Abstract
RAD51 and other members of the RecA family of strand exchange proteins assemble on ssDNA to form presynaptic filaments, which carry out the central steps of homologous recombination. A microplate-based assay was developed for high-throughput measurement of hRAD51 filament formation on ssDNA. With this method, a 10,000 compound library was screened, leading to the identification of a small molecule (RS-1) that enhances hRAD51 binding in a wide range of biochemical conditions. Salt titration experiments showed that RS-1 can enhance filament stability. Ultrastructural analysis of filaments formed on ssDNA showed that RS-1 can increase both protein–DNA complex lengths and the pitch of helical filament turns. RS-1 stimulated hRAD51-mediated homologous strand assimilation (D-loop) activity by at least 5- to 11-fold, depending on the condition. This D-loop stimulation occurred even in the presence of Ca2+ or adenylyl-imidodiphosphate, indicating that the mechanism of stimulation was distinct from that conferred by Ca2+ and/or inhibition of ATPase. No D-loop activity was observed in the absence of a nucleotide triphosphate cofactor, indicating that the compound does not substitute for this requirement. These results indicate that RS-1 enhances the homologous recombination activity of hRAD51 by promoting the formation of active presynaptic filaments. Cell survival assays in normal neonatal human dermal fibroblasts demonstrated that RS-1 promotes a dose-dependent resistance to the cross-linking chemotherapeutic drug cisplatin. Given that RAD51-dependent recombination is a major determinant of cisplatin resistance, RS-1 seems to function in vivo to stimulate homologous recombination repair proficiency. RS-1 has many potential applications in both research and medical settings.
Keywords: cross-linking chemotherapy, DNA repair, high-throughput screen, recombinase, strand exchange
Homologous recombination (HR) has multiple roles in DNA repair, including the repair of double strand breaks (DSBs) and recovery from the replication-blocking lesions formed by DNA cross-linking agents. HR repairs DSBs by locating a homologous stretch of DNA and replicating the missing genetic information from this homologous template. In contrast to DSB repair by nonhomologous end joining, HR repair generally occurs without mutations. Because of this, HR repair is critically important in the maintenance of genomic stability (reviewed in ref. 1). The proposed mechanism for this pathway begins with 5′ to 3′ nuclease activity at the DSB, resulting in a 3′ single-stranded tail. The tail is coated by replication protein A, which is subsequently replaced by a helical filament of RAD51 protein. This displacement of replication protein A by RAD51 seems to be controlled by a number of mediator proteins, which include BRCA2, RAD52, and RAD51 paralogue complexes (2–5). The RAD51-coated 3′ tail then locates and invades a homologous template of dsDNA. After invasion, templated DNA synthesis initiated at 3′ ends leads to formation of branched DNA intermediates, which are thought to be resolved by structure-specific endonucleases or by a process referred to as synthesis-dependent strand annealing.
Several studies have shown that HR can be upregulated by alterations in RAD51. For example, RAD51 overexpression in chicken DT40 cells can partially compensate for the loss of filament assembly mediator proteins, including RAD51 paralogues and other HR-related proteins (6–8). Furthermore, Fortin and Symington (9) have generated and characterized several gain-of-function mutations in yeast RAD51. These point mutations in scRAD51 are located in regions responsible for DNA binding and/or ATP-dependent conformational changes. One such mutant RAD51 protein had elevated DNA binding activity that could displace replication protein A from ssDNA in the absence of mediator proteins. Expression of these mutant proteins in yeast partially bypassed the requirement for the assembly mediators RAD55 and RAD57 in DNA repair. These examples demonstrate that physical changes to RAD51 protein can upregulate HR in vivo. These findings provided the rationale to search for small molecules that stimulate RAD51's DNA binding activity in vitro.
RAD51 filament formation is a well-accepted critical step in HR repair. Several biochemical assays have been developed to measure RAD51 protein assembly on ssDNA. One such method uses oligonucleotides that are end-labeled with a fluorescent tag. Binding of proteins to this substrate DNA can be measured according to fluorescence polarization (FP) of the tag (10). We modified this method so that small sample volumes could be monitored on a high-throughput scale. A naïve library of 10,000 small molecules was screened in search of compounds that modulate the formation of hRAD51 filaments on ssDNA. Using this approach, we identified a compound that specifically stimulates the formation of active hRAD51 filaments and enhances hRAD51's recombination activity.
Results
Identification of a Compound That Stimulates hRAD51.
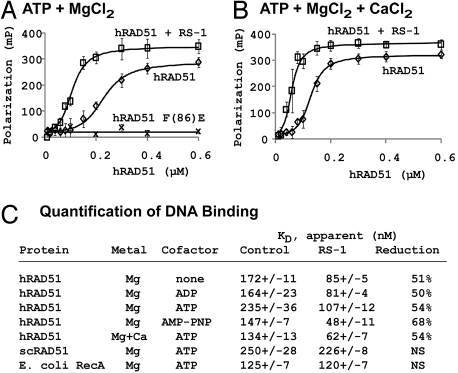
To identify small molecules that modify the DNA binding properties of hRAD51, we used an FP-based method that involves incubation of hRAD51 protein with oligonucleotides that are end-labeled with a fluorescent tag. The binding of hRAD51 to this substrate DNA increases polarization of the fluorescent tag. This assay was modified such that small-volume reactions (30 μl) could be assayed in 384-well microplates using a fluorescent plate reader. The method was validated using both wild-type hRAD51 protein and hRAD51 carrying an F86E mutation, which is known to impair the ability of hRAD51 protein to bind DNA (11, 12). A titration of wild-type hRAD51 protein in 2 mM ATP yielded a sigmoidal binding curve with good agreement with published values (10); RAD51-F86E gave almost no signal, as expected (Fig. 1A).
Fig. 1.
Effects of RS-1 on DNA binding. Various concentrations of DNA strand exchange proteins were incubated with 10 nM polydT containing a 5′ OG tag. Reactions were performed in Buffer B in the absence (diamonds) or presence of 20 μM RS-1 (squares). (A) hRAD51 in ATP and 10 mM MgCl2. (B) hRAD51 in ATP and 1 mM CaCl2 + 10 mM MgCl2. (C) Quantification of DNA binding by DNA strand exchange proteins under various conditions. NS, no statistically significant reduction.
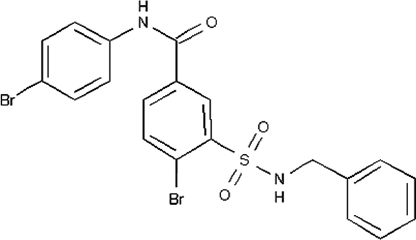
Under conditions described in the supporting information (SI) Methods, this assay was tested with a method that evaluates its robustness for high-throughput screening and yielded a Z factor of 0.82, validating it as an excellent assay (13). A naïve 10,000-compound library (Chembridge DIVERSet) was screened using a “mixed compound” strategy, wherein each reaction contained a mixture of eight compounds. When an FP elevation of >30% was observed for any given compound mixture, the eight compounds from that mixture were individually tested using identical conditions. Compound mixtures were deemed false positives when epifluorescence measurements showed them to be intrinsically fluorescent (17 mixtures) or fluorescein-quenching (4 mixtures). After omission of these, the screen yielded 15 “hit” mixtures, and the subsequent screen of individual compounds identified three small molecules that increased FP by ≥45%. This represents a “hit rate” of 0.03%. The most active compound, 3-[(benzylamino)sulfonyl]-4-bromo-N-(4-bromophenyl)benzamide, increased FP signal in excess of twofold. This compound will hereafter be referred to as RS-1 (for RAD51-stimulatory compound 1). RS-1's chemical structure is shown in Fig. 2. Physiochemical properties of RS-1 include four H-bond acceptors, two H-bond donors, a partition coefficient log P of 5.19, and a molecular mass of 524.23 Da. Therefore, RS-1 satisfies two of the four Lipinski rules that predict “drug-likeness,” and it nearly satisfies all four (14). The other two stimulatory compounds, RS-2 and RS-3, have yet to be characterized in detail and will be described in a separate article.
Fig. 2.
The chemical structure of RS-1 (3-[(benzylamino)sulfonyl]-4-bromo-N-(4-bromophenyl)benzamide).
The high-throughput screen also yielded 106 compound mixtures that reduced FP by at least 70%, and the subsequent screen of individual compounds identified 76 small molecules that reduced FP by ≥50%; these too will be described separately.
Effects of RS-1 on Binding of hRAD51 to DNA.
To quantify effects of RS-1, the FP-based assay was used to measure binding of hRAD51 to an Oregon Green (OG)-conjugated 45-mer polydT substrate. RS-1 stimulated binding in the presence of ATP or ADP and in the absence of a nucleotide cofactor (Fig. 1 A and C).
RS-1's ability to stimulate hRAD51 was not due to direct interaction with DNA. Previous studies showed that intercalating agents like ethidium bromide can promote the assembly of DNA strand exchange proteins on dsDNA (15). For this reason it was important to show that RS-1 stimulatory activity did not result from altering DNA conformation. The following three experiments were performed to eliminate this possibility. First, RS-1 was shown to have no detectable effect on the FP-based assay in the absence of RAD51 protein, suggesting that it did not interact directly with polydT. Second, relaxed and supercoiled dsDNA circular plasmids (ϕX174 RF) were electrophoresed through agarose gels containing 1 μM ethidium bromide, 10 μM RS-1, or neither. As expected, ethidium bromide induced coiling of the relaxed plasmid, resulting in an increased electrophoretic mobility; this effect was not seen in the gels containing RS-1 or no agent, suggesting that RS-1 is not a DNA intercalator (data not shown). Finally, RS-1 had no stimulatory activity when tested on related DNA strand exchange proteins, including Escherichia coli RecA, scRAD51, and scDMC1 (Fig. 1C). The specificity of RS-1 for hRAD51 indicates that it likely functions by altering contacts and/or conformations of amino acid residues in hRAD51 that are not conserved in other proteins, rather than by interacting with DNA.
RS-1 Stimulates RAD51 by a Mechanism That Does Not Require Inhibition of ATP Hydrolysis.
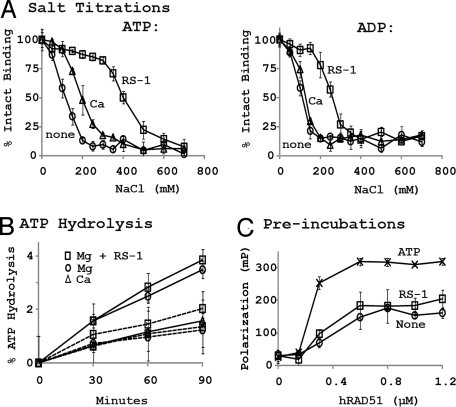
Previous work showed that hRAD51 filament formation and recombinase activities are enhanced under conditions that suppress or prevent hydrolysis of ATP. These stimulatory conditions included the use of Ca2+ in buffers or the use of adenylyl-imidodiphosphate (AMP-PNP) in place of ATP (16–18). As expected, our FP assay showed higher levels of RAD51–DNA binding in AMP-PNP or Ca2+ ions relative to standard conditions containing ATP and Mg2+ ions (Fig. 1C). Three experiments were performed to determine that the stimulation promoted by RS-1 is not similarly related to ATP hydrolysis. First, RS-1 was found to stimulate DNA binding in the presence of Ca2+ or AMP-PNP. This was true even when Ca2+ and AMP-PNP were present at or above concentrations sufficient for their maximum effect (Fig. 1 B and C). The elevation in binding affinity caused by addition of RS-1 was even greater in AMP-PNP than in standard conditions containing ATP and Mg2+. Second, the stability of filaments was tested by challenging hRAD51–DNA complexes with NaCl (Fig. 3A). Ca2+ is known to stabilize hRAD51 filaments formed in the presence of ATP but not those formed in ADP (16). We repeated these findings and also found that RS-1 stabilized hRAD51–DNA complexes formed in the presence of either ATP or ADP. Third, DNA-dependent ATP hydrolysis rates of hRAD51 were measured in the presence of Ca2+ or RS-1 (Fig. 3B). Consistent with a previous report (16), 5 mM Ca2+ inhibited ATP hydrolysis activity of hRAD51, relative to hydrolysis in 10 mM Mg2+ (kcat = 0.18 ± 0.1 vs. 0.39 ± 0.4 min−1). In contrast, RS-1 had little, if any, reproducible impact on ATPase activity (kcat = 0.43 ± 0.05 min−1). When performed in the absence of DNA, neither Ca2+ nor RS-1 had a significant effect on ATPase. Taken together, this series of experiments suggests that RS-1 does not stimulate RAD51 by inhibiting its ATPase activity.
Fig. 3.
RS-1 stimulates hRAD51 via a novel mechanism. (A) RS-1, but not CaCl2, stabilizes hRAD51 nucleoprotein filaments formed in the presence of ADP. hRAD51 (500 nM) was incubated for 30 min with 10 nM OG-tagged polydT in Buffer B containing various combinations of 2 mM nucleotide cofactor, 1 mM CaCl2 (in addition to 10 mM MgCl2), and/or 20 μM RS-1, as indicated. Protein–DNA complexes were subsequently challenged by the addition of various concentrations of NaCl. (B) RS-1 does not inhibit hRAD51's ATPase activity. DNA-dependent ATP hydrolysis (shown by solid lines) was measured after incubation of 2 μM hRAD51 with 6 μM M13 ssDNA under conditions described in SI Methods. Buffers contained either 10 mM MgCl2, 5 mM CaCl2, or 10 mM MgCl2 + 20 μM RS-1, as indicated. Dotted lines show repeat experiments performed without DNA. (C) ATP but not RS-1 allows the DNA binding activity of hRAD51 to tolerate pretreatment with MgCl2. hRAD51 was preincubated at for 30 min at 37°C in Buffer B in combination with either 2 mM ATP, 20 μM RS-1 with no nucleotide cofactor, or neither, as indicated. OG-tagged polydT (10 nM) and 2 mM ATP were subsequently added, and the mixtures were incubated for an additional 30 min before FP was measured.
Because nucleotide cofactor binding can alter RAD51–DNA interactions, the possibility that RS-1 functions by mimicking a nucleotide cofactor was considered. Prior studies demonstrated that hRAD51's ability to bind DNA is reduced by preincubation of the protein with Mg2+ unless a nucleotide cofactor is also present. This loss seems to result from hRAD51 protein aggregation (19). We took advantage of this effect to ask whether RS-1 mimics ATP. We found that RS-1 could not prevent the reduction of RAD51 binding activity caused by incubation with Mg2+ in the absence of nucleotide (Fig. 3C).
RS-1 Alters the Length and Pitch of RAD51 Protein–DNA Complexes.
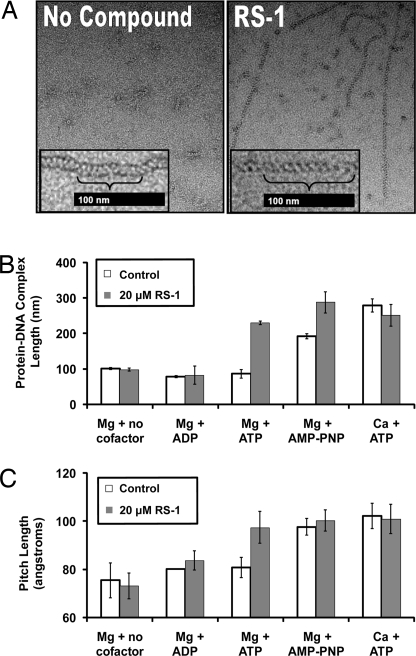
Transmission electron microscopy (TEM) imaging was performed using a 1,000-nucleotide ssDNA substrate to determine whether RS-1 influences hRAD51 filaments. Addition of RS-1 to reactions containing ATP and Mg2+ resulted in longer protein–DNA complexes than those seen without RS-1 addition (Fig. 4 A and B). The protein–DNA complex length distribution for this experiment is reported in Fig. S1. When repeated with ADP in place of ATP or in the absence of a nucleotide cofactor, RS-1 had no detectable effect on length (Fig. 4B). Substituting AMP-PNP for ATP or substituting Ca2+ for Mg2+ increased the average lengths by 2.2- and 3.2-fold, respectively. In the case of AMP-PNP, the addition of RS-1 resulted in even further stimulation of length. RS-1 had no additional effect on length beyond that observed with Ca2+.
Fig. 4.
RS-1 affects the average length and helical pitch of hRAD51 protein–DNA complexes. Electron microscopy images of hRAD51 filaments formed on ssDNA were obtained at a magnification of ×25,000. Fifty randomly selected filaments were analyzed for each reaction condition. (A) RS-1 alters protein–DNA complex length and appearance. Representative images for filaments formed in ATP and Mg2+ plus the presence (Right) or absence (Left) of 20 μM RS-1. Insets at higher magnification demonstrate differences in length (brackets indicate 10 striations and bars denotes 100 nm). (B) Forty to fifty unselected protein–DNA complexes were measured. Cofactor requirements for RS-1 stimulation are displayed, and error bars represent SD. (C) RS-1 has a similar effect as AMP-PNP or Ca2+ on filament pitch (magnification was ×39,000).
Filaments were also examined to determine their helical pitch. Previous studies with RecA and RAD51 showed that active filaments formed in the presence of poorly hydrolyzable NTP analogues have pitch lengths of 90–100 Å, whereas inactive filaments have an average pitch length of 70–80 Å (18, 20–23). In the presence of ATP and Mg2+, hRAD51 filaments had a pitch consistent with the inactive form. This finding was expected because filament-bound ATP is expected to be hydrolyzed to yield ADP and hence form inactive filaments (18, 22, 23). In contrast, addition of RS-1 to ATP yielded an average pitch length of 98 ± 7 Å. Thus, the majority of protomers in each filament seem to be in the active conformation (Fig. 4 A Left Inset and Right Inset and C). When repeated in ADP or in the absence of a nucleotide cofactor, filaments had an average pitch of approximately 70–80 Å, regardless of whether RS-1 was present. These findings suggest that stabilization of hRAD51 filaments in the active conformation by RS-1 requires an NTP. Given that RS-1 does not inhibit RAD51's ATPase activity, this finding also suggests that, so long as ATP is present initially, RS-1 can lock filaments in an active conformation that is maintained even when hydrolysis converts ATP to ADP.
RS-1 Stimulates the Strand Assimilation Activity of hRAD51.
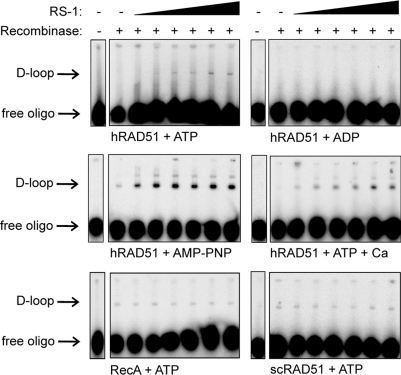
Strand assimilation assays were performed to assess the effect of RS-1 on hRAD51's recombination function in vitro. In this assay, hRAD51 is first allowed to assemble on a 32P-labeled ssDNA oligonucleotide in the presence or absence of RS-1. A homology-containing supercoiled target plasmid is then added, and the two are allowed to form a homology-dependent joint molecule referred to as a “D-loop.” RS-1 supported the formation of D-loops (by at least fivefold) in a concentration-dependent manner in the presence of ATP and Mg2+ (Fig. 5). D-loop formation was not observed under these conditions in the absence of RS-1. When repeated in the presence of ADP, no D-loops were observed regardless of RS-1 concentration, indicating a requirement of NTP for this activity. This is consistent with the NTP requirement for the formation of filaments with approximately 100-Å pitch lengths in the TEM experiments (Fig. 4C). D-loop formation was strongly stimulated by buffers containing AMP-PNP or Ca2+ ions, as expected (16). Addition of RS-1 resulted in further stimulation of D-loop formation to levels that were 5.7- or 11-fold higher, respectively, than those seen with AMP-PNP or Ca2+ alone (see quantifications of gel images in Fig. S2). RS-1 had no effect when hRAD51 was replaced with E. coli RecA or scRAD51 using standard buffer conditions; the D-loop activities for both of these proteins could be stimulated by Ca2+ ions, but no additional stimulation was observed with the addition of RS-1 to Ca2+-containing buffers (Fig. S2). Furthermore, no stimulatory activity was observed with scDMC1 (data not shown). These findings are consistent with the FP assay results, and taken together, these results suggest that the stimulatory effects of RS-1 require contacts with residues in hRAD51 that are not conserved in these homologous DNA strand exchange proteins.
Fig. 5.
RS-1 stimulates strand assimilation activity of hRAD51. Various DNA strand exchange proteins were allowed to form joint molecules (“D-loops”) in the presence of RS-1 (concentration range: 0, 1, 5, 10, 15, 20, and 25 μM). Reactions were performed as described in Methods, and phosphorimages of agarose gels are displayed for various conditions, as indicated. The positions of free oligo and oligo associated with D-loops are indicated.
RS-1 Promotes Resistance of Primary Human Fibroblasts to the Chemotherapeutic Agent Cisplatin.
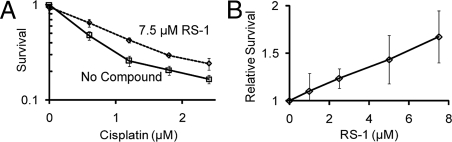
The HR pathway is critically important for cellular repair of DNA damage induced by cross-linking chemotherapies (6, 24). Mutations that eliminate RAD51-dependent recombination show profound sensitivity to these agents. For this reason, cell survival analyses were performed to determine whether RS-1 could stimulate hRAD51 in vivo and increase cellular resistance to cisplatin. Cells were incubated for 24 h in cisplatin with or without RS-1 and were subsequently incubated for an additional 6 days in normal media lacking both agents. Normal nonimmortalized early-passage neonatal human dermal fibroblasts demonstrated significantly greater survival after receiving cisplatin and RS-1, relative to those receiving cisplatin alone (Fig. 6A). Furthermore, this protective effect was dose dependent in the range of 1–7.5 μM RS-1 (Fig. 6B), which is identical to the concentration range required for biochemical stimulation of DNA binding or strand assimilation. It should be noted that the 24-h time of incubation with RS-1 represents less than one doubling time for this cell type, and 24-h incubations in RS-1 at concentrations of ≤7.5 μM did not significantly affect cell counts (data not shown). These considerations make it quite unlikely that the observed effect of RS-1 on cisplatin sensitivity is the result of a nonspecific effect on cell growth. Taken together, this series of experiments suggests that RS-1 is capable of gaining intracellular access and increasing the efficiency with which potentially lethal cisplatin-induced DNA damage is repaired.
Fig. 6.
RS-1 promotes resistance of human cells to cross-linking chemotherapy. Survival analyses were performed on neonatal human dermal fibroblasts. Cells were incubated for 24 h in media containing varying concentrations of cisplatin in the presence or absence of RS-1. Drugs were then removed, and cells were allowed to grow in complete media for an additional 6 days. Survival was measured with an sulforhodamine B method, as described in Methods. (A) Cisplatin titration shows that RS-1 at 7.5 μM promotes a resistance to cisplatin. (B) RS-1 titration at 1.2 μM cisplatin shows a concentration-dependent RS-1 effect.
Discussion
We identified RS-1 on the basis of its ability to stimulate binding of hRAD51 to ssDNA. Further work has demonstrated that low-micromolar concentrations of this small molecule enhance DNA binding and result in longer protein–DNA complex lengths. In addition, RS-1 stabilizes the active form of hRAD51 filaments, in which each helical turn has a pitch length of approximately 100 Å, and this is reflected in an enhanced strand assimilation activity.
RS-1 seems to modulate hRAD51 interaction with DNA in two distinct ways: one that is NTP-dependent and another that is NTP-independent. The FP-based assay showed that RS-1 can stimulate hRAD51 binding in the absence of a nucleotide cofactor. However, it should be noted that this assay, like most DNA binding assays, does not distinguish active DNA filaments from other types of protein–DNA complexes. RS-1's NTP-independent stimulatory activity leads to hRAD51–DNA complexes that are not well visualized on TEM and lack the ability to form D-loops. In the presence of an NTP, on the other hand, RS-1 exerts a functional stimulation that promotes active filament formation. Assays that are sensitive to filament length, filament quality, or strand exchange activity have consistently demonstrated that an NTP cofactor is required for optimal hRAD51 activity (10, 17, 25–27). One common spectroscopic assay uses the change in fluorescence of etheno-DNA, which is caused by the unstacking of bases that is associated with filament formation. It was not possible to use this etheno-DNA method in our case because the spectroscopic properties of RS-1 were incompatible (data not shown). However, our TEM-based filament imaging and D-loop experiments provide clear evidence that ATP or an NTP analogue is required for RS-1 to stimulate the formation of active functional hRAD51 filaments.
A striking finding was that RS-1 stabilized the active hRAD51 filaments formed on ssDNA in the presence of ATP. Active hRAD51 filaments are known to be favored under conditions that inhibit hRAD51's ATPase activity, which is thought to lock hRAD51-DNA complexes in their active ATP-bound state (16–18). RS-1's mechanism of action is clearly distinct from the effects associated with ATPase inhibition because it (i) does not inhibit ATP hydrolysis, and (ii) stimulates HR activity above that induced by ATPase inhibition. These results suggest that RS-1 acts as an allosteric regulator that locks hRAD51 in an active conformation and does so without influencing the active site for ATP hydrolysis.
Recent studies identified two peptide fragments of BRCA2 that can act to promote RAD51 filaments (28, 29). These peptides seem to bind an interface created by two adjacent RAD51 monomers. One of the peptides, derived from the Caenorhabditis elegans BRCA2 orthologue Ce-Brc2, was found to stimulate assembly of RAD51 at damaged sites in vivo. It is possible that RS-1's mechanism of action shares properties with one or both of these peptides and that further study of RS-1's activity will prove instructive for understanding BRCA2 function. On the other hand, it is clear that the Ce-Brc2–derived peptide differs in activity from RS-1, because the peptide inhibits RAD51's ATPase activity. Structural characterization of the interaction between RS-1 and RAD51 should provide better insights into its mechanism of action.
Cell survival assays show RS-1 can increase cellular resistance of neonatal human dermal fibroblasts to cisplatin. This activity is likely the result of RS-1's ability to stimulate HR. If HR can be effectively stimulated in various cell types, one can envision a number of potential applications for RS-1 (or RS-1–derived compounds) that could be far-reaching in significance. Such an agent could potentially be used to temporarily elevate cellular resistance to DNA-damaging agents, thereby increasing cell survival and perhaps reducing mutagenesis. This would be an attractive approach for protecting the normal tissues of patients receiving chemotherapy and/or radiotherapy. After treatment with cisplatin, for example, injuries to the kidneys and auditory organs are common and remain dose-limiting toxic events (30). Such an agent could also be used to protect healthy individuals from the effects of acute exposure to DNA-damaging agents in the environment. Another potential use of an HR-stimulating compound such as RS-1 would be as a reagent to enhance HR-mediated gene targeting. If such activity is found, the implications for controlled genetic modification of human cells could be far reaching. Furthermore, related compounds could greatly improve genetic analysis of many model systems, including C. elegans and Arabidopsis thaliana, for which gene targeting is currently either very inefficient or not possible. Finally, RS-1 is likely to be generally useful in characterization of RAD51's activity in purified systems.
In summary, we have identified a chemical compound that stimulates the DNA binding and recombination activities of human RAD51 protein. RS-1 seems to promote the recombinogenic activity of RAD51 by acting as an allosteric effector of active filament structure. RS-1 or related compounds could potentially have many important applications in both research and medical settings.
Materials and Methods
Plasmids and Proteins.
Methods for preparing hRAD51 are detailed in the SI Methods. A plasmid encoding scRAD51 was provided by Dr. Phoebe Rice, and the protein was purified by a previously described method (31). RecA was purchased from New England Biolabs. scDMC1 was purified as described previously (32). Patrick Sung and Wolf-Dietrich Heyer generously provided scRAD54.
DNA Binding Assays.
All reactions were performed in black polystyrene 384-well plates with reaction volumes of 30–100 μl. Purified DNA strand exchange proteins and chemical compounds were incubated at 37°C for 30 min with 5′ terminal fluorescently tagged oligonucleotide substrates (synthesized and purified by Integrated DNA Technologies). DNA binding was measured as a function of FP. Concentrations are reported as nucleotide concentrations for ssDNA and base pair concentrations for dsDNA.
A fluorescein-conjugated 45-mer polydT substrate at 100 nM was used for the small molecule screen, and methodologic details of the screen are provided in the SI Methods. All other FP-based experiments used an OG-conjugated 45-mer polydT substrate at 10 nM, because this fluorescent tag is more photo-stable. Reactions were performed in 1× Buffer B [40 mM Hepes (pH 7.5), 10 mM MgCl2, 0.1 mM DTT, 1 μM BSA, 2% glycerol, 30 mM NaCl, and 2 mM nucleotide cofactor] and other components as indicated. FP was measured with a Safire2 plate reader (Tecan), using the following settings: excitation 470 ± 5 nm, emission 530 ± 5 nm, 10 reads per well, with Z height and G factor autocalibrated from control wells. Data were fit to the cooperativity equation (33) using KaleidaGraph 4.0 software (Synergy), and KD apparent values were calculated. All conditions were performed at least in triplicate; mean values are reported and errors represent SD.
ATPase Assays.
These were performed as previously described (16, 34), with some modifications detailed in the SI Methods.
Imaging with Electron Microscopy.
A 1,000-nucleotide ssDNA substrate was generated as described in the SI Methods. RAD51 protein–DNA complexes were prepared by incubating 15 μM 1,000-nucleotide ssDNA with 5 μM hRAD51 protein at 37°C for 10 min in buffer containing 20 mM Hepes pH 7.5, 1 mM DTT, 0.5 mM nucleotide cofactor, and 5 mM MgCl2. Some conditions contained 2 mM CaCl2 in place of MgCl2 and/or contained 20 μM RS-1 as indicated. Samples were spread on carbon-coated grids and negatively stained with 1% uranyl acetate. Images were taken with a Gatan digital camera on a Tecnai F30 transmission electron microscope operated at 300 kV. The collection method consisted of an initial scan at ×19,500 to locate a region of the grid with several visible filaments. Starting from that location, the grid was then continuously scanned at ×25,000 to collect images of 50 unselected complexes. Protein–DNA complexes less than 50 nm in length were omitted in the analyses of contour lengths; final sample sizes contained 40–50 protein–DNA complexes. Pitch lengths were measured only from filaments with clear striations using ×39,000 images and DigitalMicrograph 3.10.1 software (Gatan). For each filament, a stretch of three striations was measured, and pitch length was determined as that value divided by 3.
D-Loop Assays.
The ability of various DNA strand exchange proteins to promote homologous strand assimilation was tested with methods previously described (34), with some modifications described in the SI Methods. The scRAD51 accessory protein scRAD54 was added to reactions containing scRAD51, because scRAD51 is incapable of D-loop production by itself (35).
Cell Survival Assays.
Early-passage neonatal human dermal fibroblasts were maintained in Medium 106 supplemented with low serum growth supplement (cells and media from Cascade Biologics). Cell-survival experiments were performed using a sulforhodamine B assay as previously described (36). Nine hundred cells were plated per well in 96-well plates per treatment condition, and each condition was performed in quintuplicate. Sulforhodamine B–stained cells were quantified with a Synergy (BioTek) plate reader. Survival was reported as OD at 564 nm of experimental wells divided by that of control wells that received no treatment with cisplatin (American Pharmaceutical Partners) or small molecule. Error bars denote SD.
Supplementary Material
Acknowledgments.
We thank Geoff Greene, Wolf-Dietrich Heyer, Brian Kay, Stephen Kowalczykowski, Phoebe Rice, and Patrick Sung for helpful conversations and/or materials; and Jennifer Grubb for assistance with D-loop and ATPase assays. This work was supported by funding from the American Society for Therapeutic Radiology and Oncology (to P.P.C.); and by GM50936 (to D.K.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808046105/DCSupplemental.
References
- 1.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 2.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 3.Gasior SL, et al. Assembly of RecA-like recombinases: Distinct roles for mediator proteins in mitosis and meiosis. Proc Natl Acad Sci USA. 2001;98:8411–8418. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masson JY, et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu N, Schild D, Thelen MP, Thompson LH. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res. 2002;30:1009–1015. doi: 10.1093/nar/30.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takata M, et al. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatanaka A, et al. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol Cell Biol. 2005;25:1124–1134. doi: 10.1128/MCB.25.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin RW, et al. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007;67:9658–9665. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
- 9.Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HK, Morimatsu K, Norden B, Ardhammar M, Takahashi M. ADP stabilizes the human Rad51-single stranded DNA complex and promotes its DNA annealing activity. Genes Cells. 2002;7:1125–1134. doi: 10.1046/j.1365-2443.2002.00588.x. [DOI] [PubMed] [Google Scholar]
- 11.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 12.Yu DS, et al. Dynamic control of Rad51 recombinase by self-association and interaction with BRCA2. Mol Cell. 2003;12:1029–1041. doi: 10.1016/s1097-2765(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 14.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 15.Thresher RJ, Griffith JD. Intercalators promote the binding of RecA protein to double-stranded DNA. Proc Natl Acad Sci USA. 1990;87:5056–5060. doi: 10.1073/pnas.87.13.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasad TK, Yeykal CC, Greene EC. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J Mol Biol. 2006;363:713–728. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 18.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc Natl Acad Sci USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tombline G, Heinen CD, Shim KS, Fishel R. Biochemical characterization of the human RAD51 protein. III. Modulation of DNA binding by adenosine nucleotides. J Biol Chem. 2002;277:14434–14442. doi: 10.1074/jbc.M109917200. [DOI] [PubMed] [Google Scholar]
- 20.Stasiak A, Di Capua E. The helicity of DNA in complexes with recA protein. Nature. 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Egelman EH. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J Mol Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- 22.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, et al. Conformational changes modulate the activity of human RAD51 protein. J Mol Biol. 2004;337:817–827. doi: 10.1016/j.jmb.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 24.Tebbs RS, et al. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc Natl Acad Sci USA. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta RC, Bazemore LR, Golub EI, Radding CM. Activities of human recombination protein Rad51. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 27.Morrison C, et al. The essential functions of human Rad51 are independent of ATP hydrolysis. Mol Cell Biol. 1999;19:6891–6897. doi: 10.1128/mcb.19.10.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 29.Petalcorin MI, Galkin VE, Yu X, Egelman EH, Boulton SJ. Stabilization of RAD-51-DNA filaments via an interaction domain in Caenorhabditis elegans BRCA2. Proc Natl Acad Sci USA. 2007;104:8299–8304. doi: 10.1073/pnas.0702805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeVita V, Hellman S, Rosenberg S. Cancer: Principle and Practice of Oncology. Philadelphia: Lippincott Raven; 1997. [Google Scholar]
- 31.Conway AB, et al. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 32.Hong EL, Shinohara A, Bishop DK. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem. 2001;276:41906–41912. doi: 10.1074/jbc.M105563200. [DOI] [PubMed] [Google Scholar]
- 33.Kowalczykowski SC, et al. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: Experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- 34.Lee MH, et al. Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J Biol Chem. 2005;280:40980–40984. doi: 10.1074/jbc.M505896200. [DOI] [PubMed] [Google Scholar]
- 35.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 36.Pauwels B, et al. Comparison of the sulforhodamine B assay and the clonogenic assay for in vitro chemoradiation studies. Cancer Chemother Pharmacol. 2003;51:221–226. doi: 10.1007/s00280-002-0557-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.