Abstract
The factors that mediate chromosomal rearrangement remain incompletely defined. Among regions prone to structural variant formation, chromosome 6p25 is one of the few in which disease-associated segmental duplications and segmental deletions have been identified, primarily through gene dosage attributable ocular phenotypes. Using array comparative genome hybridization, we studied ten 6p25 duplication and deletion pedigrees and amplified junction fragments from each. Analysis of the breakpoint architecture revealed that all the rearrangements were non-recurrent, and in contrast to most previous examples the majority of the segmental duplications and deletions utilized coupled homologous and non-homologous recombination mechanisms. One junction fragment exhibited an unprecedented 367 bp insert derived from tandemly arranged breakpoint elements. While this accorded with a recently described replication-based mechanism, it differed from the previous example in being unassociated with template switching, and occurring in a segmental deletion. These results extend the mechanisms involved in structural variant formation, provide strong evidence that a spectrum of recombination, DNA repair and replication underlie 6p25 rearrangements, and have implications for genesis of copy number variations in other genomic regions. These findings highlight the benefits of undertaking the extensive studies necessary to characterize structural variants at the base pair level.
INTRODUCTION
The contribution that chromosomal anomalies make to human disease was, until relatively recently, under-appreciated. Introduction of array technologies has revealed that rearrangements below the resolution of standard karyotyping are exceedingly common (1,2), with sub-microscopic structural genomic variants underlying a substantial proportion of human genetic variation (3,4) and disease (5–8). Estimates that such variants account for 12% or more of our genomes (2,9) are likely conservative due to the resolution of the arrays used, and these technologies' general inability to detect most balanced rearrangements (9). Despite their importance to a wide range of human diseases (5–8), structural variants have been defined at the base pair level in only a small number of cases, thereby restricting our understanding of the underlying mechanisms involved. As the majority of characterized rearrangements were identified prior to the advent of comparative genomic hybridization (CGH) and single-nucleotide polymorphism (SNP) arrays, it remains to be determined whether the mechanisms involved in their formation are representative of the generally smaller genomic variants being detected with array approaches.
Since copy number alteration predominates among the mechanisms by which genomic rearrangements induce phenotypes (10), copy number variation (CNV) is becoming the preferred descriptor for segmental gains or losses of chromosomal material, and as such will be used in this paper. CNVs have particular relevance in ocular genetics due to the eye's well-documented sensitivity to the effects of altered gene dosage (11–19). Combined with the organ's accessibility to detailed phenotyping, these factors provide unique opportunities to use CNV-induced phenotypes to elucidate the functions of dosage-sensitive genes (12,16,18,20). Among dosage-sensitive ocular developmental genes, the forkhead box transcription factor FOXC1, located on chromosome 6p25, is noteworthy for the frequency of its involvement in segmental and telomeric chromosomal rearrangements (16,18,21–24). Increased or decreased FOXC1 copy number causes the Axenfeld–Rieger Syndrome (ARS) spectrum of glaucoma-associated ocular anomalies (16,18,22,24), and occurs at a similar prevalence to FOXC1 mutation (25). Although previous studies localized 6p25 segmental anomalies using microsatellite marker genotyping or large-insert bacterial clones as probes for FISH, the resulting broad intervals (150–200 kb) precluded determining the causes of these rearrangements (22,26,27). Further investigation was warranted by the unique architecture of 6p25, which contains a triplet of forkhead genes (FOXC1, FOXF2 and FOXQ1). Such tandemly arranged paralogs are not observed in the other five regions where segmental duplications and deletions cause human disease {Charcot–Marie–Tooth disease type 1A23 (28) [dup (17)(p12p12)] and Hereditary Neuropathy with liability to pressure palsies (29) [del (17)(p12p12)]; Potocki–Lupski [dup (17)(p11.2p11.2)] and Smith Magenis syndromes (30) [del (17)(p11.2p11.2)]; dup (22)(q11.2q11.2) and Velocardiofacial syndrome (31,32); dup (7)(q11.23q11.23) and Williams–Beuren syndrome (33); dup (15)(q11q13) (34) and Prader–Willi/Angelman syndromes (35)}.
In order to characterize the 6p25 rearrangements at the base pair level, amplicon-based, and subsequently commercial oligonucleotide CGH arrays were used to comprehensively analyze the 6p25 region in the largest ARS pedigree collection worldwide (16,22,24–26). The findings revealed a novel mechanistic spectrum involved in the genesis of these CNVs, including junction fragment architecture incompatible with the two primary mechanisms [non-allelic homologous recombination (NAHR) and non-homologous end joining (NHEJ)] that underlie characterized genomic rearrangements. These results extend the mechanisms that generate CNV with implications for CNV formation in other parts of the genome.
RESULTS
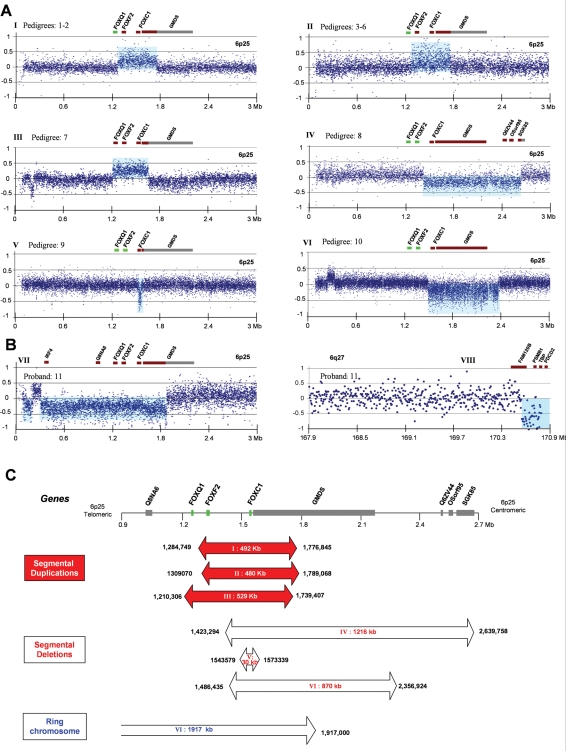
In order to determine the extent of the 6p25 structural variants (pedigrees #1–7, 6p25 segmental duplication; #8–10, 6p25 segmental deletion; #11, ring chromosome 6), DNA samples from a subset of the probands were initially hybridized to an in-house PCR amplicon-based array. However, the limited extent of this array, not encompassing both breakpoints in pedigrees #7 and #10 (data not shown), was a factor in subsequent adoption of a commercial array. The latter accurately defined the extent of the rearrangements in eight of the nine pedigrees (duplication: #1, 3–5; deletion: #8–10; ring: #11) in which it was used (Fig.1). These data facilitated amplification of junction fragments with long-range (but not Alu-based) PCR, in six of the seven segmental duplication pedigrees (#1–6) as well as in all deletion pedigrees (#8–10). The identical junction fragment sizes observed in the duplication pedigrees #1 and 2 (2.5 kb) and #3–6 (2.1 kb) were compatible with either recurrent rearrangements or founder effects. Bioinformatic studies, including Pipmaker analysis of the telomeric 2.5 Mb of chromosome 6p25 did not reveal a significant level of low-copy repeats (LCRs) (data not shown).
Figure 1.
(A and B) Array CGH results from patients with Axenfeld–Rieger phenotypes illustrating the extent of the structural genomic variants relative to the position of the forkhead paralogs and neighboring genes. Duplicated or deleted genes are depicted in brown (others gray): to emphasize the constant (FOXC1) and varying (FOXF2/FOXQ1) involvement in the rearrangements, forkhead genes present at normal copy number are shown in green. The segmental duplications extend 492 (A, I pedigrees #1–2), 480 (A, II #3–6) and 529 kb (A, III #7), respectively, the segmental deletions (A, IV #8, V #9 and VI #10) 1216, 30 and 870 kb, respectively, while the ring chromosome (B) lacks ∼2452 kb of sequence from 6p25 and 6q27. (C) Schematic overview of the extent of each 6p25 chromosomal anomaly (red and open arrows) [x-axis chromosome position, NCBI build 35, y-axis log 2 Cy3:Cy5 ratio), the copy number variant region located at 0.2 Mb represents a recognized polymorphism].
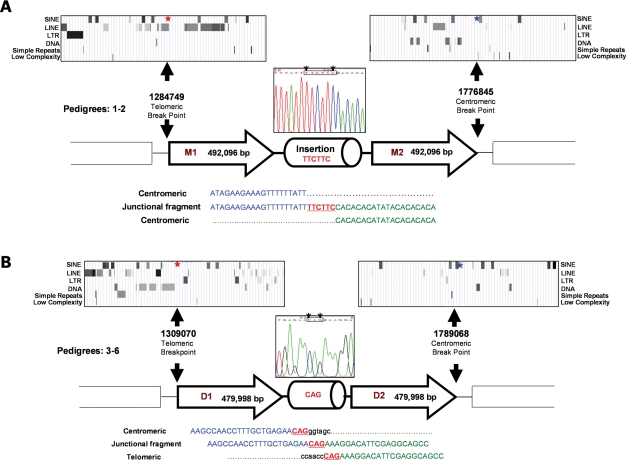
Sequence analysis of the junction fragment from pedigrees #1 and #2 revealed identical head to tail arranged ∼492 kb duplicons, separated by a 6 bp insertion (Fig. 2A). The telomeric breakpoint lies within a long interspersed nuclear element (LINE), located within 3 kb of near-contiguous repetitive elements, whereas the centromeric breakpoint lies 381 bp away from a low complexity repeat that is located within a 10 kb region of repetitive sequence. The maximum repetitive element composition in a 10 kb region encompassing the telomeric and centromeric breakpoints is 71 and 45%, respectively, compared with ∼35% genome-wide (36). Four pedigrees (#3–6) share similarly oriented, but smaller 480 kb duplicated segments, with 3 bp of microhomology at the junction (Fig. 2B). The breakpoints, located ∼25 kb from those in #1 and 2, lie close to repetitive elements [telomeric: MER1_type repeat (191 bp); centromeric: between two short interspersed nuclear elements (SINE), AluSx and MIRb, respectively]. The 10 kb region adjacent to the telomeric breakpoint is noteworthy for the high (94%) repetitive element density (compared with centromeric 28%), indeed a near contiguous (97%) 13.8 kb array of repeats (including 6 SINES, 10 LINES, 1 LTR and other elements) lies adjacent to the telomeric breakpoint.
Figure 2.
Summary of genomic architecture in 6p25 segmental duplication pedigrees (#1–6): the genomic architecture of the 20 kb region flanking the breakpoints is shown above the respective breakpoints, with the distribution of repeat elements inset above (UCSC Genome Browser). (A) Pedigrees (#1–2) have an identical head to tail arrangement of a 492 096 bp duplicated segment separated by a 6 bp insertion. The telomeric breakpoint (red star) lies within a LINE, whereas the centromeric breakpoint (blue star) lies 381 bp from a low complexity repeat. (B) Pedigrees (#3–6) share similarly oriented 479 998 bp duplicated motifs: note the 3 bp microhomology (CAG) present at the junction between the centromeric (blue) and telomeric (green) sequence. The telomeric breakpoint lies 191 bp from a MER1_type repeat, while the centromeric breakpoint lies between two SINES (For clarity, descriptions of sequence location in all figures are made relative to the telomere/centromere as opposed to the telomeric/centromeric end of each duplicon).
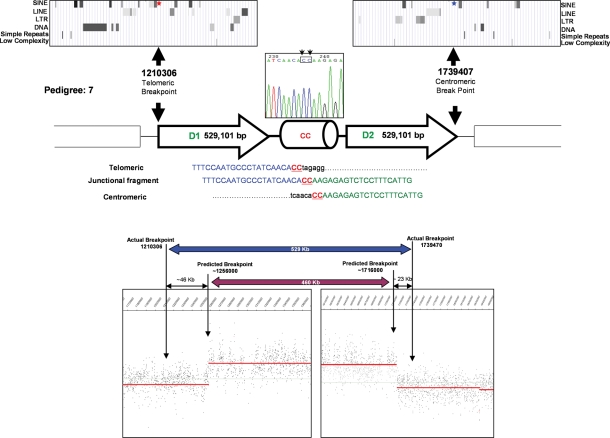
Despite clear prediction of the breakpoint positions from a mean 100 bp probe density customized CGH array, no junction fragment was initially amplified in #7 (Fig. 3). To assess whether the duplicons were separated by >20 kb, additional primers extending up to 70 kb from the predicted breakpoints were used. One primer combination yielded a junction fragment, with subsequent sequencing revealing significant discrepancies (telomeric 46 kb and centromeric 23 kb) between the actual and predicted breakpoints (Fig. 3). Quantitative PCR (qPCR) with multiple amplicons spanning these two intervals accorded with the array CGH result (data not shown). While these intervals had no significant homology to other chromosomal regions, the telomeric interval exhibited a 40% repetitive element composition (centromeric 15%). The head to tail orientated 529 kb duplicated segment (Fig. 3) of pedigree #7 is larger than the previous examples, and exhibits a 2 bp microhomology at the junction and the breakpoints lie adjacent to AluJo and AluJb repeat elements (telomeric 47 bp, centromeric 735 bp) that share 90% (135/150) sequence homology. The prevalence of repeat sequences in the telomeric and centromeric 10 kb regions encompassing the breakpoints is 59 and 42%, respectively.
Figure 3.
Montage illustrating the genomic architecture of the seventh segmental duplication, with the actual breakpoints, and those predicted by array CGH, shown below. Note the: head to tail orientated 529 101 bp duplicated segments; location of breakpoints adjacent to Alu elements; and 2 bp microhomology (CC) at the junction. Appreciable discrepancies exist between the positions of the actual and predicted breakpoints, despite clear CGH data from closely spaced probes. The variation in signal intensity observed is representative of the 6p25 hybridization results achieved with other samples and reflects natural differences in hybridization efficiency.
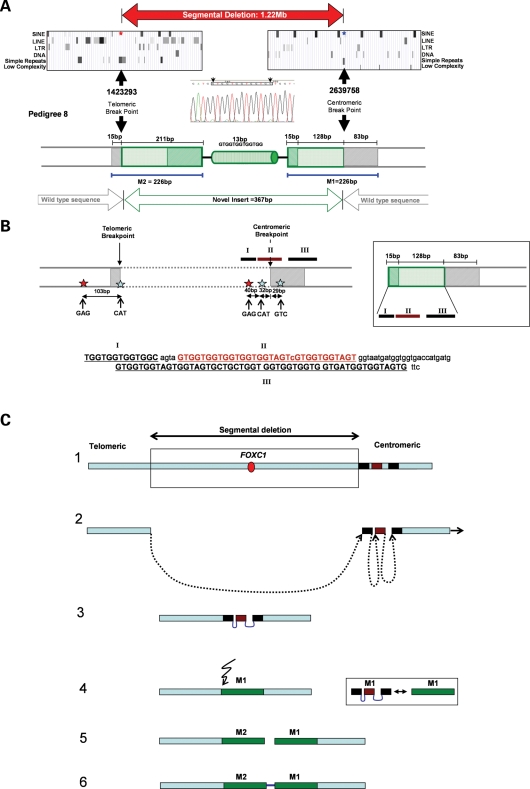
In contrast to the 480–529 kb duplications (#1–7), segmental deletion #8 is 1.22 Mb in size with breakpoints separated by insertion of an unexpectedly large 367 bp of novel sequence. This insert comprises two 100% homologous motifs (M1 and M2), separated by a 13 bp DNA segment (Fig. 4A). Motif M1 consists of three portions, the largest of which at 128 bp of primarily (GTG)n repeats is revealed by BLAT analysis to be homologous to three adjacent segments of sequence spanning the centromeric breakpoint (Fig. 4BI–III). The remaining motif portions of 15 and 83 bp are wild-type sequence that respectively lie at the telomeric and centromeric breakpoints (Fig. 4A). Motif M2 comprises 211 bp that is identical to M1 together with the same 15 bp of telomeric wild-type sequence (gray, Fig. 4A and C). The breakpoints are located within similarly sized (GTG)n simple repeats (telomeric 183 bp, centromeric 180 bp) with the former adjacent to three (GTG)n or (ATGGTG)n elements that extend for 889 bp. In addition, topoisomerase I consensus cleavage (CAT/GTC), and DNA polymerase α pause sites (GAG) are present at or adjacent to both regions (Fig. 4B).
Figure 4.
(A) Summary of the segmental deletion's (#8) genomic architecture illustrating the composition of the 226 bp duplicated motifs (M1, M2), with the 20 kb region flanking each breakpoint (filled arrows) shown above. The motifs comprise novel (green) and wild-type sequence (gray), with the pattern of shading used to highlight the 100% homology of these motifs. (B) Illustration of the 100% sequence homology between motif segments and regions adjacent to both breakpoints: the location of DNA polymerase α pause sites and consensus cleavage sites for Topo-isomerase I are depicted with red and blue stars, respectively. Note the corresponding composition, order and orientation of the 128 bp motif portion and segments of sequence (I–III) that straddle the centromeric breakpoint (identical bases in capitals). (C) Diagrammatic representation of the 6p25 region (1) and the proposed steps in generating this rearrangement: fork stalling (2), formation of motif M1 (3), double strand break and strand dependent synthesis generating motif M2 (4–6).
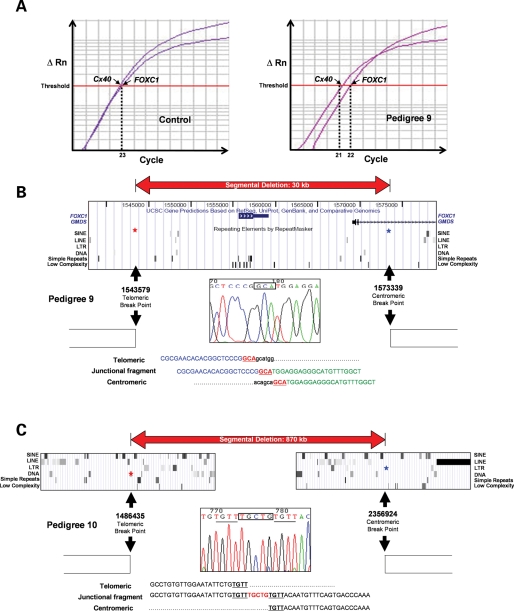
qPCR screening of an ARS patient panel (n = 32) unattributable to FOXC1 or PITX2 mutation, identified one sample (#9) with reduced FOXC1 copy number. Array CGH confirmed this, defining a small segmental deletion (29.76 kb) (Fig. 5) in contrast to the previous much larger CNVs (∼480–1220 kb). Sequencing of the junction fragment demonstrated 3 bp of microhomology, with minimal repetitive elements present in the 10 kb peri-breakpoint regions (2.2 and 0%, respectively) (Fig. 5). This deletion encompasses FOXC1 together with a small part of GDP-mannose-4,6-dehydratase (GMDS). Sequence analysis of the junction fragment from the final segmental deletion pedigree (#10) demonstrated the breakpoints of this 870 kb deletion to be separated by a 5 bp insertion (Fig. 5C). Four base pairs of microhomology exist between the sequences adjacent to the breakpoints, which lie within (telomeric) or adjacent (centromeric) to repetitive elements.
Figure 5.
Summary of genomic architecture of #9 and #10. (A) Schematic representation of qPCR-identified segmental deletion (#9), note the lag in the qPCR's exponential phase, with FOXC1 reaching threshold one cycle after the Connexin40 (Cx40) control. (B) Architecture of #9 segmental deletion breakpoint, illustrating 3 bp microhomology at junction. (C) Genomic architecture of #10 segmental deletion illustrating the sequence homology (underlined) at both breakpoints and the 5 bp insertion (TGCTG). Note that part of this insert (CTG) and the contiguous 4 bp (TGTT) are identical to the 7 bp (CTGTGTT) present at the telomeric breakpoint.
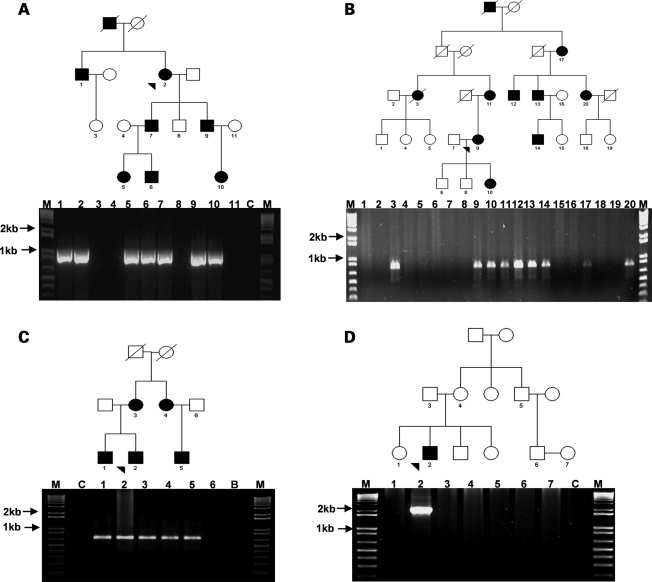
The junction fragments segregated with the disease phenotype in all the segmental anomaly pedigrees (Fig. 6A–C and unpublished data), and the absence of a junction fragment in either parent of the proband (#9) is compatible with gonadosomal mosaicism (Fig. 6D). Five of 19 microsatellite markers or SNPs studied were informative (Table 1). This demonstrated that pedigrees #1 and 2, and separately #3–6 share common haplotypes across the duplicated segment, in addition to identically sized segmental duplications and breakpoint architectures (#1 and 2: 492 096 bp with a 6 bp insertion; #3–6: 479 998 bp with a 3 bp microhomology), confirming that they are ancestrally related. Array CGH of #11 revealed three regions of CNV, including loss of chromosomal material from the telomeres of 6p and 6q (Fig. 1B). The inability to amplify a junction fragment from #11 may be attributable to this rearrangement's complexity. While the number of forkhead genes encompassed by each rearrangement differ [duplication (FOXC1 and FOXF2: 7/7; FOXQ1: 1/7), deletion (FOXC1: 3/3), ring (FOXC1, FOXF2 and FOXQ1)], FOXC1 is the only gene whose copy number is altered in all the rearrangements. Clinically, the seven duplication pedigrees, and separately the three deletion pedigrees exhibit similar ocular structural phenotypes (Table 2).
Figure 6.
Segregation of junction fragments with disease phenotype in representative pedigrees: (A and B) Segmental duplications (#1 and #4, respectively), (C and D) Segmental deletion (#8 and #9, respectively); note the unaffected parents of individuals 2 and 3 (D), a finding indicative of germline mosaicism (probands denoted by arrowhead).
Table 1.
Haplotype of segmental duplication pedigrees
| Pedigrees | Microsatellite markers |
SNPs |
Architecture |
||||
|---|---|---|---|---|---|---|---|
| D6S967 | BA121 | FF2 | SNP-1 | SNP-2 | Extent (bp) | Insertion/deletion | |
| 1 | 318 | 318 | 160 | CCC | GGG | 492 096 | 6 bp insertion |
| 2 | 318 | 318 | 160 | GCC | TGG | 492 096 | 6 bp insertion |
| 3 | 314 | 302 | 163 | TTC | AAG | 479 998 | 3 bp deletion |
| 4 | 314 | 302 | 163 | TTC | AAG | 479 998 | 3 bp deletion |
| 5 | 314 | 302 | 163 | CTC | GAG | 479 998 | 3 bp deletion |
| 6 | 314 | 302 | 163 | TTC | AAG | 479 998 | 3 bp deletion |
The duplication genotype showing the duplicated base pair in bold (and the unduplicated in standard font).
Table 2.
Ocular phenotypes of 6p25 copy number variation families
| Pedigree no. | Phenotype | Location and reference(s) |
|---|---|---|
| 1 | Iris hypoplasia and glaucoma | Canada (66,68,69) |
| 2 | Iris hypoplasia and glaucoma | Canada |
| 3 | Iris hypoplasia and glaucoma | UK (27) |
| 4 | Iris hypoplasia and glaucoma | UK |
| 5 | Iris hypoplasia and glaucoma | UK (16) |
| 6 | Iris hypoplasia and glaucoma | Canada (68,69) |
| 7 | Iris hypoplasia and glaucoma | UK (27,66) |
| 8 | Axenfeld–Rieger Syndrome and glaucoma | UK (22,27) |
| 9 | Axenfeld–Rieger Syndrome and glaucoma | Canada |
| 10 | Ocular developmental anomalies | USA |
| 11 | Anterior segment dysgenesis | UK |
The ancestral origin of #1, 2 and 6 is from the UK; #10’s ocular phenotype cannot be fully determined at present due to this infant's age.
DISCUSSION
Elucidating the genesis of CNVs is important in view of the major contributions that they make to human disease and the fact that despite recent advances (37), the mechanisms mediating their formation remain incompletely defined. The recurrent nature of most characterized disease-associated segmental rearrangements illustrates that they arise due to the neighboring genomic architecture. Such (recurrent) structural variants, which have common breakpoints in unrelated individuals, are usually generated by LCRs that induce misalignment during meiosis and thus provide the substrate for NAHR (38–41). Less frequently, rearrangements occur in which the breakpoint positions vary, and these non-recurrent rearrangements are generally attributable to NHEJ. The latter is an error-prone repair process with breakpoints located in non-homologous regions (42–48) and characterized by either microhomology, insertion or deletion of nucleotides at the junction. One novel aspect of the present study stems from the broader mechanistic spectrum observed and is evident from the following features.
First, previous examples of disease-associated duplication and deletion in the same genomic area predominantly have common breakpoints (49–51), whereas all six ancestral 6p25 segmental duplications and deletions are non-recurrent (Fig.1AI–VI). This indication that 6p25 may differ from most previously studied genomic regions, is supported by the substantially broader than previously described segmental anomaly size spectrum (∼30–1220 kb) observed with these CNVs. The second feature relates to the sequences that mediate the rearrangements, which have generally been LCRs, comprising elements some 10–500 kb in size that share ≥95% sequence homology (39). Although no LCRs are present in this region, evidence for a homologous error-free repair mechanism is provided by the normal reference genomic sequence present at the end of each duplicated or deleted sequence. In this context, location of breakpoints in five of the six ancestral rearrangements within (#1, 2 and 8) or adjacent (#3–7 and 10) to repetitive elements that are themselves located within large repeat blocks, indicates that the blocks of repetitive sequence contribute to the rearrangements. It is therefore noteworthy that in #3–6 (Fig. 2) the extent (13.8 kb) of the adjacent repeat block, which is 97% composed of repetitive elements, exceeds the minimum size of an LCR. Furthermore, in pedigrees #7 and 8 (Figs 3 and 4), location of members of the same repeat element class at or adjacent to each breakpoint provides putative recombination substrates. Taken together with the microhomology observed in the peri-breakpoint sequence (#10), these data indicate that 6p25 segmental rearrangements are frequently mediated by sequences smaller than LCRs.
In addition, all six ancestral rearrangements exhibit NHEJ with microhomology (#3–7 and 9) or base pair insertion (#1, 2, 8 and 10) at the junction. Multiple rearrangements exhibiting both NAHR and NHEJ have only been previously reported in Pelizaeus–Merzbacher disease, a disorder primarily characterized by X-chromosomal duplication (48). Concurrent involvement of NAHR and NHEJ on an autosome, and with both segmental duplication and deletion, convincingly demonstrates that a greater proportion of rearrangements use mixed mechanisms. The differing degrees of NAHR and NHEJ evident in the six rearrangements, suggest that these may represent components of a mechanistic spectrum. It is interesting to contrast our findings with the X-chromosomal duplications (and less frequent deletions) underlying Pelizaeus–Merzbacher disease, where co-existing NAHR and NHEJ were first observed (48). While a broad CNV size spectrum is a feature of Pelizaeus–Merzbacher disease, the abundant LCRs of up to 122 kb in size associated with the underlying rearrangements are not evident in the 6p25 region.
The architecture of rearrangement #8 (Fig. 4) is unprecedented, with a 60-fold larger insert at the junction than previously observed in NHEJ (6 bp) (48), as well as features of NAHR [breakpoints within (GTG)n repeats]. The size of the insert (367 bp), identical motifs and particularly homology to sequence adjacent to both breakpoints (Fig. 4A) are incompatible with NAHR and NHEJ being the sole mechanisms involved. Indeed, existence of DNA polymerase α pause and topo-isomerase I consensus cleavage sites at, or adjacent to, both breakpoints (Fig. 4B) accord with a more complex mechanism, notably the replication-based mechanism recently described in two non-recurrent Pelizaeus–Merzbacher disease cases (37). The DNA polymerase pause sites and extensive (GTG)n at the breakpoints represent recognized impediments to polymerase progression (52,53), capable of causing the same successive replication fork stalling that characterized the Pelizaeus–Merzbacher cases (37). Stalled forks are recombinogenic (54), in part due to the potential for the increased length of single-strand DNA to lead to double strand break and subsequent template switching (37).
Hence the most parsimonious explanation for the observed architecture of motif M1 is recurrent stalling followed by re-initiation of strand synthesis ahead of the sequence that induced the fork to stall. This is in agreement with the extensive homology between M1 and the three sequence blocks (labeled I–III, Fig. 4B) that straddle the centromeric breakpoint, which lie in the correct order and orientation to yield this 128 bp motif portion. The imperfect sequence identity at the junctions of blocks ‘I’ and ‘II’, and ‘II’ and ‘III’ is explicable by error-prone repair of the resultant single strand gaps using either the sister chromatid, or other sequence, as template (55). The key differences with the Fork Stalling and Template Switching Mechanism (37) concern the absence of evidence of template switching in #8 and the higher order symmetry of the identical motifs. The latter is compatible with a double strand break at the telomeric breakpoint—the location of a topoisomerase I cleavage site (CAT)—followed by single strand-dependent synthesis using the antisense strand of M1 as template to generate M2. Such findings, albeit based upon a single complex rearrangement, accord with the central tenet of the replication-based mechanism (37), while illustrating potential for additional intricacies and demonstrating involvement of this process in segmental deletion in addition to the previously described two duplications (37).
The unique architecture of 6p25 among regions prone to segmental duplication and deletion (28–35) with three tandemly arranged paralogs (FOXC1, FOXF2 and FOXQ1), heightens interest in these rearrangements. CNVs in regions containing paralogs triplets cause both human (color blindness: opsin cluster, Xq28) (56,57) and canine phenotypes (ridge and dermal sinus: FGF3/4/19 cluster, chr. 18) (58). Since homology between the parologous forkhead domains is at a level sufficient to promote strand exchange (59,60), their presence is compatible with a role in CNV formation, possibly by promoting misalignment or stabilizing intermediate structures. CNV prevalence is increased in genomic regions characterized by segmental duplication and the available evidence, including the identical zebrafish forkhead triplet, indicate that the 6p25 (FOXC1, FOXF2 and FOXQ1) and 16q24 clusters (FOXC2, FOXF1 and FOXL1) were generated by ancestral segmental duplication events (26,61). This raises the intriguing possibility that the segmental duplications that generated the 6p25 forkhead triplet may predispose to subsequent CNV.
Defining the breakpoint architecture of each of the six ancestral segmental rearrangements, a particular strength of this study, revealed a discrepancy between array CGH-predicted and actual breakpoints in one pedigree (#7; Fig. 3). This observation is of relevance due to increasing transition of array CGH from the research laboratory into clinical testing. Two possible explanations can be proposed for the discordant result in this well-validated 6p25 segmental duplication pedigree (22). The first is masking by paralogs test or reference sequence, since array CGH copy number data reflect the overall level of hybridization to individual probes. The second is that use of an extensive panel of primers to amplify the junction fragment has detected a more complex rearrangement than defined by array CGH alone. At the present time we are unable to differentiate between these possibilities.
In summary, our findings illustrate the value of undertaking the extensive experiments necessary to characterize individual CNVs at the base pair level, identifying the novel spectrum of mechanisms that mediate 6p25 rearrangements. This combination of recombination, DNA repair and replication-based mechanisms, together with the CNV size spectrum and 100% non-recurrence rate, highlight unique features of this genomic region. Use of increasingly higher resolution techniques to interrogate 6p25 has detected progressively smaller CNVs, providing translational opportunities for determining the molecular basis of an appreciable proportion of pediatric glaucoma. Although CNVs smaller than the current chromosome 6p25 ∼30 kb minimum are present genome-wide, their prevalence is incompletely defined due to the ≥5 kb probe density of most current arrays. If our hypothesis is correct that formation of small structural variants may be mediated by smaller sequence elements than larger CNVs (as in 6p25), analysis of regions containing multiple paralogs dosage sensitive genes that exhibit a particularly broad spectrum of rearrangements, may be especially fruitful in determining novel causes of CNV.
MATERIALS AND METHODS
Patients
Genomic DNA from 148 individuals with ARS phenotypes secondary to 6p25 rearrangements (Table 2), were analyzed after isolation from venous blood samples using conventional techniques (62,63). These samples were derived from seven segmental duplication, and two segmental deletion pedigrees, as well as a single ring chromosome proband. The ancestral UK origin of the duplication pedigrees, recruited from Canada (n = 3) and the UK (n = 4), was compatible with the existence of founder effects (Table 2). A panel of 32 mutation-negative ARS cases, previously screened by microsatellite marker genotyping for copy number changes involving the known ARS-associated genes FOXC1 and PITX2, were also studied with FOXC1 qPCR (TaqMan, ABI). The study was approved by the Health Research Ethics Board of the University of Alberta.
Analysis of 6p25 copy number variants
DNA from four segmental duplication patients were initially hybridized to CGH arrays comprising 544 contiguous 744–10 289 bp PCR amplicon-based probes from a 680 kb region encompassing the segmental duplications. Amplicon design and hybridizations were performed as previously described (64). Subsequently, a customized commercial array (Nimblegen) comprising isothermal, oligonucleotide probes tiled at a high (∼100 bp) density, across the telomeric 5 Mb of 6p25 was employed using Cy3-labeled patient and Cy5-labeled reference DNA as described elsewhere (65). DNA samples from a single affected individual from eight pedigrees [#1 (66), 3 (27), 4, 5 (16) and 7 (22,66) (duplication); #8 (22,27) and #10 (deletion); #11 (ring chromosome 6)] plus an ARS case (#9) in which qPCR was compatible with a CNV, were hybridized using standard techniques to determine the extent of the 6p25 rearrangements. After an Alu-based PCR approach (67) (n = 20 primer pairs) proved unsuccessful at amplifying breakpoint-spanning junction fragments, additional primer pairs (n = 116) extending 20 kb from the predicted breakpoints were designed. Since array CGH cannot define the relative positions or orientation of the duplicated and unduplicated segments, long-range PCR (Elongase, Invitrogen) was next performed with multiple primer permutations (n = 336). Amplification of junction fragments in duplication pedigrees #1–6 with the primer pairs listed (Table 3), obviated the need for array CGH in #2 and #6; while in #7, supplementary primers (n = 48) extending up to 70 kb from the predicted breakpoints, were required. Junction fragments from all segmental duplication and deletion pedigrees (#1–10) were visualized on ethidium bromide stained 1% agarose gel and sequenced with internal primers using a BigDye (v3.1) terminator kit and 3100 DNA sequencer (Applied Biosystems). In pedigree #7, SYBR Green qPCR was performed in quadruplicate with nine PCR amplicons to validate the array CGH data. In pedigree #8, the amplicon's architecture necessitated cloning into a vector (pCR®4-TOPO®, Invitrogen) prior to sequencing.
Table 3.
Primer pairs used for junction fragment, qPCR and haplotype analyses
| Forward | Reverse | Annealing temperature (°C) | |
|---|---|---|---|
| Junction fragment | |||
| #1 and 2 | GAACTTGTAACATTCCCAACAGTTCT | CATGTGTGACAAGTGATATAAGCAAG | 60 |
| #3–6 | TACAGAACAGACAGTACAGATTATGAGG | GGAGATGGGTCATAAACATTTGAACTATAT | 60 |
| #7 | TGCTTAGGTCTATAGGACAGAGTCG | CCAAGAGAGTCTCCTTTCATTGTTA | 60 |
| #8 | TCAAACATTGAGGGTAGTGTTTTGG | CCATGTATTGTGCACACACA | 60 |
| #9 | AGAGCCTCTCTTCTGTTTAAGACATC | CCAGAAACTCTTCCTAAGAGAAAGAA | 62 |
| #10 | AAAAATACAAAAATTAGCTGGGTGTG | TAGAATTGTACTTGATCCACCACTTC | |
| FOXC1 qPCR | |||
| #9 | CACTCGGTGCGGGAGATG Probe: TCGAGTCACAGAGGAT |
TGAACAACTCTCCAGTGAACGG | 60 |
| Microsatellite markers/SNPs | |||
| BA121 | CAGGAGGCAGAGGTCACAGT | ATAGGGTTGGTTCCCAAAGG | 60 |
| FF2 | CGGGTTCCACCTACATGGC | CACTCCAGCATGTCCTCCTACT | 58 |
| SNP-1 and SNP-2 | GTGGCTAGATTTGTGAGTATCACTTC | CGAGTTATCGGTGGTAGTCATACA | 58 |
Bioinformatic and haplotype analysis
In silico analyses of sequences adjacent to the breakpoints were performed with Ensembl, University of California–Santa Cruz (UCSC) and National Center for Biotechnology Information (NCBI) Entrez genome browsers. Sequence composition was determined with BLASTN (NCBI Blast) and BLAT multi-species comparison (UCSC), with the boundaries of repeat elements defined using Ensembl and UCSC. Analysis of the repeat masked telomeric 2.5 Mb of chromosome 6p25 was undertaken with Pipmaker to determine if LCRs were present. To determine whether the duplication pedigrees shared common haplotypes, genotyping was performed with known (D6S967, RH122719, SHGC-82115, SHGC-53095 and SHGC-112337) or predicted (BA121, BA118 and FF2) fluorescent labeled microsatellite markers identified by analysis of repeat elements in the duplicated segments, supplemented by 11 SNPs (rs12524120/4544/5352/5740; rs1961687; rs1785778/5836/6028/6106) together with 2 SNPs identified within an E74-like factor pseudogene.
WEB RESOURCES
The accession number and URLs for data presented herein are as follows:
Ensembl Genome Browser: http://www.ensembl.org/index.html (for version 36).
NCBI BLAST: http://www.ncbi.nlm.nih.gov/blast/.
NCBI Entrez: http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi.
UCSC Genome Browser: http://genome.ucsc.edu/.
UniGene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene.
Pip Maker: http://pipmaker.bx.psu.edu/pipmaker/.
Repeat Masker: http://repeatmasker.org/cgi-bin/WEBRepeatMasker.
Palindrome: http://bioweb.pasteur.fr/seqanal/interfaces/palindrome.html.
FUNDING
This work was initially supported by the Birth Defects Foundation and the Wellcome Trust, and subsequently by the Canadian Institutes of Health Research, Alberta Heritage Foundation for Medical Research and Canada Research Chair program.
ACKNOWLEDGEMENTS
We are grateful to Dr Cordelia Langford for provision of the amplicon-based 6p25 arrays and assisting this study. We thank Drs Peter Carlsson and Andrew Waskiewicz for helpful advice, Dr Shomi Bhattacharya for his longstanding support, Dr Kathleen Millen and Kimberly Aldinger for provision of samples, and the many clinicians who contributed patients to these studies, particularly Professor Peng Khaw and the late Dr Ron Casey.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 2.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–754. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perry G.H., Dominy N.J., Claw K.G., Lee A.S., Fiegler H., Redon R., Werner J., Villanea F.A., Mountain J.L., Misra R., et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollox E.J., Armour J.A., Barber J.C. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am. J. Hum. Genet. 2003;73:591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moessner R., Marshall C.R., Sutcliffe J.S., Skaug J., Pinto D., Vincent J., Zwaigenbaum L., Fernandez B., Roberts W., Szatmari P., et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am. J. Hum. Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y., Chung E.K., Wu Y.L., Savelli S.L., Nagaraja H.N., Zhou B., Hebert M., Jones K.N., Shu Y., Kitzmiller K., et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am. J. Hum. Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollox E.J., Huffmeier U., Zeeuwen P.L., Palla R., Lascorz J., Rodijk-Olthuis D., van de Kerkhof P.C., Traupe H., de Jongh G., den Heijer M., et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat. Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scherer S.W., Lee C., Birney E., Altshuler D.M., Eichler E.E., Carter N.P., Hurles M.E., Feuk L. Challenges and standards in integrating surveys of structural variation. Nat. Genet. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinjan D.A., van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDermid H.E., Morrow B.E. Genomic disorders on 22q11. Am. J. Hum. Genet. 2002;70:1077–1088. doi: 10.1086/340363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fantes J., Ragge N.K., Lynch S.A., McGill N.I., Collin J.R., Howard-Peebles P.N., Hayward C., Vivian A.J., Williamson K., van Heyningen V., et al. Mutations in SOX2 cause anophthalmia. Nat. Genet. 2003;33:461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 13.Lines M.A., Kozlowski K., Kulak S.C., Allingham R.R., Heon E., Ritch R., Levin A.V., Shields M.B., Damji K.F., Newlin A., et al. Characterization and prevalence of PITX2 microdeletions and mutations in Axenfeld–Rieger malformations. Invest. Ophthalmol. Vis. Sci. 2004;45:828–833. doi: 10.1167/iovs.03-0309. [DOI] [PubMed] [Google Scholar]
- 14.Chang B., Smith R.S., Peters M., Savinova O.V., Hawes N.L., Zabaleta A., Nusinowitz S., Martin J.E., Davisson M.L., Cepko C.L., et al. Haploinsufficient Bmp4 ocular phenotypes include anterior segment dysgenesis with elevated intraocular pressure. BMC Genet. 2001;2:18. doi: 10.1186/1471-2156-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crisponi L., Deiana M., Loi A., Chiappe F., Uda M., Amati P., Bisceglia L., Zelante L., Nagaraja R., Porcu S., et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat. Genet. 2001;27:159–166. doi: 10.1038/84781. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann O.J., Ebenezer N.D., Jordan T., Fox M., Ocaka L., Payne A., Leroy B.P., Clark B.J., Hitchings R.A., Povey S., et al. Chromosomal duplication involving the forkhead transcription factor gene FOXC1 causes iris hypoplasia and glaucoma. Am. J. Hum. Genet. 2000;67:1129–1135. doi: 10.1016/s0002-9297(07)62943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schedl A., Ross A., Lee M., Engelkamp D., Rashbass P., van Heyningen V., Hastie N.D. Influence of PAX6 gene dosage on development: overexpression causes severe eye abnormalities. Cell. 1996;86:71–82. doi: 10.1016/s0092-8674(00)80078-1. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura D.Y., Searby C.C., Alward W.L., Walton D., Craig J.E., Mackey D.A., Kawase K., Kanis A.B., Patil S.R., Stone E.M., et al. A spectrum of FOXC1 mutations suggests gene dosage as a mechanism for developmental defects of the anterior chamber of the eye. Am. J. Hum. Genet. 2001;68:364–372. doi: 10.1086/318183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Baere E., Van Roy N., Speleman F., Fukushima Y., De Paepe A., Messiaen L. Closing in on the BPES gene on 3q23: mapping of a de Novo reciprocal translocation t(3;4)(q23;p15.2) breakpoint within a 45-kb cosmid and mapping of three candidate genes, RBP1, RBP2, and beta’-COP, distal to the breakpoint. Genomics. 1999;57:70–78. doi: 10.1006/geno.1999.5747. [DOI] [PubMed] [Google Scholar]
- 20.Asai-Coakwell M., French C.R., Berry K.M., Ye M., Koss R., Somerville M., Mueller R., van Heyningen V., Waskiewicz A.J., Lehmann O.J. GDF6, a Novel Locus for a Spectrum of Ocular Developmental Anomalies. Am. J. Hum. Genet. 2007;80:306–315. doi: 10.1086/511280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kume T., Deng K.Y., Winfrey V., Gould D.B., Walter M.A., Hogan B.L. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell. 1998;93:985–996. doi: 10.1016/s0092-8674(00)81204-0. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann O.J., Ebenezer N.D., Ekong R., Ocaka L., Mungall A.J., Fraser S., McGill J.I., Hitchings R.A., Khaw P.T., Sowden J.C., et al. Ocular developmental abnormalities and glaucoma associated with interstitial 6p25 duplications and deletions. Invest. Ophthalmol. Vis. Sci. 2002;43:1843–1849. [PubMed] [Google Scholar]
- 23.Maclean K., Smith J., St Heaps L., Chia N., Williams R., Peters G.B., Onikul E., McCrossin T., Lehmann O.J., Ades L.C. Axenfeld–Rieger malformation and distinctive facial features: Clues to a recognizable 6p25 microdeletion syndrome. Am. J. Med. Genet. A. 2005;132:381–385. doi: 10.1002/ajmg.a.30274. [DOI] [PubMed] [Google Scholar]
- 24.Gould D.B., Jaafar M.S., Addison M.K., Munier F., Ritch R., MacDonald I.M., Walter M.A. Phenotypic and molecular assessment of seven patients with 6p25 deletion syndrome: relevance to ocular dysgenesis and hearing impairment. BMC Med. Genet. 2004;5:17. doi: 10.1186/1471-2350-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strungaru M.H., Dinu I., Walter M.A. Genotype-phenotype correlations in Axenfeld–Rieger malformation and glaucoma patients with FOXC1 and PITX2 mutations. Invest. Ophthalmol. Vis. Sci. 2007;48:228–237. doi: 10.1167/iovs.06-0472. [DOI] [PubMed] [Google Scholar]
- 26.Lehmann O.J., Sowden J.C., Carlsson P., Jordan T., Bhattacharya S.S. Fox's in development and disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann O.J., Tuft S., Brice G., Smith R., Blixt A., Bell R., Johansson B., Jordan T., Hitchings R.A., Khaw P.T., et al. Novel anterior segment phenotypes resulting from forkhead gene alterations: evidence for cross-species conservation of function. Invest. Ophthalmol. Vis. Sci. 2003;44:2627–2633. doi: 10.1167/iovs.02-0609. [DOI] [PubMed] [Google Scholar]
- 28.Lupski J.R., de Oca-Luna R.M., Slaugenhaupt S., Pentao L., Guzzetta V., Trask B.J., Saucedo-Cardenas O., Barker D.F., Killian J.M., Garcia C.A., et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 29.Chance P.F., Alderson M.K., Leppig K.A., Lensch M.W., Matsunami N., Smith B., Swanson P.D., Odelberg S.J., Disteche C.M., Bird T.D. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 30.Potocki L., Chen K.S., Park S.S., Osterholm D.E., Withers M.A., Kimonis V., Summers A.M., Meschino W.S., Anyane-Yeboa K., Kashork C.D., et al. Molecular mechanism for duplication 17p11.2- the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat. Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- 31.Yobb T.M., Somerville M.J., Willatt L., Firth H.V., Harrison K., MacKenzie J., Gallo N., Morrow B.E., Shaffer L.G., Babcock M., et al. Microduplication and triplication of 22q11.2: a highly variable syndrome. Am. J. Hum. Genet. 2005;76:865–876. doi: 10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edelmann L., Pandita R.K., Spiteri E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum. Mol. Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- 33.Somerville M.J., Mervis C.B., Young E.J., Seo E.J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., et al. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N. Engl. J. Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long F.L., Duckett D.P., Billam L.J., Williams D.K., Crolla J.A. Triplication of 15q11-q13 with inv dup(15) in a female with developmental delay. J. Med. Genet. 1998;35:425–428. doi: 10.1136/jmg.35.5.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ledbetter D.H., Riccardi V.M., Airhart S.D., Strobel R.J., Keenan B.S., Crawford J.D. Deletions of chromosome 15 as a cause of the Prader-Willi syndrome. N. Engl. J. Med. 1981;304:325–329. doi: 10.1056/NEJM198102053040604. [DOI] [PubMed] [Google Scholar]
- 36.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 37.Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 38.Kanaar R., Hoeijmakers J.H., van Gent D.C. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 39.Stankiewicz P., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 40.Shaw C.J., Lupski J.R. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum. Mol. Genet. 2004;13(Spec 1):R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- 41.Lupski J.R., Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raghavan S.C., Tong J., Lieber M.R. Hybrid joint formation in human V(D)J recombination requires nonhomologous DNA end joining. DNA Repair (Amst) 2006;5:278–285. doi: 10.1016/j.dnarep.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz M., Zlotorynski E., Goldberg M., Ozeri E., Rahat A., le Sage C., Chen B.P., Chen D.J., Agami R., Kerem B. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 2005;19:2715–2726. doi: 10.1101/gad.340905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonoda E., Hochegger H., Saberi A., Taniguchi Y., Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Lieber M.R., Ma Y., Pannicke U., Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 46.Tsukamoto Y., Ikeda H. Double-strand break repair mediated by DNA end-joining. Genes Cells. 1998;3:135–144. doi: 10.1046/j.1365-2443.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- 47.Roth D.B., Porter T.N., Wilson J.H. Mechanisms of nonhomologous recombination in mammalian cells. Mol. Cell. Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodward K.J., Cundall M., Sperle K., Sistermans E.A., Ross M., Howell G., Gribble S.M., Burford D.C., Carter N.P., Hobson D.L., et al. Heterogeneous duplications in patients with Pelizaeus–Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am. J. Hum. Genet. 2005;77:966–987. doi: 10.1086/498048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pentao L., Wise C.A., Chinault A.C., Patel P.I., Lupski J.R. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat. Genet. 1992;2:292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- 50.Juyal R.C., Figuera L.E., Hauge X., Elsea S.H., Lupski J.R., Greenberg F., Baldini A., Patel P.I. Molecular analyses of 17p11.2 deletions in 62 Smith-Magenis syndrome patients. Am. J. Hum. Genet. 1996;58:998–1007. [PMC free article] [PubMed] [Google Scholar]
- 51.Park S.S., Stankiewicz P., Bi W., Shaw C., Lehoczky J., Dewar K., Birren B., Lupski J.R. Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res. 2002;12:729–738. doi: 10.1101/gr.82802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirkin E.V., Mirkin S.M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fouche N., Ozgur S., Roy D., Griffith J.D. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 2006;34:6044–6050. doi: 10.1093/nar/gkl757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambert S., Watson A., Sheedy D.M., Martin B., Carr A.M. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 55.Langston L.D., O'Donnell M. DNA replication: keep moving and don't mind the gap. Mol. Cell. 2006;23:155–160. doi: 10.1016/j.molcel.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 56.Nathans J., Merbs S.L., Sung C.H., Weitz C.J., Wang Y. Molecular genetics of human visual pigments. Annu. Rev. Genet. 1992;26:403–424. doi: 10.1146/annurev.ge.26.120192.002155. [DOI] [PubMed] [Google Scholar]
- 57.Deeb S.S., Motulsky A.G. Molecular genetics of human color vision. Behav. Genet. 1996;26:195–207. doi: 10.1007/BF02359380. [DOI] [PubMed] [Google Scholar]
- 58.Salmon Hillbertz N.H., Isaksson M., Karlsson E.K., Hellmen E., Pielberg G.R., Savolainen P., Wade C.M., von Euler H., Gustafson U., Hedhammar A., et al. Duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat. Genet. 2007;39:1318–1320. doi: 10.1038/ng.2007.4. [DOI] [PubMed] [Google Scholar]
- 59.Waldman A.S., Liskay R.M. Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol. Cell. Biol. 1988;8:5350–5357. doi: 10.1128/mcb.8.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lam K.W., Jeffreys A.J. Processes of copy-number change in human DNA: the dynamics of {alpha}-globin gene deletion. Proc. Natl. Acad. Sci. USA. 2006;103:8921–8927. doi: 10.1073/pnas.0602690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carlsson P., Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 62.Beutler E., Gelbart T., Kuhl W. Interference of heparin with the polymerase chain reaction. Biotechniques. 1990;9:166. [PubMed] [Google Scholar]
- 63.Tsai M., Miyamoto M., Tam S.Y., Wang Z.S., Galli S.J. Detection of mouse mast cell-associated protease mRNA. Heparinase treatment greatly improves RT-PCR of tissues containing mast cell heparin. Am. J. Pathol. 1995;146:335–343. [PMC free article] [PubMed] [Google Scholar]
- 64.Dhami P., Coffey A.J., Abbs S., Vermeesch J.R., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D. Exon array CGH: detection of copy-number changes at the resolution of individual exons in the human genome. Am. J. Hum. Genet. 2005;76:750–762. doi: 10.1086/429588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Selzer R.R., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 66.Mears A.J., Jordan T., Mirzayans F., Dubois S., Kume T., Parlee M., Ritch R., Koop B., Kuo W.L., Collins C., et al. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld–Rieger anomaly. Am. J. Hum. Genet. 1998;63:1316–1328. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meese E.U., Meltzer P.S., Ferguson P.W., Trent J.M. Alu-PCR: characterization of a chromosome 6-specific hybrid mapping panel and cloning of chromosome-specific markers. Genomics. 1992;12:549–554. doi: 10.1016/0888-7543(92)90447-z. [DOI] [PubMed] [Google Scholar]
- 68.Walter M.A., Mirzayans F., Mears A.J., Hickey K., Pearce W.G. Autosomal-dominant iridogoniodysgenesis and Axenfeld–Rieger syndrome are genetically distinct. Ophthalmology. 1996;103:1907–1915. doi: 10.1016/s0161-6420(96)30408-9. [DOI] [PubMed] [Google Scholar]
- 69.Mears A.J., Mirzayans F., Gould D.B., Pearce W.G., Walter M.A. Autosomal dominant iridogoniodysgenesis anomaly maps to 6p25. Am. J. Hum. Genet. 1996;59:1321–1327. [PMC free article] [PubMed] [Google Scholar]