Abstract
Cyclic GMP (cGMP) is an important second messenger in eukaryotes. It is formed by guanylyl cyclases (GCs), members of the nucleotidyl cyclases class III, which also comprises adenylyl cyclases (ACs) from most organisms. To date, no structures of eukaryotic GCs are available, and all bacterial class III proteins were found to be ACs. Here we describe the biochemical and structural characterization of the class III cyclase Cya2 from cyanobacterium Synechocystis PCC6803. Cya2 shows high specificity for GTP versus ATP, revealing it to be the first bacterial GC, and sequence similarity searches indicate that GCs are also present in other bacteria. The crystal structure of Cya2 provides first structural insights into the universal GC family. Structure and mutagenesis studies show that a conserved glutamate, assisted by an interacting lysine, dominates substrate selection by forming hydrogen bonds to the substrate base. We find, however, that a second residue involved in substrate selection has an unexpected sterical role in GCs, different from its hydrogen bonding function in the related ACs. The structure identifies a tyrosine that lines the guanine binding pocket as additional residue contributing to substrate specificity. Furthermore, we find that substrate specificity stems from faster turnover of GTP, rather than different affinities for GTP and ATP, implying that the specificity-determining interactions are established after the binding step.
Keywords: bacterial, cGMP
The second messenger cGMP plays a central role in regulating bodily function such as the cardiovascular system and vision (1). In mammals, cGMP is formed by a family of guanylyl cyclases comprising a soluble GC (sGC) and several transmembrane receptor enzymes (rGCs). Mammalian GCs have a catalytic domain fused, via a central domain, to either an extracellular ligand binding domain (rGCs) or a nitric oxide binding heme domain (sGC) (1–3). In lower eukaryotes, guanylyl cyclase domains are fused to a variety of regulatory domains and function in processes such as cell motility control or chemotaxis (4).
All known guanylyl cyclases belong to class III of the purine nucleotidyl cyclase family, which was subdivided in six phylogenetically separated classes, I–VI, based on sequence homologies within the catalytic cores (5, 6). Class III also comprises adenylyl cyclases (ACs) from almost all domains of life (no member has been confirmed in plants), including most bacteria (7). GCs, in contrast, have been described only in Dictyostelium and higher organisms, and their existence in prokaryotes has only been speculated upon (8). Despite their physiological importance, no structural data on GC enzymes are available to date. For class III ACs, in contrast, several crystal structures have been reported, revealing a conserved basic architecture of the catalytic cores (6, 9–11), which is assumed to be shared by GCs (1, 12).
Class III cyclases require dimerization of two catalytic domains for activity, which leads to formation of shared active sites in ACs (6, 7). Modeling studies indicate that several features of these AC active sites also apply to GCs (13). Two conserved acidic residues bind the nucleoside triphosphate via two divalent ions, normally magnesium (9, 14). One ion contributes to catalysis (catalytic ion A), whereas the second ion (ion B) interacts with the nucleotide β- and γ-phosphates for substrate binding and possibly for leaving group stabilization. With few deviations, mainly in microbial cyclases, two conserved active-site residues in ACs form hydrogen bonds to the adenine base of the substrate and confer specificity for ATP (12): A lysine interacts with N1, and an aspartate or a threonine acts as hydrogen bond acceptor for the 6-amino group. In GC enzymes, these positions are also conserved and contribute to GTP specificity (12): A glutamate replaces the lysine and is supposed to interact with N1 and the 2-amino group. The second position is occupied by either a cysteine or a serine, possibly for interaction with the guanosine 1-keto group. Despite these similarities, GCs often show lower substrate specificity than ACs (9, 12), and mutagenesis studies on these key residues often enable only the partial conversion of a GC to an AC and vice versa (12, 15, 16). Furthermore, class III genes have been identified that do not carry one of the generic AC or GC residues at these positions (9, 12). The Rhizobium meliloti AC Cya1, e.g., carries a glutamine/threonine pair (12), and Cya2 from Synechocystis PCC6803 has the GC characteristic glutamate but carries a glycine at the second position (12, 17). Knocking out the cya2 gene caused a loss in cGMP levels in Synechocystis (17), suggesting that Cya2 could be the first bacterial GC enzyme to be identified or, alternatively, that Cya2 is an AC with high GC side activity due to the lack of one of the specificity-determining side chains.
Here we present the biochemical and structural characterization of Cya2, revealing the first bacterial GC enzyme and the first crystal structure of a GC catalytic domain. The enzyme shows high specificity for GTP, and crystal structure and mutagenesis results indicate that the conserved glutamate, assisted by a lysine, dominates substrate selection through formation of hydrogen bonds. The second position, in contrast, influences substrate specificity indirectly by providing space for proper orientation of the substrate. Furthermore, we find that the specificity-mediating interactions are formed after an initial binding step, which does not discriminate between ATP and GTP.
Results
Cya2 Is a Bacterial GC with High Substrate Specificity.
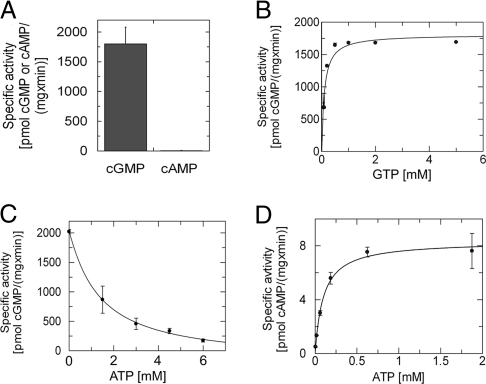
Cya2 from Synechocystis PCC6803 (Swiss-Prot entry P72951) (17) is a class III cyclase connected through putative transmembrane segments to a regulatory CHASE2 domain (18). The Cya2 catalytic domain carries a GC characteristic glutamate (Glu-488), but the second conserved position is Gly-562 instead of the residues typically found in ACs (Asp or Thr and sometimes Ser) or GCs (Cys or Ser) (12). Cya2 thus might either be the first bacterial GC to be described or an AC with high GTP-converting side activity. To answer this question, we cloned, expressed, and purified the catalytic domain of Cya2 (residues 434–635). From gel filtration, a Cya2 dimer was obtained, as expected for an active class III cyclase. Testing Cya2 in activity assays revealed a specific GC activity of ≈2 nmol/(mg × min), whereas its AC activity corresponds to <1% of the GC activity (Fig. 1A and Table 1). Thus, Cya2 qualifies as the first bacterial guanylyl cyclase to be identified and, surprisingly, shows higher substrate specificity than most cyclases, which carry two putative hydrogen bonding partners for the substrate base (12).
Fig. 1.
Cyclase activities of the recombinant Cya2 catalytic domain. (A) Cyclase activity of Cya2 with 3 mM of either GTP or ATP as substrate nucleotide. (B) Substrate saturation curve for Cya2. GC activities assayed at various GTP concentrations were fitted assuming Michaelis–Menten kinetics, yielding a Km value of 0.11 mM and a Vmax of 1.8 nmol/(mg × min). (C) Inhibition of Cya2 activity, at a fixed GTP concentration of 3 mM, by increasing concentrations of ATP/Mg2+. (D) Substrate saturation curve for Cya2 with ATP as substrate determined by using a RIA. The Michaelis–Menten fit yielded a Km of 0.10 mM for this substrate.
Table 1.
Vmax and Km values for Cya2 wild type and variants
| Protein/substrate | ELISA |
Radioimmunoassay |
||
|---|---|---|---|---|
| Km, mM | Vmax, nmol/(min × mg) | Km, mM | Vmax, nmol/(min × mg) | |
| wt/GTP-Mn2+Mg2+ | 0.11 ± 0.03 | 1.81 ± 0.09 | 0.22 ± 0.06 | 1.62 ± 0.11 |
| wt/ATP-Mn2+Mg2+ | ND | ND | 0.10 ± 0.01 | 0.008 ± 0.001 |
| wt/GTP-Mn2+ | 0.48 ± 0.02 | 1.48 ± 0.22 | ND | ND |
| wt/GTP-Mg2+ | 2.1 ± 0.7 | 0.64 ± 0.10 | ND | ND |
| Glu488Lys/GTP-Mn2+Mg2+ | ND | ND | 0.14 ± 0.06 | 0.007 ± 0.001 |
| Glu488Lys/ATP-Mn2+Mg2+ | 0.10 ± 0.05 | 1.50 ± 0.23 | ND | ND |
| Gly562Ser/ATP-Mn2+Mg2+ | 0.45 ± 0.17 | 0.42 ± 0.04 | ND | ND |
| Tyr572Ala/ATP-Mn2+Mg2+ | 0.09 ± 0.08 | 0.50 ± 0.07 | ND | ND |
ND, not determined.
The >100-fold-higher GC activity over AC activity could either stem from a higher affinity for GTP or be caused by its faster turnover. We thus determined Km values for ATP and GTP (Table 1). Saturation curves with the substrate GTP yielded a Km of 0.1 mM (Fig. 1B). Experiments with ATP as substrate indicated a similar affinity, but measurements were unreliable because of the low AC activity. We thus tested ATP affinity in competition experiments. Adding between 0 and 6 mM Mg2+-ATP in assays with 3 mM GTP as substrate lowered GC activity in a dose-dependent manner (Fig. 1C). From the ATP concentration resulting in half-maximal inhibition (IC50), we calculated the affinity for ATP, with the equation Ki = IC50/[1 + ([S]/Km)] for competitive binding (19), to be 0.05 mM, comparable to the affinity for GTP. To confirm this surprising result, we used a more sensitive radioimmunoassay (RIA) for measuring saturation curves with either GTP or ATP as substrate. Again, comparable Km values were obtained for GTP (0.22 mM; Table 1) and ATP (0.10 mM; Fig. 1D). These results show that both nucleotides bind to the active site, that they do so with comparable affinities, and that GC specificity primarily stems from different turnover efficiencies.
Dependency of Cya2 Activity on Ionic Strength and Divalent Ions.
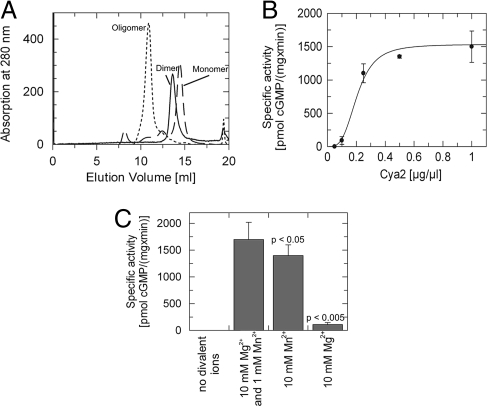
The initially purified Cya2 homodimer exhibited a strong dependency of its oligomerization state on ionic strength. In gel filtration experiments (Fig. 2A), the protein behaved as a monomer at high salt concentrations (20 mM Tris buffer plus 200 mM NaCl), as a higher oligomer (trimer or tetramer) with no NaCl added, and as a dimer at a concentration of 50 mM NaCl. Consistent with a homodimer as active Cya2 species, conditions favoring Cya2 dimerization yielded the highest activity (data not shown), and specific Cya2 activity depended on protein concentration (Fig. 2B). Assuming a homodimerization equilibrium, we determined a Kd of 8 μM in the presence of 50 mM NaCl. This value is consistent with the weak dimerization affinity observed for a mammalian AC (11) and the working model that dimer rearrangements and stabilization contribute to class III regulation (11, 20).
Fig. 2.
Influence of ions and protein concentration on Cya2 oligomerization and activity. (A) Gel filtration profiles of recombinant Cya2 catalytic domain. Different oligomeric states are obtained depending on the ionic strengths of the buffer. Dotted line, 0 mM NaCl in the gel filtration buffer; solid line, 50 mM NaCl; dashed line, 200 mM NaCl. (B) Specific GC activity of Cya2 increases with increasing enzyme concentration, consistent with the assumption that the active species is dimeric. (C) GC activity of Cya2 in presence of 5 mM GTP and various combinations of divalent ions, showing that Cya2 activity depends on the availability of either Mg2+ or Mn2+. The highest activity is observed when both ions are present; using either ion alone resulted in lower activity. P values were determined by using Student's t test.
In class III cyclases, two divalent cations contribute to substrate binding (ion B) and catalytic turnover (ion A) (6, 9). Consistently, we found that either Mg2+ or Mn2+ is essential for Cya2 activity, and Mn2+ alone led to a higher specific activity than Mg2+ alone (Fig. 2C). The highest activity, however, was found when using both ions, with Mg2+ exceeding Mn2+ in a ratio range of 2:1 to 10:1 [Fig. 2C and supporting information (SI) Fig. S1]. The Km value for GTP (Table 1) was higher with Mn2+ alone (0.5 mM) compared with the value obtained with the Mg2+/Mn2+ combination (0.1 mM), but even higher with Mg2+ alone (2.1 mM). The Vmax value with Mn2+ alone, in contrast, was only slightly reduced, indicating that magnesium addition to manganese increases activity mainly through improved substrate binding. The observation that magnesium increases substrate affinity only if manganese is also present suggests a complex interplay between the two ions, but more extensive studies will be required to reveal the details of this interaction. Like most class III cyclases (6, 21), however, Cya2 could be inhibited by adding micromolar concentrations of calcium or zinc (data not shown), which can compete for the ion A and B sites (9).
The Crystal Structure of Cya2 Confirms a Close Similarity Between GCs and Class III ACs.
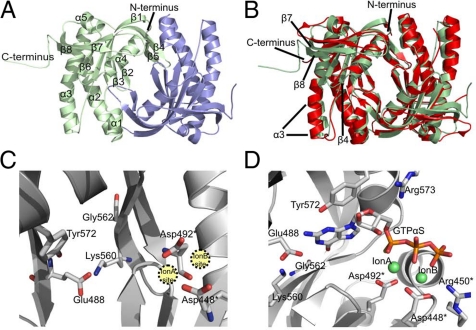
To provide a first experimental structure of a GC enzyme and to examine the molecular details of a GC active site, we solved the crystal structure of the homodimeric Cya2 catalytic core (Fig. 3A). The structure was refined at 2.3-Å resolution to Rcryst and Rfree values of 19.8% and 28.2%, respectively (Table 2), with no residues in the disallowed region of the Ramachandran plot and 92.3% of the residues in the most favorable areas. The Cya2 monomer within the tightly packed homodimer (dimerization interface area of 1,578 Å2) carries a central seven-stranded β-sheet (Fig. 3A), and the monomers dimerize through a clamp formed by β4 and β5. Although Cya2 uses a different substrate and carries unique regulatory domains, the overall fold of the catalytic core resembles the known structures of AC catalytic dimers from bacteria (Fig. 3B) and higher organisms (6, 9). Differences compared to the AC enzyme CyaC include an extended C terminus in Cya2 that folds back on strand β8 and helix α3. Furthermore, β7 and β8 are slightly extended, whereas α3 is truncated and the loop between α3 and β4 is considerably shortened. Despite these differences, the Cya2 structure experimentally confirms that the conserved general architecture of class III catalytic domains now includes GCs, as suggested by AC structures and the low but significant sequence similarities in this enzyme family (6, 11, 12).
Fig. 3.
Crystal structure of Cya2 catalytic domain. (A) Overall structure of the Cya2 homodimer. The two monomers are colored green and blue, and the secondary structure elements are labeled in one monomer. (B) Overlay of the Cya2 catalytic domain (green) with the AC enzyme CyaC (red). Secondary structure elements displaying differences between the two cyclases are labeled. (C) Active site of the Cya2 dimer. Residues mentioned in the text are labeled; residues supplied by the second monomer of the dimer are marked with an asterisk. (D) Cya2 active site with a modeled GTPαS ligand. The nucleotide was positioned in Cya2 after superposition with the structure of a CyaC/ATPαS complex; the nucleotide base was then modified manually to guanosine.
Table 2.
X-ray data and refinement statistics
| Cya2 | |
|---|---|
| Space group | P21 |
| Unit cell constants | a = 62.4 Å, b = 84.1 Å, c = 115.3 Å; β = 97.4 |
| Resolution, Å | 50.0–2.3 |
| Unique reflections | 50,776 |
| 〈I/σ〉 | 8.4 |
| Completeness, % | 97.3 (85.0) |
| Rmerge,* % | 8.0 (37.9) |
| Refinement resolution, Å | 20.0–2.3 |
| Total reflections used | 48,079 |
| Final Rcryst/Rfree,†‡ % | 19.4/28.2 |
| Atoms: protein/solvent | 8,911/468 |
| rmsd bond lengths, Å | 0.02 |
| rmsd bond angles, ° | 1.9 |
| rmsd bonded B, Å2 | 1.6 |
| Average B-factor: protein /solvent, Å2 | 41/48 |
Numbers in parentheses are for the outermost shell.
*Rmerge = Σ I − 〈I〉/Σ I; I is the intensity of an individual measurement, and 〈I〉 is the corresponding mean value.
†R-factor = Σ||Fobs| − k|Fcalc||/Σ|Fobs|; |Fobs| is the observed and |Fcalc| the calculated structure factor amplitude.
‡Rfree was calculated from 5% of measured reflections omitted from refinement.
The GC Active Site.
The GC active site of Cya2 comprises several features known from AC catalytic cores (6, 9). It is located at the dimer interface and is formed by residues from both monomers (Fig. 3C). The conserved aspartates, Asp-448 and Asp-492, are oriented toward each other to form the ion A and ion B sites for the two divalent cations. Interestingly, all six monomers of the three dimers in the asymmetric unit have very similar overall conformations, but only one active site of each dimer has these residues oriented in a manner that both ions would be coordinated by both aspartates. In the second active site, Asp-448 is turned around and provides only one oxygen for the ion sites. This conformation is influenced through an interaction with Arg-450 and appears to be an inactive state but might also be an open form ready to accommodate ions and substrate. Notably, “half-of-the-sites” activity was previously proposed for the bacterial AC enzyme Rv1900c based on asymmetry in its crystal structure and the observation that heterodimers of mutants reconstituting a single active site displayed wild-type activity (9, 22). Similar experiments with Cya2 resulted in no increase in activity when Cya2-Asp492Ala was mixed with Cya2-Lys560Ala, which would reconstitute one active site in heterodimers (Fig. S2). Combining either mutant with Cya2 wild type revealed a dominant negative effect, as previously reported for mammalian rGC mutants (23, 24). Thus, Cya2 shows an active-site communication that appears to be typical for homodimeric GCs, but its details remain to be revealed.
Using the structural conservation between Cya2 and the AC enzyme CyaC, we generated a model of a nucleotide in the Cya2 active site. From superposition of Cya2 with a CyaC/ATPαS complex, the nucleotide was transferred and then modified into GTPαS (Fig. 3D). The model illustrates that the conserved Arg-573 contributes to the phosphate binding site, whereas the conserved Lys-608 points away but could reach a substrate through a local conformational change. In the hydrophobic pocket responsible for binding of the substrate base, the Glu-488 characteristic for GCs has a position and an orientation identical to those of the conserved lysine in ACs, which contributes to adenine base recognition (12). Glu-488 is well oriented for interaction with the guanine base, especially at the 2-amino group and possibly also at position N1. The orientation of the Glu-488 side chain is stabilized through Lys-560. This interaction was previously speculated on based on sequence alignments [“supporting residue” hypothesis (12)]. Our structure confirms this interaction, which is likely to be a general feature of GCs.
Comparison of the Cya2 structure to ACs confirms the sequence-based assumption (12, 17) that Gly-562 occupies the second position involved in base recognition in AC enzymes. ACs typically carry an adenine-binding aspartate or threonine at this position (12). Having a glycine at this site means that one of the assumed hydrogen bonding interactions with the base cannot be present, and it also means that more space is available at this side of the base binding pocket. Close to the opposite edge of the base, Cya2 carries a tyrosine (Tyr-572) that replaces Ala-1149 in CyaC and that would be positioned properly for a hydrogen bond to the guanine 2-amino group (Fig. 3D). Most of the space needed for this replacement is provided by replacement of CyaC Phe-1015 by Cya2 Thr-446, but Tyr-572 is still closer to the base, which requires the base to shift toward Gly-562 for proper binding. In this way, Gly-562 appears to contribute to base recognition by providing space that enables an optimized interaction between guanine and other active-site residues such as Glu-488 and Tyr-572.
The Active-Site Glutamate Conserved in GCs Dominates Cya2 Substrate Specificity.
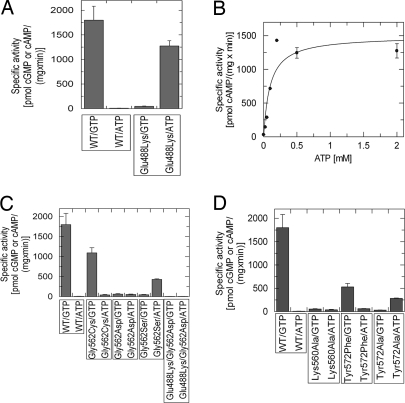
To further analyze the role of active-site residues we tested Cya2 variants for their activity and specificity. First, we changed the conserved GC residue and hydrogen bond acceptor Glu-488 into the AC-characteristic hydrogen bond donor lysine (12). Consistently, the Cya2-Glu488Lys variant showed clearly reduced GC activity in comparison to the wild type and a 100-fold increased AC activity (Fig. 4A). Like for wild-type protein, we found that the Km values for ATP (0.10 mM) and GTP (0.14 mM; Fig. 4B and Table 1) are comparable. Thus, this conserved residue strongly influences substrate selection in Cya2 and presumably in most GCs, likely by forming hydrogen bonds (see above), but only after an initial, nonspecific binding step has taken place.
Fig. 4.
The conserved active-site glutamate dominates substrate specificity. Shown are activities of Cya2 wild type and mutants in presence of 10 mM Mg2+ and 1 mM Mn2+. (A) Specific cyclase activity of purified Cya2 wild type and Cya2-Glu488Lys with either ATP or GTP as substrate. (B) Saturation curve for the AC activity of Cya2-Glu488Lys, yielding a Km value for ATP of 0.1 mM. (C) Specific activities of Cya2 wild type and the variants Cya2-Gly562Cys, Cya2-Gly562Asp, Cya2-Lys560Ala, and Cya2-Glu488Lys/Gly562Asp at a concentration of 5 mM ATP or GTP, respectively. (D) Specific AC and GC activities of Cya2 wild type and the variants Cya2-Lys560Ala, Cya2-Tyr572Phe, and Cya2-Tyr572Ala.
We next investigated the role of the unusual glycine (Gly-562) of Cya2. The corresponding position is typically occupied by a cysteine in GCs (assumed to act as hydrogen bond donor) and by an aspartate in ACs (assumed to act as hydrogen bond acceptor) (12). Changing Gly-562 to aspartate indeed increased AC activity and reduced GC activity (Fig. 4C). However, the mutant's AC activity did not exceed its GC activity and also stayed low compared with the Glu-488/Lys mutant, indicating that this residue is not as strong a determinant for substrate specificity as Glu-488. Surprisingly, changing the glycine to cysteine (Cya2-Gly562Cys), i.e., generating the generic GC pair glutamate/cysteine, diminished GC activity to approximately half of wild-type activity, whereas AC activity was slightly increased (Fig. 4C). Thereby, GTP specificity was lowered to values observed for GCs natively carrying the glutamate/cysteine pair, such as mammalian sGC, which shows 5% AC side activity (25). Thus, adding a second putative H-bond partner increases only AC activity, whereas GC activity is even lowered, and we conclude that this residue serves a different function in GCs than forming a hydrogen bond to guanine. Consistently, a Cya2-Gly562Ser mutant carrying a side chain that can act as hydrogen bond donor and acceptor also showed increased ATP turnover but dramatically decreased GC activity (Fig. 4C and Table 1).
Based on our structural data, we suggest a sterical function for Gly-562, i.e., to provide space for proper orientation and binding of GTP to Glu-488 and additional residues such as Tyr-572, which lines the base binding pocket opposite from Gly-562. Contribution of additional residues to specificity is indicated by the observation that the same glutamate/serine pair can lead either to the GC activity of a GC from Paramecium (26) or to the AC activity of Cya2-Gly562Ser (Fig. 4C). This notion is further supported by results we obtained with a double mutant Glu488Lys/Gly562Asp. Based on hydrogen bonding residues, increased AC activity would have been expected. However, AC and GC activity were both decreased compared with wild-type Cya2 and the single-site mutants Gly562Asp (Fig. 4C) and Glu488Lys (Fig. 4A). We thus analyzed the contribution of additional residues to specificity. Changing the newly identified guanine interaction partner Tyr-572 to phenylalanine lowered the GC activity (Fig. 4D), consistent with a hydrogen bond from the tyrosine hydroxyl group to guanine. However, a Cya2-Tyr572Ala mutant showed further decreased GC activity and an increase in ATP turnover (Fig. 4D and Table 1), indicating that this residue also contributes sterically to specificity by restricting the base binding site opposite to Gly-562. We also tested the “supporting residue” Lys-560, which we found to interact with Glu-488 (see above) and which thus should influence specificity by fine-tuning the orientation of this guanine-recognizing residue. Consistent with such a function, exchange of Lys-560 to alanine significantly lowered the GC activity of Cya2 and slightly increased its AC activity (Fig. 4D). We conclude that the conserved Glu-488 dominates substrate specificity, supported by Lys-560, and that Gly-562 influences specificity in a more subtle way than hydrogen bonding, together with Tyr-572.
Cya2-Like Bacterial Cyclases Are Present in a Wide Range of Prokaryotes.
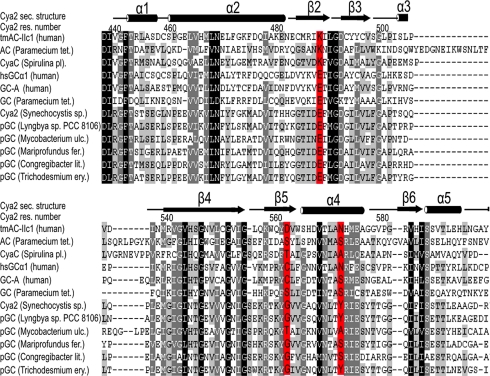
No function for cGMP and thus for GCs has yet been established in bacteria. The discovery of the bacterial GC enzyme Cya2 raises the question of whether GCs exist in a wider range of prokaryotes. We thus did a BLAST search of the UniProt database for bacterial Cya2 homologs and analyzed hits for sequences carrying the glutamate/glycine combination of the highly GTP-specific Cya2 wild type. Thereby, we identified several putative class III GCs from a variety of bacteria of the phyla cyanobacteria, actinobacteria, and proteobacteria (Fig. 5). Comparison with confirmed bacterial adenylyl cyclase in a phylogenetic tree reveals that the putative bacterial GCs cluster together, indicating that they form a defined cyclase subfamily (Fig. S3). Interestingly, a subset of the putative GCs carries a tyrosine at the position equivalent to Tyr-572 of Cya2 (Fig. 5), reinforcing their classification as GCs; other sequences carry a serine or alanine also found in ACs, which might indicate that these enzymes have a decreased specificity compared with the highly specific Cya2, or that the function of Tyr-572 is exerted by other positions in other cyclases. Putative GC containing organisms also comprise pathogens such as Mycobacterium ulcerans, which causes the common human mycobacteriosis Buruli ulcer (27), and Lyngbya sp., which causes swimmer's itch (28). We thus assume that Cya2 is a representative of a widespread bacterial subfamily of the class III GCs, and further studying the physiological functions of these enzymes might also be of medical interest.
Fig. 5.
Cya2-like cyclases in other prokaryotes. Shown is a sequence alignment of eukaryotic and mammalian AC and GC domains, Cya2, and several prokaryotic sequences obtained from a BLAST search of the UniProt database. Shown are the regions containing the specificity-determining residues, which are highlighted in red. pGCk, putative guanylyl cyclase.
Discussion
Class III is the largest and most prevalent nucleotidyl cyclases family (5, 6, 9). All known eukaryotic ACs belong to class III, and most prokaryotes possess class III ACs (7, 29). Furthermore, all known GCs are grouped into class III (2, 4). GCs so far have been identified only in eukaryotes, whereas all cyclases from prokaryotes tested biochemically were found to be ACs (12). Consistently, no function is known for cGMP in bacteria, although the related cyclic dinucleotide c-di-GMP was found to be important for motility and biofilm formation in several prokaryotes (30). Our finding that Cya2 from Synechocystis PCC6803 is a highly specific GC shows, however, that GCs are not restricted to eukaryotes. The identification of Cya2-like cyclase sequences in other prokaryotes in fact indicates that Cya2 is a representative of a larger bacterial GC subfamily. Cya2 appears to be a receptor GC (17) functioning as osmosensor (18), but the exact physiological functions of Cya2 and of bacterial GCs in general remain to be elucidated and will be interesting targets for future research.
In both ACs and GCs, magnesium ions are assumed to be the physiological active-site ions, but for most class III cyclases nonphysiological concentrations of manganese lead to higher activities (31). Manganese is a trace element in eukaryotes but appears to be present in higher concentrations in cyanobacteria (32) and could be a physiological cofactor for Cya2. The increased activity of eukaryotic cyclases in presence of Mn2+ could be an evolutionary remnant or simply be due to the larger size and more flexible coordination geometry of manganese (9, 33).
A major reason that contributed to the late discovery of a bacterial GC is our still-incomplete understanding of substrate selection and the incomplete conservation of substrate-recognizing residues (12). Based on the current model of substrate selection through hydrogen bond formation (12), a low specificity would have been expected for Cya2 with its glutamate/glycine pair because of the missing second side chain. However, our results show that the GTP specificity of Cya2 even exceeds that of many other GCs, which tend to display considerable AC side activity (9, 12). We find that the GC-characteristic glutamate dominates this substrate specificity. The first crystal structure of a GC presented here shows that this residue is positioned analogous to the conserved lysine in ACs (9, 11), apparently for the formation of hydrogen bonds to the substrate base. However, changing in Cya2 the second position assumed to interact with the base into a residue capable of hydrogen bonding increased AC activity but decreased GC activity, even when introducing the cysteine found in many GCs (12). We thus assume that the second position forms hydrogen bonds in ACs but has a different function in GCs. Consistently, similar exchange experiments in other cyclases most times did not result in a complete switch of specificity, especially when trying to convert an AC into a GC (12, 15). Introducing the AC-characteristic hydrogen bond partner in the second position can strongly contribute to ATP specificity (16) and in the case of sGC was even sufficient to switch its specificity (15), but a more subtle role in GCs would make it more difficult to introduce the correct interactions through a single mutation. The Cya2 structure indicates that the second residue, Gly-562, contributes sterically to substrate selection. It reveals Tyr-572 as novel substrate binding residue, and this interaction requires a base orientation best enabled by a small residue in position 562. The lack of strict conservation of Tyr-572 means that another residue can assume this function in other GCs, indicating that features slightly varying in detail have developed during evolution for substrate recognition by GCs. Our results further show how additional residues, which do not directly contact the base, can contribute to specificity and activity: The conserved, positively charged “assisting” residue Lys-560 (12, 16) acts by positioning and neutralizing the conserved glutamate.
Our data show that GTP selection takes place only after the initial binding step. Similar affinities for ATP and GTP have also been reported for a mammalian soluble GC (25), and our results on a receptor GC indicate substrate selection after binding to be a general feature of GCs. It even appears necessary to fully explain mammalian AC specificity (34). This finding could be important for the physiological regulation of GCs, because ATP is present in comparable or even higher concentrations as GTP in most cells, and it explains the long-known inhibitory effect of ATP on mammalian GCs (35). Mechanistically, it implies that specific interactions to the nucleotide base, such as the hydrogen bonds assumed in Cya2 based on a GTP complex model, and observed in structures of AC/nucleotide complexes (9, 36), are established or optimized after the initial binding step. The Cya2 structure described here is one of few structures of a class III cyclase without a ligand, either nucleotide analog or regulator (9). Comparison to different states of AC enzymes (10, 11) shows that it acquires a closed conformation previously proposed to be induced by substrate binding (9, 36). Thus, Cya2 has to open up for uptake of the nucleotide before it can close again, either induced by substrate binding or as part of substrate conversion as predicted in an alternative model (6, 10). We assume that this closure movement enables the specificity-determining interactions to the nucleotide in the active site. It remains to be elucidated whether the open–closed transition is also the step facilitated by activators such as Gsα for ACs (11, 21) and p21-activated kinase for GCs (37).
Methods
Cloning of Cya2 and Site-Directed Mutagenesis.
Cya2 catalytic domain (protein residues 434–635) was PCR-amplified from Synechocystis genomic DNA and cloned into pET151/D-TOPO (Invitrogen), resulting in a N-terminally his-tag construct with a TEV (tobacco etch virus) protease cleavage site. Cya2 mutants were generated by using the QuikChange site-directed mutagenesis (Stratagene) protocol.
Protein Expression and Purification.
Catalytic domains of Cya2 and Cya2 mutants were expressed in Escherichia coli BL21(DE3) at 37°C in Luria–Bertani broth containing 100 μg/ml ampicillin. Expression was induced at OD600 of 0.8 by adding 0.5 mM isopropyl-thiogalactoside, and culturing continued at 20°C overnight. Cells were disrupted in a French press, and cell debris was removed by centrifugation at 18,000 rpm for 45 min at 4°C in a HFA 22.50 rotor. The supernatant was supplemented with 10 mM imidazole and incubated with Talon resin (Clontech) for 1 h at 4°C. The resin was washed with 10 volumes of buffer A (50 mM Tris·HCl, pH 7.8/500 mM NaCl) and 10 volumes of buffer B (50 mM Tris·HCl, pH 7.8/200 mM NaCl), each supplemented with 10 mM imidazole. Protein was eluted with buffer B containing 200 mM imidazole. For activity assays, the protein was concentrated by using a Centriprep-10 (Millipore) and further purified by gel filtration on a S12 column (GE Healthcare) in buffer C (20 mM Tris, pH 7.8/50 mM NaCl/3 mM TCEP). For crystallization, the affinity chromatography elution buffer was exchanged to buffer B with 7 mM 2-mercaptoethanol by using a NAP25 column (GE Healthcare), and the his-tag cleaved off by incubation with TEV protease overnight at 10°C. Cleaved protein was incubated with Talon resin for 1 h at 4°C, and the flow-through was subjected to gel filtration as described above. The purified protein was concentrated and stored at −80°C.
Activity Assays.
GC and AC activity assays were done in a 30-μl volume of 20 mM Tris·HCl (pH 7.8), 50 mM NaCl, 5 mM GTP or ATP, 10 mM MgCl2, 1 mM MnCl2, and 0.5–2.5 μg/μl protein. Deviations from this standard setup are noted in the description of the respective experiment. Samples were incubated at 30°C for 30 min, and cAMP or cGMP was determined by using ELISA kits (Biomol) according to the instructions of the manufacturer.
For the RIA, 4.2–30 μg of Cya2-wt or Cya2-E488K was incubated for 10 min at 30°C in a 10-μl total volume of 3 mM DTT, 0.25 mg/mg BSA, 10 mM MgCl2, 1 mM MnCl2, 50 mM triethalolamine hydrochloride (pH 7.4), and 6.25–3,000 μM GTP or ATP as indicated. Reactions were stopped by addition of 90 μl of ice-cold 100 mM sodium acetate (pH 6), put on ice, and 3 μl of a 2:1 mixture of triethylamine and acetic anhydride were added. cGMP or cAMP were then determined by using a RIA as described (38, 39).
Crystallization, Data Collection, Structure Solution, and Refinement.
Crystallization experiments were carried out at 20°C with 30 mg/ml Cya2. Initial crystallization conditions from factorial screens (Hampton Research) were refined to 16% PEG 3350 and 20 mM MgCl2, yielding well diffracting crystals in space group P21. For data collection, crystals were soaked with reservoir solution supplemented with 30% (vol/vol) glycerol. A complete dataset was collected at 2.3-Å resolution at SLS beamline X10SA. Data integration and reduction were done with HKL2000 (40).
The structure of Cya2 was solved by using Patterson search techniques using Phaser (41) with a structure of CyaC [Protein Data Bank ID code 1WC1 (10)] as search model. The structure was rebuilt and refined with Coot (42) and Refmac (43), respectively. Weak to medium noncrystallographic symmetry restraints between all six monomers in the asymmetric unit were applied, except for loops. Individual isotropic Debye–Waller factors were refined for all atoms, and, in later stages of refinement, chains were refined as individual TLS groups (44) and solvent atoms were added. The refined structure was analyzed by using ProCheck (45), the protein/protein interaction server (www.bioinformatics.sussex.ac.uk/protorp/), and the quality check functions of Coot.
Supplementary Material
Acknowledgments.
We thank Nadine Waschewski (Department of Plant Biochemistry, Ruhr-University Bochum) for Synechocystis PCC6803 genomic DNA and Arkadius Pacha (Department of Pharmacology and Toxicology, Ruhr-University Bochum) for technical assistance. We further thank the beamline staff of X10SA at the Swiss Light Source (Villigen, Switzerland) for support and our colleagues from Max Planck Institute (Dortmund, Germany) for help with data collection. This work was supported by Deutsche Forschungsgemeinschaft SFB642 (to C.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors for Cya2 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2W01).
This article contains supporting information online at www.pnas.org/cgi/content/full/0808473105/DCSupplemental.
References
- 1.Lucas KA, et al. Guanylyl cyclases and signaling by cyclic GMP. Pharmacol Rev. 2000;52:375–414. [PubMed] [Google Scholar]
- 2.Padayatti PS, Pattanaik P, Ma X, van den Akker F. Structural insights into the regulation and the activation mechanism of mammalian guanylyl cyclases. Pharmacol Ther. 2004;104:83–99. doi: 10.1016/j.pharmthera.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Wedel B, Garbers D. The guanylyl cyclase family at Y2K. Annu Rev Physiol. 2001;63:215–233. doi: 10.1146/annurev.physiol.63.1.215. [DOI] [PubMed] [Google Scholar]
- 4.Schaap P. Guanylyl cyclases across the tree of life. Front Biosci. 2005;10:1485–1498. doi: 10.2741/1633. [DOI] [PubMed] [Google Scholar]
- 5.Danchin A. Phylogeny of adenylyl cyclases. Adv Second Messenger Phosphoprotein Res. 1993;27:109–162. [PubMed] [Google Scholar]
- 6.Kamenetsky M, et al. Molecular details of cAMP generation in mammalian cells: A tale of two systems. J Mol Biol. 2006;362:623–639. doi: 10.1016/j.jmb.2006.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linder JU, Schultz JE. The class III adenylyl cyclases: Multi-purpose signalling modules. Cell Signal. 2003;15:1081–1089. doi: 10.1016/s0898-6568(03)00130-x. [DOI] [PubMed] [Google Scholar]
- 8.Baker DA, Kelly JM. Structure, function and evolution of microbial adenylyl and guanylyl cyclases. Mol Microbiol. 2004;52:1229–1242. doi: 10.1111/j.1365-2958.2004.04067.x. [DOI] [PubMed] [Google Scholar]
- 9.Sinha SC, Sprang SR. Structures, mechanism, regulation and evolution of class III nucleotidyl cyclases. Rev Physiol Biochem Pharmacol. 2006;157:105–140. doi: 10.1007/112_0603. [DOI] [PubMed] [Google Scholar]
- 10.Steegborn C, Litvin TN, Levin LR, Buck J, Wu H. Bicarbonate activation of adenylyl cyclase via promotion of catalytic active site closure and metal recruitment. Nat Struct Mol Biol. 2005;12:32–37. doi: 10.1038/nsmb880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsα. GTPγS. Science. 1997;278:1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- 12.Linder JU. Substrate selection by class III adenylyl cyclases and guanylyl cyclases. IUBMB Life. 2005;57:797–803. doi: 10.1080/15216540500415636. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Ruoho AE, Rao VD, Hurley JH. Catalytic mechanism of the adenylyl and guanylyl cyclases: Modeling and mutational analysis. Proc Natl Acad Sci USA. 1997;94:13414–13419. doi: 10.1073/pnas.94.25.13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linder JU. Class III adenylyl cyclases: Molecular mechanisms of catalysis and regulation. Cell Mol Life Sci. 2006;63:1736–1751. doi: 10.1007/s00018-006-6072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunahara RK, et al. Exchange of substrate and inhibitor specificities between adenylyl and guanylyl cyclases. J Biol Chem. 1998;273:16332–16338. doi: 10.1074/jbc.273.26.16332. [DOI] [PubMed] [Google Scholar]
- 16.Tucker CL, Hurley JH, Miller TR, Hurley JB. Two amino acid substitutions convert a guanylyl cyclase, RetGC-1, into an adenylyl cyclase. Proc Natl Acad Sci USA. 1998;95:5993–5997. doi: 10.1073/pnas.95.11.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochoa De Alda JA, Ajlani G, Houmard J. Synechocystis strain PCC 6803 cya2, a prokaryotic gene that encodes a guanylyl cyclase. J Bacteriol. 2000;182:3839–3842. doi: 10.1128/jb.182.13.3839-3842.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhulin IB, Nikolskaya AN, Galperin MY. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J Bacteriol. 2003;185:285–294. doi: 10.1128/JB.185.1.285-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Copeland RA. Enzymes—A Practical Introduction to Structure, Mechanism, and Data Analysis. 2nd Ed. New York: Wiley-VCH; 2000. [Google Scholar]
- 20.Tews I, et al. The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme. Science. 2005;308:1020–1023. doi: 10.1126/science.1107642. [DOI] [PubMed] [Google Scholar]
- 21.Sunahara RK, Taussig R. Isoforms of mammalian adenylyl cyclase: Multiplicities of signaling. Mol Interv. 2002;2:168–184. doi: 10.1124/mi.2.3.168. [DOI] [PubMed] [Google Scholar]
- 22.Sinha SC, Wetterer M, Sprang SR, Schultz JE, Linder JU. Origin of asymmetry in adenylyl cyclases: Structures of Mycobacterium tuberculosis Rv1900c. EMBO J. 2005;24:663–673. doi: 10.1038/sj.emboj.7600573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudner XL, Mandal KK, de Sauvage FJ, Kindman LA, Almenoff JS. Regulation of cell signaling by the cytoplasmic domains of the heat-stable enterotoxin receptor: Identification of autoinhibitory and activating motifs. Proc Natl Acad Sci USA. 1995;92:5169–5173. doi: 10.1073/pnas.92.11.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson DK, Garbers DL. Dominant negative mutations of the guanylyl cyclase-A receptor. Extracellular domain deletion and catalytic domain point mutations. J Biol Chem. 1995;270:425–430. doi: 10.1074/jbc.270.1.425. [DOI] [PubMed] [Google Scholar]
- 25.Gille A, et al. Differential inhibition of adenylyl cyclase isoforms and soluble guanylyl cyclase by purine and pyrimidine nucleotides. J Biol Chem. 2004;279:19955–19969. doi: 10.1074/jbc.M312560200. [DOI] [PubMed] [Google Scholar]
- 26.Linder JU, Hoffmann T, Kurz U, Schultz JE. A guanylyl cyclase from Paramecium with 22 transmembrane spans. Expression of the catalytic domains and formation of chimeras with the catalytic domains of mammalian adenylyl cyclases. J Biol Chem. 2000;275:11235–11240. doi: 10.1074/jbc.275.15.11235. [DOI] [PubMed] [Google Scholar]
- 27.Wansbrough-Jones M, Phillips R. Buruli ulcer: Emerging from obscurity. Lancet. 2006;367:1849–1858. doi: 10.1016/S0140-6736(06)68807-7. [DOI] [PubMed] [Google Scholar]
- 28.Osborne NJ, Webb PM, Shaw GR. The toxins of Lyngbya majuscula and their human and ecological health effects. Environ Int. 2001;27:381–392. doi: 10.1016/s0160-4120(01)00098-8. [DOI] [PubMed] [Google Scholar]
- 29.Shenroy AR, Visweswariah SS. Class III nucleotide cyclases in bacteria and archaebacteria: Lineage-specific expansion of adenylyl cyclases and a dearth of guanylyl cyclases. FEBS Lett. 2004;561:11–21. doi: 10.1016/s0014-5793(04)00128-0. [DOI] [PubMed] [Google Scholar]
- 30.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurley JH. Structure, mechanism, and regulation of mammalian adenylyl cyclase. J Biol Chem. 1999;274:7599–7602. doi: 10.1074/jbc.274.12.7599. [DOI] [PubMed] [Google Scholar]
- 32.Keren N, Kidd MJ, Penner-Hahn JE, Pakrasi HB. A light-dependent mechanism for massive accumulation of manganese in the photosynthetic bacterium Synechocystis sp. PCC 6803. Biochemistry. 2002;41:15085–15092. doi: 10.1021/bi026892s. [DOI] [PubMed] [Google Scholar]
- 33.Seebeck B, Reulecke I, Kamper A, Rarey M. Modeling of metal interaction geometries for protein-ligand docking. Proteins. 2008;71:1237–1254. doi: 10.1002/prot.21818. [DOI] [PubMed] [Google Scholar]
- 34.Gille A, et al. Differential interactions of G-proteins and adenylyl cyclase with nucleoside 5′-triphosphates, nucleoside 5′-[gamma-thio]triphosphates and nucleoside 5′-[beta,gamma-imido]triphosphates. Biochem Pharmacol. 2005;71:89–97. doi: 10.1016/j.bcp.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Kimura H, Murad F. Two forms of guanylate cyclase in mammalian tissues and possible mechanisms for their regulation. Metabolism. 1975;24:439–445. doi: 10.1016/0026-0495(75)90123-7. [DOI] [PubMed] [Google Scholar]
- 36.Tesmer JJ, et al. Two-metal-ion catalysis in adenylyl cyclase. Science. 1999;285:756–760. doi: 10.1126/science.285.5428.756. [DOI] [PubMed] [Google Scholar]
- 37.Guo D, et al. A Rac-cGMP signaling pathway. Cell. 2007;128:341–355. doi: 10.1016/j.cell.2006.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooker G, Harper JF, Terasaki WL, Moylan RD. Radioimmunoassay of cyclic AMP and cyclic GMP. Adv Cyclic Nucleotide Res. 1979;10:1–33. [PubMed] [Google Scholar]
- 39.Rosenthal W. Giessen, Germany: Universität Giessen; 1983. Pharmakologische Beeinflussung der zyklischen Nukleotide in der Meerschweinchenlunge. PhD thesis. [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276A:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.