mutations are highlighted in gold. The selectivity filter is enclosed in the red rectangle. The kink near V85 in WSK3 is marked with an asterisk. The underlined residues are non-α-helix in some structures.
In this work, we report an experimental solution structure of WSK3 determined by NMR. Our study provides direct experimental evidence for the high fidelity with which WSK3 reproduces KcsA. The NMR structure of WSK3 shows similarity to the native KcsA crystal structures (PDB ID codes 1K4C and 1K4D), validating the design of the water-soluble analogue. By several more-stringent criteria, however, the designed protein is not identical to the native version. Our results have valuable implications not only for the design of water-soluble analogues of other membrane proteins but also for the understanding of the determinants of intrinsic protein structure and dynamics.
Results
WSK3 Forms Helical Tetramers in Water.
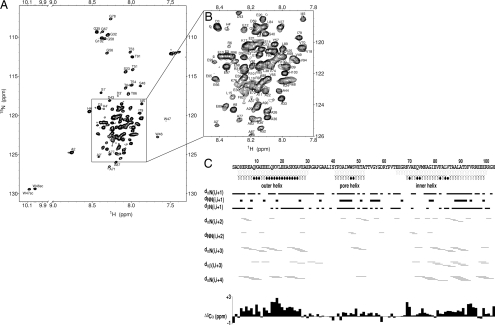
Previously, it was shown by size-exclusion chromatography (SEC) and analytical ultracentrifugation (AUC) (4) that WSK3 at neutral pH predominantly forms tetramers that can further loosely associate into higher-order aggregates, most probably consisting of trimers of tetramers. At high protein concentrations appropriate for NMR, the high-order aggregates led to poor spectral quality. Because KcsA is activated under acidic conditions, we explored lower-pH conditions. A combination of low pH (≈4.5) and low ionic strength gave rise to well-resolved NMR spectra. The tetrameric structure under the NMR sample condition was confirmed by dynamic light scattering (DLS) and NMR diffusion measurements [see supporting information (SI) Text and Figs. S1 and S2], which showed that 85–88% of the protein had a hydrodynamic volume consistent with that of a tetramer, with high-order aggregates comprising the remaining minor species. A single set of resonances was observed in the 2D and 3D NMR spectra. A typical [15N,1H]-heteronuclear single quantum coherence (HSQC) spectrum is shown in Fig. 2A and expanded in Fig. 2B, exhibiting spectral characteristics of a water-soluble helical protein. The high quality of this and the 3D spectra of the 15N,13C-uniformly labeled protein allowed nearly complete backbone assignment and ≈90% side-chain assignment. The observation of a single set of resonances suggests that the small fraction of high-order oligomers was either too broad to detect or in rapid exchange with the tetramers.
Fig. 2.
NMR structural analysis of WSK3. (A) Representative [1H,15N]-HSQC spectrum of WSK3 in 95% H2O and 5% D2O acquired at 293 K and 16.5 T. (B) A zoom-in view of the crowded region in A. (C) NOESY connectivity with line thickness proportional to cross-peak intensities. Also shown are the Cα chemical shift index and helical regions in KcsA (light gray) and in WSK3 (black). The filled circles below the sequence mark the residues where backbone hydrogen-bond restraints were imposed on the basis of unambiguous temperature dependence of exchangeable amide protons.
Circular dichroism (CD) confirmed that WSK3 was predominantly helical, having a helical content of 75–80% (Fig. S3). Most importantly, the backbone Cα, C′ and N chemical shifts correlate well with those of native KcsA in detergent (Fig. S4) (9). This finding indicates that the secondary structure of native KcsA is retained in its water-soluble counterpart and suggests that the tertiary structure is similar.
NMR measurements provided further evidence that WSK3 adopted a reasonably well-defined conformation. Specifically, the indole amide protons of both W46 and W47 were protected from rapid exchange with water; the temperature dependence of the chemical shift changes was −3.1 and −4.2 ppb/K, respectively. As will be seen in the solution NMR structure below, the W46 side chain is fully exposed and not protected by an adjacent subunit in a monomer, being thus more exposed to water than W47. The fact that the W46 indole amide proton has a lower water exchange rate than the W47 indole amide proton, which is protected within the hydrophobic region of a monomer, suggests that WSK3 is in an oligomer state in the NMR sample. Moreover, by purging the intrasubunit cross-peaks using 13C- and 15N-filtered 3D NOESY on a mixture of labeled and unlabeled WSK3, we observed many intersubunit cross-peaks between the labeled and unlabeled proteins (see structure determination below), again supporting the oligomeric state of WSK3. As pointed out, SEC, AUC, and toxin binding assays from previous studies and the data from this study all support the conclusion that WSK3 is predominantly tetramer in the NMR samples.
NMR Structure of WSK3 Mimics, But Is Not Identical to, Low-K+ KcsA Crystal Structure.
The solution NMR structure of WSK3 was determined by using established methods. The average and a bundle of the 20 lowest-energy structures [PDB ID code 2K1E, Biological Magnetic Resonance Bank (BMRB): 15677] are shown superimposed onto the low-K+ KcsA crystal structure (PDB ID code 1K4D) in Fig. 1 A and B, respectively. The mutations for water solubility and agitoxin-2 binding are highlighted. Fig. 2C shows the NOE connectivity (see also Fig. S5) along with the chemical shift index (CSI) and backbone hydrogen bonding restraints, which were imposed conservatively only for those residues with small temperature coefficient of the amide proton chemical shift (10). A total of 69 pairs of intersubunit long-range NOEs between the labeled and unlabeled subunits from the isotope-filtered 3D NOESY and additional 346 pairs of intrasubunit long-range NOEs are identified, assigned, and summarized in Fig. S6. These long-range NOEs define the tertiary folding and quaternary association of the tetramer structure. The structure statistics for the entire protein, including the flexible loops, are summarized in Table S1. The number of restraints by distance range or by individual residues is depicted in Fig. S7. The averaged pairwise r.m.s.d. calculated for backbone-only and for all heavy atoms are 1.4 and 2.2 Å, respectively. Adding 41 N-H residual dipole coupling restraints (11) during structural refinement in the torsion angle space did not improve the statistics, probably because of the highly-dynamic nature of the WSK3 tetramer (see Discussion below).
Given that there are 33 mutations in WSK3 relative to KcsA (68% sequence identity) and that a dramatic difference exists between water and lipid as a solvent, the agreement between the WSK3 and KcsA structures is remarkable. The outer, inner, and pore helices and the selectivity filter of WSK3 are well-defined, with the corresponding N and C termini of various regions nearly perfectly matching those in KcsA. This underscores the importance of short-range backbone interactions in controlling secondary structure and the propensity of side-chain packing in determining the tertiary structure (12).
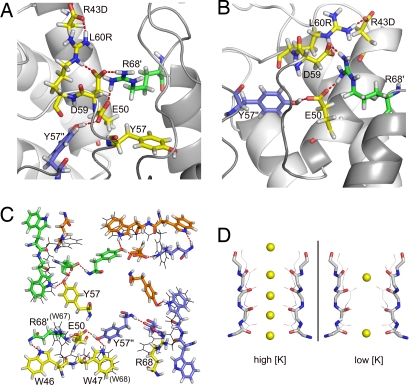
Close inspections of Fig. 1B indicate that the most structurally variable region encompasses residues 31–42, corresponding to the loop between the outer and pore helices. This is also the region where the WSK3 and KcsA structures deviate greatly from each other (Fig. 1A), suggesting a highly dynamic segment. There is also a packing discrepancy between the two structures at the selectivity filter and the pore helix. This discrepancy seems to be a direct consequence of the R43D mutation, which was introduced to provide an artificial agitoxin-2-binding site rather than to solubilize the protein in water. R43D is located near the beginning of the pore helix; the refined structures suggest the formation of a salt bridge between R43D and L60R, which is located immediately after the selectivity filter. As shown in Fig. 3 A and B, L60R also forms a salt bridge with the adjacent D59. The L60R's interaction with R43D alters D59 side-chain orientation, twisting the top of the selectivity filter and placing the D59 side chain near R68 of an adjacent subunit, where an intersubunit salt bridge is likely formed. E50 near the end of the pore helix also forms a salt bridge with the repositioned R68 of the adjacent subunit. Additionally, E50 also forms a hydrogen bond either with the Y57 hydroxyl group from the selectivity filter of a third adjacent subunit or with the Y57 backbone from the same subunit. The key experimental distance restraints supporting the possible formation of these salt bridges are summarized in Tables S2 and S3. The 3-point interaction between E50 and D59 of one subunit and R68 of the adjacent subunit fills the space normally occupied by the W46 side chain, which instead flips to the outside of the inner helix (Fig. 3B) and forms two hydrogen bonds with the backbone amide proton of R68 and the backbone carbonyl of T64. This network of interactions differs from that in KcsA. The corresponding residues in KcsA are E71, W67, W68, Y78, D80, and R89. The KcsA selectivity filter is positioned primarily by carboxyl–carboxylate hydrogen bonding from E71 in the pore helix directly to D80 at the end of the selectivity filter, without the intermediating R89, and by the hydrogen bonding and van der Waals contact among residues Y78 in the selectivity filter, W67 on the exterior face of the pore helix of the same subunit, and W68 on the interior face of the pore helix of the adjacent subunit (5). As pointed out above, the displacement of W46 in the WSK3 structure resulted in Y57 forming a hydrogen bond with E50 while maintaining most of its van der Waals contact with W47. This shift in Y57, combined with the repositioning of D59, slightly changed the orientation of the carbonyls lining the selectivity filter, thereby increasing the radius of the selectivity filter pore (Fig. 3D).
Fig. 3.
Stabilization of the selectivity filter and tetramer conformation in WSK3 by a network of salt bridges and hydrogen bonds. Important side chains are depicted in the licorice representation, with hydrogen bonds indicated by red dashed lines. (A and B) The same region is depicted from different viewing angles. (C) Quaternary relationship among W46, W47, E50, Y57, and R68 in WSK3. W67 and W68 in KcsA are shown in black lines for comparison. (D) Comparison of the selectivity filter between WSK3 structure and KcsA crystal structures obtained in the presence of high (1K4C) and low (1K4D) K+ concentrations. WSK3 is depicted in thick sticks and KcsA in thin lines. Element colors: C, gray; N, blue; O, red; and K, yellow. K+ locations are taken from the crystal structures and not from the NMR data.
Another clear difference between the WSK3 structure and the closed-channel KcsA crystal structure is a kink in the inner helix, which splays the C-terminus away from the pore. Fig. S8 provides a pore-profile analysis, showing a larger pore radius in WSK3 than in KcsA after the hydrophobic gate near the C-terminus. The WSK3 structure shows a larger inner helix bending (≈17.6° compared with ≈8.4° in PBD ID code 1K4D) centered at the helical turn from V85 to A88. This conformational difference from KcsA is probably caused by the low pH, which is known to activate KcsA. Indeed, the kink location near V85 in the WSK3 structure (V106 in KcsA) agrees well with the proposed pivot point near T107 in the “scissoring-type motion” in the pH-dependent opening (13). Consequently, the F93 side chain is packed within the pore in WSK3 instead of between the inner and outer helices of two adjacent subunits as shown in PDB ID code 1K4D. In excellent agreement with the WSK3 structure, a recent NMR study of KcsA in detergents (14) revealed that F93 is involved in the pH-dependent gating.
Backbone Dynamics Reflect an Unusually Dynamic Structure.
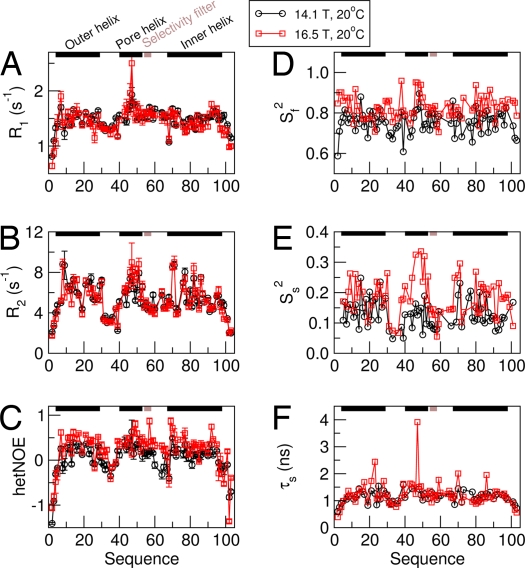
Despite high helical content as determined by CD (Fig. S3) and by the NMR CSI and distance restraints, WSK3 exhibits an unusual and interesting dynamic profile. 15N R1, R2, and 15N-{1H} NOE acquired at 14.1 and 16.5 T and 20 °C are shown in Fig. 4 A–C. The most conspicuous features include relatively high R1, and relatively low R2 and NOE values, as compared with relaxation data from typical globular proteins with molecular mass of ≈45 kDa. Low 15N-{1H} hetNOE values constitute a fingerprint of internal flexibility, notably nanosecond (ns) motions. For detergent-solubilized KcsA (15), comparable R1 and R2 and hetNOE values have been obtained for the C-terminal intracellular helix segment. In contrast, the TM region of KcsA in detergent micelles is associated with the relatively low R1, and high R2 and hetNOEs typical of regular globular proteins. The reduced spectral density mapping analysis of the data in Fig. 4 A–C is provided in Fig. S9.
Fig. 4.
NMR dynamics analysis of WSK3. (A–C) Experimental amide 15N relaxation rate constants and 15N-{1H} hetNOE of WSK3 at 20 °C and at 14.1 T (circles) and 16.5 T (squares). (D–F) Extended model-free analysis of the experimental data in A–C.
Based on the qualitative analysis detailed in SI Text, we estimated a τm of 25 ns for WSK3 at 20 °C. By using this estimate, a more quantitative analysis was performed with the extended model-free approach (16) using DYNAMICS (17). An extensive search in the range of 15–35 ns based on minimum reduced χ2 value and minimum number of nonfitted residues yielded a τm of 20 ns at 20 °C. Using this value, all of the residues have been fit with the so-called “model 5,” in which the squared generalized order parameter, Ss2, and the effective correlation time τs characterize the slow internal motion, and the squared generalized order parameter Sf2 characterizes the fast internal motion.
Fig. 4 D–F depicts Sf2, Ss2, and τs as a function of residue number. Sf2 is ≈0.8 in the helical regions, typical for globular proteins. Ss2 is on the order of 0.1–0.3, which is unusually low. τs is on the order of 1–2 ns, which is not unusual. The loop regions comprising residues 30–42 and 55–69 are associated with relatively low Ss2 and τs values, i.e., higher flexibility than the 3 helical regions. These results indicate that WSK3 is similar to regular globular proteins except for low Ss2 values, which most likely reflect collective domain motions of loosely interacting helices or subunits with low local ordering. This is interesting information that is rarely obtained with regular proteins. A noteworthy difference between WSK3 and detergent-solubilized KcsA is that in KcsA, the selectivity filter appears as rigid as the TM helices (15), whereas in WSK3, the selectivity filter exhibits relatively higher flexibility. A recent structural analysis of the KcsA slow-inactivation mutants (18) seems to suggest that the selectivity filter is “intrinsically unstable” and thus prone to structural rearrangement leading to an inactivated open state. Molecular-dynamics simulations seem to support this conclusion (19). A NMR study of conformational dynamics of KcsA based on NMR-measurable changes of the residues in the selectivity filter and the C-terminus also suggests that slow dynamics in these regions are crucial for ion selection and gating (14).
K+ and Na+ Affect WSK3 Dynamics.
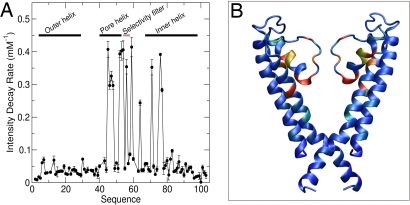
By using NMR spectral quality as a sensitive measure of specific ion effects, a series of HSQC spectra were recorded with increasing concentrations of KCl or NaCl. We found that increasing ionic strength up to 50 mM produced only small chemical shift changes (<10 Hz) at a few residues, most of which were located within or adjacent to the selectivity filter, with Y57 and G58 being particularly sensitive. Other residues showed insignificant chemical shift changes, suggesting that the overall secondary structure of WSK3 is insensitive to the ionic strength in the range examined. In contrast, the intensities of several peaks decreased exponentially with increasing ionic strength, ultimately becoming undetectable. The rate of the intensity decay for each residue, measured by titrating with 0–50 mM NaCl, is plotted in Fig. 5A. Note that the most pronounced intensity decreases occur in the pore helix, the selectivity filter, and several residues of the inner helix (Fig. 5B). The clustering of ion-sensitive residues in this region further confirms a channel-type conformation of WSK3 in water. Although our current structure resolution precludes a detailed analysis of ion binding to WSK3, the decrease in NMR peak intensity without large corresponding chemical shift changes is a strong indicator of changes in dynamics, which might be expected if K+ binds to this region and dynamically hops from the multiple, partially occupied sites observed in crystal structures of KcsA. Intriguingly, K+ is ≈5 times more effective than Na+, with the intensity decay being nearly the same for 10 mM KCl and 50 mM NaCl. This finding is consistent with WSK3 being a soluble analogue of a K+ channel, albeit the selectivity in KcsA is much greater. It should be appreciated, however, that the selectivity filter and pore cavity have many binding sites, some of which can interact with Na+ nonelectrostatically (20). Further studies will be needed to differentiate the highly-selective from the less-selective K+-binding sites in WSK3.
Fig. 5.
Effects of ionic strength on WSK3. The rate of NMR intensity decay with increasing ion concentration is plotted as a function of sequence number (A) and color-coded onto the WSK3 structure (B). In B, two subunits are removed for display clarity. Most- to least-affected residues are color coded from red to blue.
Discussion
To the best of our knowledge, a solution-state NMR structural study of a polytopic ion channel converted from a membrane protein into a water-soluble analogue has not been presented before. WSK3 successfully mimics most aspects of the KcsA crystal structure (5), on which the design of the water-soluble analogue was based. However, a few detailed structural features and some important dynamic features are not the same. These results have revealed some fundamental properties of water-soluble analogues of membrane proteins and provide valuable information useful for the future design of this class of proteins.
Mutation Effects on the WSK3 Structure.
Although a solution structure of KcsA is not yet available, the existing NMR data seem to suggest that KcsA structure in detergent is comparable to the closed-pore crystal structure (14, 21). The WSK3 structure determined in this study resembles the low-K+ KcsA crystal structure except for the kink in the inner helix causing a larger vestibule at the C-terminus and the shifted carbonyls lining in the selectivity filter, resulting in a slightly larger pore passage. Given the overall similarity in folding, it is tempting to presume that the mechanism of pore opening probability is properly represented in the WSK3 construct. Recently, H25 (H4 in WSK3) was reported to act as the pH sensor in opening the hydrophobic gate at the cytoplasmic side of KcsA (22). H25 lies in a hydrophobic intersubunit region in both the WSK3 and KcsA structures. Protonation of H25 in KcsA would likely change the intersubunit contacts, similar to what was observed for WSK3 as a consequence of mutations in this region (L3D, W5E, and W92D). H4 in WSK3 is indeed protonated at pH ≈4.5 used in our studies, as confirmed by the chemical shifts of the imidazole Hδ2 and Hε1 (7.22 and 8.53 ppm, respectively). Similarly, the carboxy-terminal segment, when present, also modulates pore opening in response to decreasing pH, apparently by weakening the stability of the closed state at low pH (23). Thus, the cytoplasmic vestibule of the WSK3 pore, lacking the stability provided by a membrane environment, assumes a configuration that perhaps bears more resemblance to the state of transition to an open channel.
As discussed earlier, the R43D mutation plays an important role in the structural change in WSK3 relative to KcsA. This mutation, combined with L60R, brings about an altered hydrogen-bonding network and packing geometry near the selectivity filter and the pore helix, leading to slight shift in the placement of the carbonyls within the selectivity filter. Without attempting to analyze the ion binding but simply superimposing the selectivity filter of WSK3 to that of KcsA, we found that the selectivity filter in the WSK3 structure is more comparable to the conducting-state crystal structure (PDB ID code 1K4C) than to the nonconducting-state crystal structure (PDB ID code 1K4D) (see Fig. 3D). The distortion in the WSK3 selectivity filter is sufficient to result in a different radius profile within the selectivity filter. In KcsA, the selectivity filter is stabilized by a hydrogen-bond network between E71 and D80, W67 and D80, and a water molecule bound to the backbone nitrogens of G79 and L81 (6). It has been proposed recently that the conformational change in the inner helix during gating propagates to the selectivity filter to trigger the slow inactivation by altering the interaction between E71 and D80 (18). In WSK3, the tertiary contact corresponding to E71 and D80 (E50 and D59) is replaced by a quaternary 3-residue contact among E50 and D59 of one subunit and R68 of the adjacent subunit. This change apparently makes the selectivity filter less stable, in agreement with the dynamics data. It is well known that very small variations in the orientation of the backbone carbonyls in the selectivity filter have dramatic effects on its function. Thus, mutations to charged residues in WSK3 aimed at solubilizing the protein have some unintended implications. It is clear from the WSK3 structure that polar amino acids might be better choices than charged amino acids in the future design of soluble membrane protein analogues, unless the charged amino acids can be accurately paired in such a way as to prevent unintended alternate contacts.
High Flexibility of WSK3.
Little is known about the extent to which lipids contribute to the structure and stability of TM proteins. The absence of a bilayer may be the reason for the NMR-detected slow (τs ≈ 1–3 ns) motions with unusually low local ordering (Ss2 ≈ 0.1–0.3). These motions might represent helix reorientation, domain wobbling, twisting, and rotating or even the entire subunit wobbling within the confine of the tetramer association.
It is not common for globular proteins to have the entire protein backbone experience ns motions with Ss2 as low as 0.1–0.3. However, several cases showing similar behavior have been reported. In the studies of detergent-solubilized KcsA (15, 21), the intracellular C-terminal helix, which extends into the aqueous phase, exhibits low R2 and low hetNOE similar to WSK3 helices (see Table S4). In a model-free analysis, the entire backbone of Ca2+-loaded calmodulin was found to experience ns motions (24). These motions have been attributed to “wobble-in-a-cone-type” domain reorientation. The 15N spin relaxation of diubiquitin (25) was analyzed in terms of domain reorientation between two distinct conformations. The correlation time was found to be 9.3 ns at pH 4.5 and 31.9 ns at pH 6.8. The results were correlated with a tighter interface with observable interdomain NOEs at low pH and loss of these contacts at high pH. These scenarios are analogous to the detergent-solubilized KcsA and WSK3, respectively. Indeed, the corresponding intersubunit contacts in KcsA (15) might have been lost in WSK3 because of mutations at the intersubunit interface, enhancing subunit flexibility. By inference, mutations at interhelix interfaces might have enhanced the flexibility of the TM helices and loops within a given WSK3 subunit. The latter feature is of interest in its own right and might be useful in the design of future water-soluble membrane protein constructs.
The unique dynamics characteristics of WSK3 also contribute to the high spectral resolution observed for the tetramers. The WSK3 NMR peaks as seen in Fig. 2 are broader than what would be expected for a ≈11-kDa monomer at 200 μM (50 μM for a tetramer concentration) but are unusually narrow for non-TROSY spectra of a rigid nondeuterated protein of ≈45 kDa. At least two factors explain the well-resolved narrow peaks. WSK3 is a homotetramer, making the overlapping issue less severe. Furthermore, the dynamics analysis suggests that the 4 subunits have a significant degree of freedom to move relative to each other. Although our quantitative analysis is within the scope of model-free formulism, which applies separately to individual N-H sites, a posteriori one can infer from the parameter commonality on collective subunit motions. A similar situation was identified for the 47-kDa tetrameric yeast elongin C (Elc1) (26), which yielded HSQC spectra with good resolution without 2H labeling. The high resolution was ascribed to tetramerization by flexible monomer association. The WSK3 monomers might associate in a similar manner.
Functional Implications.
The biological function of proteins is determined by both their static 3D structure and their motional state. The fact that the NMR signal intensities are 5 times more sensitive to K+ than to Na+ for residues in and near the selectivity filter and the pore helix (Fig. 5) is a strong indication of the functional relevance of WSK3. Although it is inappropriate to speak of channel function in terms of ion conductance without the membrane, one may envision an aqueous space enclosed within the WSK3 structure distinguishable from the rest of the (bulk) aqueous phase, revealed by the selective NMR signal sensitivity to ionic strength. At the very least, this suggests that key structural aspects of KcsA function are preserved in the current WSK design. Dynamically, WSK3 behaves differently from KcsA embedded in micelles. Note that the intrinsic dynamics of KcsA, without the effect of detergent micelles or lipids, is currently unknown. When the bare WSK3 and KcsA structures were subjected to normal mode analysis, the global mode was found to be essentially identical for the two proteins (data not shown). The global mode manifests as a quaternary twist (tilt) of both the inner and outer helices with relatively large amplitude, along with a twist of the pore region in the opposite direction with smaller amplitude. Thus, it is possible that KcsA, when detached from the membrane, is intrinsically as flexible as WSK3. An advantage of TM channel proteins being intrinsically flexible is that their functional motions can be easily controlled by the dynamic characteristics of the membrane. Studying water-soluble analogues of membrane proteins can potentially reveal such intrinsic protein dynamics independent of the influence of the membrane.
In conclusion, using a water-soluble analogue of a channel protein with known crystal structure, we demonstrated an NMR approach to investigating the structure and dynamics of membrane proteins. Because sequence identity between the original membrane proteins and the water-soluble analogues can be optimized to reach 70–90%, this method will permit atom-resolution structural templates to be generated at a fast pace for superfamilies of receptor proteins that are otherwise not amenable to high-resolution structural analyses. The approach also opens avenues to a better understanding of both membrane protein folding properties and the functionally important intrinsic slow dynamics without the confounding effects of a membrane.
Methods
Protein Expression and NMR Sample Preparation.
The design of WSK3 and its expression plasmid have been published elsewhere (4). For the present studies, the published expression and purification procedures were modified to increase the yield and to control the aggregation of the purified WSK3. Detailed procedures are given in SI Text. To prepare NMR samples, lyophilized WSK3 was dissolved in 95% H2O and 5% D2O to a monomer concentration of ≈200 μM (i.e., ≈50 μM for tetramers). The pH was measured to be ≈4.5. The WSK3 NMR samples prepared in this manner were stable for many weeks.
Experimental Procedures.
Details of experimental procedures measurements, including CD, DLS, and the parameters of various NMR pulse sequences, are given in SI Text. NMR spectra were recorded on Bruker 600, 700, and 800 MHz spectrometers with Topspin 1.3 software using cryogenic triple-resonance probes. NMR pulse sequences from the Topspin sequence library, including HNCO, HNCA, HNCACB, and CBCACONH for sequential assignment, HCCH-TOCSY for side-chain assignment, 3D 15N- or 13C-filtered NOESY for distance restraints, TROSY-IPAP for residual dipole coupling (RDC), HSQC-based R1 and R2 sequences for 15N relaxation parameters, HSQC-based CPMG method for R2 dispersion measurements, and DOSY for translational diffusion constant, were used either with standard setting or with minor modifications. Except as otherwise noted, most of the NMR experiments were carried out at 20 °C.
Data Processing and Analysis.
NMR spectral processing, structure calculation, and dynamics analysis are detailed in SI Text. A total of 100 structures were calculated and refined. The 20 lowest-energy structures were further energy-minimized inside a water box to remove bad contacts between proton atoms and are presented here. The structure statistics with and without RDC restraints are given in Table S1. Without RDCs, the Ramachandran statistics are 87.4%, 9.3%, 1.9%, and 1.4% in most favored, additionally allowed, generously allowed, and disallowed regions, respectively. With RDCs, these statistics are 86.3%, 10.2%, 1.9%, and 1.6%.
Supplementary Material
Acknowledgments.
We thank Profs. Jeff Saven and Rieko Ishima for stimulating discussions and Dr. Filippo Pullara and Prof. Guillermo Calero for help with DLS measurements. This work was supported in part by National Institutes of Health Grants R01GM056257 (to P.T.), P01GM055876 (to R.E. and Y.X.), and R37GM049202 (to Y.X. and P.T.). E.M. acknowledges Israel Science Foundation Grant 279/03, Israel–USA Binational Science Foundation Grant 2000399), and the Damadian Center for Magnetic Resonance at Bar-Ilan University.
Footnotes
The authors declare no conflict of interest.
Data deposition: The NMR chemical shifts have been deposited in the Biological Magnetic Resonance Bank, www.bmrb.wisc.edu (accession no. 15677). The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2K1E).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805501105/DCSupplemental.
References
- 1.Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 2.DeGrado WF, Gratkowski H, Lear JD. How do helix–helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. Protein Sci. 2003;12:647–665. doi: 10.1110/ps.0236503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rees DC, DeAntonio L, Eisenberg D. Hydrophobic organization of membrane proteins. Science. 1989;245:510–513. doi: 10.1126/science.2667138. [DOI] [PubMed] [Google Scholar]
- 4.Slovic AM, Kono H, Lear JD, Saven JG, DeGrado WF. Computational design of water-soluble analogues of the potassium channel KcsA. Proc Natl Acad Sci USA. 2004;101:1828–1833. doi: 10.1073/pnas.0306417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 7.Zou J, Saven JG. Statistical theory of combinatorial libraries of folding proteins: Energetic discrimination of a target structure. J Mol Biol. 2000;296:281–294. doi: 10.1006/jmbi.1999.3426. [DOI] [PubMed] [Google Scholar]
- 8.MacKinnon R, Cohen SL, Kuo A, Lee A, Chait BT. Structural conservation in prokaryotic and eukaryotic potassium channels. Science. 1998;280:106–109. doi: 10.1126/science.280.5360.106. [DOI] [PubMed] [Google Scholar]
- 9.Chill JH, Louis JM, Delaglio F, Bax A. Local and global structure of the monomeric subunit of the potassium channel KcsA probed by NMR. Biochim Biophys Acta. 2007;1768:3260–3270. doi: 10.1016/j.bbamem.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baxter NJ, Williamson MP. Temperature dependence of 1H chemical shifts in proteins. J Biomol NMR. 1997;9:359–369. doi: 10.1023/a:1018334207887. [DOI] [PubMed] [Google Scholar]
- 11.Bryce DL, Bax A. Application of correlated residual dipolar couplings to the determination of the molecular alignment tensor magnitude of oriented proteins and nucleic acids. J Biomol NMR. 2004;28:273–287. doi: 10.1023/B:JNMR.0000013701.16162.0c. [DOI] [PubMed] [Google Scholar]
- 12.Bhutani K, Tang P, Xu Y. Helical pairing propensity of amino acids in membrane proteins. Biophys J. 2005;88:47a. [Google Scholar]
- 13.Liu YS, Sompornpisut P, Perozo E. Structure of the KcsA channel intracellular gate in the open state. Nat Struct Biol. 2001;8:883–887. doi: 10.1038/nsb1001-883. [DOI] [PubMed] [Google Scholar]
- 14.Baker KA, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R. Conformational dynamics of the KcsA potassium channel governs gating properties. Nat Struct Mol Biol. 2007;14:1089–1095. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chill JH, Louis JM, Baber JL, Bax A. Measurement of 15N relaxation in the detergent-solubilized tetrameric KcsA potassium channel. J Biomol NMR. 2006;36:123–136. doi: 10.1007/s10858-006-9071-4. [DOI] [PubMed] [Google Scholar]
- 16.Clore GM, et al. Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen-15 nuclear magnetic relaxation of proteins. J Am Chem Soc. 1990;112:4989–4991. [Google Scholar]
- 17.Fushman D, Cahill S, Cowburn D. The main-chain dynamics of the dynamin pleckstrin homology (PH) domain in solution: Analysis of 15N relaxation with monomer/dimer equilibration. J Mol Biol. 1997;266:173–194. doi: 10.1006/jmbi.1996.0771. [DOI] [PubMed] [Google Scholar]
- 18.Cordero-Morales JF, et al. Molecular driving forces determining potassium channel slow inactivation. Nat Struct Mol Biol. 2007;14:1062–1069. doi: 10.1038/nsmb1309. [DOI] [PubMed] [Google Scholar]
- 19.Domene C, Vemparala S, Furini S, Sharp K, Klein ML. The role of conformation in ion permeation in a K+ channel. J Am Chem Soc. 2008;130:3389–3398. doi: 10.1021/ja075164v. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, MacKinnon R. Ion binding affinity in the cavity of the KcsA potassium channel. Biochemistry. 2004;43:4978–4982. doi: 10.1021/bi049876z. [DOI] [PubMed] [Google Scholar]
- 21.Chill JH, Louis JM, Miller C, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeuchi K, Takahashi H, Kawano S, Shimada I. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J Biol Chem. 2007;282:15179–15186. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]
- 23.Pau VP, Zhu Y, Yuchi Z, Hoang QQ, Yang DS. Characterization of the C-terminal domain of a potassium channel from Streptomyces lividans (KcsA) J Biol Chem. 2007;282:29163–29169. doi: 10.1074/jbc.M703277200. [DOI] [PubMed] [Google Scholar]
- 24.Baber JL, Szabo A, Tjandra N. Analysis of slow interdomain motion of macromolecules using NMR relaxation data. J Am Chem Soc. 2001;123:3953–3959. doi: 10.1021/ja0041876. [DOI] [PubMed] [Google Scholar]
- 25.Ryabov YE, Fushman D. A model of interdomain mobility in a multidomain protein. J Am Chem Soc. 2007;129:3315–3327. doi: 10.1021/ja067667r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Botuyan MV, et al. Binding of elongin A or a von Hippel-Lindau peptide stabilizes the structure of yeast elongin C. Proc Natl Acad Sci USA. 1999;96:9033–9038. doi: 10.1073/pnas.96.16.9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.