Abstract
Dyskeratosis congenita (DC) is a multisystem bone marrow failure syndrome characterized by a triad of mucocutaneous abnormalities and a predisposition to cancer. The genetic basis of DC remains unknown in more than 60% of patients. Mutations have been identified in components of the telomerase complex (dyskerin, TERC, TERT, NOP10, and NHP2), and recently in one component of the shelterin complex TIN2 (gene TINF2). To establish the role of TINF2 mutations, we screened DNA from 175 uncharacterised patients with DC as well as 244 patients with other bone marrow failure disorders. Heterozygous coding mutations were found in 33 of 175 previously uncharacterized DC index patients and 3 of 244 other patients. A total of 21 of the mutations affected amino acid 282, changing arginine to histidine (n = 14) or cysteine (n = 7). A total of 32 of 33 patients with DC with TINF2 mutations have severe disease, with most developing aplastic anaemia by the age of 10 years. Telomere lengths in patients with TINF2 mutations were the shortest compared with other DC subtypes, but TERC levels were normal. In this large series, TINF2 mutations account for approximately 11% of all DC, but they do not play a significant role in patients with related disorders. This study emphasises the role of defective telomere maintenance on human disease.
Introduction
Dyskeratosis congenita (DC) is a bone marrow failure syndrome classically associated with a triad of mucocutaneous features: nail dystrophy, oral leukoplakia, and abnormal skin pigmentation. A variety of other (dental, gastrointestinal, neurological, ophthalmic, pulmonary, and skeletal) abnormalities have also been described.1 Genetically, DC is heterogeneous, with 3 forms identified: X-linked recessive, autosomal dominant, and autosomal recessive. Although causative gene mutations have been identified in some of the families, a significant number remain uncharacterized. DC displays several features that overlap other bone marrow failure syndromes such as aplastic anemia (AA), both constitutional and idiopathic (CAA and IAA, respectively), and myelodysplastic syndromes (MDSs). Of these, CAA is most similar in presentation to DC, as it describes patients in whom there is a family history of AA or myelodysplasia, or patients with AA with one or more additional physical abnormality but not enough to formally diagnose as DC. MDS again covers a spectrum of disorders; the main features are cytopenia and dysplastic bone marrow. IAA is unexplained AA with usually no other physical feature or family history.2
Mutations in DC have been identified in 5 genes encoding components of the telomerase complex. DKC1 encoding dyskerin was the first causative gene to be identified and is the main gene responsible for the X-linked recessive form of the disease.3 Autosomal-dominant DC is caused by heterozygous mutations in the core components of telomerase, namely TERC (the RNA component)4 and TERT (the enzymatic component).5 Autosomal recessive DC has been shown to be caused by biallelic NOP10, NHP2 (components of the small nucleolar ribonucleoprotein particle)6,7 and TERT mutations.8,9 Heterozygous mutations in both TERC and TERT have been identified in patients with CAA, IAA, and MDS.10–12 Very recently, heterozygous mutations in a sixth gene, TINF2, which encodes TIN2, a component of the shelterin telomere protection complex, have been described in some patients with DC.13
The underlying causes of DC are now evolving. Until the identification of mutations in TIN2 of the shelterin complex, all mutations centred on telomerase and although DC is accepted as a disease of defective telomere maintenance, it had been via defective telomere elongation. As the shelterin complex is involved in telomere protection (for reviews see de Lange14 and Gilson and Geli15), and now with the identification of mutations in TIN2, the previous observation of defective telomere maintenance in DC is now in terms of telomere elongation and protection. Shelterin has high sensitivity for the telomeric TTAGGG repeats, which are added by telomerase, and TIN2 is a core component of this complex. Without the protective activity of shelterin, telomeres are no longer hidden from DNA damage repair mechanisms; thus, chromosome ends can be incorrectly processed by the DNA repair pathways.16 It is therefore a combination of the activities of telomerase and shelterin that correctly protects and processes the telomeres.
Recently, Savage et al described 3 mutations in the shelterin component TINF2 in 5 of 9 genetically uncharacterized index patients with DC and short telomeres.13 Currently, more than 60% of patients entered on the Dyskeratosis Congenita Registry (DCR) remain genetically uncharacterized.17 Most of these patients are sporadic cases. This study was designed to determine whether TINF2 mutations could account for the genetic basis of this group. By screening the uncharacterized patients we aimed to answer this question, which in turn could give another valuable biomarker for the diagnosis of DC. This may allow discrimination between ambiguous cases of DC and many of the overlapping diseases such as AA (idiopathic and constitutional), myelodysplasia, and related disorders.
Methods
Patient selection
All uncharacterized index patients in the DCR were screened for mutations in TINF2 (median age, 9 years; range, 0-58 years; n = 175). In addition, 244 patients with related diseases that were part of a separate collection held at The Royal London Hospital (United Kingdom) were screened for mutations in exon 6 only. Of the 244 patients with related diseases, there were 111 patients with IAA (median age, 28 years; range, 1-71 years), 77 patients with CAA (median age, 14.5 years; range, 0-81 years), 15 patients with MDS (median age, 26.5 years; range, 4-54 years), and 41 patients with one or more characteristics shared by DC/Hoyeraal-Hreidarsson syndrome (HH) but limited clinical information available (median age, 11.5 years; range, 0-48 years). Where available, family members of patients with known mutations were screened for segregation, and an additional 91 healthy individuals were screened for polymorphisms in exon 6. All samples were prepared from peripheral blood leukocytes and were obtained with informed consent and with the approval of our local ethics committee, the Barts and The London School of Medicine and Dentistry, and in accordance with the Declaration of Helsinki.
Mutation detection by heteroduplex analysis
The coding region of TINF2 was amplified from genomic DNA by PCR on 6 fragments covering coding sequences described as TINF2 isoform 1 (NM_001099274; primers given in Table 1). After checking the products on 1.8% agarose gel, they were heated to 95°C and allowed to cool slowly to allow the formation of heteroduplexes. They were then analyzed by denaturing high-performance liquid chromatography (dHPLC) on a Wave DNA fragment analysis system (Transgenomic, San Jose, CA) at a temperature at which the fragment was approximately 75% helical. Any fragments displaying abnormal elution patterns were reamplified and sequenced using the Big Dye (Applied Biosystems, Foster City, CA) chain termination chemistry.
Table 1.
Primer sequences for PCR amplification of TINF2
| Fragment | Exon | Forward | Reverse | Product size, bp |
|---|---|---|---|---|
| 1 | 1 | CCTCTTACCGCCCTTTTCC | CTTGTCAGGTGCTCGCATC | 600 |
| 2 | 2 + 3 | AGACTAGTTGGAAAAGGTCAGC | GGGCGACAGAGCAAGATTC | 463 |
| 3 | 4 + 5 | CAAATGGCCAGGATTACAGG | TTCTTATGCCCGGAGCCC | 455 |
| 4 | 6 | GGCTCCGGGCATAAGAAAC | TGAGGTGAGAGCAAG CAAAG | 617 |
| 5 | CAGAGCAAAAGG AGTGAGTG | TACAGTCACAGGAAGAAACAG | 611 | |
| 6 | GGAATCTCTGGAAAACTATCAG | ACACCCAGATAATCTGGCAG | 552 |
Primers are designed to genomic accession number AL096870. Fragment 1 includes the 5′ untranslated region (UTR). Fragments 4, 5, and 6 overlap and cover the alternative splice regions detailed in the 3′ UTR and described as 2 different isoforms (isoform 1, nm_001099274; isoform 2, nm_012461). All PCR amplifications were performed using 2 mM magnesium with an annealing temperature of 58°C for 30 cycles. Exceptions to this were for fragments 2 and 3, where 10% DMSO was used and the annealing temperature was increased to 60°C.
Detection of mutations occurring at amino acid 282
It was observed that arginine 282 was commonly mutated to either cysteine or histidine. The sequence spanning this amino acid GCGC incorporated a HhaI restriction site that is lost when the amino acid is mutated. After standard polymerase chain reaction (PCR) amplification of the appropriate fragment, the products were digested overnight with 2 U of HhaI (New England Biolabs, Ipswich, MA) and the results were scored after visualization on a 2% agarose gel.
Telomere length measurements
Genomic DNA, extracted from whole blood, was digested with BamHI and analyzed by Southern blot analysis using 0.75% agarose gels and the subtelomeric probe pTelBam8.18,19 Telomere lengths were measured as the size of the fragment of peak signal intensity, which includes approximately 8 kb of subtelomeric DNA. Sizes were determined with reference to the same standards run on each gel using Image Quant software (GE Healthcare, Sunnyvale, CA). Data from healthy control samples, patients with DKC1 mutations, and age-adjusted telomere length (delta Tel) values were obtained as described previously.6 Differences between the various groups were tested using the Mann-Whitney U test.
TERC quantitation by real-time PCR
Where samples were available, TERC accumulation was determined as previously described.6 Briefly, cDNA was prepared from RNA extracted from whole blood. Absolute TERC and ABL expression levels were measured by quantitative reverse transcription–PCR (Q-RT/PCR) using the ABI PRISM 7700 sequence detection system (Applied Biosystems). These ratios were compared with those from healthy individuals and patients with known DKC1 mutations. Statistical differences were tested as in the previous paragraph.
Results
Mutation analysis of TINF2 in patients with genetically uncharacterized DC
As previously described mutations in TINF2 affected amino acids (aa) 280 and aa 282 in exon 6, we initially screened this exon in all uncharacterized index patients present in the DCR. Of the 292 families available in the registry, 175 remained genetically uncharacterized. Any individual that gave an abnormal trace was sequenced. In this initial screen, abnormal wave patterns were observed in 33 (18.9%) of 175 samples. Mutations were identified in these samples by direct sequencing and were verified by sequencing the reverse strand or by HhaI digestion for Arg282 mutations.
Of these 33 samples, 21 were found to have a mutation in Arg282. A total of 14 had a mutation where the arginine was mutated to a histidine (c.845G>A, Arg282His); in the remaining 7, the arginine was mutated to cysteine (c.844C>T, Arg282Cys). Of the remaining 12 mutations, 3 were insertion or deletion of a single base and the other 9 were missense. Table 2 details all the relevant clinical information as well as the genetic mutation for all affected index patients. All the mutations seen were clustered in an 18-aa segment, and all the mutations affected highly conserved residues (Figure 1). In silico analysis was performed for predicting possible functional effects of the TINF2 missense mutations with sorting intolerant from tolerant (SIFT). SIFT predicted none of the mutations seen would be tolerated,21 which is consistent with them being pathogenic. None of these changes were observed in a screen of 91 ethnically matched healthy individuals, nor in a previously reported screen of 298 healthy control individuals.13 We also performed a complete TINF2 screen on 98 index patients who did not have mutations in exon 6 and found no other coding mutations.
Table 2.
Characteristics of patients with TINF2 mutations
| DCRno. | Diagnosis | Age, y/sex | Nail dystrophy | Skin abnormality | Leukoplakia | Other features | AA | Hb, g/L | WBC, ×109/L | Platelets, ×109/L | Base change* | Amino acid change |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 45 | DC/HH/RS | 1/M | + | − | − | Retinopathy, intercranial calcification | + | NA | NA | NA | c.838A>T | Lys280X |
| 282 | HH/RS | 3/M | + | − | + | Microcephaly, short stature, retinopathy, intracerebral calcifications, BMT age 3 y | + | 95 | 1.3 | 15 | c.838A>T | Lys280X |
| 19 I1 | DC | 37/M | + | + | + | Bronchitis | + | 129 | 8.1 | 142 | c.844C>T | Arg282Cys |
| II2 | AA | 4/F | − | − | − | + | 87 | 3.1 | 26 | c.844C>T | Arg282Cys | |
| II3 | AA | 7/NA | − | − | − | + | NA | NA | NA | c.844C>T | Arg282Cys | |
| 24 I1 | DC | 24/M | + | − | + | Post-BMT lung disease, died 36 y | + | NA | NA | NA | c.844C>T | Arg282Cys |
| I2 | DC | 2/F | + | − | − | Died AA 9 y | + | NA | NA | NA | NA | NA |
| 36 | DC | 12/M | + | + | − | No family history | + | 58 | 8.2 | 40 | c.844C>T | Arg282Cys |
| 139 | DC | 9/F | + | − | − | Short, fine hair, lacrimal duct stenosis | + | 100 | 2.3 | 15 | c.844C>T | Arg282Cys |
| 181 | DC | 41/M | + | − | − | Pulmonary fibrosis, squamous Ca, alopecia | − | 1250 | 10.8 | 106 | c.844C>T | Arg282Cys |
| 189 | DC | 11/M | + | + | − | Short stature | + | 125 | 5.4 | 30 | c.844C>T | Arg282Cys |
| 195 | DC | 14/M | + | + | + | Osteoporosis, SAA-BMT age 5 y | + | NA | NA | NA | c.844C>T | Arg282Cys |
| 18 | DC | NA/M | NA | NA | NA | DC with AA | + | NA | NA | NA | c.845G>A | Arg282His |
| 40 | DC | 13/M | + | + | − | Phimosis, osteoporosis | + | 113 | 3.9 | 8 | c.845G>A | Arg282His |
| 54 | DC | NA/M | + | ? | ? | + | NA | NA | NA | c.845G>A | Arg282His | |
| 85 | DC | 4/M | + | − | − | AA–transfusion dependent | + | NA | NA | NA | c.845G>A | Arg282His |
| 90 | DC/HH/RS | 3/M | + | − | − | Retinopathy, deaf, microcephaly, learning problems | + | 87 | 4.1 | 14 | c.845G>A | Arg282His |
| 95 | DC | 4/M | + | + | + | Epiphora, osteoporosis | + | 73 | 1.5 | 20 | c.845G>A | Arg282His |
| 130 | DC/RS | 4/M | + | + | + | Retinopathy, heart defect, SAA | + | 75 | 1.3 | 24 | c.845G>A | Arg282His |
| 220 | DC | 9/F | + | + | + | Hair loss, osteoporosis, BMT age 1 y for SAA | + | 72 | NA | 19 | c.845G>A | Arg282His |
| 250 | DC | 7/M | + | − | + | Thin skin on trunk | + | 95 | 2.1 | 20 | c.845G>A | Arg282His |
| 253 | HH | 2/F | − | − | + | Microcephaly, cerebellar hypoplasia, developmental delay, low NK cells | + | 107 | 4.3 | 15 | c.845G>A | Arg282His |
| 258 | DC | 5/M | + | + | + | Epiphora, hyperhiderosis, undescended testes | + | 85 | 2.4 | 36 | c.845G>A | Arg282His |
| 265 | HH | 1/F | − | − | + | Developmental delay, cerebellar atrophy, hypotonia, short stature | + | 80 | 0.5-1.0 | <10 | c.845G>A | Arg282His |
| 269 | DC | 5/M | + | − | − | Alopecia, dental loss, short stature, skeletal defects | + | 105 | 3.8 | 25 | c.845G>A | Arg282His |
| 283 | DC | 4/M | + | + | + | Alopecia, osteoporosis, BMT for SAA | + | 60 | 3.4 | 2.0 | c.845G>A | Arg282His |
| 126 | DC/HH | 4/M | + | − | + | Ataxia, BMT age 4 y, pulmonary disease after BMT | + | 86 | 5.0 | 10 | c.847C>T | Pro283Ser |
| 211 | DC | 9/F | + | + | + | Alopecia, hyperhiderosis, dental loss, short stature, BMT age 3 y | + | 86 | 4.7 | 28 | c.847C>G | Pro283Ala |
| 276 | DC | 2/M | + | + | − | AA at age 1 y | + | 93 | 1.3 | 10 | c.848C>A | Pro283His |
| 89 | DC | 6/M | + | + | + | Response to oxymetholone | + | 106 | 3.9 | 10 | c.850A>G | Thr284Ala |
| 273 | DC/AA | 4/M | + | + | − | AA at age 1 y | + | 105 | 3.5 | 30 | c.849 850insC | Thr284HisfsX8 |
| 206 | DC | 10/F | + | + | − | Osteoporosis, BMT for SAA at 6 y | + | 120 | <2 | 20 | c.860T>C | Leu287Pro |
| 69 | DC | 12/M | + | + | + | DC with SAA | + | 114 | 3.9 | 12 | c.865 866 delinsAG | Pro289Ser |
| 133 | DC | 7/M | − | + | + | Microcephaly, short stature, learning difficulties | + | 70 | 2.0 | 10 | c.867 868insC | Phe290LeufsX2 |
| 94 | DC | 29/F | + | + | + | DC with AA, therapy with androgens | + | 72 | 2.21 | 39 | c.871A>G | Arg291Gly |
| 14 | DC | 15/F | + | + | + | Lacrimal duct stenosis, BMT for SAA age 11 y | + | 71 | 2.3 | 10 | c.892delC | Gln298Arg fsX19 |
| Non-DCR patients | ||||||||||||
| 1293 | AA | 4/M | − | − | − | AA at age 3 y | + | 82 | 6.1 | 26 | c.706C>T | Pro236Ser |
| 1771 | AA | 50/M | − | − | − | AA age 50 y | + | NA | NA | NA | c.734C>A | Ser245Tyr |
| 14433 | Low WBC | 40/M | − | − | − | Low WBC at age ∼30 y | + | 154 | 2.7 | 194 | c.841G>A | Glu281Lys |
Age indicates age when sample was obtained; BMT, bone marrow transplantation; Ca, carcinoma; DCR no., Dyskeratosis Congenita Registry number; F, female; Hb, hemoglobin; M, male; NA, not available; NK, natural killer cells; SAA, severe AA; WBC, white blood cells; +, present; −, absent; and ?, diagnosis unclear.
Nomenclature as recommended by the Human Genome Variation Society (http://www.hgvs.org/rec.html).
Figure 1.
Conservation of amino acids in a small region of TIN2 between different species. Close-up of the residues mutated in the patients with DC. Arrows indicate the sites of mutations. Changes are K280X (n = 2), R282C (n = 7), R282H (n = 14), P283A, P283H, P283S, T284A, T284Hfs8X, L287P, P289S, F290LfsX2, R291G, and Q298RfsX19. Red indicates highly conserved; blue, limited conservation; black, no conservation. Alignment obtained using MultAlin.20
Mutations in TINF2 are largely de novo
Parental samples were available for 17 of the index patients; these were screened for mutation segregation either by HhaI digest (Arg282 mutations; 11 families) or by dHPLC (6 families). A total of 16 of 17 mutations were de novo. In family DCR19, the Arg282Cys mutation is inherited from an affected father by his 2 children with AA. There was a slight anomaly in another family (DCR24). Both parents did not carry the Arg282Cys mutation seen in the index case, yet there were 2 affected children (1 with Arg282Cys; sample unavailable for the other).
Telomere lengths are extremely short
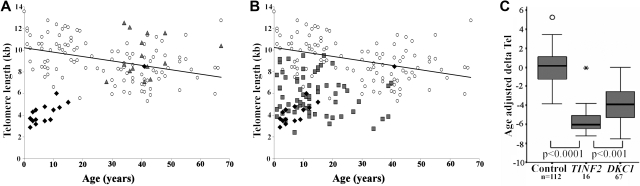
Telomere length measurements for the patients with TINF2 mutations and their parents (which, allowing for sample availability, were run on the same gel), are shown in Figure 2A. It is clear that patients with TINF2 mutations have significantly shorter telomeres than both their healthy parents and healthy individuals. When telomere lengths are compared directly between patients with TINF2 and DKC1 mutations, the age of presentation of TINF2 group is younger than the DKC1 group, and the telomere lengths are shorter as shown in Figure 2B. When age-adjusted delta Tel values are compared, the extent of the difference between healthy controls, patients with DC with TINF2 mutations, and patients with DC with DKC1 mutations becomes apparent (TINF2 vs controls, P < .0001; TINF2 vs DKC1, P < .001; Figure 2C). When an age-matched analysis is performed between the 2 patient groups, the difference is still maintained (P < .001, Mann-Whitney U test; data not shown). Interestingly there is one patient that appears to have normal telomere lengths. This individual is DCR181, who has a less severe phenotype and is the only patient in this group not to have AA.
Figure 2.
Telomere lengths in patients with DC with TINF2 mutations are the shortest compared with other DC subtypes. (A) Telomere lengths in healthy controls (○, n = 112), patients (♦, n = 16) and parents ( , n = 15). (B) Telomere length compassion between DC patients with TINF2 mutations (♦, n = 16), DKC1 mutations (
, n = 15). (B) Telomere length compassion between DC patients with TINF2 mutations (♦, n = 16), DKC1 mutations ( , n = 67) and healthy controls (○, n = 112). The line of best fit for the healthy controls is shown in panels A and B. (C) Comparison of age-adjusted delta Tel in healthy controls and patients with DKC1 and TINF2 mutations. The box represents the interquartile range which contains the 50% of values. The whiskers are lines that extend from the box to the highest and lowest values, excluding outliers and extreme outliers (○ and
, n = 67) and healthy controls (○, n = 112). The line of best fit for the healthy controls is shown in panels A and B. (C) Comparison of age-adjusted delta Tel in healthy controls and patients with DKC1 and TINF2 mutations. The box represents the interquartile range which contains the 50% of values. The whiskers are lines that extend from the box to the highest and lowest values, excluding outliers and extreme outliers (○ and  , respectively). A line across the box indicates the median.
, respectively). A line across the box indicates the median.
TERC levels are not reduced in patients with DC with TINF2 mutations
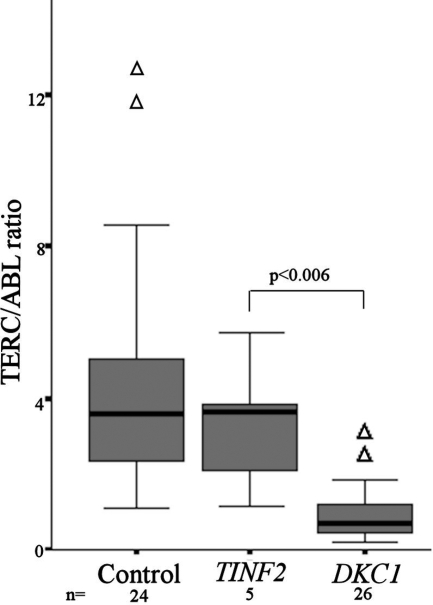
Samples were available for TERC quantitation in 5 of the patients with DC with TINF2 mutations. From previous studies we had demonstrated a range of normal TERC/ABL expression and showed that patients with DKC1 mutations have significantly lower TERC levels.6 Comparing TERC levels seen in patients with TINF2 mutations, there is no difference between the normal and the TINF2 patient groups, while the DKC1 patient group is significantly lower than the TINF2 group (Mann-Whitney P < .006; Figure 3).
Figure 3.
TERC/ABL levels are not reduced in patients with DC with TINF2 mutations. Comparison of TERC/ABL levels between healthy controls (n = 24), patients with DC with TINF2 mutations (n = 5), and patients with DC with DKC1 mutations (n = 26). The P value between TERC/ABL levels for TINF2 and DKC1 patients is given. The box represents the interquartile range which contains the 50% of values. The whiskers are lines that extend from the box to the highest and lowest values, excluding outliers (△). A line across the box indicates the median.
Patients with DC with TINF2 mutations are clinically severe
Clinically, most patients with DC with a mutation in TINF2 have severe disease. Compared with the normal view of DC pathology, AA is extremely prevalent in this group of patients, with 32 of 33 patients presenting with AA, often as a very early feature, with 23 of 33 patients developing AA before the age of 10 years. Table 3 summarizes the key differences between DC in the general patient population and those with TINF2 mutations. Differential diagnoses of HH and Revesz syndrome (RS) in combination with DC or each other have been made in a subgroup of 7 patients. This is dependent on their clinical presentations: features of microcephaly and developmental delay are associated with HH, and retinopathy is associated with RS. The mutations associated with the RS diagnosis are either Lys280X or Arg282His (2 each). The mutations associated with a diagnosis of pure HH is Arg282His (x2) or as an overlap in Lys280X (x2), Arg282His (1), and Pro283Ser (1).
Table 3.
Summary of the main characteristics of patients (with TINF2 mutations) in this study group compared with patients with DC in general
| Clinical feature | No. index patients in this study | Percentage of patients in this study | Percentage of patients reported in DC16 |
|---|---|---|---|
| AA at any age | 32 | 97 | 86 |
| AA at < 10 y | 21 | 63 | 51 |
| Nail dystrophy | 29 | 88 | 88 |
| Abnormal skin pigmentation | 19 | 58 | 89 |
| Leukoplakia | 19 | 58 | 78 |
| Classical triad | 12 | 36 | NA |
| Features of HH | 6 | 18 | NA |
| Osteoporosis | 6 | 18 | NA |
| Retinopathy (Revesz syndrome) | 4 | 12 | NA |
Classical triad indicates combination of nail dystrophy, abnormal skin pigmentation, and leukoplakia; NA, data not available.
TINF2 mutations are not a major cause of disease in the non-DC group
We also screened TINF2 exon 6 for mutation in 244 patients with different clinical presentations that have some overlap with DC. In this screen, we identified missense sequence changes in 8 individuals. Of these, 5 shared the polymorphism c.710G>A, Gly237Asp (rs17102313). The other 3 were unique missense mutations: c.706C>T, Pro236Ser, c.734C>A, Ser245Tyr and c.841G>A, Glu281Lys (Table 2). Due to lack of parental material, it was not possible to determine the segregation of these mutations.
Discussion
In this study, we undertook a screen of the uncharacterized index patients for mutations in TINF2 in the largest collection of patients with DC internationally. Of the 175 DC families screened, we identified coding mutations in 33 index patients. A total of 21 of these mutations affected residue 282, mutating this arginine to either cysteine or histidine. The remaining 12 mutations were all in a very tight cluster between aa 280 and aa 298. No additional mutations were found elsewhere in the gene. None of these changes are thought to be rare polymorphisms due to the number of healthy individuals that had been screened in this and other studies.13,22 All the mutated aa's were highly conserved across different mammalian species (Figure 1), and this combined with the lack of variation data suggest that these mutations are likely to be pathogenic. The significance of this tight clustering is unclear at present, as these aa's have not been associated with known interactions, but this could form a new unidentified binding site that could have a critical role in telomere protection. However, only 3 private missense mutations were found in 244 patients who did not have DC (including those with AA and myelodysplasia), with 2 of 3 lying outside the cluster associated with DC. This suggests TINF2 mutations do not have major role in patients who do not have DC.
Clinically, all the patients with DC presenting with a TINF2 mutation have severe disease, including some with features of the severe variant HH and the rare RS (Table 2). The mutations in patients with HH and/or RS are clustered at aa 280, 282, and 283 and overlap with the location of the mutation identified by Savage for RS, Arg282His.13 As highlighted in Table 3, the overriding feature of patients with TINF2 mutations is the high prevalence and early presentation of AA, often arising before the development of the more classical DC abnormalities. Osteoporosis is also more prevalent in this subgroup of patients with DC. These data suggest that any patients with genetically uncharacterized DC presenting with AA should be screened for mutations in exon 6 of TINF2, particularly if the presentation appears to be sporadic rather than having a family history. Interestingly, there also appears to be a bias toward males (approximately 3:1; Table 2). The precise explanation for this is unclear because of the 175 uncharacterized index patients, the male-to-female ratio is 1.7:1 (111:64).
The telomere length analysis shows that, as observed previously in DC, patients with TINF2 mutations have significantly shorter telomeres compared with controls. However, what is striking is that patients with TINF2 mutations have even shorter telomeres compared with patients with DKC1 mutations, which are usually the more severe patient group (Figure 2B,C). Thus, in the TINF2 group there appears to be correlation between disease severity, telomere length, and age; specifically, the severest disease appears to be associated with the shortest telomeres in the youngest patients. This is in contrast to the TERC/ABL ratios observed in the different groups, where patients with TINF2 mutations have similar ratios to those seen in the control population, and are significantly higher than those of the DKC1 mutation group (P < .006). This supports our earlier findings that reduced TERC accumulation levels are not a universal feature of DC, and appears to be mutation dependent.6
In families where parental samples were available, 16 of 17 appeared to be de novo. In DCR19, the Arg282Cys mutation is inherited from the father by 2 of his children. There are 2 possible explanations for this. It is either another example of disease anticipation as previously observed in families with TERC mutations,23 as the 2 children have developed disease at a younger age than their father, or Arg282Cys is a milder mutation, and individuals with this variant survive long enough to have children. DCR24 has an unusual presentation relative to the parental status. In this family the disease appears to be familial, as there were 2 affected children (1 deceased, no sample available). However, the Arg282Cys is not detected in either parent. This may be an example of gonadal mosaicism, but this cannot be explored further due to a lack of appropriate material.
The high prevalence of the Arg282 mutations in patients with DC is interesting and broadly agrees with the findings of Savage et al, where in 4 out of 5 families identified with TINF2 mutations, the mutation affected Arg 282.13 A possible explanation is the spontaneous deamination of a CpG site resulting in C>T transitions of both the forward and reverse strands. This gives rise to the 2 mutations (Arg>Cys and Arg>His) seen repeatedly in this study. Although this mechanism may increase the mutation rate, it cannot explain the very tight cluster of mutations in this region of the protein to the exclusion of the rest of the protein.
Considering all the mutations arising in patients in the DCR, the 2 most commonly mutated genes, DKC1 and TINF2, account for 119 index patients. Ala353Val in dyskerin has been observed in 32 index patients, and mutation of Arg282 in TIN2 has been seen in 21 index patients. Together, these 2 changes account for approximately 35% of the genetic changes in the DCR. As both of these mutations can be identified by simple tests, it would be appropriate to screen all newly diagnosed patients for these before more extensive genetic analysis is undertaken.
The precise effect of these mutations is unknown. As a group, individuals with a TINF2 mutation have shorter telomeres than others with known mutations, but their TERC accumulation levels appear normal. This observation gives rise to a potential mechanism of defective telomere protection. TIN2 is considered to be the linchpin in the shelterin complex. It has been shown to tether the other components of the shelterin complex together and thereby allow their correct interaction with telomeric DNA.24,25 If mutations in TIN2 change the conformation of the protein sufficiently, then these interactions may not be as tight and the protection afforded by the shelterin complex is reduced. This potentially could allow inappropriate DNA-processing mechanisms access to the telomeric DNA, thereby reducing telomere length. This idea is strengthened by recent work involving fusing TRF1 to POT1. In this study telomere lengths were shortened, whereas when POT1 and TFR1 are expressed, normally telomere lengths were lengthened,26 suggesting a link between shelterin interactions and telomere length. What is striking, however, is the similarity in the phenotypes produced by defects in telomere elongation and protection mechanisms both in terms of telomere lengths and clinical phenotypes, suggesting that however telomeres are inappropriately shortened, the clinical outcome is the same.
In conclusion, the findings from this study show that TINF2 mutations account for approximately 11% of patients with DC, but that they do not have a major role in related disorders such as AA. As a group, patients with TINF2 mutations have severe disease and the shortest telomeres compared with other DC subgroups. These findings further support the idea that DC is principally a disorder of defective telomere maintenance and highlight the severe consequences in humans of telomere dysfunction. Finally, these findings will facilitate early diagnosis and appropriate management in a significant subset of patients with DC.
Acknowledgments
We thank families and their clinicians for patient samples, which make this work possible.
This work was supported by The Wellcome Trust and The Medical Research Council (MRC).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.J.W. performed dHPLC analysis, sequence analysis, and QT-PCR analysis, and is the main author; T.V. performed dHPLC analysis, telomere length analysis, restriction digest analysis, and is an assisting author; R.B. performed additional experimental analysis; M.K. performed figure preparation and additional experimental analysis; and I.D. performed clinical data analysis, heads the laboratory, and is an assisting author.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Amanda J. Walne, Centre for Paediatrics, Institute of Cell and Molecular Science, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Barts and The London Children's Hospital, 4 Newark Street, London, E1 2AT, United Kingdom; e-mail: a.walne@qmul.ac.uk.
References
- 1.Vulliamy TJ, Marrone A, Knight SW, Walne A, Mason PJ, Dokal I. Mutations in dyskeratosis congenita: their impact on telomere length and the diversity of clinical presentation. Blood. 2006;107:2680–2685. doi: 10.1182/blood-2005-07-2622. [DOI] [PubMed] [Google Scholar]
- 2.Savage SA, Alter BP. The role of telomere biology in bone marrow failure and other disorders. Mech Ageing Dev. 2008;129:35–47. doi: 10.1016/j.mad.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heiss NS, Knight SW, Vulliamy TJ, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 4.Vulliamy T, Marrone A, Goldman F, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 5.Armanios M, Chen JL, Chang YP, et al. Haplo-insufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci U S A. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walne AJ, Vulliamy T, Marrone A, et al. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum Mol Genet. 2007;16:1619–1629. doi: 10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vulliamy T, Beswick R, Kirwan M, et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc Natl Acad Sci U S A. 2008;105:8073–8078. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marrone A, Walne A, Tamary H, et al. Telomerase reverse-transcriptase homozygous mutations in autosomal recessive dyskeratosis congenita and Hoyeraal-Hreidarsson syndrome. Blood. 2007;110:4198–4205. doi: 10.1182/blood-2006-12-062851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du HY, Pumbo E, Manley P, et al. Complex inheritance pattern of dyskeratosis congenita in two families with 2 different mutations in the telomerase reverse transcriptase gene. Blood. 2008;111:1128–1130. doi: 10.1182/blood-2007-10-120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vulliamy T, Marrone A, Dokal I, Mason PJ. Association between aplastic anaemia and mutations in telomerase RNA. Lancet. 2002;359:2168–2170. doi: 10.1016/S0140-6736(02)09087-6. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi H, Baerlocher GM, Lansdorp PM, et al. Mutations of the human telomerase RNA gene (TERC) in aplastic anemia and myelodysplastic syndrome. Blood. 2003;102:916–918. doi: 10.1182/blood-2003-01-0335. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi H, Calado RT, Ly H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 13.Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 15.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 16.Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem. 2004;279:43799–43804. doi: 10.1074/jbc.M408650200. [DOI] [PubMed] [Google Scholar]
- 17.Walne AJ, Dokal I. Dyskeratosis Congenita: a historical perspective. Mech Ageing Dev. 2008;129:48–59. doi: 10.1016/j.mad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Brown WR, MacKinnon PJ, Villasante A, Spurr N, Buckle VJ, Dobson MJ. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- 19.Notaro R, Cimmino A, Tabarini D, Rotoli B, Luzzatto L. In vivo telomere dynamics of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 1997;94:13782–13785. doi: 10.1073/pnas.94.25.13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savage SA, Stewart BJ, Eckert A, Kiley M, Liao JS, Chanock SJ. Genetic variation, nucleotide diversity, and linkage disequilibrium in seven telomere stability genes suggest that these genes may be under constraint. Hum Mutat. 2005;26:343–350. doi: 10.1002/humu.20226. [DOI] [PubMed] [Google Scholar]
- 23.Vulliamy T, Marrone A, Szydlo R, Walne A, Mason PJ, Dokal I. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet. 2004;36:447–449. doi: 10.1038/ng1346. [DOI] [PubMed] [Google Scholar]
- 24.Ye JZ, Donigian JR, van Overbeek M, et al. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Yang Y, van Overbeek M, et al. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 26.Etheridge KT, Compton SA, Barrientos KS, Ozgur S, Griffith JD, Counter CM. Tethering telomeric double- and single-stranded DNA-binding proteins inhibits telomere elongation. J Biol Chem. 2008;283:6935–6941. doi: 10.1074/jbc.M708711200. [DOI] [PubMed] [Google Scholar]