Abstract
Mitogen-activated protein (MAP) kinases are pivotal components of eukaryotic signaling cascades. Phosphorylation of tyrosine and threonine residues activates MAP kinases, but either dual-specificity or monospecificity phosphatases can inactivate them. The Candida albicans CPP1 gene, a structural member of the VH1 family of dual- specificity phosphatases, was previously cloned by its ability to block the pheromone response MAP kinase cascade in Saccharomyces cerevisiae. Cpp1p inactivated mammalian MAP kinases in vitro and acted as a tyrosine-specific enzyme. In C. albicans a MAP kinase cascade can trigger the transition from the budding yeast form to a more invasive filamentous form. Disruption of the CPP1 gene in C. albicans derepressed the yeast to hyphal transition at ambient temperatures, on solid surfaces. A hyphal growth rate defect under physiological conditions in vitro was also observed and could explain a reduction in virulence associated with reduced fungal burden in the kidneys seen in a systemic mouse model. A hyper-hyphal pathway may thus have some detrimental effects on C. albicans cells. Disruption of the MAP kinase homologue CEK1 suppressed the morphological effects of the CPP1 disruption in C. albicans. The results presented here demonstrate the biological importance of a tyrosine phosphatase in cell-fate decisions and virulence in C. albicans.
INTRODUCTION
The developmental transition from the predominant yeast form (blastospores) to the hyphal form of the opportunistic pathogen Candida albicans is thought to contribute to early steps in invasion of epithelial tissues (reviewed in Fidel and Sobel, 1994); however, both forms can be found in infected human tissues (Odds, 1988). Elucidation of the signaling events that lead to germ tube formation and subsequent hyphal development will help us to understand both the initial steps in the encounter of C. albicans cells with host epithelia and the contribution of the yeast and hyphal forms to pathogenesis. Such studies may reveal new targets for antimycotic drugs.
Many environmental factors induce the yeast form of C. albicans to form germ tubes which develop into hyphae (Odds, 1988; reviewed in Soll, 1986), suggesting the involvement of multiple signal transduction pathways or multiple inputs into the same pathway. Conditions that mimic those found in the blood, including neutral pH (Buffo et al., 1984) and physiological temperature (Lee et al., 1975; Buffo et al., 1984) induce the yeast to hyphal switch, whereas ambient temperatures or acid pH favor the budding yeast form. In combination with these conditions, serum (Gow and Gooday, 1982), or certain synthetic defined and complex media, allows hyphal development in vitro either in liquid culture (Lee et al., 1975; Buffo et al., 1984) or on solid media (Gow and Gooday, 1982; Liu et al., 1994; Leberer et al., 1996). A conserved protein kinase cascade in Saccharomyces cerevisiae drives several cellular responses including pheromone response, yeast to pseudohyphal switching, and agar invasion (Gimeno et al., 1992; Liu et al., 1993; Roberts and Fink, 1994; Herskowitz, 1995). Components of this cascade have recently been shown to stimulate yeast to hyphal switching in C. albicans (Liu et al., 1994; Malathi et al., 1994; Clark et al., 1995; Kohler and Fink, 1996; Leberer et al., 1996). The C. albicans genes CST20, HST7, and CPH1 are homologues of S. cerevisiae genes encoding a mitogen-activated protein (MAP) kinase kinase kinase kinase (STE20), a MAP kinase kinase (STE7), and a transcription factor (STE12) involved in this signaling pathway (Liu et al., 1994; Malathi et al., 1994; Clark et al., 1995; Kohler and Fink, 1996; Leberer et al., 1996). In C. albicans, these genes function in glucose-independent in vitro hyphal formation on solid surfaces, but not in serum-dependent or in vivo hyphal formation (Liu et al., 1994; Kohler and Fink, 1996; Leberer et al., 1996). These results point toward distinct signaling mechanisms for hyphal development in response to different stimuli, including solid surfaces. Contact sensing at solid surfaces could be involved in stimulating hyphal development as it is in guiding hyphae toward pores (Sherwood et al., 1992). Despite the absence of an effect on hyphal formation in mice, deletion of CST20 from C. albicans results in a minor but significant reduction in virulence in mice (Leberer et al., 1996).
A balance between MAP kinase activation by kinases and their inactivation by phosphatases is likely to be important for decisions which govern developmental processes and virulence in C. albicans. We have previously screened a C. albicans genomic library in an attempt to find genes which interfere with pheromone-mediated cell cycle arrest in S. cerevisiae (Whiteway et al., 1992). One of the genes (CEK1) isolated during this screen encodes a C. albicans homologue of the S. cerevisiae Fus3p and Kss1p MAP kinases involved in pheromone response (Whiteway et al., 1992).This screen also identified a gene called CPP1 for Candida protein phosphatase with similarities to protein tyrosine phosphatases (PTPs) (Whiteway et al., 1993). CPP1 expression was found, using a series of epistasis experiments, to block the S. cerevisiae pheromone response pathway at the level of the Fus3p MAP kinase (Whiteway et al., 1993). We show here that Cpp1p is a member of the VH1 family of dual-specificity phosphatases and is most similar to the S. cerevisiae MAP kinase phosphatase, Msg5p, which is involved in adaptation to pheromone (Doi et al., 1994). Although deletion of the mammalian VH1 phosphatase (MKP1) gene has no effect on mouse development (Dorfman et al., 1996) and deletion of the two S. cerevisiae VH1-phosphatase genes (MSG5 and YVH1) have only subtle effects on S. cerevisiae cells (Guan et al., 1992; Doi et al., 1994), deletion of the CPP1 gene in C. albicans derepresses the yeast to hyphal transition at ambient temperatures on solid surfaces under normally noninducing conditions. This result suggests that Cpp1p is required for repression of the yeast to hyphal switch. In addition, under hyphal-stimulating physiological conditions in vitro which inhibit the initial growth of the yeast form, deletion of the CPP1 gene results in a hyphal growth rate defect. In vivo, this latter effect may be of significance because virulence is greatly reduced in a mouse model for systemic candidiasis when mice are infected with CPP1-defective cells.
MATERIALS AND METHODS
DNA Manipulations and Analysis
DNA manipulations were carried out according to standard procedures (Sambrooke et al., 1989). Southern blot analysis was performed with a nonradioactive labeling and detection kit (Boerhinger Mannheim, Canada, Laval, QC) according to the manufacturer’s recommendations. Both strands of C. albicans sequences in the plasmid M195p4 were sequenced.
Blast similarity searches were done with the GenBank and SwissProt databases. A protein motif alignment was done using Protomat (Henikoff et al., 1995). Alignments of the active site domains of protein sequences were done with the program GeneWorks (IntelliGenetics) using a modified FASTA algorithm as described in the GeneWorks Reference Manual.
Plasmid Constructions
Plasmid pCCY10.2 was constructed by digestion of plasmid M195p4 with BglII and SmaI, generation of blunt ends and religation, leaving the vector YEP352 (Hill et al., 1986) with a 4.5-kb fragment containing the CPP1 gene and flanking DNA. M195p4, itself, was derived from the original plasmid M153p11 (Whiteway et al., 1992) by partial SpeI digestion and religation to the YEP352 XbaI site. Plasmid pGALCPP1-M178 expresses an active truncated version of CPP1 starting at amino acid 178. Synthetic oligonucleotides with 5′ EcoRI sites were used to amplify DNA spanning nucleotides 506 to 1886 (nucleotide 1 refers to the first nucleotide of the open reading frame). Polymerase cahin reaction (PCR) products were inserted into the EcoRI site of an S. cerevisiae centromeric TRP1-based shuttle vector pRS314GAL containing the GAL1 S. cerevisiae promoter on a BamHI-EcoRI fragment modified from the vector pRS314 (Sikorski and Hieter, 1989). The conserved cysteine (residue 516) of the active site region of the CPP1 polypeptide in pGALCPP1-M178 was changed to a serine residue by site-directed mutagenesis (Kunkel et al., 1987) creating pGALCPP1-M178-C516S. The synthetic oligonucleotide used (5′-GACGTAAAATATTGATTCATTCACAATGTGGAGTATCG3-′) replaces cysteine 516 with a serine, while introducing a DraIII site (underlined). Mutants were verified by sequence analysis. pGST-ERK1 was created by subcloning the EcoRI fragment encoding hamster Erk1 from plasmid pCMV/HAPMK (Meloche et al., 1992) into the EcoRI site of pGEX-KG (Guan and Dixon, 1991). pGST-CPP1 was created by amplifying nucleotides 767 to 1886 of CPP1 by PCR using synthetic oligonucleotides with 5′ EcoRI sites and then cloning the amplified DNA fragment into pGEX-KT (Hakes and Dixon, 1992). This same fragment, when placed under the control of the S. cerevisiae GAL1 promoter, interfered with pheromone-induced cell cycle arrest in yeast. For gene disruptions pCCB201 was created in two steps. First, PCR was used to amplify the CPP1 gene and flanking sequences in pCCY10.2 and to add SacI sites using the oligodeoxynucleotide primers O27 (5′-GAACAACCAGGAGAGCTCTTTCCAACTGATTTAATTTG-3′) and O26 (5′-GTTGTCTTTAGTTGGAGCTCCTTATTTTATATAATAGATG3′) (SacI sites are underlined). After digestion, this 2.3-kb SacI fragment was inserted into the Bluescript KS(+) vector (Stratagene, La Jolla, CA) to yield plasmid pCCB200. Oligodeoxynucleotide primers O24 (5′-GAAGATCTGATATCTATTTTCCCTTGATCTGGATCTG3′) and O25 (5′-GAAGATCTGTTGTAGCATTTTATATGAAGAAATTCCAATTGGGAG-3′) were then used to delete the PTP-active site region in pCCB200 using divergent PCR while adding BglII sites (underlined). The amplified fragment was cut with BglII and joined to a 4-kb BamHI-BglII hisG-URA3-hisG fragment from the plasmid p5921 (Fonzi and Irwin, 1993) to create pCCB201. pCCa2 was constructed by subcloning a 4.5-kb PstI-KpnI fragment from the plasmid pCCY10.2 into pBS-cURA3 (Leberer et al., 1996) containing the C. albicans URA3 gene.
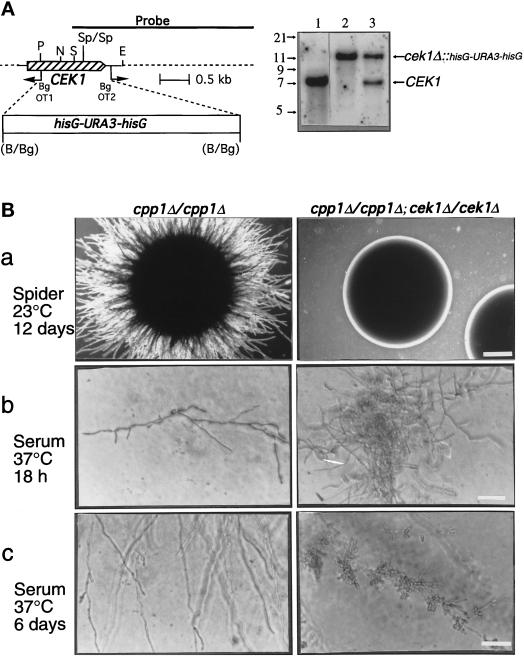
Construction of plasmids for disruption of the CEK1 gene (Whiteway et al., 1992) will be presented in greater detail elsewhere. Briefly, the plasmid pMO3 contains an 8-kb KpnI-XbaI DNA fragment in the KpnI-XbaI sites of the Bluescript KS(+) vector (Stratagene). This insert contains a C. albicans genomic DNA fragment in which a 1.2-kb portion of the CEK1 gene was replaced by a 4-kb BamHI-BglII hisG-URA3-hisG fragment (Fonzi and Irwin, 1993). Deletion of the 1.2-kb portion of the CEK1 gene was accomplished by reverse PCR with the oligonucleotide primers OT1 and OT2 (positions shown in Figure 8A) which were flanked by BglII sites for ligation to the hisG-URA3-hisG blaster cassette.
Figure 8.
Construction and phenotypic analysis of cek1/cpp1 double null mutants. (A) Deletion of CEK1 in C. albicans. Left, Deletion strategy and restriction map of the CEK1 gene. PCR with the divergent oligodeoxynucleotides OT1 and OT2 was used to delete a 1.2-kb fragment of the CEK1 gene. A 4.0-kb hisG-URA3-hisG cassette was then inserted. Restriction sites are as follows: B, BamHI; Bg, BglII; P, PstI; S, SacI; N, NsiI; E, EcoRI; and Sp, SpeI. Right, Southern blot analysis of CEK1 disruptions with a 3.2-kb KpnI-SacI fragment containing the CEK1 gene as a probe. Genomic DNA samples were digested with SpeI (absent from the hisG-URA3-hisG cassette) from the following strains: lane 1, CP29–1-7 L4 (ura3/ura3 cpp1Δ::hisG/cpp1Δ::hisG; CEK1/CEK1); lane 2, CP29-1-7 CK14 (ura3/ ura3cpp1Δ::hisG/cpp1Δ::hisG; cek1Δ::hisG-URA3hisG/cek1Δ::hisG-URA3-hisG); and lane 3, CP29-1-7 CK13 (ura3/ura3cpp1Δ::hisG/cpp1Δ::hisG; CEK1/cek1Δ::hisG-URA3-hisG). Hybridization of a very small part (between the SacI and SpeI sites) of the probe to a 1.4-kb SpeI fragment, present only in the wild-type CEK1 gene, was not detectable in these Southern blots and was not used for diagnostic purposes. (B) Colonies of cpp1 null mutants (cpp1Δ/cpp1Δ) or cpp1/cek1 double null mutants (cpp1Δ/cpp1Δ; cek1Δ/cek1Δ) grown at room temperature (a) on solid Spider medium containing mannitol (2× objective; bar, 1.4 mm) and under physiological conditions (b and c) on solid serum medium (40× objective; bar, 70 μm). In a and b colonies are shown. In c hyphae (and lateral blastospores) from peripheries of mycelial colonies are shown. Our unpublished data demonstrated that the cpp1/cek1 double null mutant phenotypes resembled those of the wild-type strain SC5314.
S. cerevisiae Pheromone Response
Pheromone response was assessed by transforming plasmids (Rose et al., 1990) into a supersensitive S. cerevisiae strain HC1–4D (Whiteway et al., 1993). For examination of growth in the presence of pheromone on solid media, patches of HC1–4D cells transformed with pGALCPP1-M178, pGALCPP1-M178-C516S, and the control vector pRS314GAL were grown on selective solid medium (SC) without tryptophan using 2% galactose as a carbon source (Rose et al., 1990) and were replica plated onto selective medium spread with 5 μg of the yeast mating pheromone α-factor (Sigma, St. Louis, MO).
Biochemical Assays
Recombinant glutathione S-transferase (GST) fusion proteins were produced as follows: Plasmids pGST-CPP1, pGST-ERK1, and pGEX-KT (GST) were expressed in Escherichia coli, and the GST fusion proteins or GST were purified as described (Guan and Dixon, 1991). The approximate concentration of recombinant proteins was determined by SDS-PAGE using bovine serum albumin as a protein standard.
The enzymatic activity of Cpp1p was assayed using Erk1 as a substrate. For these experiments, recombinant GST-Erk1 immobilized on glutathione-agarose beads was first activated by incubation with a cytosolic extract of serum-stimulated Rat 1 cells in the presence of 50 μM ATP at 30°C for 30 min (Meloche, 1995). The beads were then washed in phosphatase buffer containing 25 mM HEPES (pH 7.4), 100 mM NaCl, and 1 mM dithiothreitol prior to incubation with GST-Cpp1p at 37°C for 30 min in phosphatase buffer. Myelin basic protein (MBP) phosphotransferase assays were done as described (Meloche, 1995). The tyrosine phosphorylation of Erk1 was evaluated by antiphosphotyrosine immunoblotting using the monoclonal antibody PY20 (ICN). The bands were visualized by chemiluminescence (ECL, Amersham Canada Ltd., Oakville, Ontario) according to the manufacturer’s instructions. For phosphoamino acid analysis, Erk1 and Erk2 were immunoprecipitated from 32P-labeled serum-stimulated Rat 1 fibroblasts using the specific Erk1 antiserum SM1 or the specific Erk2 antiserum aIIcp42 and analyzed by two-dimensional phosphoamino acid analysis as described (Meloche, 1995).
Candida Strains and Growth Conditions
All strains are listed in Table 1. To induce germ tube formation in liquid culture, cells were diluted 10-fold from overnight cultures into fresh Spider medium (Liu et al., 1994), Lee’s medium (Lee et al., 1975), or 10% fetal bovine serum (Intergen Co., Purchase, NY) and incubated for 3 h at 37°C (Lee et al., 1975).
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype | Origin or source |
|---|---|---|
| S. cerevisiae | ||
| HC1-4D | MATa sst1 sst2 ura3 trp1 leu2 ade-kex2::ura3 | Whiteway et al., 1993 |
| C. albicans | ||
| SC5314 | URA3/URA3 CPP1/CPP1 CEK1/CEK1 | Gillum et al., 1984 |
| CAI4 | ura3 CPP1/CPP1 CEK1/CEK1 | Fonzi and Irwin, 1993 |
| CP29 | ura3/ura3 CPP1/cpp1Δ::hisG-URA3-hisG | CAI4 |
| CP29-1 | ura3/ura3 CPP1/cpp1Δ::hisG | CP29 |
| CP29-1-7 | ura3/ura3 cpp1Δ::hisG-URA3-hisG/cpp1Δ::hisG | CP29-1 |
| CP29-1-7L4 | ura3/ura3 cpp1Δ::hisG/cpp1Δ::hisG | CP29-1-7 |
| CP29-1-7RI | ura3/ura3 cpp1Δ::hisG/cpp1Δ::hisG::CPP1-URA3 | CP29-1-7L4 |
| CP29-1-7CK13 | ura3/ura3 cpp1Δ::hisG/cpp1Δ::hisG; | CP29-1-7L4 |
| CEK1/cek1Δ::hisG-URA3-hisG | ||
| CP29-1-7CK14 | ura3/ura3 cpp1Δ::hisG/cpp1Δ::hisG; | CP29-1-7L4 |
| cek1Δ::hisG-URA3-hisG/cek1Δ::hisG-URA3-hisG |
To induce hyphal growth on solid media, budding C. albicans were grown overnight at 30°C with vigorous shaking in YP medium (2% yeast extract, 4% Bacto Peptone) supplemented with either 2% glucose for YPD (Rose et al., 1990) or 2% mannitol for YPM. Twenty-five to 100 cells per plate were then incubated for indicated times at 23°C or 37°C on different media. Solid Spider medium contains 1% nutrient broth, 0.2% K2HPO4, 1.4% agar, and 1% of either glucose or mannitol (Liu et al., 1994). Lee’s medium is as described (Lee et al., 1975). For serum plates, 10% fetal bovine serum was added to 1.4% agar at 50°C after autoclaving.
To examine agar-penetrating growth beneath colonies, cells were gently scraped off plates with a plastic inoculating loop and plates were then washed several times with sterile water. Photomicroscopy of colonies and invasive growth was done with a Nikon TMS inverted microscope and photographed with Kodak TMAX film.
Nomarski optics was used to photograph germ tubes and to monitor for septa formation and branching of cells scraped from agar surfaces. A 100× objective and a Leitz Aristoplan microscope were used.
Candida Gene Disruptions
Homologous recombination was used for gene disruptions by a sequential gene disruption strategy using the selectable marker URA3 flanked by hisG direct repeats (Fonzi and Irwin, 1993). The hisG repeats facilitate the removal of the URA3 gene and allow the use of the same selectable marker for disruption of the second allele. This hisG-URA3-hisG cassette was used to replace parts of the CPP1 and CEK1 genes. For CPP1 disruptions, the active site region of the CPP1 gene was deleted to allow a maximum number of defined sequences flanking the disruption cassette and because previous experiments showed that removal of the active site was sufficient to totally inactivate Cpp1p function. pCCB201 was digested with SacI and the isolated fragment was used to transform spheroplasts (Kurtz et al., 1986) of the strain CAI4 (Table 1). Transformants able to grow in the absence of uracil were selected and replacement of the chromosomal gene with the fragment containing a hisG-URA3-hisG cassette in place of the active site region was verified by Southern blot analysis using a HindIII-SpeI fragment from the CPP1 gene as a probe. Transformants with a disruption in the CPP1 gene were chosen for selection of Ura3− derivatives on 5-fluoroorotic acid, which kills Ura3+ prototrophs. Ura3− auxotrophs were examined using Southern blot analysis to identify excisions of the URA3 repeat leaving behind one copy of hisG. These steps were repeated to obtain cells with disruptions in both alleles of CPP1. For reintegration of CPP1 into the genome, the C. albicans reintegration plasmid containing the CPP1 gene and flanking sequences, pCCa2, was linearized with NsiI to target integration to the NsiI site of cpp1Δ::hisG and transformed into Ura− C. albicans containing the double deletion of the CPP1 gene cpp1Δccp1Δ::hisG. Most in vitro studies were done with two completely independent, but phenotypically identical, homozygous mutants: CP29–1-7 (shown in this study) and CP27–1-1.
Disruption of the CEK1 (GenBank M76585) gene from the cpp1 null mutant strain was achieved in one step. An 8-kb KpnI-NotI insert from the plasmid pMO3 was used to transform CP29–1-7L4 (Table 1) as described above. This fragment contains a 4.0-kb hisG-URA3-hisG cassette in place of a 1.2-kb portion of the open reading frame of the CEK1 gene. Replacement of the chromosomal gene with the exogenously provided fragment was verified by Southern blot analysis using a 3.2-kb KpnI-SacI probe containing the CEK1 gene and genomic DNA digested with SpeI. Of 14 transformants examined using Southern blot analysis, 12 contained a disruption of one allele of CEK1 and one had both alleles of the CEK1 gene disrupted (Figure 8A). This transformant was called CP29–1-7CK14 (cpp1Δcpp1Δ/cek1Δcek1Δ) and was used for subsequent experiments.
Virulence Studies
Inbred female BALB/c mice were obtained from Charles Rivers Breeding Laboratories (Sulzfeld, Germany) and used for infection at 8 to 10 wk of age.
C. albicans in vivo virulence testing and colony-forming unit (CFU) enumeration was done as follows: Strains for infection were routinely grown at 30°C in YPD medium and kept at stationary phase for 48 h prior to infection. Aliquots of approximately 2 × 108 cells were harvested and washed three timed in phosphate-buffered saline, and adjusted to the desired density to be used for in vivo virulence testing. C. albicans blastospores were injected i.v. into the tail vein in a final volume of 200 μl. Three to four mice per group were killed 2 and 5 d after infection, and the number of CFUs was quantified using a plate dilution method of homogenized organs on modified Lee’s medium agar plates (Soll et al., 1981). Results are expressed as the log CFU per g organ wet weight.
For histological analysis of C. albicans cell morphology in vivo, five infected animals were killed 2 d after infection, and the kidneys were solubilized in 20 ml of 10% potassium hydroxide (KOH) solution at 50°C for 3 h. Alkaline-resistant particles, including yeast cells, were sedimented at 1500 × g, resuspended in 50 μl of KOH solution containing 5 μg/ml Calcofluor white (Sigma, Deisenhofen, Germany). Slides were directly mounted without further manipulations and screened for C. albicans cells with a Zeiss Axiophot microscope under fluorescent light (365-nm filter for excitation and 420-nm filter for emission).
RESULTS
Sequence Analysis of the CPP1 Gene
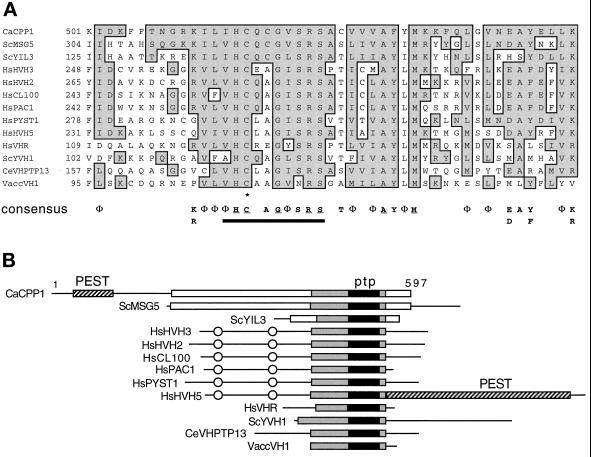
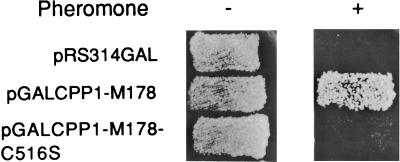
The CPP1 gene (GenBank L01038) codes for a 597-amino acid polypeptide with similarity to the dual- specificity (serine/threonine and tyrosine) phosphatases of the Vaccinia VH1 subfamily of PTPs (Figure 1) (Guan et al., 1991; Keyse, 1995; Zolnierowicz and Hemmings, 1996) and maps to C. albicans chromosome 1 (Magee, personal communication). A 140-amino acid region of the CPP1 polypeptide, which contains a core PTP-active site signature sequence (V/I)HCxAGxxR(S/T) (Fischer et al., 1991; Zolnierowicz and Hemmings, 1996), aligns with other members of the VH1 phosphatase family (Figure 1). An active site cysteine residue is required for catalysis by PTPs (Keyse, 1995). Mutation of the equivalent cysteine residue of Cpp1p to a serine, expressed in plasmid pGALCPP1-M178 in S. cerevisiae, destroyed its capacity to block pheromone-induced cell cycle arrest (Figure 2), demonstrating that phosphatase activity was required for biological activity.
Figure 1.
Structural similarity of CPP1 with VH1 family phosphatases. (A) Similarity of the CPP1 amino acid sequence to a conserved region surrounding the active-site signature sequence of the VH1 family phosphatases. Aligned sequences are C. albicans (Ca) CPP1, S. cerevisiae (Sc) MSG5 (Doi et al., 1994), YIL3 (SWISS-PROT P40558, YIL003W, putative phosphatase), and YVH1 (Guan et al., 1992), human (Hs) HVH3 (Kwak and Dixon, 1995), HVH2 (Guan and Butch, 1995), CL100 (MKP1) (Alessi et al., 1993), PAC1 (Rohan et al., 1993), PYST1 (Groom et al., 1996), HVH5 (Martell et al., 1995), VHR (Ishibashi et al., 1992), Chlamydomonas eugametos (Ce) VHPTP13 (Haring et al., 1995), and Vaccinia (Vacc) VH1 (Guan et al., 1991). Conserved residues (ILVM =Φ; KR; ED; NQ; YF) and identical residues present in the CPP1 polypeptide and one or more of the other polypeptides are boxed and shaded. A VH1 family consensus is presented below the alignment for residues present in 8 or more of the 13 sequences. Residues identical in all 13 sequences are underlined. The PTP-active site motif is underlined with a black bar and the active site essential cysteine is shown with an asterisk. Numbers at the left of the margin specify the amino acid positions within each polypeptide. (B) Schematic alignment of the VH1 family phosphatases. Black boxes represent the highly conserved active site region (PTP) aligned in A around which the alignment is centered. The gray boxes represent an extended homology region with lower identities between sequences. White boxes represent additional regions of alignment among CPP1, MSG5, and YIL3. PEST sequences are shown. CH2 domains are circled. Numbers indicate the size of the CPP1 polypeptide.
Figure 2.
Site-directed mutagenesis of the conserved PTP-active site cysteine stops Cpp1p from interfering with the S. cerevisiae pheromone response pathway. Growth of S. cerevisiae cells on selective media with or without pheromone is shown. Cells contain the plasmids pRS314GAL (vector), pGALCPP1-M178 containing the extended region of CPP1 homology with MSG5, or pGALCPP1-M178 with the essential PTP-active site cysteine changed to a serine (pGALCPP1-M178-C516S).
CPP1 is most similar to two S. cerevisiae genes: MSG5, which encodes a protein which dephosphorylates the Fus3p MAP kinase (Doi et al., 1994), and YIL3, an open reading frame in the databases (SWISS-PROT P40558; YIL003W; putative phosphatase). CPP1 has 44% and 40% amino acid identity with MSG5 and YIL3, respectively, in the region surrounding the active site (Figure 1B; gray boxes). All three have a cysteine instead of an alanine at position five of the PTP signature sequence. The CPP1, MSG5, and YIL3 polypeptides do not contain CH2 domains (Figure 1B), which are domains shared by several mammalian VH1 family phosphatases (many of which dephosphorylate MAP kinases) and the dual- specificity cdc25 phosphatases (Keyse and Ginsburg, 1993). In addition, Cpp1p contains a putative PEST motif (Rogers et al., 1986) rich in serine (25%) and threonine (15%) residues in its amino terminal domain (Figure 1B), which suggests that Cpp1p may have a short half-life (Rogers et al., 1986).
Biochemical Characterization of Cpp1p Activity
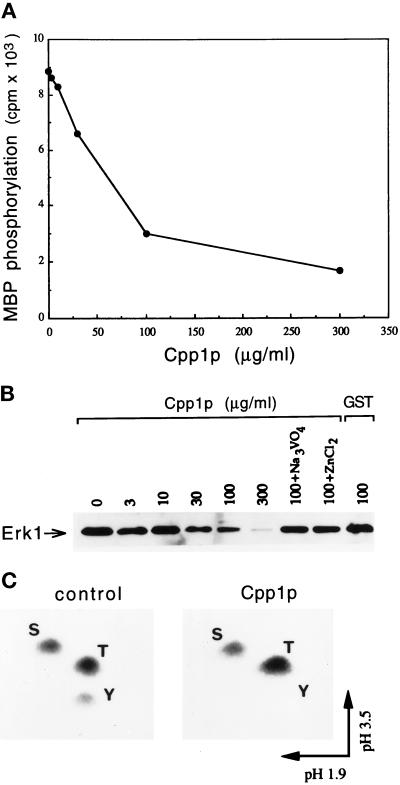
The indications that Cpp1p might act as a MAP kinase phosphatase prompted us to examine the ability of Cpp1p to dephosphorylate and inhibit the enzymatic activity of the mammalian MAP kinase Erk1 in vitro. Cpp1p was found to inhibit the phosphotransferase activity of Erk1 toward its substrate MBP in a dose-dependent manner (Figure 3A). The inactivation of the enzyme correlated with the dephosphorylation of the phosphotyrosine residue as shown by immunoblotting with an antiphosphotyrosine antibody (Figure 3B). The dephosphorylation of the tyrosine in Erk1 was inhibited by vanadate and Zn2+, which are common inhibitors of tyrosine phosphatases (Walton and Dixon, 1993). Our unpublished observations showed that the Cpp1p-active site mutant Cpp1pC516S could not dephosphorylate Erk1, but was still able to inhibit Erk1 activity. Similarly, site-directed mutants of the equivalent active site cysteine residue of the Chlamydomonas eugametos VHPTP13 phosphatase or the S. cerevisiae MSG5 phosphatase retain some ability to inactivate MAP kinases in vitro (Haring et al., 1995).
Figure 3.
Cpp1p dephosphorylates and inactivates MAP kinase in vitro. (A) Cpp1p inhibits the MBP kinase activity of Erk1. Purified active GST-Erk1 on beads was incubated with indicated concentrations of GST-Cpp1p. Following incubation, the beads were washed and assayed for MBP kinase activity. (B) Cpp1p dephosphorylates Erk1. Purified active GST-Erk1 was incubated in the absence or presence of indicated concentrations of GST-Cpp1p or GST, or with 100 μg/ml GST-Cpp1p in the presence of the phosphatase inhibitors sodium vanadate and zinc chloride. Proteins were resolved by SDS-PAGE and immunoblotted with antiphosphotyrosine antibody. (C) Cpp1p specifically dephosphorylates Erk1 on tyrosine. Erk1 was immunoprecipitated from 32P-labeled serum-stimulated Rat 1 cells and incubated in the absence (control) or presence (Cpp1p) of soluble GST-Cpp1p. Two-dimensional phosphoamino acid analysis of the 32P-labeled Erk1 is shown. S, phosphoserine; T, phosphothreonine; Y, phosphotyrosine.
Interestingly, Cpp1p was able to dephosphorylate phosphotyrosine residues but not phosphoserine or phosphothreonine residues of 32P-labeled activated-Erk1 (and Erk2) as determined by phosphoamino acid analysis (Figure 3C).
Derepression of the Yeast to Hyphal Switch in CPP1 Null Mutants
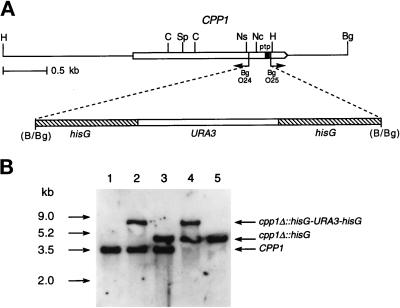
We constructed null mutants of the CPP1 gene by sequential gene disruption using a URA3 gene, flanked by hisG repeats, as a selectable marker (Figure 4).
Figure 4.
Deletion of CPP1 in C. albicans. (A) Deletion strategy and restriction map of the CPP1 gene. PCR with the divergent oligodeoxynucleotides O24 and O25 was used to delete the PTP-active site region (ptp) of CPP1. A hisG-URA3-hisG cassette was then inserted. Restriction sites are as follows: B, BamHI; Bg, BglII; C, ClaI; H, HindIII; Nc, NcoI; Ns, NsiI; Sp, SpeI. (B) Southern blot analysis of CPP1 disruptions with a HindIII-SpeI CPP1 fragment as a probe. Genomic DNA samples were digested with HindIII from the following strains: lane 1, CAI4 (ura3/ura3 CPP1/CPP1); lane 2, CP29 (ura3/ura3 CPP1/cpp1Δ::hisG-URA3-hisG); lane 3, CP29–1(ura3/ura3 CPP1/cpp1Δ::hisG): lane 4, CP29–1-7 (ura3/ura3 cpp1Δ::hisG-URA3hisG/cpp1Δ::hisG), and lane 5, CP29–1-7 L4 (ura3/ura3 cpp1Δ::hisG/ cpp1Δ::hisG).
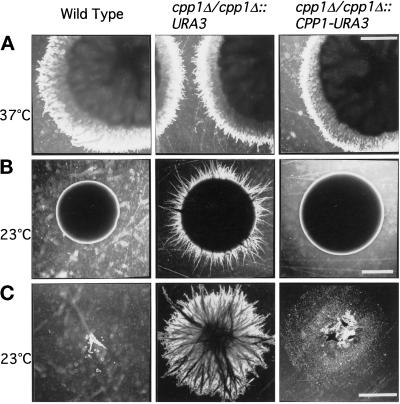
Previous work had shown that null mutants of the CST20, CPH1, and HST7 genes encoding C. albicans MAP kinase cascade components are defective in normal hyphal outgrowth at 37°C (Liu et al., 1994; Kohler and Fink, 1996; Leberer et al., 1996); this was assessed as the absence of filamentous growth from mature colony borders on solid spider agar in which mannitol, but not glucose, is used as a carbon source. We reasoned that if C. albicans Cpp1p has a target in this kinase pathway, it might also modulate hyphal development under these conditions. We examined whether Ura+ cpp1 null mutants would either derepress filamentous growth under hypha-inhibiting conditions (23°C) or hyperactivate filamentous growth under hypha-inducing conditions (37°C). Under hypha-inhibiting conditions (23°C), cpp1 null mutants formed hyphae which invaded the agar beneath colonies with either glucose (Figure 5, B and C) or mannitol as carbon sources, and on a wide variety of rich and defined solid media including Lee’s medium, YPD, YPM, and 10% serum. Our unpublished observations show that Ura− cpp1 null mutants also demonstrate invasive hyphal growth not seen in the Ura− CAI4 parent. On the other hand, cpp1 null mutants did not hyperactivate hyphal growth at 37°C on solid Spider medium containing mannitol or glucose (Figure 5A). In liquid culture, cpp1 null mutants had no effect on germ tube formation at 37°C and little effect was seen at room temperature. Identical phenotypes were obtained with two independent double disruptions, and phenotypes were reversed either by site-directed reintegration of the CPP1 gene (cpp1Δ/cpp1Δ::CPP1-URA3; Figure 5) or, as our unpublished results show, by high-copy expression of CPP1 from an ADH1 promoter. Phenotypes were also reversed by overexpression of the CPP1 gene with a deletion of the PEST region (Cpp1pΔPEST), or a deletion of a larger portion of the amino terminus, but were not reversed by overexpression of an active site mutant Cpp1pC516S. This suggests that the mutant phenotypes were the result of the loss of phosphatase activity. Taken together, these results indicate an important role of the Cpp1p phosphatase in repressing the yeast to hyphal transition from stationary C. albicans cells in contact with solid surfaces.
Figure 5.
Colonies of C. albicans cells grown on solid Spider medium containing glucose. Strain SC5314 (wild type); cpp1 null mutant CP29–1-7 (cpp1Δ/cpp1Δ::URA3); cpp1 null mutant into which the CPP1 gene has been replaced by homologous recombination CP29–1-7RI (cpp1Δ/cpp1Δ::CPP1-URA3). (A) Cells grown for 5 d at 37°C. (B) Cells grown for 8 d at 23°C. (C) Agar-penetrating growth remaining after surface washing of colonies grown for 8 d at 23°C from B (2× objective). Bars, 2 mm.
A Role of Cpp1p in Radial Mycelial Growth
In contrast to the derepression of hyphal growth under noninducing conditions, we observed, that when compared with controls, cpp1 null mutants formed a smaller zone of lateral hyphae when grown on carbon sources such as mannitol or raffinose which normally stimulated extensive radial hyphal growth from colony borders. This raised the possibility that absence of Cpp1p was detrimental to extended radial growth of mycelial colonies.
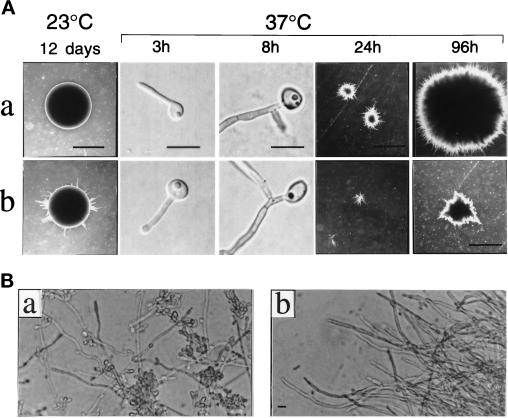
To test this hypothesis, we examined the growth of cells on solid serum media at 37°C (Figure 6). Wild-type C. albicans are stimulated on solid serum media to form germ tubes which develop into invasive hyphal colonies (Gow and Gooday, 1982). In addition, serum may provide an in vitro environment more closely related to that of animal hosts. Wild-type cells, cpp1 null mutants, and CPP1-reintegrants resembled each other in the extent of germ tube and hyphal elongation and septum formation up to 8 h after plating at 37°C on serum (Figure 6A; 3 and 8 h). Subsequent growth of the agar-imbedded mycelial colonies was reduced in cpp1 null mutants when compared with the CPP1 reintegrants (Figure 6A; 24 and 96 h) or the wild-type strain SC5314 (which our unpublished data demonstrated was phenotypically identical to the CPP1 reintegrants). In an independent experiment, we counted the number of hyphal tips in a colony (Gow and Gooday, 1982) to estimate the mycelial growth rates of cpp1 null mutants and wild-type SC5314 strains. Between 6 and 18 h, the rates of growth (μ) were 0.062 h−1 and 0.12 h−1 for the cpp1 null mutant and the wild type, respectively. Although the structure of cpp1 null hyphae appears to be normal, we observed fewer lateral buds on cpp1 null hyphae than on hyphae from CPP1 reintegrants (Figure 6B) or the wild-type strain SC5314 (which our unpublished data demonstrated was phenotypically identical to the CPP1 reintegrants), suggesting that lateral bud formation or accumulation is suppressed in cpp1 null mutants under these conditions. On the other hand, cpp1 null mutants were able to make lateral buds from hyphae growing at room temperature. This suggests that a cellular threshold above which hyphal development occurs and below which the yeast form is favored may be lowered in the cpp1 null mutant strain, and that lowering of this threshold may in addition have some detrimental effects on normal hyphal growth rates.
Figure 6.
Growth of C. albicans colonies on solid 10% serum medium at 23 and 37°C. a, CPP1 reintegrant strain CP29–1-7RI (cpp1Δ/cpp1Δ:: CPP1-URA3) and b, cpp1 null mutant strain CP29-1-7 (cpp1Δ/cpp1Δ::URA3). (A, 23°C) Yeast-form colonies were grown for 12 d and examined for hyphal outgrowth from colony borders (2× objective; bar, 2 mm). (A, 37°C) Single yeast-form cells spread on agar plates were monitored for germ tube formation and the onset of branching (3 and 8 h, Nomarski optics, 100× objective; bar, 15 μm), and for subsequent formation of invasive hyphal colonies (24 and 96 h; 2× objective; bar, 2 mm). (B) Mycelia from 96-h hyphal colony peripheries grown at 37°C and excised from agar for microscopy (40× objective; bar, 15 μm). Our unpublished observations showed that the wild-type strain SC5314 and the CPP1 reintegrant gave identical results.
Virulence Studies
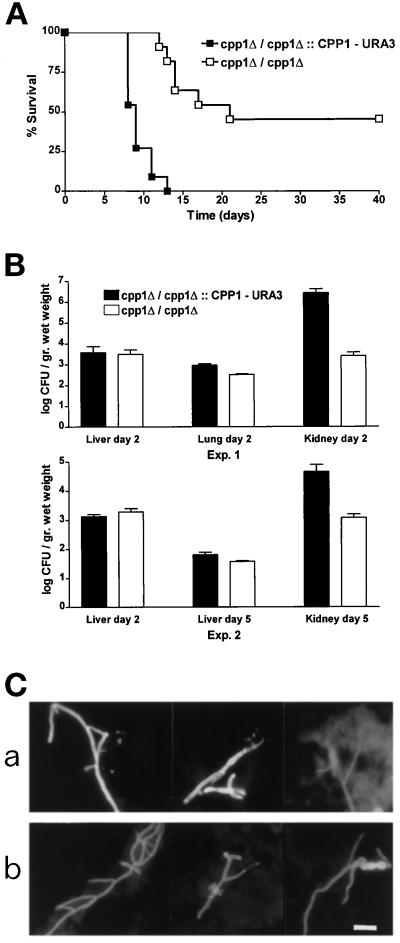
To determine the role of Cpp1p in virulence, mice were injected i.v. with cpp1 null mutants (cpp1Δ/ cpp1Δ::URA3) and cpp1 null mutants into which the CPP1 gene had been reintroduced (cpp1Δ/ cpp1Δ::CPP1-URA3), which in vitro and in mice acted like the wild-type strain. Mice were monitored for survival and for fungal burden of the liver, lungs, and kidneys. Infection with 5 × 105 stationary phase cells resulted in complete mortality by d 14 for strains containing the wild-type CPP1 gene (Figure 7A). In contrast, 45% of mice injected with an equal inoculum of cpp1 null mutant cells were still alive after 40 d. We thought it unlikely that the reduction in virulence of cpp1 null mutants was the result of a generalized growth defect because cpp1 null mutants in vitro have only a very minor (8%) difference in yeast form growth rates (71 min for the cpp1Δ/cpp1Δ::CPP1-URA3 strain used as a control and 65 min for the cpp1 null mutant strain (cpp1Δ/cpp1Δ::URA3); however, because of the striking mycelial growth defect seen on serum, we decided to examine the fungal burden of infected tissues.
Figure 7.
Virulence assays of BALB/c mice infected i.v. with either cpp1 null mutant (cpp1Δ/cpp1Δ::URA3) or CPP1 reintegrant (cpp1Δ/cpp1Δ::CPP1-URA3) C. albicans strains. (A) Survival curves of mice (n = 11) injected with 5 × 105 cells. (B) Fungal burden of infected tissues. (C) Fluorescence micrographs of Calcofluor white-stained hypha of C. albicans purified from KOH-solubilized kidneys 2 d after infection. a, CP29–1-7RI (cpp1Δ/cpp1Δ::CPP1-URA3); b, CP29–1-7 (cpp1Δ/cpp1Δ::URA3). Bar, 15 μm. Our unpublished results showed that survival curves of mice injected with the wild-type strain SC5314 and the CPP1 reintegrant were identical.
When C. albicans gains access to the blood stream it disseminates to all major organs including the liver, lungs, spleen, brain, and kidneys (Odds, 1988; Anaiffie et al., 1993). To determine whether reduced virulence of mice infected with cpp1 null mutants occurred because of decreased levels of tissue infection, we determined the fungal burden of the lungs, liver, and the kidneys in two independent experiments (both shown in Figure 7B). By 2 d, the kidneys were much more heavily infected (100- to 1000-fold greater CFUs) than the liver or lungs in mice infected with the CPP1 reintegrant as a wild-type control strain (Figure 7B, experiment 1, top graph), consistent with previous reports of the fungal burdens in these tissues 2 d after experimental infections (reviewed by Odds, 1988). On the other hand, the fungal burden of kidneys from mice infected with the cpp1 null mutant was two to three orders of magnitude lower than the fungal burden of kidneys from mice infected with the reintegrant. The filamentous form, and not the yeast form, of C. albicans predominated in kidneys infected with both strains (Figure 7C). Surprisingly, livers and lungs from animals infected with either strain showed no significant difference in fungal burden (Figure 7B) at d 2 (lungs and liver) or d 5 (liver). We did not examine fungal burden in the first few hours after infection during which the lungs and the liver typically have the highest fungal counts of any infected organ (Odds, 1988), but we did see the predicted (Odds, 1988) decline in fungal burden in the liver during the course of infection in mice infected with both strains (Figure 7B, experiment two, lower graph), suggesting that the mutant strain was being cleared normally from these organs. From these studies, we suggest that the kidney-specific reduction in fungal burden accounts for the greater number of mice surviving infections with the cpp1 null mutant strain.
Chromosomal Disruption of the MAP Kinase Homologue CEK1 Suppresses the cpp1 Null Mutant Phenotypes
To verify our assumption that Cpp1p acts on a MAP kinase affecting hyphal growth in C. albicans, we constructed a double mutant (Figure 8A) of CPP1 and the MAP kinase homologue CEK1 (Whiteway et al., 1992). CEK1 is a potential member of the MAP kinase cascade involved in C. albicans hyphal growth. Like Cst20p, Hst7p, and Cph1p, Cek1p has S. cerevisiae homologues (Fus3p and Kss1p) which function in the pheromone response MAP kinase cascade. We found that deletion of CEK1 completely suppressed the phenotypes of the cpp1 null mutants (Figure 8B), strengthening our hypothesis that Cpp1p functions as part of the MAP kinase cascade involved in C. albicans hyphal growth. Cpp1 null mutant phenotypes, including: derepressed hyphal development at ambient temperature; reduced growth rates of serum-induced mycelia, and the absence of lateral buds on serum at 37°C, were all reversed by deletion of CEK1 from the strain (Figure 8B, a–c).
DISCUSSION
Uncovering the genes involved in C. albicans pathogenicity is important for the discovery of new therapeutic strategies. Through functional complementation of S. cerevisiae genes, C. albicans homologues of components of a MAP kinase cascade involved in pheromone response and invasiveness in haploids and pseudohyphal growth in diploids (Gimeno et al., 1992; Liu et al., 1993; Roberts and Fink, 1994; Herskowitz, 1995) have been found which trigger the C. albicans yeast to hyphal switch in vitro (Liu et al., 1994; Malathi et al., 1994; Clark et al., 1995; Kohler and Fink, 1996; Leberer et al., 1996). The pheromone response pathway of S. cerevisiae is an extensively characterized and genetically tractable system in which to study signal transduction (reviewed in Herskowitz, 1995 and Leberer et al., 1997a). We previously identified C. albicans genes that when overexpressed blocked the function of the pheromone-response MAP kinase cascade in S. cerevisiae (Whiteway et al., 1992, 1993). Among the genes identified were CEK1, a MAP kinase homologue and CPP1, a gene with structural similarities to the VH1 family of protein tyrosine phosphatases. The VH1 family contains a growing number of dual- specificity MAP kinase-specific enzymes (reviewed in Keyse, 1995 and Zolnierowicz and Hemmings, 1996). We show here that Cpp1p is a putative MAP kinase phosphatase involved in determining cell-fate decisions in C. albicans by blocking the yeast to hyphal transition in the absence of a temperature cue. Genetic evidence in C. albicans suggests that Cpp1p mediates this repression by blocking activation of a MAP kinase cascade involved in hyphal growth. The Cpp1p MAP kinase phosphatase, like some components of the MAP kinase pathway that trigger hyphal growth, is involved in C. albicans virulence.
The net level and duration of MAP kinase activity in cells depends on the phosphorylation of critical tyrosine and threonine residues on MAP kinases by dual-specificity MAP kinase kinases and on the dephosphorylation of at least one of these residues by protein phosphatases (Keyse, 1995). Both dual- specificity and monospecificity phosphatases have been found to be involved in MAP kinase inactivation (Zhan et al., 1997; Millar et al., 1995), and both types of phosphatases together can coordinate the regulation of a single MAP kinase (Zhan et al., 1997). Several lines of evidence using heterologous systems in vivo or in vitro point to a role of Cpp1p as a MAP kinase phosphatase. First, overproduction of wild-type, but not an active site mutant of Cpp1p, interferes with pheromone-mediated cell-cycle arrest in S. cerevisiae. Genetic evidence suggests that Cpp1p acts at the level of the Fus3p (and Kss1p) MAP kinases of this pathway (Whiteway et al., 1993). These MAP kinases convey the signal from the pheromone receptor to a transcription factor and to the cell cycle machinery and, unlike other components of the cascade, require tyrosine phosphorylation for activity (Gartner et al., 1992). Indeed, Cpp1p is most closely related to the S. cerevisiae MAP kinase phosphatase, Msg5p, which acts in synergy with two other tyrosine-specific phosphatases, on the MAP kinases of this cascade (Doi et al., 1994; Zhan et al., 1997). Although Cpp1p is a member of the dual- specificity VH1 phosphatases, we found that under the conditions used, Cpp1p inactivated and dephosphorylated phosphotyrosine residues of the mammalian MAP kinases Erk1 and Erk2 in vitro, but did not dephosphorylate their phosphothreonine or phosphoserine residues. This is not entirely surprising in view of the finding that tyrosine can be the preferred in vitro substrate of other VH1 phosphatases (Denu et al., 1995; Groom et al., 1996). Because dephosphorylation of either residue of a MAP kinase can result in its inactivation, the tyrosine specificity of Cpp1p is consistent with it having a role in MAP kinase inactivation in C. albicans.
Our experiments in heterologous systems suggested to us that Cpp1p functions as a MAP kinase phosphatase in C. albicans, and that its physiological target could be the MAP kinase cascade that triggers C. albicans hyphal development on solid surfaces. Strains containing deletions of genes encoding the MAP kinase kinase kinase kinase, CST20, the MAP kinase kinase, HST7, and the transcription factor, CPH1, are defective in hyphal growth on solid substrata at physiological temperatures (Liu et al., 1994; Kohler and Fink, 1996; Leberer et al., 1996). Our unpublished results reveal the same phenotype for a strain with a deletion of the CEK1 MAP kinase gene (Whiteway et al., 1992). Cpp1 null mutants have an opposite phenotype: they form hyphae from mature colonies on solid substrata at ambient temperatures, and our unpublished observations also demonstrate the same phenotype with cells constitutively overexpressing the Cph1p transcription factor. Both of these strains derepress the yeast to hyphal transition under noninducing conditions and do not hyperactivate invasive hyphal growth at physiological temperatures. However, the strongest evidence that Cpp1p acts on this MAP kinase pathway is that disruption of the CEK1 MAP kinase gene completely suppresses the phenotypes of the cpp1 null mutant. These observations support a role of Cpp1p in determining cell fate through the inactivation of a Cek1p MAP kinase cascade, and by inference, through the control of the duration or intensity of MAP kinase response.
The potential importance of controlling the duration of MAP kinase activity for cellular responses is illustrated by the differential roles of transient versus sustained MAP kinase activities in proliferation and differentiation in rat pheochromocytoma PC12 cells (Wu et al., 1994; Fukuda et al., 1995; Marshall, 1995) and in transcriptional induction and cell cycle arrest of S. cerevisiae cells exposed to pheromone (Couvé and Hirsch, 1996). In addition, adaptation to pheromone, in the absence of a mating partner, involves shutting off MAP kinase activity to resume the cell cycle (Moore, 1984; Doi et al., 1994). Several phosphatases, including the tyrosine phosphatases Ptp2p and Ptp3p and the dual-specificity phosphatase Msg5p, are involved in this adaptive response (Doi et al., 1994; Zhan et al., 1997). Therefore, by controlling the duration of MAP kinase activity, MAP kinase-specific phosphatases have the potential to determine the end result of a MAP kinase cascade. In addition, MAP kinase phosphatases may also be key modulators of responses by helping to maintain low basal levels of MAP kinase activity (Zhan et al., 1997).
Because of the variety of responses in which components of MAP kinase cascades can function, it is possible that Cpp1p affects the same MAP kinase cascade or other similar MAP kinase cascades in other developmental processes or cellular responses in C. albicans. We have observed that although cpp1 null mutants were able to form germ tubes normally, they had a hyphal growth rate defect and formed few lateral buds under some hyphal-inducing conditions (at physiological rather than ambient temperature). The cpp1 null mutant hyphae may have a growth defect under these conditions because of the detrimental effects of higher than normal levels of hyphal-inducing cellular activities resulting from the Cek1p MAP kinase cascade being inappropriately hyperactive. Removal of the CEK1 gene suppresses the hyphal growth rate defect of cpp1 null mutants, suggesting that this is a plausible situation. Because cpp1 null mutant hyphae appear to be less capable of differentiating or accumulating new yeast cells, under these same conditions, it is also possible that the development of lateral buds from hyphae could represent an adaptive response to hyphal-inducing stimuli. Once again, this phenotype is suppressed in the cpp1/cek1 double null mutants. In cpp1 null mutants the hyphal-inducing signals may remain too high for cells to resume growing in the yeast form. Indeed, overexpression of either Msg5p or Cpp1p in S. cerevisiae promotes adaptation and budding growth by shutting off the pheromone response pathway (Doi et al., 1994; Whiteway et al., 1993). Reentry into the budding cell cycle in C. albicans could parallel the process of adaptation to pheromone in S. cerevisiae.
Tyrosine phosphatases have been shown to play a role in the pathogenicity of the bacterial genus Yersinia which includes species responsible for enteric diseases, septicemia, and bubonic plague, and of the viral genus Orthopoxvirus which contains the causative agent of smallpox (Bliska et al., 1991; Hakes et al., 1993; reviewed in Ninfa, 1994). In addition, we have found that the Cpp1p MAP kinase phosphatase contributes to the pathogenicity of C. albicans. Cpp1 null mutants demonstrate a dramatic reduction in virulence and, in addition, show reduced fungal load in the kidneys, a typical secondary site of infection. The reduction in virulence may well be attributed to decreased infection of the kidneys, since during experimental candidiasis the kidneys are the most highly infected and abscessed organs in the body (Odds, 1988). What accounts for the reduction in fungal burden in the kidneys? This question is not easily answered in view of the different phenotypes of cpp1 null mutants in vitro; however, it may simply be that cpp1 null mutant mycelia have a growth rate defect specifically in the kidneys resembling that seen under physiological conditions in vitro, although one can also envision that the absence of lateral buds in cpp1 null mutants under physiological conditions could make dissemination from primary sites of infection such as the lungs and liver to secondary sites of infection such as the kidneys more difficult. Other possibilities also exist, such as the cpp1 null mutant strain being more susceptible to kidney-specific defense mechanisms. Although this study does not answer the question of what the role of the hyphal form of C. albicans plays in pathogenicity, our data are consistent with studies that report that C. albicans morphological mutant strains that exist in hyphal forms at ambient temperatures are avirulent, as are those mutant strains which are unable to undergo the yeast to hyphal switch (Sobel et al., 1984; Hubbard et al., 1986; Gil et al., 1990; Leberer et al., 1997b). One of the latter class of mutants is a null mutant of a homologue (CaCLA4) of the S. cerevisiae CLA4 gene [a relative of STE20 (CST20)]; although having no growth rate defect, Cacla4 null mutants cannot make hyphae and are completely avirulent, reinforcing the idea that hyphae are required for virulence.
Because the current state of understanding of C. albicans virulence is rudimentary, it is not yet possible to pinpoint the precise molecular basis of Cpp1p-mediated virulence; however, it is through the isolation of genes and the detailed analysis of phenotypes, coupled with virulence studies, that progress can be made in defining some of the elements that define pathogenicity. The Cpp1p phosphatase may indeed serve as a useful target for therapeutics against systemic disease, the most devastating and least treatable form of fungal infection (Odds, 1988), especially in view of its limited structural similarity to mammalian counterparts of this class of enzymes. Most important, the present study demonstrates the involvement of a tyrosine phosphatase in fungal disease and provides a demonstration of the important role of a phosphatase of the VH1 family in cell fate decisions.
ACKNOWLEDGMENTS
We thank D. Harcus for discussion and technical consultations, T. Leeuw for help with microscopy, E. Leberer, A. Nantel, and C. Wu for helpful discussions and for reviewing the manuscript, A. Mcarther for help with alignments and phylogenetic analyses, R. Swoboda, R. Barton, M. Raymond, and J.-C. Scimeca for helpful discussions, and Candida news. Thanks to W. Fonzi and E. Leberer for strains and plasmids. C.C. was supported by a Canadian Government Laboratory Visiting Fellowship with funds from Glaxo and a Medical Research Council of Canada postdoctoral fellowship. S.M. is a scholar of the Medical Research Council of Canada. K.S. was supported by grant Deutsche Forschungsgemeinschaft Schr 450/2-1. This work was supported in part by a grant from the National Cancer Institute of Canada. This is National Research Council of Canada Publication No. NRC39975.
REFERENCES
- Alessi DR, Smythe C, Keyse SM. The human CL100 gene encodes a Tyr/Thr-protein phosphatase which potently and specifically inactivates MAP kinase and suppresses its activation by oncogenic ras in Xenopus oocyte extracts. Oncogene. 1993;8:2015–2020. [PubMed] [Google Scholar]
- Anaiffie EJ, Pinczowfski H, Louria B. Candidiasis: Pathogenesis, Diagnosis and Treatment. G.P. Bodey, New York: Raven Press; 1993. Candida infections in experimental animals; pp. 43–58. [Google Scholar]
- Bliska JB, Guan KL, Dixon JE, Falkow S. Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc Natl Acad Sci USA. 1991;88:1187–1191. doi: 10.1073/pnas.88.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffo J, Herman MA, Soll DR. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia. 1984;85:21–30. doi: 10.1007/BF00436698. [DOI] [PubMed] [Google Scholar]
- Clark KL, Feldmann PJ, Dignard D, Larocque R, Brown AJ, Lee MG, Thomas DY, Whiteway M. Constitutive activation of the Saccharomyces cerevisiae mating response pathway by a MAP kinase kinase from Candida albicans. Mol Gen Genet. 1995;249:609–621. doi: 10.1007/BF00418030. [DOI] [PubMed] [Google Scholar]
- Couvé A, Hirsch JP. Loss of sustained Fus3p kinase activity and the G1 arrest response in cells expressing an inappropriate pheromone receptor. Mol Cell Biol. 1996;16:4478–4485. doi: 10.1128/mcb.16.8.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denu JM, Zhou G, Wu L, Zhao R, Yuvaniyama J, Saper MA, Dixon JE. The purification and characterization of a human dual-specific protein tyrosine phosphatase. J Biol Chem. 1995;270:3796–3803. doi: 10.1074/jbc.270.8.3796. [DOI] [PubMed] [Google Scholar]
- Doi K, Gartner A, Ammerer G, Errede B, Shinkawa H, Sugimoto K, Matsumoto K. MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 1994;13:61–70. doi: 10.1002/j.1460-2075.1994.tb06235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman K, Carrasco D, Gruda M, Ryan C, Lira SA, Bravo R. Disruption of the erp/mkp-1 gene does not affect mouse development: Normal MAP kinase activity in ERP/MKP-1-deficient fibroblasts. Oncogene. 1996;13:925–931. [PubMed] [Google Scholar]
- Fidel P, Jr, Sobel JD. The role of cell-mediated immunity in candidiasis. Trends Microbiol. 1994;2:202–206. doi: 10.1016/0966-842x(94)90112-i. [DOI] [PubMed] [Google Scholar]
- Fischer EH, Charbonneau H, Tonks NK. Protein tyrosine phosphatases: a diverse family of intracellular and transmembrane enzymes. Science. 1991;253:401–406. doi: 10.1126/science.1650499. [DOI] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Gotoh Y, Tachibana T, Dell K, Hattori S, Yoneda Y, Nishida E. Induction of neurite outgrowth by MAP kinase in PC12 cells. Oncogene. 1995;11:239–244. [PubMed] [Google Scholar]
- Gartner A, Nasmyth K, Ammerer G. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev. 1992;6:1280–1292. doi: 10.1101/gad.6.7.1280. [DOI] [PubMed] [Google Scholar]
- Gil C, Pomes R, Nombela C. Isolation and characterization of Candida albicans morphological mutants derepressed for the formation of filamentous hypha-type structures. J Bacteriol. 1990;172:2384–2391. doi: 10.1128/jb.172.5.2384-2391.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum AM, Tsay EY, H, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Gow NA, Gooday GW. Growth kinetics and morphology of colonies of the filamentous form of Candida albicans. J Gen Microbiol. 1982;128:2187–2194. doi: 10.1099/00221287-128-9-2187. [DOI] [PubMed] [Google Scholar]
- Groom LA, Sneddon AA, Alessi DR, Dowd S, Keyse SM. Differential regulation of the MAP, SAP, and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J. 1996;15:3621–3632. [PMC free article] [PubMed] [Google Scholar]
- Guan K, Broyles SS, Dixon JE. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature. 1991;350:359–362. doi: 10.1038/350359a0. [DOI] [PubMed] [Google Scholar]
- Guan K, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Guan K, Hakes DJ, Wang Y, Park HD, Cooper TG, Dixon JE. A yeast protein phosphatase related to the vaccinia virus VH1 phosphatase is induced by nitrogen starvation. Proc Natl Acad Sci USA. 1992;89:12175–12179. doi: 10.1073/pnas.89.24.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Butch E. Isolation and characterization of a novel dual specific phosphatase, HVH2, which selectively dephosphorylates the mitogen-activated protein kinase. J Biol Chem. 1995;270:7197–7203. doi: 10.1074/jbc.270.13.7197. [DOI] [PubMed] [Google Scholar]
- Hakes DJ, Dixon JE. New vectors for high level expression of recombinant proteins in bacteria. Anal Biochem. 1992;202:293–298. doi: 10.1016/0003-2697(92)90108-j. [DOI] [PubMed] [Google Scholar]
- Hakes DJ, Martell KJ, Zhao W-G, Massung RF, Esposito JJ, Dixon JE. A protein phosphatase related to the Vaccinia virus VH1 is encoded in the genomes of several orthopoxviruses and a baculovirus. Proc Natl Acad Sci USA. 1993;90:4017–4021. doi: 10.1073/pnas.90.9.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring MA, Siderius M, Jonak C, Hirt H, Walton KM, Musgrave A. Tyrosine phosphatase signalling in a lower plant: cell-cycle and oxidative stress-regulated expression of the Chlamydomonas eugametos VH-PTP13 gene. Plant J. 1995;7:981–988. doi: 10.1046/j.1365-313x.1995.07060981.x. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Alford WJ, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:GC17–GC26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hubbard MJ, Markie D, Poulter RT. Isolation and morphological characterization of a mycelial mutant of Candida albicans. J Bacteriol. 1986;165:61–65. doi: 10.1128/jb.165.1.61-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Bottaro DP, Chan A, Miki T, Aaronson SA. Expression cloning of a human dual-specificity phosphatase. Proc Natl Acad Sci USA. 1992;89:12170–12174. doi: 10.1073/pnas.89.24.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyse SM. An emerging family of dual specificity MAP kinase phosphatases. Biochim Biophys Acta. 1995;1265:152–160. doi: 10.1016/0167-4889(94)00211-v. [DOI] [PubMed] [Google Scholar]
- Keyse SM, Ginsburg M. Amino acid sequence similarity between CL100, a dual-specificity MAP kinase phosphatase and cdc25. Trends Biochem Sci. 1993;18:377–378. doi: 10.1016/0968-0004(93)90092-2. [DOI] [PubMed] [Google Scholar]
- Kohler JR, Fink GR. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TD, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kurtz MB, Cortelyou MW, Kirsch DR. Integrative transformation of Candida albicans, using a cloned Candida ADE2 gene. Mol Cell Biol. 1986;6:142–149. doi: 10.1128/mcb.6.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak SP, Dixon JE. Multiple dual specificity protein tyrosine phosphatases are expressed and regulated differentially in liver cell lines. J Biol Chem. 1995;270:1156–1160. doi: 10.1074/jbc.270.3.1156. [DOI] [PubMed] [Google Scholar]
- Leberer E, Harcus D, Broadbent ID, Clark KL, Dignard D, Ziegelbauer K, Schmidt A, Gow NA, Brown AJ, Thomas DY. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Thomas DY, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997a;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas DY. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr Biol. 1997b;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- Lee KL, Buckley HR, Campbell CC. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Malathi K, Ganesan K, Datta A. Identification of a putative transcription factor in Candida albicans that can complement the mating defect of Saccharomyces cerevisiae ste12 mutants. J Biol Chem. 1994;269:22945–22951. [PubMed] [Google Scholar]
- Marshall CJ. Specificity of tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Martell KJ, Seasholtz AF, Kwak SP, Clemens KK, Dixon JE. hVH-5: a protein tyrosine phosphatase abundant in brain that inactivates mitogen-activated protein kinase. J Neurochem. 1995;65:1823–1833. doi: 10.1046/j.1471-4159.1995.65041823.x. [DOI] [PubMed] [Google Scholar]
- Meloche S. Cell cycle reentry of mammalian fibroblasts is accompanied by the sustained activation of p44mapk and p42mapk isoforms in the G1 phase and their inactivation at the G1/S transition. J Cell Physiol. 1995;163:577–588. doi: 10.1002/jcp.1041630319. [DOI] [PubMed] [Google Scholar]
- Meloche S, Pagès G, Pouysségur J. Functional expression and growth factor activation of an epitope-tagged p44 Mitogen-activated protein kinase, p44mapk. Mol Biol Cell. 1992;3:63–71. doi: 10.1091/mbc.3.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JB, Buck V, Wilkinson MG. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- Moore SA. Yeast cells recover from mating pheromone alpha factor-induced division arrest by desensitization in the absence of alpha-factor destruction. J Biol Chem. 1984;259:1004–1010. [PubMed] [Google Scholar]
- Ninfa EG. Protein tyrosine phosphatases in disease processes. Trends Cell Biol. 1994;4:427–430. doi: 10.1016/0962-8924(94)90103-1. [DOI] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis: A Review and Bibliography. 2nd ed. London: Bailliere Tindall; 1988. [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Rohan PJ, Davis P, Moskaluk CA, Kearns M, Krutzsc H, Siebenlist U, Kelly K. PAC-1: a mitogen-induced nuclear protein tyrosine phosphatase. Science. 1993;259:1763–1766. doi: 10.1126/science.7681221. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sambrooke J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sherwood J, Gow NA, Gooday GW, Gregory DW, Marshall D. Contact sensing in Candida albicans: a possible aid to epithelial penetration. J Med Vet Mycol. 1992;30:461–469. doi: 10.1080/02681219280000621. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JD, Muller G, Buckley HR. Critical role of germ tube formation in the pathogenesis of candidal vaginitis. Infect Immun. 1984;44:576–580. doi: 10.1128/iai.44.3.576-580.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR. The regulation of cellular differentiation in the dimorphic yeast Candida albicans. Bioessays. 1986;5:5–11. doi: 10.1002/bies.950050103. [DOI] [PubMed] [Google Scholar]
- Soll DR, Bedell GW, Brummel M. Zinc and regulation of growth and phenotype in the infectious yeast Candida albicans. Infect Immun. 1981;32:1139–1147. doi: 10.1128/iai.32.3.1139-1147.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KM, Dixon JE. Protein tyrosine phosphatases. Annu Rev Biochem. 1993;62:101–120. doi: 10.1146/annurev.bi.62.070193.000533. [DOI] [PubMed] [Google Scholar]
- Whiteway M, Csank C, Thomas DY. Dominant negative selection of heterologous genes in yeast. Methods. 1993;5:110–115. [Google Scholar]
- Whiteway M, Dignard D, Thomas DY. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc Natl Acad Sci USA. 1992;89:9410–9414. doi: 10.1073/pnas.89.20.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Lau LF, Sturgill TW. Rapid deactivation of MAP kinase in PC12 cells occurs independently of induction of phosphatase MKP-1. FEBS Lett. 1994;353:9–12. doi: 10.1016/0014-5793(94)01000-5. [DOI] [PubMed] [Google Scholar]
- Zhan X-L, Deschenes RJ, Guan K-L. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 1997;11:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]
- Zolnierowicz S, Hemmings BA. Protein phosphatases on the piste. Trends Cell Biol. 1996;6:359–362. doi: 10.1016/0962-8924(94)90012-4. [DOI] [PubMed] [Google Scholar]