Abstract
We explored the role of microRNAs (miRNAs) in acquiring resistance to tamoxifen, a drug successfully used to treat women with estrogen receptor-positive breast cancer. miRNA microarray analysis of MCF-7 cell lines that are either sensitive (parental) or resistant (4-hydroxytamoxifen-resistant (OHTR)) to tamoxifen showed significant (>1.8-fold) up-regulation of eight miRNAs and marked down-regulation (>50%) of seven miRNAs in OHTR cells compared with parental MCF-7 cells. Increased expression of three of the most promising up-regulated (miR-221, miR-222, and miR-181) and down-regulated (miR-21, miR-342, and miR-489) miRNAs was validated by real-time reverse transcription-PCR. The expression of miR-221 and miR-222 was also significantly (2-fold) elevated in HER2/neu-positive primary human breast cancer tissues that are known to be resistant to endocrine therapy compared with HER2/neu-negative tissue samples. Ectopic expression of miR-221/222 rendered the parental MCF-7 cells resistant to tamoxifen. The protein level of the cell cycle inhibitor p27Kip1, a known target of miR-221/222, was reduced by 50% in OHTR cells and by 28–50% in miR-221/222-overexpressing MCF-7 cells. Furthermore, overexpression of p27Kip1 in the resistant OHTR cells caused enhanced cell death when exposed to tamoxifen. This is the first study demonstrating a relationship between miR-221/222 expression and HER2/neu overexpression in primary breast tumors that are generally resistant to tamoxifen therapy. This finding also provides the rationale for the application of altered expression of specific miRNAs as a predictive tamoxifen-resistant breast cancer marker.
Breast cancer is the most common malignancy in women, accounting for 31% of all female cancers. An estimated 178,480 new cases of invasive breast cancer was diagnosed in the United States in 2007, and 40,460 women will die of this cancer. Over two-thirds of breast cancers exhibit high concentrations of estrogen receptor, which contribute to tumor growth and progression. Blocking the steroid hormone pathway with tamoxifen and/or oophorectomy has been shown to be effective in this patient population. The Early Breast Cancer Trialists' Collaborative Group overview demonstrated a significant improvement in 15-year survival with the addition of adjuvant tamoxifen for 5 years following surgery (1). Furthermore, tamoxifen can also reduce the incidence of contralateral breast cancer and has been approved as a prophylactic agent to prevent breast cancer. Despite this accomplishment in the management of women with potentially endocrine-responsive breast cancers, a significant proportion of these women will experience disease progression due to either an intrinsic or acquired resistance to tamoxifen.
Nongenomic activation of epidermal growth factor receptor/HER2 signaling by tamoxifen is an important factor contributing to tamoxifen resistance. This leads to activation of both the p42/44 mitogen-activated protein kinase (MAPK) and Akt signaling pathways, which favor cell proliferation and survival. These changes could be blocked by the selective epidermal growth factor receptor tyrosine kinase inhibitor gefitnib, suggesting that epidermal growth factor receptor/HER2 signaling is directly involved in tamoxifen resistance (2). The preclinical data are corroborated by clinical observations that tumors expressing HER2 exhibit poor outcome when treated with tamoxifen (3). None of the molecular mechanisms proposed for tamoxifen resistance (for review, see Ref. 4) have led to the development of a gene expression profile that can consistently identify resistant tumors and benefit these patients from upfront use of alternative drugs such as aromatase inhibitors.
Recent studies have highlighted the key regulatory roles of microRNAs (miRNAs)3 in all fundamental cellular processes in animals and plants. Altered expression of miRNAs in primary human cancers has been used for tumor diagnosis, classification, staging, and prognosis (5). These small noncoding RNAs regulate expression of their target proteins primarily by inhibiting translation of the target mRNA and in some cases by inducing rapid decay of the message (6). A study with 76 neoplastic and 34 normal breast tissue samples revealed altered expression of several miRNAs that could correctly predict the nature of the tumor analyzed (7). This study did not, however, attempt to profile miRNA expression in drug resistance.
Here, we used a cell culture model to determine the miRNA expression profile of a tamoxifen-resistant cell line that was subsequently validated in primary human breast cancers. We further identified a target protein of the highly overexpressed miRNAs in tamoxifen-resistant breast cancer and established its regulatory role in conferring tamoxifen resistance.
EXPERIMENTAL PROCEDURES
Cell Culture and Tissue Procurement—Tamoxifen-sensitive MCF-7 cells and 4-hydroxytamoxifen-resistant (OHTR) cells were obtained from Dr. Kenneth P. Nephew (Indiana University) and maintained as described (8). Before all experiments, MCF-7 and OHTR cells were cultured in growth medium (minimum essential medium with 2 mmol/liter l-glutamine, 0.1 mmol/liter nonessential amino acids, 50 units/ml penicillin, 50 mg/ml streptomycin, 6 ng/ml insulin, and 10% fetal bovine serum) in the absence of 4-hydroxytamoxifen for 7 days. ZR75.1 cells were obtained from Dr. Brett Hall (Ohio State University) and maintained in RPMI 1640 medium containing 10% fetal bovine serum. Primary human breast samples were obtained from the Stephanie Spielman Tissue Bank collected under Protocol 2003C0036.
miRNA Microarray—The miRNA microarray was performed at the Ohio State University Comprehensive Cancer Center Microarray Core Facility (see supplemental “Experimental Procedures” for details).
TaqMan Reverse Transcription (RT)-PCR for miRNA Quantification—Total RNA was isolated from formaldehyde-fixed paraffin-embedded unsectioned primary human breast cancer tissue cores with a RecoverAll™ total nucleic acid isolation kit (Ambion) and from the cell lines with TRIzol™ (Invitrogen), reverse-transcribed using a TaqMan™ microRNA reverse transcription kit, and subjected to real-time PCR using a TaqMan™ microRNA assay kit (Applied Biosystems). Reactions were performed using a Stratagene Mx3000 instrument in triplicate. miRNA expression was normalized to small nuclear RNA (snRNA) RNU6B as described (9).
Cell Proliferation Assay—Cells (3000/well) were seeded in 96-well plates and treated with 5 μm tamoxifen in complete medium after overnight serum starvation. Cell proliferation was documented using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay kit (Roche Applied Science).
Apoptosis Assay—Tamoxifen-induced apoptosis was monitored by Western blotting (10) and cell cycle analysis (see supplemental “Experimental Procedures” for details).
Statistical Analysis—Statistical significance was analyzed by unpaired Student's t test, and p ≤ 0.05 was considered to be statistically significant.
RESULTS
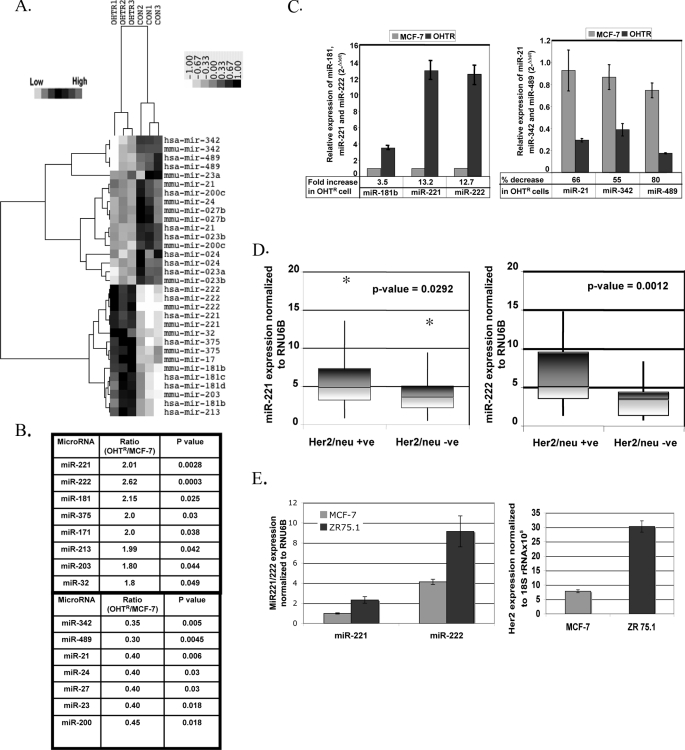
Distinct miRNA Expression Profile Is Associated with Tamoxifen Resistance in Breast Cancer Cell Lines—As a first step to identify differentially expressed miRNAs in tamoxifen-resistant breast cancer, we compared the miRNA profiles of the OHTR and tamoxifen-sensitive MCF-7 breast cancer cell lines by microarray analysis. Both cell lines originated from a single estrogen-responsive MCF-7 clone (8), and any alteration in the miRNA expression profile is therefore expected to result from prolonged tamoxifen treatment. To analyze the microarray data, the expression of each miRNA was normalized to the average median of all the genes and compared between the two cell lines. Using Student's t test, we obtained a list of differentially expressed miRNAs (p ≤ 0.05) in the OHTR and MCF-7 cells. Unsupervised clustering of significantly deregulated miRNAs is presented in Fig. 1A. Eight miRNA genes, miR-221, miR-222, miR-181, miR-375, miR-32, miR-171, miR-213, and miR-203, were significantly overexpressed (>1.5-fold) in OHTR cells compared with MCF-7 cells. In addition, seven miRNAs, miR-342, miR-489, miR-21, miR-24, miR-27, miR-23, and miR-200, were underexpressed (<50%) in OHTR cells (Fig. 1B).
FIGURE 1.
miRNA expression profile of tamoxifen-sensitive and tamoxifen-resistant MCF-7 cells. A, shown are the results from cluster analysis. Total RNAs isolated from three biological replicates of tamoxifen-sensitive MCF-7 cells and tamoxifen-resistant MCF-7 cells (OHTR) were subjected to miRNA microarray analysis. miRNA expression data were normalized to the average median of all the genes present in the array. miRNAs expressed at least 1.5-fold higher or 50% lower in OHTR cells compared with MCF-7 cells were considered for cluster analysis. B, shown are the miRNAs that are up-regulated and down-regulated in OHTR cells. C, validation is shown. Total RNAs isolated from three biological replicates of MCF-7 and OHTR cells were subjected to real-time RT-PCR to validate differential expression of miR-221, miR-222, miR-181b, miR-21, miR-342, and miR-489. Each assay was done in triplicate, and expression of miRNA was normalized to snRNA RNU6B. D, the expression of miR-221 and miR-222 was analyzed in HER2/neu-positive and HER2/neu-negative primary human breast cancer tissues. Total RNAs isolated from formalin-fixed paraffin-embedded tissue sections were subjected to real-time RT-PCR with specific miRNA assay kits. Expression of snRNA RNU6B was used as normalizer. Normalized expression of the miRNAs was compared between the HER2/neu-positive (HER2/Neu +ve) and HER2/neu-negative (HER2/Neu -ve) samples using a Whisker plot. The asterisk indicate the outliers. E, total RNAs isolated from MCF-7 and ZR75.1 cells were subjected to real-time RT-PCR to analyze the expression levels of miR-221, miR-222, and HER2. miRNA levels were normalized to snRNA RNU6B, and HER2 levels were normalized to 18 S rRNA.
On the basis of maximal alteration in the expression levels in OHTR cells, we next validated differential expression of three up-regulated miRNAs (miR-221, miR-222, and miR-181) and three down-regulated miRNAs (miR-489, miR-342, and miR-21) by real-time RT-PCR. Normalization of the miRNA expression levels to snRNA RNU6B revealed 3.5-, 13.2-, and 12.7-fold increases in the miR-181b, miR-221 and miR-222 levels, respectively, in OHTR cells compared with MCF-7 cells (Fig. 1C). In contrast, the miR-21, miR-342, and miR-489 expression levels in OHTR cells were reduced by 66, 55, and 80% respectively, compared with MCF-7 cells (Fig. 1C).
miR-221 and miR-222 Are Up-regulated in HER2/neu-positive Primary Human Breast Cancer—The next obvious question was whether similar deregulation in miRNA expression observed in a cell culture model also occurs in primary breast cancer samples. To address this issue, we studied a panel of primary human breast cancer samples that were either HER2/neu-positive or HER2/neu-negative. Because increased expression and signaling via HER2/neu in breast cancer patients are associated with poor outcome of endocrine therapy (11), we used HER2/neu expression as a marker for tamoxifen resistance. Real-time RT-PCR and normalization to RNU6B expression revealed significant increases in miR-221 (p = 0.029) (Fig. 1D, left panel) and miR-222 (p = 0.0012) (right panel) expression in the HER2/neu-positive tumor samples compared with the HER2/neu-negative samples (n = 24). The data obtained from the patient samples corroborated the data obtained from the cell culture model of tamoxifen resistance.
We also tested miR-221 and miR-222 expression levels in similar breast cancer cell lines with a differential expression of HER2 to support these patient data. A 2.3-fold increase in both miR-221 and miR-222 expression was seen in ZR75.1 cells (high HER2 expression) compared with MCF-7 cells (low HER2 expression) (Fig. 1E).
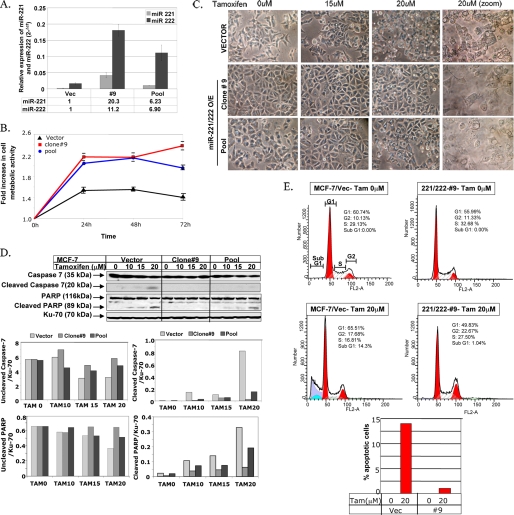
Overexpression of miR-221 and miR-222 in MCF-7 Cells Confers Resistance to Tamoxifen—Previous studies have reported increased expression of miR-221 and miR-222 in primary glioblastoma (12), papillary thyroid carcinoma (13), and pancreatic cancer (14). Because the levels of these two miRNAs were also significantly elevated in HER2/neu-positive breast tumors, we further explored their role in acquiring resistance to endocrine therapy. miR-221 and miR-222 were ectopically expressed in tamoxifen-sensitive MCF-7 cells (MCF-7/221/222), and miR-221/222-overexpressing cells were selected using G418. We used the G418-selected pool and a clone (clone 9) derived from these cells for further studies. Increased expression of miR-221/222 in the G418-selected pool and clone 9 was confirmed by real-time RT-PCR (Fig. 2A). MCF-7 cells transfected with the empty vector (MCF-7/Vec) and miR-221/222-overexpressing cells were treated with 5 μm tamoxifen, and cell viability was monitored using the MTT assay. The viability of both the miR-221/222-overexpressing clone and pool in the presence of tamoxifen was significantly higher than that of the MCF-7/Vec pool (p ≤ 0.005), demonstrating that miR-221/222 can confer resistance to tamoxifen in the breast cancer cell line (Fig. 2B).
FIGURE 2.
Ectopic expression of miR-221/222 in MCF-7 cells results in increased tamoxifen resistance. A, expression of miR-221 and miR-222 was analyzed using total RNA isolated from miR-221/222-transfected MCF-7 cells by real-time RT-PCR and normalized to snRNA RNU6B. -Fold increase is shown below the bars. B, the G418-selected pool of miR-221/222-expressing MCF-7 cells, G418-selected clone 9 (MCF-7/221/222), and the empty vector-transfected MCF-7 cell pool were treated with 5 μm tamoxifen for 72 h. Cell metabolic activity in the presence of the drug was measured every 24 h using the MTT assay. The metabolic activity of cells at 0 h was taken as 1. The results are the means ± S.D. of triplicate assays. C, vector- and miR-221/222-expressing MCF-7 cells were treated with 0, 15, and 20 μm tamoxifen for 16 h. The cells were photographed using a phase-contrast microscope. O/E, overexpressing. D, whole cell extracts from the vector- and miR-221/222-transfected MCF-7 cells were separated by SDS-PAGE and probed with antibodies against PARP and caspase-7 that detect respective intact and cleaved products. The blot was reprobed with anti-Ku-70 antibody to ensure equal protein loading. The signal in each band was quantified using Kodak Imaging software. Quantification of both caspase-7 and PARP (uncleaved and cleaved) was plotted after normalization to Ku-70. The results are representative of two independent experiments. E, the vector- and miR-221/222-expressing MCF-7 cells were treated with 20 μm tamoxifen (Tam) for 24 h. The cells were fixed and stained with propidium iodide, and cell cycle distribution was monitored by flow cytometry in a FACSCalibur. The percentage of apoptotic cells (sub-G1 peak) in vector (Vec)- and miR-221/222-expressing cells is represented in the bar diagram. Similar results were obtained from three independent experiments.
Because tamoxifen is known to induce apoptosis in breast cancer cells (15), we explored the potential role of miR-221/222 in inhibiting tamoxifen-mediated apoptosis. For this purpose, we treated miR-221/222-overexpressing MCF-7 cells with increasing concentrations of the drug and monitored its effect on cell morphology under a phase-contrast microscope. The MCF-7/Vec cells exhibited signs of cellular damage in the presence of 15 μm tamoxifen that were more pronounced upon treatment with 20 μm tamoxifen (Fig. 2C). In contrast, both the miR-221/222-expressing clone and pool showed minimal signs of cellular damage when treated with 15 μm tamoxifen (Fig. 2C). At 20 μm, the drug-induced cellular damage was significantly less in these cells compared with MCF-7/Vec cells (Fig. 2C). These observations can be considered biologically relevant, as the median in vivo concentration of tamoxifen attained in breast cancer patients treated with a standard 20 mg/day dose of tamoxifen was found to be 83.6 μm (16). To prove further the differential response of the vector- and miR-221/222-transfected cells to tamoxifen-induced apoptosis, we monitored caspase-7 and poly(ADP-ribose) polymerase (PARP) cleavage by Western blot analysis. Although both caspase-7 and PARP cleavage increased in MCF-7/Vec cells with increasing concentrations of tamoxifen (0–20 μm), the cleaved products were barely detectable in tamoxifen-treated MCF-7/221/222 clone 9 (Fig. 2D). The extent of PARP degradation in the miR-221/222-overexpressing pool was also reduced by 40% relative to the MCF-7/Vec cells. It is noteworthy that the level of miR-221/222 expression in the MCF-7/221/222 pool was considerably diminished compared with that in clone 9, and a dose-dependent effect of miR-221/222 on tamoxifen-induced PARP cleavage in MCF-7 cells was observed upon ectopic expression of miR-221/222 (Fig. 2D).
We then quantified the extent of tamoxifen-induced apoptosis in these cell lines by flow cytometry (Fig. 2E). Although 14.3% of the MCF-7/Vec cells were apoptotic (sub-G1 peak) upon tamoxifen treatment, only 1% of clone 9 ((Fig. 2E) and 2.9% of the miR-221/222-overexpressing pool (data not shown) were apoptotic under the same condition. This series of studies demonstrated that the miR-221/222 cluster indeed plays a key role in conferring tamoxifen resistance to MCF-7 cells.
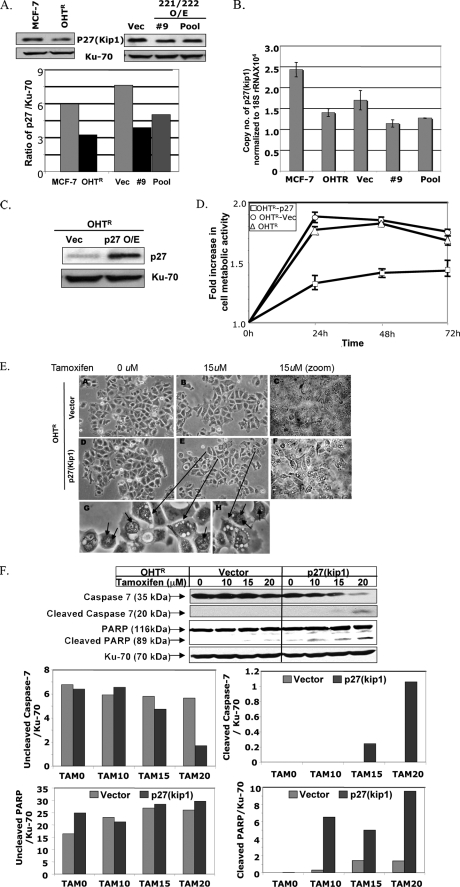
p27Kip1 Protein, a Target of miR-221/222, Is the Effector Molecule That Regulates Tamoxifen Sensitivity—The cell cycle inhibitor p27Kip1 is a known target of miR-221/222 (17). Recent studies have shown that sequestration of p27Kip1 in the cytoplasm by Akt-mediated phosphorylation correlates with poor prognosis of breast cancer (reviewed in Ref. 18). Based on this information, we hypothesized that the effect of miR-221/222 in augmenting tamoxifen resistance is mediated by the loss of p27Kip1 protein in breast cancer. Indeed, Western blot analysis revealed 50% reduction in the level of p27Kip1 in MCF-7 cells compared with OHTR cells (Fig. 3A). Furthermore, the level of p27Kip1 was reduced by 50 and 28% in miR-221/222-expressing clone 9 and the G418-selected pool, respectively, compared with the vector-transfected cells (Fig. 3A). Real-time RT-PCR analysis showed that the mRNA level of p27Kip1 was also reduced in these cell lines (Fig. 3B), indicating that miR-221/222 regulates p27Kip1 at the level of RNA stability and its translation.
FIGURE 3.
p27Kip1, a target of miR-221/222, imparts tamoxifen sensitivity to MCF-7 cells. A, whole cell extracts from MCF-7 cells, OHTR cells, and miR-221/222-transfected and vector (Vec)-transfected MCF-7 cells were subjected to SDS-PAGE and probed with anti-p27Kip1 antibody. The membrane was reprobed with anti-Ku-70 antibody, and p27Kip1 expression was normalized to Ku-70 protein. O/E, overexpressing. B, total RNAs from MCF-7 cells, OHTR cells, and miR-221/222-transfected and vector-transfected MCF-7 cells were analyzed by real-time RT-PCR with primers specific for p27Kip1 and 18 S rRNA. C, whole cell extract from the p27Kip1-transfected OHTR cell pool was subjected to Western blot analysis with anti-p27Kip1 antibody and reprobed with anti-Ku-70 antibody. D, OHTR cells transfected with empty vector and p27 expression vector were treated with 5 μm tamoxifen for 72 h. Cell metabolic activity was measured every 24 h using the MTT assay. The metabolic activity of cells at 0 h was taken as 1. The results are the means ± S.D. of triplicate assays. E, vector-overexpressing (panels A–C) and p27Kip1-overexpressing (D–F) OHTR cells were treated with 0 and 15 μm tamoxifen for 16 h. The cells were photographed using a phase-contrast microscope. The small arrows in panels G and H indicate autophagosome-like bodies. F, whole cell extracts from the vector-transfected and p27Kip1-transfected OHTR cells were subjected to SDS-PAGE and probed with antibodies against caspase-7 and PARP. The blot was reprobed with anti-Ku-70 antibody to normalize protein loading. The signal in each band was quantified using Kodak Imaging software. Quantification of uncleaved and cleaved caspase-7 and PARP is presented in the bar diagrams. The results are representative of three independent experiments. TAM, tamoxifen.
We next determined whether the increased expression of p27Kip1 in OHTR cells can sensitize the OHTR cells to tamoxifen. To test this possibility, we transfected a p27Kip1 expression vector into OHTR cells and selected a G418-resistant pool for further studies. The ectopic expression of p27Kip1 was monitored by Western blot analysis with anti-p27 antibody (Fig. 3C). The viability of untransfected OHTR cells and the vector- and p27Kip1-transfected cell pools in the presence of tamoxifen was analyzed by the MTT assay. A significant loss of cell viability was observed when p27Kip1 was ectopically expressed in OHTR cells (p ≤ 0.002) (Fig. 3D).
To investigate further the effect of p27Kip1 in tamoxifen-induced cell death, we examined potential morphological changes in the vector-transfected and p27Kip1-overexpressing OHTR cells in the presence of tamoxifen. The cellular damage of tamoxifen-treated p27Kip1-expressing cells was obvious as observed under a microscope, whereas such damage was minimal in vector-transfected cells (Fig. 3E). To authenticate this observation, we monitored caspase-7 and PARP cleavage in these cell lines. Cleaved caspase-7 was detectable only in the tamoxifen-treated p27Kip1-transfected cells but not in the vector control (Fig. 3F). Under the same treatment conditions, PARP cleavage was significantly more pronounced in the p27Kip1-expressing cells than in the vector-transfected cells at all concentrations of tamoxifen tested (Fig. 3F). These data demonstrate that the maintenance of high p27Kip1 levels may play a key role in the successful endocrine therapy of breast cancer.
DISCUSSION
Tamoxifen is the current standard adjuvant therapy in women with estrogen receptor-positive breast cancer. Because ∼30% of these tumors are resistant to tamoxifen, there is considerable interest in elucidating the molecular mechanisms of acquiring resistance to this important anticancer drug. Although altered expression of miRNAs in primary human cancers has been used for tumor diagnosis and prognosis, the potential involvement of miRNAs in induction of drug resistance, particularly tamoxifen resistance, has not been explored. Recent studies have demonstrated the role of miR-214 in conferring cisplatin resistance in ovarian cancer by targeting PTEN (19). Similar studies with gemcitabine-resistant cholangiocarcinoma showed that inhibition of miR-21 and miR-200b sensitizes cholangiocytes to gemcitabine (20). The present study showed significantly increased expression of eight miRNAs and down-regulation of seven miRNAs in a tamoxifen-resistant breast cancer cell line compared with a tamoxifen-sensitive cell line. A finding of considerable clinical significance was the increased expression of miR-221 and miR-222 in HER2/neu-positive compared with HER2/neu-negative primary human breast tumors that are resistant to endocrine therapy. Earlier studies identified increased expression of miR-221 and miR-222 as a signature of primary glioblastoma (12), papillary thyroid carcinoma (13), and pancreatic cancer (14). Here, we report for the first time increased expression of miR-221/222 as a potential signature of tamoxifen resistance in breast cancer.
miR-221/222 functions as an oncogene by targeting the cell cycle inhibitor p27Kip1, thereby controlling cell proliferation (17, 21). In addition, increased expression of these two miRNAs facilitates transit from quiescence to proliferation by targeting p27 proteins (22). The present study has revealed a relationship between increased tamoxifen resistance and reduced levels of p27Kip1 by augmenting miR-221/222 expression, further emphasizing the importance of this cell cycle inhibitor in multifarious biological processes. Interestingly, the elevated expression of this cell cycle inhibitor has been associated with diminished apoptosis in small cell lung cancer (23). A small but reproducible increase in caspase-7 and PARP cleavage in tamoxifen-treated p27Kip1-overexpressing OHTR cells demonstrates that apoptosis is involved in the drug-induced death of these cells. The p27-mediated cell death could also occur by autophagy (24). Indeed, the presence of autophagosome-like bodies in the tamoxifen-treated cells (Fig. 3E, panels E, G, and H) suggests that the pathways leading to both apoptosis and autophagy may be involved in this process. Although the tumor suppressor function of p27Kip1 is regulated largely by post-translational mechanisms rather than by mutation or loss of heterozygosity, segregating p27Kip1 from the nuclear compartment of carcinomas, including those of breast, thyroid, esophagus, and colon, may lead to an additional growth advantage (reviewed in Ref. 18). It would be of interest to study potential alterations in the localization of the low level of p27Kip1 in tamoxifen-resistant cells, further facilitating their growth in the presence of this drug.
We have identified other miRNAs that are differentially expressed in tamoxifen-resistant cell lines. Some of these miRNAs could emerge as potential biomarkers of tamoxifen-resistant tumors. Identification of target genes of these deregulated miRNAs will further enhance our knowledge of tamoxifen resistance and facilitate design of new therapeutic agents targeting these proteins.
Supplementary Material
Acknowledgments
We sincerely thank Dr. Kenneth P. Nephew for the OHTR and MCF-7 cells and Drs. Brett Hall, Carlo M. Croce, and Robert T. Pilarski (Ohio State University) for the ZR75.1 cells, miR-221/222 plasmid, and breast cancer tissues, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grants CA101956, CA122523, and CA086978. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and additional references.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: miRNA, microRNA; OHTR, 4-hydroxytamoxifen-resistant; RT, reverse transcription; snRNA, small nuclear RNA; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; PARP, poly(ADP-ribose) polymerase.
References
- 1.Early Breast Cancer Trialists' Collaborative Group (2005) Lancet 365 1687-1717 [DOI] [PubMed] [Google Scholar]
- 2.Shou, J., Massarweh, S., Osborne, C. K., Wakeling, A. E., Ali, S., Weiss, H., and Schiff, R. (2004) J. Natl. Cancer Inst. 96 926-935 [DOI] [PubMed] [Google Scholar]
- 3.Carlomagno, C., Perrone, F., Gallo, C., De Laurentiis, M., Lauria, R., Morabito, A., Pettinato, G., Panico, L., D'Antonio, A., Bianco, A. R., and De Placido, S. (1996) J. Clin. Oncol. 14 2702-2708 [DOI] [PubMed] [Google Scholar]
- 4.Lewis, J. S., and Jordan, V. C. (2005) Mutat. Res. 591 247-263 [DOI] [PubMed] [Google Scholar]
- 5.Croce, C. M. (2008) N. Engl. J. Med. 358 502-511 [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008) Nat. Rev. Genet. 9 102-114 [DOI] [PubMed] [Google Scholar]
- 7.Iorio, M. V., Ferracin, M., Liu, C. G., Veronese, A., Spizzo, R., Sabbioni, S., Magri, E., Pedriali, M., Fabbri, M., Campiglio, M., Menard, S., Palazzo, J. P., Rosenberg, A., Musiani, P., Volinia, S., Nenci, I., Calin, G. A., Querzoli, P., Negrini, M., and Croce, C. M. (2005) Cancer Res. 65 7065-7070 [DOI] [PubMed] [Google Scholar]
- 8.Fan, M., Yan, P. S., Hartman-Frey, C., Chen, L., Paik, H., Oyer, S. L., Salisbury, J. D., Cheng, A. S., Li, L., Abbosh, P. H., Huang, T. H., and Nephew, K. P. (2006) Cancer Res. 66 11954-11966 [DOI] [PubMed] [Google Scholar]
- 9.Livak, K. J., and Schmittgen, T. D. (2001) Methods (Amst.) 25 402-408 [DOI] [PubMed] [Google Scholar]
- 10.Datta, J., Majumder, S., Kutay, H., Motiwala, T., Frankel, W., Costa, R., Cha, H. C., MacDougald, O. A., Jacob, S. T., and Ghoshal, K. (2007) Cancer Res. 67 2736-2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu, L., Chow, L. W., Loo, W. T., Guan, X. Y., and Toi, M. (2004) Clin. Cancer Res. 10 4639-4644 [DOI] [PubMed] [Google Scholar]
- 12.Ciafre, S. A., Galardi, S., Mangiola, A., Ferracin, M., Liu, C. G., Sabatino, G., Negrini, M., Maira, G., Croce, C. M., and Farace, M. G. (2005) Biochem. Biophys. Res. Commun. 334 1351-1358 [DOI] [PubMed] [Google Scholar]
- 13.Pallante, P., Visone, R., Ferracin, M., Ferraro, A., Berlingieri, M. T., Troncone, G., Chiappetta, G., Liu, C. G., Santoro, M., Negrini, M., Croce, C. M., and Fusco, A. (2006) Endocr.-Relat. Cancer 13 497-508 [DOI] [PubMed] [Google Scholar]
- 14.Lee, E. J., Gusev, Y., Jiang, J., Nuovo, G. J., Lerner, M. R., Frankel, W. L., Morgan, D. L., Postier, R. G., Brackett, D. J., and Schmittgen, T. D. (2007) Int. J. Cancer 120 1046-1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obrero, M., Yu, D. V., and Shapiro, D. J. (2002) J. Biol. Chem. 277 45695-45703 [DOI] [PubMed] [Google Scholar]
- 16.Kisanga, E. R., Gjerde, J., Guerrieri-Gonzaga, A., Pigatto, F., Pesci-Feltri, A., Robertson, C., Serrano, D., Pelosi, G., Decensi, A., and Lien, E. A. (2004) Clin. Cancer Res. 10 2336-2343 [DOI] [PubMed] [Google Scholar]
- 17.le Sage, C., Nagel, R., Egan, D. A., Schrier, M., Mesman, E., Mangiola, A., Anile, C., Maira, G., Mercatelli, N., Ciafre, S. A., Farace, M. G., and Agami, R. (2007) EMBO J. 26 3699-3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blain, S. W., and Massagué, J. (2002) Nat. Med. 8 1076-1078 [DOI] [PubMed] [Google Scholar]
- 19.Yang, H., Kong, W., He, L., Zhao, J. J., O'Donnell, J. D., Wang, J., Wenham, R. M., Coppola, D., Kruk, P. A., Nicosia, S. V., and Cheng, J. Q. (2008) Cancer Res. 68 425-433 [DOI] [PubMed] [Google Scholar]
- 20.Meng, F., Henson, R., Lang, M., Wehbe, H., Maheshwari, S., Mendell, J. T., Jiang, J., Schmittgen, T. D., and Patel, T. (2006) Gastroenterology 130 2113-2129 [DOI] [PubMed] [Google Scholar]
- 21.Visone, R., Russo, L., Pallante, P., De Martino, I., Ferraro, A., Leone, V., Borbone, E., Petrocca, F., Alder, H., Croce, C. M., and Fusco, A. (2007) Endocr.-Relat. Cancer 14 791-798 [DOI] [PubMed] [Google Scholar]
- 22.Medina, R., Zaidi, S. K., Liu, C. G., Stein, J. L., van Wijnen, A. J., Croce, C. M., and Stein, G. S. (2008) Cancer Res. 68 2773-2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuda, A., Osada, H., Yatabe, Y., Kozaki, K., Tatematsu, Y., Takahashi, T., and Hida, T. (2001) Am. J. Pathol. 158 87-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, J., Shao, S. H., Xu, Z. X., Hennessy, B., Ding, Z., Larrea, M., Kondo, S., Dumont, D. J., Gutterman, J. U., Walker, C. L., Slingerland, J. M., and Mills, G. B. (2007) Nat. Cell Biol. 9 218-224 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.