Abstract
Several isoforms of phospholipase C (PLC) are regulated through interactions with Ras superfamily GTPases, including Rac proteins. Interestingly, of two closely related PLCγ isoforms, only PLCγ2 has previously been shown to be activated by Rac. Here, we explore the molecular basis of this interaction as well as the structural properties of PLCγ2 required for activation. Based on reconstitution experiments with isolated PLCγ variants and Rac2, we show that an unusual pleckstrin homology (PH) domain, designated as the split PH domain (spPH), is both necessary and sufficient to effect activation of PLCγ2 by Rac2. We also demonstrate that Rac2 directly binds to PLCγ2 as well as to the isolated spPH of this isoform. Furthermore, through the use of NMR spectroscopy and mutational analysis, we determine the structure of spPH, define the structural features of spPH required for Rac interaction, and identify critical amino acid residues at the interaction interface. We further discuss parallels and differences between PLCγ1 and PLCγ2 and the implications of our findings for their respective signaling roles.
Phosphoinositide-specific phospholipase C (PLC)3 enzymes have been established as crucial signaling nodes involved in regulation of a variety of cellular functions via hydrolysis of the membrane lipid phosphatidylinositol 4,5-bisphosphate. There are six major families of PLC enzymes (PLCβ,-γ,-δ,-ε,-ζ, and -η) that share a common core of domains related to catalysis and are distinguished by family-specific regulatory regions (1-3). The two isoforms of the PLCγ family, PLCγ1 and PLCγ2, uniquely incorporate an array of domains comprising two SH2 domains, an SH3 domain, and an internal or “split” PH domain (spPH). spPHs represent a unique subclass of PH domains that are characterized by insertions of one or several autonomously folded protein modules encoded within the boundaries of PH domain sequences (4). This array also contains sites for phosphorylation by several receptor (e.g. epidermal growth factor and platelet-derived growth factor receptors) and nonreceptor tyrosine kinases. In addition to tyrosine phosphorylation, multiple protein-protein interactions (mainly mediated by SH2 and SH3 domains) contribute to PLCγ activation and have an important role in localizing the enzyme to protein complexes in different cellular compartments (5, 6). However, the elucidation at the molecular level of how PLCγ isoforms are regulated remains an area of intense study.
Despite the common domain organization shared by the PLCγ1 and PLCγ2 isoforms, studies using gene-targeting approaches demonstrated that each has a distinct biological role (7, 8). Different functions of PLCγ1 (essential role in embryonic development) and PLCγ2 (requirement for development and function of hematopoietic cells) to some degree reflect their different expression patterns and, in particular, the abundance of PLCγ2 in hematopoietic cells. However, studies of different cell types where both isoforms are present (e.g. platelets, macrophages/monocytes, granulocytes, and NK cells) have shown that one isoform can be preferentially activated over the other, suggesting that additional mechanisms must exist to determine the distinct roles of PLCγ1 and PLCγ2 (9-11). Overall, studies of proteins that bind to SH2 and SH3 domains and target PLCγ1 and PLCγ2 to signaling complexes suggest that these binding partners are not specific to either PLCγ isoform (11). However, a recent analysis of the two PLCγ isoforms has shown that only PLCγ2 can be activated by the Rho family GTPase, Rac (12). Importantly, this was the first report to identify a signaling component that could provide a basis for differential regulation of these two closely related PLCγ isoforms.
The report of activation of PLCγ2 by Rac has also expanded the scope of potential regulators of the PLCγ family and is in line with the interconnection between other Ras superfamily GTPases and PLC isoforms. Thus, although the possible role of small GTPases in the activation of phosphoinositide-specific PLC was noted over 20 years ago, it is only recently that progress has been made in uncovering the identity of the interacting protein components (13, 14). Initially, it was reported that Rac GTPases and Cdc42 specifically activate the PLCβ2 isoform (15, 16). The recently discovered PLCε isoform was reported to be regulated by specific Ras and Rho family GTPases (17-20). In addition, the PLCδ1 isoform has been implicated in binding to Ral GTPases, leading to subsequent activation (21). Furthermore, recent studies have defined the structural basis for select examples of these interactions. For example, the crystal structure of activated H-Ras bound to the isolated PLCε RA2 domain revealed an interaction surface that is distinctly different from those of other known Ras effectors (c-Raf, RalGDS, and phosphatidylinositol 3-kinase) that contain the same RA/RBD fold (22). More recently, the crystal structure of activated Rac1 bound to a C-terminally truncated PLCβ2 has been reported (23), in which the interaction interface is restricted to the N-terminal PH domain, a region previously implicated as a key structural determinant for Rac-dependent PLCβ2 activation (24-26). This Rac-PH domain complex has expanded the structural diversity of domain types involved in binding Rho family GTPases and highlighted the potential role of PH domains as a site for either protein-lipid and protein-protein interactions (27).
Here, we report an investigation of the structural basis of the Rac binding specificity for PLCγ2 over PLCγ1 and how Rac-dependent activation of PLCγ2 compares with that found for PLCβ2. We uncover a specific mode of interaction with PLCγ2 that involves the spPH rather than the N-terminal PH domain common to PLCβ2. Determination of the three-dimensional structure of the PLCγ2 spPH and identification of residues critical for Rac binding further identify relatively subtle differences between highly similar PLCγ1 and PLCγ2 isoforms, resulting in distinct selectivity for Rac regulatory proteins, important for their function in cellular signaling.
EXPERIMENTAL PROCEDURES
Construction of Vectors—Complementary DNAs encoding c-myc epitope-tagged human PLCγ1 (1291 aa, accession number ABB84466) and human PLCγ2 (1265 aa, accession number NP_002652) were inserted into pcDNA3.1(-) and pVL1393 or pcDNA3.1(+) and pVL1392, respectively. The epitope was attached to the carboxyl termini ((L/S)EQKLISEEDL, carboxyl-terminal residues of PLCγ1 and PLCγ2 underlined).
In our discussion of the chimeric versions of PLCγ1 and PLCγ2, the following nomenclature will be used: PLCγW-XYZ, where W refers to the PLCγ isoform backbone; X, the amino-terminal PH domain; Y, the N-terminal portion of spPH; and Z, the C-terminal portion of spPH. According to this designation, for example, construct PLCγ1-122 corresponds to PLCγ1 with the amino-terminal PH domain from PLCγ1, the N-terminal portion of spPH from PLCγ2, and the C-terminal portion of spPH from PLCγ2 (Fig. 1A). For construction of the cDNAs encoding the chimeric, c-myc epitope-tagged PLCγ enzymes PLCγ1-211 and PLCγ2-122, two separate cDNA fragments, one encoding the amino-terminal PH domain of either PLCγ1 (aa 1-144) or PLCγ2 (aa 1-133) and the other encoding the remainder of PLCγ2 or PLCγ1 followed by the epitope tag, were obtained by PCR and joined together. The cDNA of c-myc epitope-tagged PLCγ1-β11 was constructed using the PCR overlap extension method (28) to join the cDNAs encoding the amino-terminal PH domain of human PLCβ2 (aa 1-137) to the cDNA encoding PLCγ1 without its amino-terminal PH domain (aa 145-1291). The cDNAs of the chimeric, c-myc epitope-tagged PLCγ isozymes PLCγ1-112, PLCγ1-121, PLCγ1-122, PLCγ2-212, PLCγ2-221, and PLCγ2-211, in which one or both portions of the split PH domain of one isozyme (aa 482-527 and 872-937 of PLCγ1; aa 468-513 and 849-914 of PLCγ2) were replaced by the corresponding regions of the other, were constructed using a two-step megaprimer PCR protocol (29). The primer sequences and PCR protocols are available from the authors upon request. The cDNA of a deletion mutant (Δ1-188) of human Vav1 was amplified by PCR and ligated into pcDNA3.1(+) already containing a DNA sequence encoding the 12CA5 hemagglutinin epitope tag (MGYPYDVPDYAGGSM; hemagglutinin epitope underlined and Met189 of Vav1 shown in italic type).
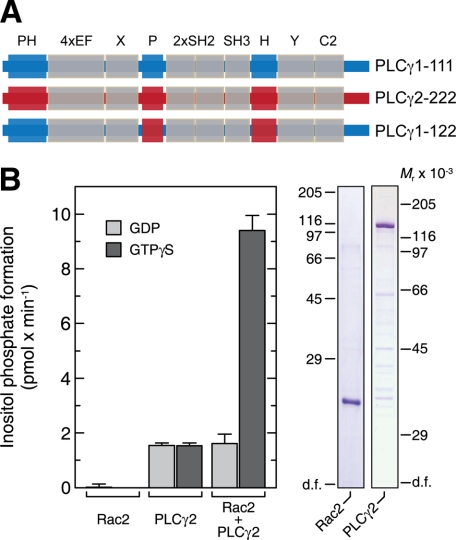
FIGURE 1.
PLCγ2 is activated by Rac2. A, domain organization of PLCγ isoforms and their chimera. The common core domains (N-PH, EF, catalytic, and C2) and unique regions are shown. The PLCγW-XYZ nomenclature, used throughout, refers to W (PLCγ isoform backbone), amino-terminal PH domain (X), N-terminal portion of spPH (Y), and C-terminal portion of spPH (Z). According to this designation, for example, construct PLCγ1-122 corresponds to PLCγ1 with the amino-terminal PH domain from PLCγ1, the N-terminal portion of spPH from PLCγ2, and C-terminal portion of spPH from PLCγ2. B, the activation of purified PLCγ2 by purified Rac2 can be reconstituted in vitro. Recombinant Rac2 and PLCγ2 were purified from baculovirus-infected insect cells and reconstituted in the presence of 100 μm GDP or 100 μm GTPγS with phospholipid vesicles containing phosphatidylinositol (4,5)-bisphosphate (left). The purity of the preparations is also shown (analysis by SDS-PAGE and Coomassie Blue staining) (right).
To prepare a baculovirus encoding GST-tagged Rac2, the cDNA of human Rac2 was inserted into the baculovirus transfer vector pAc2GT (Pharmingen). For expression of proteins in Escherichia coli (PLCγ1 spPH, aa 485-936, Δ530-864; PLCγ2 spPH, aa 471-913, Δ516-841, wild type, and mutants K862I, V893Q, and F897Q; and Rac2G12V, aa 2-177), the particular cDNAs were cloned into the pTriEx4 vector (Novagen) using the Ek/LIC methodology following the manufacturer's instructions. All expression constructs were PCR-amplified with a TeV protease recognition sequence followed by a GGSGGS linker followed by the domain open reading frame.
Expression and Purification of Proteins—For production of recombinant isoprenylated Rac2, baculovirus-infected insect (Sf9) cells (Invitrogen) were grown at 27 °C in suspension culture in TNM-FH medium containing 10% (v/v) fetal calf serum (catalog number P04-83500; PAN Biotech, Aidenbach, Germany) supplemented with 0.2% (w/v) Pluronic® F-68 (Invitrogen), 50 μg/ml gentamicin (PAA Laboratories), and 2.5 μg/ml amphotericin B (Fungizone®; Invitrogen) in a 1800-ml Fernbach culture flask. Cells (109 cells/flask) were incubated at 27 °C with recombinant baculovirus in 400 ml of medium at 80 rpm on a rotary shaker with an amplitude of 25 mm. Three days after infection, the cells were harvested at room temperature by centrifugation at 300 × g for 5 min and washed once with 100 ml of buffer A (10 mm Na2HPO4, 1.8 mm KH2PO4, 140 mm NaCl, 2.7 mm KCl, pH 7.4) per 109 intact cells at the time of cell harvesting. To obtain detergent-solubilized Rho GTPases, the cells were resuspended in 15 ml per 109 intact cells of ice-cold buffer B containing 20 mm Tris/HCl, pH 8.0, 1 mm EDTA, 1 mm dithiothreitol, 100 mm NaCl, 3.75 mm MgCl2, 0.1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 3 μm GDP and homogenized using a precooled 5-ml Teflon-glass homogenizer. Nuclei and unbroken cells were removed by centrifugation at 300 × g for 10 min at 4 °C. The membrane fraction was collected from the resulting supernatant by centrifugation at 12,000 × g for 15 min at 4 °C. Rho GTPases were solubilized by resuspending the membranes in 2 ml per 109 intact cells at the time of cell harvesting of ice-cold buffer B supplemented with 23 mm sodium cholate and incubating this mixture for 90 min at 4 °C with vigorous vortexing every 10 min. Insoluble material was removed from this suspension by centrifugation at 12,000 × g for 15 min at 4 °C. The resulting detergent extract was aliquoted, snap-frozen in liquid N2, and stored at -80 °C.
For production of recombinant PLCγ isozymes, Sf9 cells were grown at 27 °C in adherent culture in 75-cm2 flasks in TNM-FH medium (catalog number T3285; Sigma) supplemented with 10% (v/v) fetal calf serum (catalog number F7524; Sigma) and 50 μg/ml gentamicin. Cells (20 × 106 cells/flask) were incubated with recombinant baculovirus at 27 °C in 10 ml of medium/flask. Three days after infection, the cells were detached from the plastic surface, harvested by centrifugation at 300 × g for 5 min at room temperature, washed once at room temperature with 1 ml/flask of buffer A, and then resuspended in 100 μl/flask of ice-cold buffer C containing 20 mm Tris/HCl, pH 7.5, 2 mm EDTA, 2 μg/ml soybean trypsin inhibitor, 3 mm benzamidine, 0.1 mm phenylmethylsulfonyl fluoride, 1 μm pepstatin, 1 μm leupeptin, and 1 μg/ml aprotinin. The cells were homogenized by forcing the suspension ten times through a 0.45 × 25-mm needle attached to a disposable syringe. The homogenate was centrifuged at 100,000 × g for 1 h at 4 °C, and the resulting supernatant was aliquoted, snap-frozen in liquid N2, and stored at -80 °C.
c-myc epitope-tagged PLCγ1-111, PLCγ2-222, and PLCγ1-122 were purified from soluble fractions of baculovirus-infected insect cells grown in suspension culture by sequential chromatography on HiTrap™ Heparin HP and MonoQ (GE Healthcare) as described for PLCβ2Δ in Ref. 30. For purification of posttranslationally modified Rac2, the protein was expressed as a glutathione S-transferase fusion protein in baculovirus-infected insect cells and solubilized from the particulate fraction as described for wild-type Rac2 (12). The protein was purified from the detergent extract by batch adsorption to glutathione-Sepharose™ 4B (GE Healthcare); cleavage of the Rac2 portion from the resin by proteolysis with thrombin (22.5 units/ml 75% (v/v) slurry) in buffer D containing 50 mm Tris/HCl, pH 7.5, 50 mm NaCl, 2 mm MgCl2, 1 mm dithiothreitol, and 0.1% (v/v) Triton X-100; and removal of the protease by batch adsorption to p-aminobenzamidine-agarose (Sigma).
Purification of proteins from E. coli was essentially as described in Refs. 31 and 32. Recombinant proteins were expressed from pTriEx4 vectors in E. coli overnight at 25 °C in the presence of 100 μm isopropyl 1-thio-β-d-galactopyranoside (induction was carried out when the bacterial culture attained an A600 of between 0.4 and 1.0). A four-step purification procedure was then adopted. First, Ni2+-chelating chromatography utilizing 5-ml HisTrap columns (GE Healthcare) and wash buffer E (25 mm Tris/Cl, 500 mm NaCl, 40 mm imidazole, and 1 mm tris(2-carboxyethyl)phosphine hydrochloride, pH 8.0) and eluting buffer F (25 mm Tris/Cl, 500 mm NaCl, 500 mm imidazole, and 1 mm tris(2-carboxyethyl) phosphine hydrochloride, pH 8.0). Second, the His and S-tags were proteolytically cleaved overnight by TeV protease in cleavage and dialysis Buffer G (25 mm Tris/Cl, 150 mm NaCl, 1 mm tris(2-carboxyethyl) phosphine hydrochloride, pH 8.0) at 4 °C. Third, the cleaved protein mix was passed over a Ni2+-loaded 5-ml HiTrap chelating column (GE Healthcare) in Buffer G, and the flow-through was collected. Last, the flow-through fractions were loaded on a Superdex 75 26/60 gel filtration column (GE Healthcare) in Buffer G, and fractions of monomeric protein were collected and concentrated. Proteins were either used immediately or stored by snap freezing in liquid N2 and transfer to -80 °C. Labeled proteins for NMR studies were expressed essentially as outlined in Ref. 33 and purified as described above.
Measurement of PLC Activity in Vitro—Phospholipase C activity was determined as described (30, 34) with minor modifications. In brief, aliquots (10 μl) of the soluble fraction of PLCγ-baculovirus-infected insect cells appropriately diluted in buffer H, containing 60 mm Tris/maleate, pH 7.3, 84 mm KCl, 3.6 mm EGTA, 2.4 mm dithiothreitol, 2 mg/ml bovine serum albumin, were incubated for 45 min at 30 °C in a volume of 60 μl containing 50 mm Tris/maleate, pH 7.3, 70 mm KCl, 3 mm EGTA, 2 mm dithiothreitol, 536 μm phosphatidylethanolamine, 33.4 μm [3H]phosphatidylinositol (4,5)-bisphosphate (185 GBq/mmol), 0.33 mg/ml bovine serum albumin, and the concentrations of sodium deoxycholate and free Ca2+ specified in the figure legends. For reconstitution of wild-type and mutant PLCγ isozymes with Rac2, the diluted soluble fraction containing the PLC or purified PLCγ2 was reconstituted with 5 μl of detergent extract containing crude or purified isoprenylated Rac2 and incubated with the phospholipid substrate as described above. Fifty mm HEPES/NaOH, pH 7.2, was present in the incubation medium instead of 50 mm Tris/maleate, pH 7.3, when purified proteins were reconstituted. The concentration of CaCl2 required to adjust the concentration of free Ca2+ to the desired value was calculated using the program EqCal for Windows (Biosoft, Ferguson, MO). The reaction was terminated, and the samples were analyzed for inositol phosphates, as described (30).
Cell Culture and Transfection—COS-7 cells were maintained at 37 °C in a humidified atmosphere of 90% air and 10% CO2 in Dulbecco's modified Eagle's medium (catalog number 41965-039; Invitrogen) supplemented with 10% (v/v) fetal calf serum (catalog number 10270-106; Invitrogen), 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (all from PAA Laboratories, Cölbe, Germany). Prior to transfection, the cells were seeded into 12-well plates at densities of 1 × 105 cells/well, respectively, and grown for 24 h in 1 ml/well of the same medium. One hour before transfection, the medium was replaced with 1 ml/well of fresh medium. For transfection of COS-7 cells, plasmid DNA (1.0 μg DNA/well) was mixed with 2.0 μl Lipofectamine™ 2000 Reagent (Invitrogen) in 0.2 ml of Opti-MEM® I (Invitrogen) according to the manufacturer's instructions. After the addition of the DNA-Lipofectamine™ 2000-complexes to the dishes, the cells were incubated for a further 24 h at 37 °C and 10% CO2 without changing the medium.
Analysis of Inositol Phosphate Formation in Intact COS-7 Cells—Twenty-four hours after transfection, the cells were washed once with 0.5 ml/well of buffer A and then supplied with 0.4 ml/well of Dulbecco's modified Eagle's medium containing fetal calf serum and supplements as specified above, 10 μCi/ml myo-[2-3H]inositol (catalog number TRK911; GE Healthcare), and 10 mm LiCl. The cells were incubated in this medium for 20 h, washed once with 0.4 ml/well of buffer A, and then lysed by the addition of 0.2 ml/well of 10 mm ice-cold formic acid (35). After keeping the samples on ice for 30 min, 0.3 ml/well of 10 mm NH4OH was added for neutralization, and the sample was centrifuged for 5 min at 15,000 × g. The supernatant was loaded onto a column containing 0.25 ml of Dowex® 1×8-200 ion exchange resin (catalog number 217425; Sigma) that had been converted to the formate form and equilibrated with H2O as described (34). The columns were washed once with 3 ml of H2O and then twice with 3.5 ml each of 60 mm sodium formate and 5 mm sodium tetraborate, and inositol phosphates were eluted with 3 ml of 1 m ammonium formate and 100 mm formic acid. The eluate was supplemented with 15 ml of scintillation fluid (Ultima Gold™; PerkinElmer Life Sciences), and the radioactivity was quantified by liquid scintillation counting. The columns were reused after regeneration, as described (34).
NMR Spectroscopy—NMR spectra were acquired at 298 K on a Varian UnityPLUS (500 MHz), Varian Inova (600 and 800 MHz), or Bruker Avance III spectrometer (700 MHz) equipped with either a triple resonance probe or a cryogenically cooled triple resonance probe, including z axis pulse field gradient coil. Sequence-specific resonance assignments were obtained using standard triple resonance NMR spectroscopy, namely 1H-15N HSQC, 1H-13C HSQC, HNCA, HN(CO)CA, HNCO, HNCACB, CBCA(CO)NH, 1H-15N TOCSY-HSQC, HC(C)H-TOCSY. Distance restraints were derived from three-dimensional 15N- and 13C-edited NOESY-HSQC spectra with a mixing time of 100 ms. All NMR spectra were processed using NMRPipe/NMRDraw (36) and analyzed using ANSIG for OpenGL version 1.0.3 (37). 1H, 13C, and 15N chemical shifts were referenced indirectly to sodium 2,2-dimethyl-2-silanepentane-5-sulfonate, using absolute frequency ratios for the 1H signals (38).
The interaction of the PLCγ2 spPH with GppNHp-loaded Rac2G12V (aa 2-177) was performed at constant concentration of spPH protein using the method previously described (39), ranging from spPH/Rac2 molar ratios of 1:0 to 1:1.1. Protein concentrations were estimated by using predicted extinction coefficients based upon amino acid composition. The concentration of the PLCγ2 spPH was 0.5 mm. Any changes in the spectrum of labeled component during the titration can be attributed directly to an intermolecular interaction, since in each experiment both proteins are pre-exchanged into the same buffer.
Structure Calculations—Interproton distance restraints were derived from the ANSIG cross-peaks file of three-dimensional 15N NOESY-HSQC and 13C NOESY-HSQC spectra for the PLCγ2 spPH domain. A proportion of the resonances were successfully assigned in a manual fashion without ambiguity. The remaining cross-peaks appearing at positions in the spectrum with overlapping resonances were labeled with ambiguous assignments by reference to the chemical shift list obtained with through-bond correlation spectra, using the “Connect” module from the program AZARA (40). The cross-peaks were grouped into five categories according to their relative peak intensities (strong, medium, weak, very weak, and very, very weak) and were designated with the corresponding interproton distance restraint limit of 1.8-2.5, 1.8-3.0, 1.8-3.5, 1.8-4.0, and 1.8-5.0 Å, respectively. A distance of 0.5 Å per methyl group was added to the upper bound of the distance restraint for NOE cross-peaks that involved methyl groups.
All structures for spPH were calculated using an ab initio simulated annealing protocol within the CNS program (41), with PARALLHDG version 5.3 force field and PROLSQ nonbonded energy function (42). The protocol adopts a mixture of Cartesian molecular dynamics and torsion angle dynamics simulated annealing to refine structures starting from random generated conformers with good local geometry.
A total of 2487 NOE-derived interproton distance restraints for spPH were included in the final iterations of the structure calculations (see Table 1). Backbone torsion angle restraints for ϕ and ψ were derived from analysis of 1Hα, 13Cα, 13Cβ, 13C′, and 15NH chemical shift data bases as implemented in the program TALOS (43). Hydrogen bond restraints for amide protons were derived from an assessment of the regular secondary structure elements. This analysis included the overall and local patterns of NOEs and the pattern of amide proton solvent exchange rates. A total of 114 dihedral angle and 70 hydrogen bond (35 hydrogen bonds; two distance restraints per hydrogen bond) interatomic distance restraints were used for spPH.
TABLE 1.
Kinetic parameters and derived dissociation constants for the interaction between Rac2 and PLCγ isozymes determined by surface plasmon resonance measurements
Kd values were derived from the ratio ka/kd. WT, wild type.
| Analyte | Liganda | ka | kd | Kd |
|---|---|---|---|---|
| m−1 s−1 | s−1 | μm | ||
| PLCγ2-222 (WT) | His-Rac2 (GTPγS) | 806 | 3.1 × 10−3 | 3.9 |
| PLCγ1-111 (WT) | His-Rac2 (GTPγS) | Noneb | ||
| PLCγ1-122 | His-Rac2 (GTPγS) | 434 | 2.5 × 10−3 | 5.8 |
| PLCβ2 (PH-C2) | His-Rac2 (GTPγS) | 385 | 2.3 × 10−3 | 6.0 |
| γ2spPH (WT) | His-Rac2 (GTPγS) | 93.8 | 1.6 × 10−3 | 17 |
| γ2spPH (WT) | His-Rac2 (GDPβS) | None | ||
| γ1spPH (WT) | His-Rac2 (GTPγS) | None |
Rac2 protein designated as His-Rac2 contains the sequence His6-S-tag-TeV-GGS-GGS- at the N terminus.
None, no significant association signal was detected.
Biosensor Measurements—The biosensor measurements were carried out on the BIAcore 3000 system (GE Healthcare) at 25 °C. The sensor chip NTA was utilized and loaded with Ni2+ according to the manufacturer's instructions. Purified, hexahistidine-tagged Rac2G12V (aa 2-177) was loaded with GTPγS or GDPβS and immobilized in biosensor buffer (10 mm HEPES/NaOH, pH 8.0, 150 mm NaCl, 1 mm MgCl2, 5% (w/v) CM-dextran, and 0.01% (v/v) Nonidet P-40) at a flow rate of 5 μl/min for 5 min, which resulted in a deposition of ∼300 response units. Next, the purified analytes (full-length PLCγ molecules or their isolated spPHs) were injected at varying concentrations. The values for nonspecific binding measured in the reference cell were subtracted. The evaluation of kinetic parameters was performed by nonlinear fitting of binding data using BiaEvaluation 2.1 software. The apparent association (ka) and dissociation rate (kd) constants were evaluated from the differential binding curves (Fc2 - Fc1) assuming an A + B = AB association type for the protein-protein interaction. The dissociation constant KD was calculated from the equation, KD = kd/ka.
Miscellaneous Methods—Recombinant baculoviruses were produced as described (44). The mouse monoclonal antibody 9B11 reactive against the c-myc epitope EQKLISEEDL was obtained from Cell Signaling Technology. The sources of all other reagents and recombinant DNAs as well as all other experimental protocols have been described (12). All experiments were performed at least three times. Similar results and identical trends were obtained each time. Data from representative experiments are shown as means ± S.D. of triplicate determinations.
RESULTS AND DISCUSSION
The PLCγ2 Split PH Domain Is Required for Isoform-specific Regulation by Rac2—It has previously been shown with reconstitution experiments that Rac GTPases activate PLCγ2 in the presence of GTPγS (12). These experiments were conducted with cell extracts enriched in recombinant PLCγ2 and Rac2 that had been separately expressed in baculovirus-infected Sf9 cells. To exclude the possibility that other signaling proteins could mediate activation of PLCγ2 by Rac, we set out to extend these data with purified components (Fig. 1B). We noted a 7-fold activation of PLCγ2 by GTPγS-loaded Rac2. This result strongly suggests that Rac2 interacts directly with PLCγ2 and that the presence of Rac2 is both necessary and sufficient for guanine nucleoside triphosphate-dependent PLCγ2 activation. Next, we prepared a number of chimeric proteins where the N-terminal PH domains of the Rac-responsive PLCγ2 and the Rac-nonresponsive PLCγ1 were exchanged. The specific PLC activities of each of these chimeras were tested in the presence of 10 μm Ca2+ and shown to be comparable (Fig. 2A, left). However, when introduced into the reconstitution assay, it was evident that the N-terminal PH domain of PLCγ2 does not impart Rac2 activation on PLCγ1 (variant PLCγ1-211) (Fig. 2A, right). Similarly, the exchange of the PLCγ1 N-terminal PH domain into PLCγ2 (variant PLCγ2-122) did not significantly alter the propensity of this chimera to be activated by Rac2. These in vitro observations were further supported by experiments in intact COS-7 cells co-transfected with cDNAs encoding Rac2 and PLCγ isozymes to display the same basal PLC activities (Fig. 3A). We confirmed that Rac2G12V activates PLCγ2 but not PLCγ1 and that the N-terminal PH domain is not involved in the Rac2-mediated activation. In addition, we showed that the Rac-binding N-terminal PH domain of PLCβ2 is functionally interchangeable by constructing a PLCγ1 chimera that incorporates this domain (variant PLCγ1-β11) and showing its activation by Rac2G12V (Fig. 3A). Together, these experiments suggest that, unlike with PLCβ2, the N-terminal PH domain of PLCγ2 is not involved in the observed interaction of Rac2. Furthermore, the findings support the idea that Rho family GTPase effector interaction sites are not conserved and cannot easily be predicted (45).
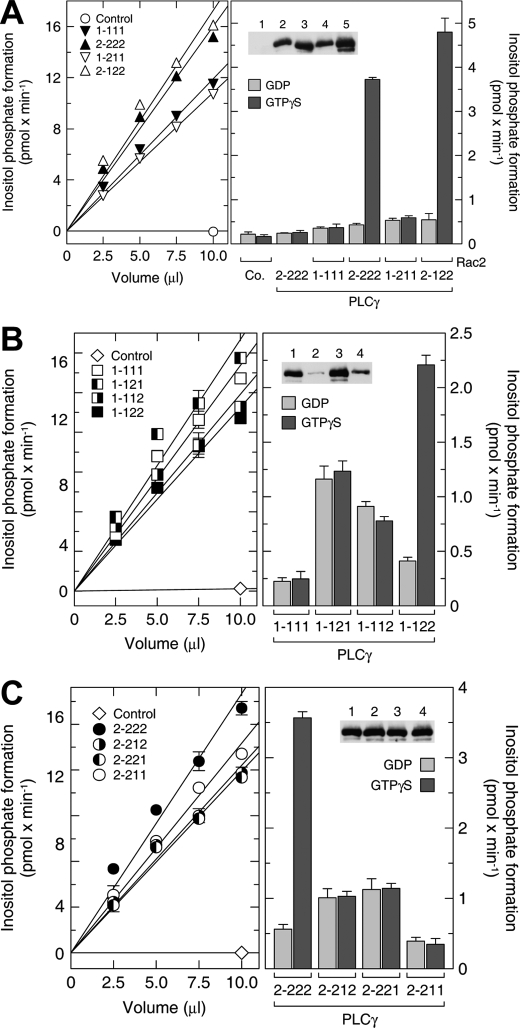
FIGURE 2.
The split PH domain of PLCγ2 is required for its regulation by Rac2 in vitro. A-C, left, soluble fractions of Sf9 cells infected with baculoviruses encoding β-galactosidase (control), wild-type PLCγ isoforms (PLCγ1-111 and PLCγ2-222), and their chimeras (PLCγ1-211, PLCγ2-122, PLCγ1-121, PLCγ1-112, PLCγ1-122, PLCγ2-212, PLCγ2-221, and PLCγ2-211) were diluted with buffer and incubated at increasing protein concentrations for 45 min at 30 °C with phospholipid vesicles containing phosphatidylinositol (4,5)-bisphosphate. The incubation was performed in the presence of 10 μm free Ca2+ and 2.5 mm sodium deoxycholate. A-C, right, the soluble fractions of Sf9 cells infected with baculoviruses encoding the indicated wild-type and mutant PLCγ isozymes were adjusted by dilution with buffer to contain similar basal PLC activity according to the results shown in the left panel. The soluble fraction of Sf9 cells infected with baculovirus encoding β-galactosidase (control) was used at the maximal protein concentration among the PLCγ-containing fractions, 1.4 mg protein/ml. Aliquots (10 μl) of these samples were reconstituted with aliquots of detergent extracts prepared from membranes of Sf9 cells infected with baculoviruses encoding β-galactosidase (no bracket) or Rac2 (brackets) and incubated for 2 h at 30 °C in the presence of 100 mm GDP or GTPγS with phospholipid vesicles containing phosphatidylinositol (4,5)-bisphosphate. The incubation was performed in the presence of 30 nm free Ca2+ and 1 mm sodium deoxycholate. Inset, aliquots (10 μl) of the samples were subjected to SDS-PAGE, and immunoblotting was performed using an antibody reactive against the c-myc epitope. In A, there are five lanes in the inset but six samples in the corresponding bar chart. The lane corresponding to PLCγ2-222 without Rac2 is not shown in the inset. Lanes 1-5, control, PLCγ1-111, PLCγ2-222, PLCγ1-211, and PLCγ2-122, respectively (all with Rac2).
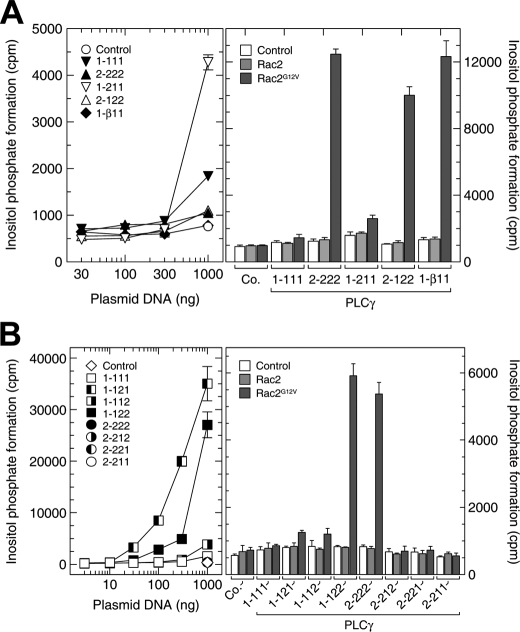
FIGURE 3.
The role of the N-terminal and split PH domains of PLCγ2 in cellular activation by Rac2. Left, COS-7 cells were transfected with increasing amounts per well of vector encoding wild-type or mutant PLCγ isozymes. The total amount of DNA was maintained constant in each transfection by adding empty vector. The empty vector (control) (A and B) and the vectors encoding PLCγ2-222, PLCγ2-212, PLCγ2-221, and PLCγ2-211 (B) were used only at 1000 ng/well, since there were only minimal changes in inositol phosphate production even at this high amount of vector DNA. Under these conditions, the inositol phosphate formation in B was as follows: control, 223 ± 30 cpm; PLCγ2-222, 436 ± 67 cpm; PLCγ2-212, 390 ± 59 cpm; PLCγ2-221, 348 ± 54 cpm; PLCγ2-211, 360 ± 6 cpm (mean ± S.D. of triplicate determinations). [3H]Inositol phosphate accumulation was measured as described under “Experimental Procedures.” Right, COS-7 cells were cotransfected as indicated with empty vector (control) and/or vectors encoding Rac2, Rac2G12V, or either wild-type or mutant PLCγ isozymes. The amounts of vectors encoding the PLCγ isozymes were adjusted according to their basal activities shown in the left panels (PLCγ1-111 and PLCγ1-211, 300 ng/well; PLCγ1-112, 100 ng/well; PLCγ1-121 and PLCγ1-122, 10 ng/well; all other vectors, 1000 ng per well). The total amount of DNA was maintained constant in each transfection by adding empty vector. In additional experiments (results not shown), we found that expression of Rac2G12V also caused only a minor (≤1.9-fold) stimulation of inositol phosphate formation in cells cotransfected with 1000 ng/well of vector encoding PLCγ1-111 or PLCγ1-211.
The PLCγ isoforms contain a second PH domain within the specific array (SA) region located between the X and Y domains of the catalytic barrel. This spPH consists of two parts separated by the tandem array insert of two SH2 domains and an SH3 domain. Although the two halves of PLCγ1 spPH can form a contiguous fold when expressed without the other domains (4), it is not known whether, in the context of the full-length PLCγ molecules, these two sections also form a contiguous PH domain or are spatially separated. Recently, there have been reports that attribute different functions to spPHs (46, 47) with the insertion of other domains between either β-strands 6 and 7 or β-strands 3 and 4, as in the case of the PLCγ1 isoform (4). Accordingly, we prepared chimeric PLCγ1 proteins that contained either one or both of the PLCγ2 spPH sections. The swapping of either the N- or C-terminal spPH subdomains (see Fig. 1A) from PLCγ2 did not confer Rac activation on PLCγ1 (Fig. 2B, right). However, the insertion of both partial spPH subdomains from PLCγ2 produced a PLCγ1 chimera that was stimulated 5.4-fold in activity by recombinant Rac2. The reverse chimera experiment was also carried out. The PLCγ1 spPH subdomains were engineered into the PLCγ2 polypeptide chain both as individual partial domains and as both halves together (Fig. 2C, right). The exchange of either or both of the partial domains abolished activation by Rac2 in reconstitution experiments. Therefore, both spPH subdomains of PLCγ2 are necessary and sufficient to impart Rac2-dependent PLCγ activation. This conclusion is supported by experiments that tested these PLCγ chimeras in transfected COS-7 cells (Fig. 3B). Of note, the two spPH subdomains of PLCγ2 also imparted on PLCγ1 a marked sensitivity to activation by exogenous Vav1 and endogenous Rac GTPases present in COS-7 cells, whereas the presence of the two spPH subdomains of PLCγ1 within the context of PLCγ2 rendered the chimeric enzyme indistinguishable in this regard from wild-type PLCγ1 (Fig. S1).
The data obtained from the analysis of PLCγ spPH (Figs. 2, B and C, and 3B), suggest that either the site of Rac2 interaction is distributed over both halves of spPH or that correct folding of each half requires the presence of the other from the same isoform. Since our further studies suggest that the first scenario is unlikely (see Fig. 7), the incorrect folding of each spPH half could be the reason for the loss of interaction with Rac. Indeed, recent studies of PLCγ1 have shown that the construct of the isolated second half of its spPH was unfolded but that the interaction with the complementary half induces the correct folding (4). Since the sequence identity (29%) of the spPH regions of the PLCγ isoforms is low, it is likely that their subdomains cannot be interchanged without losing correct spPH folding.
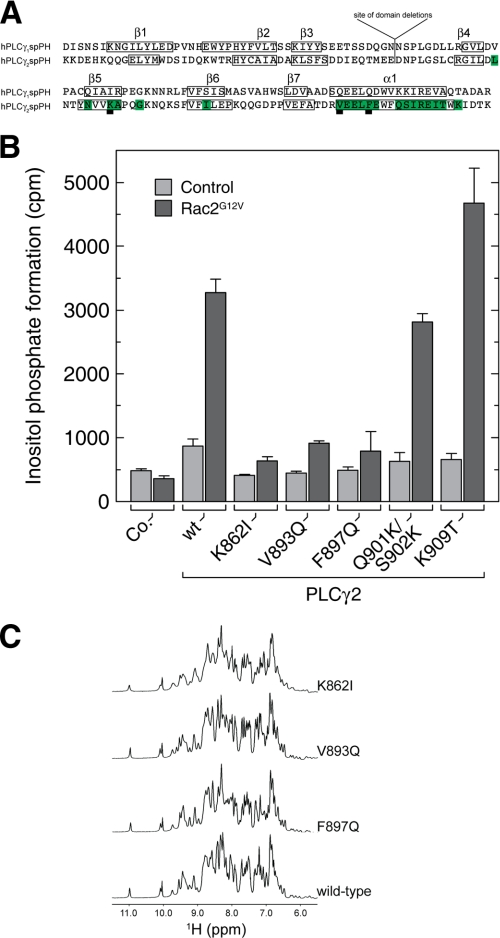
FIGURE 7.
Analysis of PLCγ2 mutants that are insensitive to Rac activation. A, alignment of the primary structures of the PLCγ1 and PLCγ2 split PH domains. Amino acid residues in PLCγ2 whose NMR resonances were perturbed in the Rac2 titration are labeled in green. Boxed elements represent regions of regular secondary structure. B, determination of PLCγ2 spPH residues important for activation by Rac2. COS-7 cells were cotransfected as indicated with empty vector (control), vector encoding Rac2G12V, and vector encoding either wild-type or mutant PLCγ2 isozymes (K862I, V893Q, F897Q, Q901K/S902K, and K909T). The total amount of DNA was maintained constant in each transfection by adding empty vector. Twenty-four h after transfection, the cells were incubated for 24 h in the presence of [3H]inositol (1.5 μCi/ml) and 10 mm LiCl, and the levels of inositol phosphates were then determined. C, one-dimensional NMR spectra of wild-type and mutant PLCγ2 spPH proteins. The indicated substitutions were introduced into the isolated spPH construct, and the corresponding encoded proteins were assessed and compared with their wild-type counterparts by one-dimensional 1H NMR spectroscopy. The downfield region encompassing resonances from the backbone NH and aromatic side chain CH protons is depicted for each variant. The maintenance of the overall chemical shift dispersion indicates that the mutants adopt a globular structure highly similar to the wild type.
The Isolated Split PH Domain from PLCγ2 Directly Binds Rac—To evaluate the role of PLCγ2 spPH as the site of Rac interaction, we purified a number of PLCγ2 and PLCγ1 variants (including the full-length and isolated spPHs) in order to carry out interaction studies in vitro. For comprehensive, quantitative analysis we used surface plasmon resonance (Table 1). Consistent with the data where we analyzed the requirements for activation of PLCγ2 by Rac2 (Figs. 1, 2, 3), PLCγ2, but not PLCγ1, selectively bound GTPγS-activated Rac2 (Table 1). Furthermore, the PLCγ1 chimera incorporating both halves of the PLCγ2 spPH (PLCγ1-122) was fully functional in Rac2-GTPγS binding. The strength of the binding of PLCγ2 and PLCγ1-122 (KD = 3.9 and 5.8 μm, respectively) was similar to that determined in this and a previous study for PLCβ2 (KD = 6.0 and 7.0 μm, respectively) (26). These data confirm PLCγ2 as a direct effector of Rac and show that its spPH determines the isoform specificity for this interaction.
We also assessed whether PLCγ2 spPH in isolation can bind GTPγS-activated Rac2. Based on the recent structural characterization of PLCγ1 spPH (4), we designed a contiguous PLCγ2 spPH construct lacking the intervening SH2 and SH3 domains and being replaced by a linker consisting of the remaining natural loop regions. In essence, a “regular” PH domain is predicted to be formed by the directly linked spPH subdomains. The corresponding domain from PLCγ1 was also constructed. Both PLCγ2 and PLCγ1 spPHs could be prepared in good yields. PLCγ2 spPH selectively bound Rac2-GTPγS, similar to the full-length protein; importantly, for PLCγ1 spPH, no interaction with Rac2 could be detected (Table 1). The binding strength for PLCγ2 spPH with Rac2 (KD = 17 μm) is in close agreement with previously reported affinities of Rac2 for the isolated PH domain of PLCβ2 (26). Subsequent structural studies have shown that the PLCβ2 isoform contacts Rac solely through its PH domain (23).
The strengths of interaction between PLCγ2 and Rac2 shown here (Table 1), in the micromolar range for KD, are generally consistent with values obtained for PLCβ2-Rac (26) and PLCε-Ras (22) complexes and more broadly with a number of other small GTPase-effector interactions (48) measured in vitro. There are, however, instances of Rac- and Cdc42-effector interactions with dissociation constants in the nanomolar range (49, 50). However, in a cellular setting, the posttranslationally modified C terminus of Rac2 or the plasma membrane could be involved in stabilizing the interaction between activated Rac2 and full-length PLCγ2. It is important to note that the spPHs of PLCγ1 and -γ2 are unlikely to bind to membrane lipids directly. Experimental (4) and molecular modeling (51) studies agree that these domains do not possess the amino acid sequence motifs typical of lipid binding modules. Accordingly, although some changes in the subcellular distribution do occur with some of the chimeras (PLCγ1-121, PLCγ1-112, and PLCγ2-221) (Fig. S2), these effects do not correlate with the variation in activity observed in Fig. 3B (left), which are also evident in the cell-free system (Fig. 2B, right).
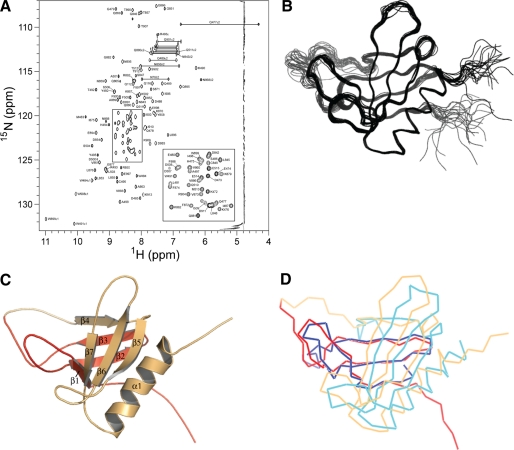
Structural Analysis of the PLCγ2 Split PH Domain Interaction Interface with Rac2—To provide a basis for further analysis of the interaction between the PLCγ2 spPH and Rac2, we determined the three-dimensional solution structure of the spPH by heteronuclear NMR spectroscopy. Single (15N) and double (13C,15N) isotope-labeled samples of PLCγ2 spPH were prepared, and nearly complete resonance assignments were obtained using standard triple resonance NMR experiments (Fig. 4A). On the basis of the analysis of three-dimensional 15N- and 13C-edited 1H NOESY spectra, 2487 interproton distance restraints were obtained and used in structure calculations along with 70 hydrogen bond restraints and 114 dihedral angle restraints. Table 2 shows the structural statistics for the bundle of 20 lowest energy conformers, each of which displays low restraint violations and good stereochemical and nonbonded interaction scores. The best fit superposition of the backbone atoms of the conformer set is shown in Fig. 4B. The lowest energy structure is shown in a ribbon representation in Fig. 4C, demonstrating the conserved core structure of a partially open two-sheet β-barrel with one end capped by the C-terminal helix. As predicted, the PLCγ2 spPH structure conforms well with the canonical PH domain architecture with seven β-strands and one α-helix: residues 478-485 (β1); 490-499 (β2) and 502-506 (β3) from the N-terminal spPH subdomain; 851-853 (β4); 860-863 (β5); 873-876 (β6); 886-889 (β7); and 893-906 (α1) from the C-terminal spPH subdomain. Omitting the loop between β3 and β4, which contains the linker between the spPH subdomains, and a small number of apparently flexible residues at the N and C termini, the refined conformer bundle provides a well defined model for the PLCγ2 spPH with coordinate root mean square difference values of 0.41 ± 0.05 Å for the backbone and 0.76 ± 0.06 Å for all heavy atoms (residues 478-508 and 849-908).
FIGURE 4.
Three-dimensional structure of the PLCγ2 split PH domain. A, heteronuclear NMR spectroscopy of PLCγ2 spPH (471-913, Δ516-841). The two-dimensional 1H-15N HSQC spectrum of 13C/15N-labeled PLCγ2 spPH (471-913, Δ516-841), recorded on a 600-MHz Varian INOVA spectrometer at 298 K. The resonance assignments for the backbone and side chain NH group cross-peaks are included. B, backbone trace of 20 lowest energy conformers of PLCγ2 spPH. C, ribbon representation of the lowest energy PLCγ2 spPH conformer with secondary structure elements labeled. Structural elements derived from the N-terminal spPH region (aa 471-515) are depicted in red, and those from the C-terminal spPH region (aa 842-913) are shown in orange. D, superposition of the backbone Cα trace of the mean solution structures of the PLCγ2 and PLCγ1 spPHs. PLCγ2 (Protein Data Bank code 2k2j; red/orange) and PLCγ1 (Protein Data Bank code 2fjl; blue/cyan) spPHs.
TABLE 2.
Summary of structure statistics for PLCγ2 spPH
<SA> represents the set of 20 selected lowest energy conformers obtained by restrained dynamical simulated annealing in CNS. SAlowest refers to the lowest energy structure of the set. There were no NOE (>0.4 Å) or dihedral (>5°) violations for any of the lowest energy conformers.
| <SA> | SAlowest | |
|---|---|---|
| Experimental restraintsa | ||
| All (Å) (2487) | 0.018 ± 0.002 | 0.014 |
| Intraresidue (786) | 0.014 ± 0.003 | 0.010 |
| Sequential (553) | 0.014 ± 0.005 | 0.010 |
| Short (373) | 0.023 ± 0.003 | 0.020 |
| Long (766) | 0.018 ± 0.001 | 0.015 |
| Ambiguous (9) | 0.009 ± 0.005 | 0.009 |
| Hydrogen bond restraints (Å) (70) | 0.031 ± 0.004 | 0.029 |
| Dihedral angle restraints (degrees) (114) | 0.26 ± 0.03 | 0.236 |
| Deviations from idealized covalent geometryb | ||
| Bonds (Å) (1930) | 0.0013 ± 0.0001 | 0.0012 |
| Angles (degrees) (3490) | 0.31 ± 0.004 | 0.31 |
| Improper dihedrals (degrees) (1020) | 0.2 ± 0.01 | 0.2 |
| Structural statistics for the ensemblec | ||
| PROCHECK parameters | ||
| Most favored region (%) | 73.1 ± 2.7 | 74.3 |
| Additionally allowed (%) | 22.5 ± 2.7 | 23.8 |
| Generously allowed (%) | 3.0 ± 1.6 | 1.0 |
| Disallowed (%) | 1.5 ± 0.7 | 1.0 |
| Number of bad contacts | 3 ± 2 | 1 |
| Root mean square difference from the average structured | ||
| Backbone (N, Cα, C) (Å) | 0.41 ± 0.05 | 0.34 |
| Heavy atoms (Å) | 0.76 ± 0.06 | 0.62 |
Sum averaging of NOE distance restraints was used for groups with degenerate proton chemical shifts. The interproton unambiguous distance restraint list comprised 786 intraresidue, 553 sequential (|i - j| = 1|), 373 short range (1 < |i - j| < 5), and 776 long range (|i - j| > 5). Hydrogen bond restraints were applied as pairs of distance restraints: HN···O, 1.2-2.2 Å; N···O, 1.2-3.2 Å. The final values for the respective force constants were as follows: NOE, 30 kcal mol−1 Å−2; hydrogen bonds, 50 kcal mol−1 Å−2; dihedral angles, 200 kcal mol−1 rad−2.
The final values for the respective force constants were as follows: bond lengths, 1000 kcal mol−1 Å−2; angles and improper torsions, 500 kcal mol−1 rad−2; the improper torsion angle restraints serve to maintain planarity and chirality.
The program PROCHECK (64) was used to assess the stereochemical parameters of the family of conformers for the spPH. The figures indicate the percentage of residues with backbone Φ and Ψ angles in separate regions of the Ramachandran plot, defined in the program. The number of bad contacts per 100 residues is expected to be in the range 0 - 30 for protein crystal structures of better than 3.0 Å resolution.
The precision of the atomic coordinates is defined as the average pairwise root mean square difference between each of the 20 conformers and a mean coordinate structure SA generated by iterative best fit of the backbone atoms (N, Cα, and C) over residues 478 - 508 and 849 - 908 of PLCγ2spPH (comprising the core secondary structure elements and omitting the flexible N and C termini and the disordered loop between β3 and β4), followed by coordinate averaging.
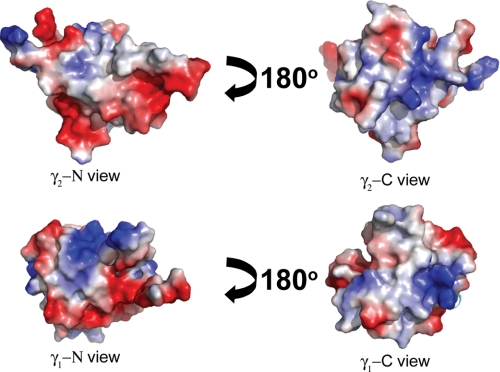
The secondary structure elements of the spPHs from PLCγ1 and PLCγ2 align reasonably well (Fig. 4D) with a backbone root mean square difference of 2.9 Å over 65 core region Cα atoms. It is noteworthy that the sequence identity of the two spPHs is considerably lower (29%) than for the intact PLCγ1 and PLCγ2 proteins (49.5%) or the respective N-terminal PH domains (48%). Despite the relatively low sequence identity, when surface representations of the PLCγ2 and PLCγ1 spPH structures are compared (Fig. 5), there are clearly similarities. Overall, there is a strong correspondence of the surface distribution of charge and hydrophobicity. Notably, a patch of negative charge visible at the lower region of the N view is common between the two domains. Given this apparent global homology, it seems likely that the differences in Rac2 interaction with these two spPHs must reside in rather specific variation in surface side chain distribution. To probe this hypothesis, the residues important for the specific Rac2-binding interface were identified through NMR titration experiments and subsequent site-directed mutagenesis.
FIGURE 5.
Surface charge distribution of PLCγ1 and PLCγ2 spPHs. Surface electrostatic potentials representations of these two spPHs were computed with PyMol (top, PLCγ2 spPH; bottom, PLCγ1 spPH). Electrostatic potentials are represented as positive (blue), negative (red), and neutral (white) charges. The large loop that links the two parts of the spPHs (which is present in the published NMR structure of the PLCγ1 spPH) is not shown. The N view notation refers to the surface derived from the amino acid residues from the N-terminal half of the domain, and the C view refers to those residues derived from the C-terminal part.
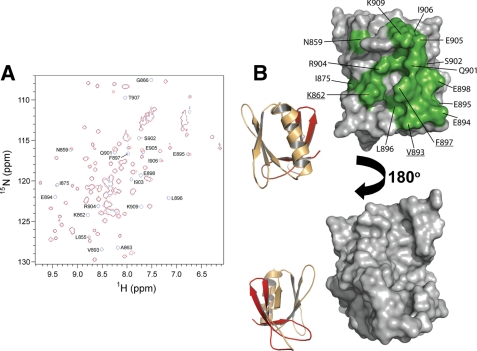
Comparison of the 1H-15N HSQC spectra of 15N-labeled PLCγ2 spPH in the presence of increasing concentrations of unlabeled GppNHp-loaded Rac2G12V (aa 2-177) reveals complex formation characterized by differential broadening and some shifting of a distinct subset of the PLCγ2 spPH cross-peaks (Fig. 6A). We observed 20 backbone NH cross-peaks for PLCγ2 spPH that shift or disappear completely upon Rac titration. The corresponding residues are highlighted in the amino acid sequence of spPH and cluster on the protein surface within the α-helix and around β-strand 5 (Figs. 6B and 7A). Interestingly, several of these residues, including Lys862, Ala863, Ile875, Val893, and Phe897, are not conserved in PLCγ1 spPH (Fig. 7A). Based on these data, we assign the site of Rac interaction to the β-strand 5 and α-helix regions in the C-terminal PLCγ2 spPH subdomain.
FIGURE 6.
The PLCγ2 split PH domain and Rac2 interaction interface. A, elucidation of PLCγ2 spPH residues involved in complex formation with Rac2. Overlay of 1H-15N HSQC spectra of PLCγ2 spPH in the absence (blue) and presence (red) of GppNHp-loaded Rac2G12V (aa 2-177) at a 1:1 molar ratio. Binding of GppNHp-loaded Rac2G12V (aa 2-177) to PLCγ2 spPH leads to a generalized broadening of the spectrum consistent with complex formation. The spectrum of the mixed proteins was plotted at slightly lower contour levels (by a factor of 0.67) in order to emphasize those spPH peaks that are differentially perturbed, thereby highlighting the specific binding site but masking the overall broadening effect. B, surface representation of the amino acid residues of PLCγ2 spPH at the interaction surface with Rac2. Amino acid residues on the surface of PLCγ2 spPH that were perturbed by the titration of Rac2 are labeled and highlighted in green. Amino acid residues that are underlined are those proposed to be important for the binding to Rac2, as presented in Fig. 7B. The figure was prepared with PyMol.
To define more rigorously the amino acid residues in PLCγ2 that are important for interaction with Rac, we carried out site-directed mutagenesis experiments and activity assays. A number of full-length PLCγ2 mutants were prepared with amino acid substitutions in its spPH subdomains. The specific mutations were designed so as to swap the PLCγ2 residue for the amino acid type in the equivalent position in PLCγ1 residue (Fig. 7A). Specifically, full-length K862I, V893Q, F897Q, Q901K/S902K, and K909T PLCγ2 variants were constructed. The wild-type and mutant PLCγ2 constructs were each co-expressed in COS-7 cells with Rac2G12V, and the PLC activities were assessed (Fig. 7B). We identified three residues as important for activation of the enzyme by Rac2. In the β-strand 5 region, the K862I mutant showed a substantially diminished activation by Rac2. The V893Q and F897Q mutations, bordering the α-helix, also yielded a substantially lower activation by Rac2. The remaining mutants demonstrated either a small reduction or even an enhancement of Rac2-dependent activation. We ruled out the possibility that the mutations perturb the spPH fold; the corresponding substitutions were introduced into the isolated spPH construct, and these variants were assessed by one-dimensional 1H NMR. All of these proteins retained a wild-type fold (Fig. 7C). Based on previous studies of PLCγ isoforms, it is also unlikely that these mutations have a direct impact on the structure and function of the catalytic domain (3). These data support the conclusion that these mutated residues (underlined in Fig. 6B) affect Rac binding directly.
Interestingly, our data also show that the surface of PLCγ2 spPH involved in the interaction with Rac2 (Fig. 6) is quite different from that described for the N-terminal PLCβ2 PH domain involved in binding of Rac1 (23), where the main contact region is located in β-strand 1 and loop regions that align with this strand. This variance is generally consistent with the observed diversity of binding sites for Rho family GTPases (45) and further shows that even when the same fold (i.e. aPH domain) is involved, the interaction surfaces engaged in Rac binding can be different.
Implications for Regulation of PLCγ Isoforms—In conjunction with the conclusions drawn from our previous study (12), the results described here reveal several aspects of the regulation of PLCγ2 activity by Rac. First, in reconstitution assays, Rac2 is sufficient to activate PLCγ2 in the absence of other protein components (Fig. 1). Second, when the proteins are co-expressed in COS-7 cells, Rac2 mediates translocation of the PLCγ2 isoform to cellular membranes (12). Third, in transfected cells, Rac2-mediated PLCγ2 activation is not dependent upon the phosphorylation of critical tyrosine residues (12) previously linked to PLCγ2 regulation in B-cells (52, 53). Taken together, these results could be taken to imply that substantial activation of PLCγ2 in vivo can be achieved through interaction with Rac alone. However, such a view potentially ignores complexity in the regulation of PLCγ isoforms that is suggested by other studies that implicate synergy of signaling inputs. Most notably, recent studies of activation of PLCγ1 by growth factor receptors have shown that Tyr783 phosphorylation is not sufficient for full PLC activation (6). The concurrent production of phosphatidylinositol 3,4,5-trisphosphate (PIP3), via epidermal growth factor-stimulated phosphatidylinositol 3-kinase activity, was reported to contribute to the activation of phosphorylated PLCγ1. Similarly, in B-cells, where signaling via PLCγ2 has been best documented, it is possible that Rac2 can contribute to full activation of this isoform together with a set of specific adapter proteins and tyrosine kinases (54-56). In the B-cell system, the evidence suggests that Rac proteins and their activators (such as the Rho guanine nucleotide exchange factors Vav) contribute to production of higher levels of IP3 and greater calcium responses rather then being essential for PLCγ2 activation (57, 58). However, the role and relative contribution of Rac GTPases in the regulation of PLCγ2 could vary between cell types. Importantly, the identification of PLCγ2 residues critical for Rac binding described here (Figs. 6, 7, and S1) provides a solid basis for the design of a Rac-insensitive PLCγ2 variant that could be exploited to dissect the roles and assess the relative importance of the Rac-dependent and other modes of PLCγ2 regulation in different cell types.
It is also interesting to ponder the mechanism by which Rac might regulate PLCγ2 activity. Here we have identified the spPH as the binding site for Rac, the domain within the γ-SA region and thus in proximity to critical tyrosine residues and the SH2 and SH3 domains that mediate interactions with a number of binding partners linked to activation (59). Some reports suggest that the unliganded γ-SA region may have an autoinhibitory role, since removal of this region appears to enhance enzymatic activity (60, 61). The observation that some of the chimeric PLCγ1 spPH mutants displayed enhanced basal activity is consistent with this concept (cf. Figs. 2B, 3B, and S1). Based on the published initial observations, it has been proposed that the γ-SA region acts as a “hinged lid” that can adopt either a closed or open state, thereby occluding or exposing the active site (62), respectively, depending upon the occupancy of the various ligand binding sites in the γ-SA region. It has recently been suggested that the inactive conformation is characterized by an intramolecular association between the N-terminal half of the split PH domain and the C-terminal SH2 domain (63). In the context of this model, the role of Rac could be to, on its own or together with other regulatory inputs, mediate the release of intramolecular constraints. However, it seems clear that more experimental data, including a greater understanding of the three-dimensional structures of holo-PLCγ enzymes, are required to critically evaluate such models for autoinhibition and activation mechanisms.
Supplementary Material
Acknowledgments
We thank N. Zanker, S. Gierschik, and A. Smith for excellent technical assistance.
The atomic coordinates and structure factors (code 2k2j) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Chemical shifts for resonance assignments for the spPH have been deposited at the BioMagResBank (accession code 15707).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: PLC, phospholipase C; SH2, Src homology 2; SH3, Src homology 3; PH, pleckstrin homology; spPH, split PH domain; aa, amino acids; NOE, nuclear Overhauser effect; GTPγS, guanosine 5′-3-O-(thio)triphosphate; GDPβS, guanyl-5′-yl thiophosphate; SA, specific array.
References
- 1.Katan, M. (2005) Biochem. J. 391 e7-e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rebecchi, M. J., and Pentyala, S. N. (2000) Physiol. Rev. 80 1291-1335 [DOI] [PubMed] [Google Scholar]
- 3.Rhee, S. G., and Bae, Y. S. (1997) J. Biol. Chem. 272 15045-15048 [DOI] [PubMed] [Google Scholar]
- 4.Wen, W., Yan, J., and Zhang, M. (2006) J. Biol. Chem. 281 12060-12068 [DOI] [PubMed] [Google Scholar]
- 5.Matsuda, M., Paterson, H. F., Rodriguez, R., Fensome, A. C., Ellis, M. V., Swann, K., and Katan, M. (2001) J. Cell Biol. 153 599-612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sekiya, F., Poulin, B., Kim, Y. J., and Rhee, S. G. (2004) J. Biol. Chem. 279 32181-32190 [DOI] [PubMed] [Google Scholar]
- 7.Ji, Q. S., Winnier, G. E., Niswender, K. D., Horstman, D., Wisdom, R., Magnuson, M. A., and Carpenter, G. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 2999-3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, D., Feng, J., Wen, R., Marine, J. C., Sangster, M. Y., Parganas, E., Hoffmeyer, A., Jackson, C. W., Cleveland, J. L., Murray, P. J., and Ihle, J. N. (2000) Immunity 13 25-35 [DOI] [PubMed] [Google Scholar]
- 9.Regunathan, J., Chen, Y., Kutlesa, S., Dai, X., Bai, L., Wen, R., Wang, D., and Malarkannan, S. (2006) J. Immunol. 177 5365-5376 [DOI] [PubMed] [Google Scholar]
- 10.Upshaw, J. L., Schoon, R. A., Dick, C. J., Billadeau, D. D., and Leibson, P. J. (2005) J. Immunol. 175 213-218 [DOI] [PubMed] [Google Scholar]
- 11.Wilde, J. I., and Watson, S. P. (2001) Cell. Signal. 13 691-701 [DOI] [PubMed] [Google Scholar]
- 12.Piechulek, T., Rehlen, T., Walliser, C., Vatter, P., Moepps, B., and Gierschik, P. (2005) J. Biol. Chem. 280 38923-38931 [DOI] [PubMed] [Google Scholar]
- 13.Bunney, T. D., and Katan, M. (2006) Trends Cell Biol. 16 640-648 [DOI] [PubMed] [Google Scholar]
- 14.Harden, T. K., and Sondek, J. (2006) Annu. Rev. Pharmacol. Toxicol. 46 355-379 [DOI] [PubMed] [Google Scholar]
- 15.Illenberger, D., Schwald, F., and Gierschik, P. (1997) Eur. J. Biochem. 246 71-77 [DOI] [PubMed] [Google Scholar]
- 16.Illenberger, D., Schwald, F., Pimmer, D., Binder, W., Maier, G., Dietrich, A., and Gierschik, P. (1998) EMBO J. 17 6241-6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley, G. G., Reks, S. E., Ondrako, J. M., and Smrcka, A. V. (2001) EMBO J. 20 743-754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song, C., Hu, C. D., Masago, M., Kariyai, K., Yamawaki-Kataoka, Y., Shibatohge, M., Wu, D., Satoh, T., and Kataoka, T. (2001) J. Biol. Chem. 276 2752-2757 [DOI] [PubMed] [Google Scholar]
- 19.Wing, M. R., Houston, D., Kelley, G. G., Der, C. J., Siderovski, D. P., and Harden, T. K. (2001) J. Biol. Chem. 276 48257-48261 [DOI] [PubMed] [Google Scholar]
- 20.Wing, M. R., Snyder, J. T., Sondek, J., and Harden, T. K. (2003) J. Biol. Chem. 278 41253-41258 [DOI] [PubMed] [Google Scholar]
- 21.Sidhu, R. S., Clough, R. R., and Bhullar, R. P. (2005) J. Biol. Chem. 280 21933-21941 [DOI] [PubMed] [Google Scholar]
- 22.Bunney, T. D., Harris, R., Gandarillas, N. L., Josephs, M. B., Roe, S. M., Sorli, S. C., Paterson, H. F., Rodrigues-Lima, F., Esposito, D., Ponting, C. P., Gierschik, P., Pearl, L. H., Driscoll, P. C., and Katan, M. (2006) Mol. Cell 21 495-507 [DOI] [PubMed] [Google Scholar]
- 23.Jezyk, M. R., Snyder, J. T., Gershberg, S., Worthylake, D. K., Harden, T. K., and Sondek, J. (2006) Nat. Struct. Mol. Biol. 13 1135-1140 [DOI] [PubMed] [Google Scholar]
- 24.Illenberger, D., Walliser, C., Nurnberg, B., Diaz Lorente, M., and Gierschik, P. (2003) J. Biol. Chem. 278 3006-3014 [DOI] [PubMed] [Google Scholar]
- 25.Illenberger, D., Walliser, C., Strobel, J., Gutman, O., Niv, H., Gaidzik, V., Kloog, Y., Gierschik, P., and Henis, Y. I. (2003) J. Biol. Chem. 278 8645-8652 [DOI] [PubMed] [Google Scholar]
- 26.Snyder, J. T., Singer, A. U., Wing, M. R., Harden, T. K., and Sondek, J. (2003) J. Biol. Chem. 278 21099-21104 [DOI] [PubMed] [Google Scholar]
- 27.Lemmon, M. A., and Ferguson, K. M. (2000) Biochem. J. 350 1-18 [PMC free article] [PubMed] [Google Scholar]
- 28.Horton, R. M., Cai, Z. L., Ho, S. N., and Pease, L. R. (1990) BioTechniques 8 528-5352357375 [Google Scholar]
- 29.Geiser, M., Cebe, R., Drewello, D., and Schmitz, R. (2001) BioTechniques 31 88-92 [DOI] [PubMed] [Google Scholar]
- 30.Illenberger, D., Stephan, I., Gierschik, P., and Schwald, F. (2000) Methods Enzymol. 325 167-177 [DOI] [PubMed] [Google Scholar]
- 31.Frankel, P., Aronheim, A., Kavanagh, E., Balda, M. S., Matter, K., Bunney, T. D., and Marshall, C. J. (2005) EMBO J. 24 54-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorli, S. C., Bunney, T. D., Sugden, P. H., Paterson, H. F., and Katan, M. (2005) Oncogene 24 90-100 [DOI] [PubMed] [Google Scholar]
- 33.Plevin, M. J., Magalhaes, B. S., Harris, R., Sankar, A., Perkins, S. J., and Driscoll, P. C. (2004) J. Mol. Biol. 341 171-184 [DOI] [PubMed] [Google Scholar]
- 34.Camps, M., Hou, C., Jakobs, K. H., and Gierschik, P. (1990) Biochem. J. 271 743-748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Offermanns, S., and Simon, M. I. (1995) J. Biol. Chem. 270 15175-15180 [DOI] [PubMed] [Google Scholar]
- 36.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 37.Kraulis, P. (1989) J. Mag. Res. 84 627-633 [Google Scholar]
- 38.Wishart, D. S., Bigam, C. G., Yao, J., Abildgaard, F., Dyson, H. J., Oldfield, E., Markley, J. L., and Sykes, B. D. (1995) J. Biomol. NMR 6 135-140 [DOI] [PubMed] [Google Scholar]
- 39.McAlister, M. S. B., Mott, H. R., vanderMerwe, P. A., Campbell, I. D., Davis, S. J., and Driscoll, P. C. (1996) Biochemistry 35 5982-5991 [DOI] [PubMed] [Google Scholar]
- 40.Boucher, W. (1996) AZARA, version 2.7, Ph.D. thesis, University of Cambridge, Cambridge, UK
- 41.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 42.Linge, J. P., and Nilges, M. (1999) J. Biomol. NMR 13 51-59 [DOI] [PubMed] [Google Scholar]
- 43.Cornilescu, G., Delaglio, F., and Bax, A. (1999) J. Biomol. NMR 13 289-302 [DOI] [PubMed] [Google Scholar]
- 44.Dietrich, A., Meister, M., Spicher, K., Schultz, G., Camps, M., and Gierschik, P. (1992) FEBS Lett. 313 220-224 [DOI] [PubMed] [Google Scholar]
- 45.Dvorsky, R., and Ahmadian, M. R. (2004) EMBO Rep. 5 1130-1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan, J., Wen, W., Xu, W., Long, J. F., Adams, M. E., Froehner, S. C., and Zhang, M. (2005) EMBO J. 24 3985-3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teo, H., Gill, D. J., Sun, J., Perisic, O., Veprintsev, D. B., Vallis, Y., Emr, S. D., and Williams, R. L. (2006) Cell 125 99-111 [DOI] [PubMed] [Google Scholar]
- 48.Wohlgemuth, S., Kiel, C., Kramer, A., Serrano, L., Wittinghofer, F., and Herrmann, C. (2005) J. Mol. Biol. 348 741-758 [DOI] [PubMed] [Google Scholar]
- 49.Mott, H. R., Owen, D., Nietlispach, D., Lowe, P. N., Manser, E., Lim, L., and Laue, E. D. (1999) Nature 399 384-388 [DOI] [PubMed] [Google Scholar]
- 50.Thompson, G., Chalk, P. A., and Lowe, P. N. (1997) Biochem. Soc. Trans. 25 509S. [DOI] [PubMed] [Google Scholar]
- 51.Singh, S. M., and Murray, D. (2003) Protein Sci 12 1934-1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim, Y. J., Sekiya, F., Poulin, B., Bae, Y. S., and Rhee, S. G. (2004) Mol. Cell. Biol. 24 9986-9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez, R., Matsuda, M., Perisic, O., Bravo, J., Paul, A., Jones, N. P., Light, Y., Swann, K., Williams, R. L., and Katan, M. (2001) J. Biol. Chem. 276 47982-47992 [DOI] [PubMed] [Google Scholar]
- 54.Inabe, K., Miyawaki, T., Longnecker, R., Matsukura, H., Tsukada, S., and Kurosaki, T. (2002) FEBS Lett. 514 260-262 [DOI] [PubMed] [Google Scholar]
- 55.Ishiai, M., Kurosaki, M., Pappu, R., Okawa, K., Ronko, I., Fu, C., Shibata, M., Iwamatsu, A., Chan, A. C., and Kurosaki, T. (1999) Immunity 10 117-125 [DOI] [PubMed] [Google Scholar]
- 56.Kurosaki, T. (2002) Nat. Rev. Immunol. 2 354-363 [DOI] [PubMed] [Google Scholar]
- 57.Inabe, K., Ishiai, M., Scharenberg, A. M., Freshney, N., Downward, J., and Kurosaki, T. (2002) J. Exp. Med. 195 189-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walmsley, M. J., Ooi, S. K., Reynolds, L. F., Smith, S. H., Ruf, S., Mathiot, A., Vanes, L., Williams, D. A., Cancro, M. P., and Tybulewicz, V. L. (2003) Science 302 459-462 [DOI] [PubMed] [Google Scholar]
- 59.Carpenter, G., and Ji, Q. (1999) Exp. Cell Res. 253 15-24 [DOI] [PubMed] [Google Scholar]
- 60.Horstman, D. A., Chattopadhyay, A., and Carpenter, G. (1999) Arch. Biochem. Biophys. 361 149-155 [DOI] [PubMed] [Google Scholar]
- 61.Horstman, D. A., DeStefano, K., and Carpenter, G. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 7518-7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou, C., Horstman, D., Carpenter, G., and Roberts, M. F. (1999) J. Biol. Chem. 274 2786-2793 [DOI] [PubMed] [Google Scholar]
- 63.DeBell, K., Graham, L., Reischl, I., Serrano, C., Bonvini, E., and Rellahan, B. (2007) Mol. Cell. Biol. 27 854-863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.