Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel is a membrane-integral protein that belongs to an ATP-binding cassette superfamily. Mutations in the CFTR gene cause cystic fibrosis in which salt, water, and protein transports are defective in various tissues. Here we expressed wild-type human CFTR as a FLAG-fused protein in HEK293 cells heterologously and purified it in three steps: anti-FLAG and wheat germ agglutinin affinity chromatographies and size exclusion chromatography. The stoichiometry of the protein was analyzed using various biochemical approaches, including chemical cross-linking, blue-native PAGE, size exclusion chromatography, and electron microscopy (EM) observation of antibody-decorated CFTR. All these data support a dimeric assembly of CFTR. Using 5,039 automatically selected particles from negatively stained EM images, the three-dimensional structure of CFTR was reconstructed at 2-nm resolution assuming a 2-fold symmetry. CFTR, presumably in a closed state, was shown to be an ellipsoidal particle with dimensions of 120 × 106 × 162Å. It comprises a small dome-shaped extracellular and membrane-spanning domain and a large cytoplasmic domain with orifices beneath the putative transmembrane domain. EM observation of CFTR·anti-regulatory domain antibody complex confirmed that two regulatory domains are located around the bottom end of the larger oval cytoplasmic domain.
The cystic fibrosis transmembrane conductance regulator (CFTR)3 (also termed ABCC7) is a unique member of the ATP-binding cassette (ABC) superfamily in that CFTR functions as an anion channel, whereas most other members function as active transporters. CFTR is expressed in the luminal membranes of secreting and absorbing epithelia and plays a critical role in transepithelial salt and water transport. Dysfunction of CFTR leads to cystic fibrosis, the most common lethal autosomal recessive disorder in Caucasians (1-3). On the other hand, extremely high activity of CFTR, usually caused by bacterial toxins, results in secretory diarrhea (4, 5), killing millions of infants in developing countries every year (6). Understanding the structure/function relationship and the underlying mechanisms of CFTR are essential for developing novel therapeutics for CFTR-mediated diseases.
Like other ABC transporters, CFTR is formed by two repeated motifs, each of which has a membrane-spanning domain (MSD) and a cytoplasmic nucleotide-binding domain (NBD). However, a regulatory domain (R domain) located between the first NBD (NBD1) and the second MSD (MSD2) is unique in CFTR among ABC transporters. This domain contains several phosphorylation sites for protein kinase A and protein kinase C, and the level of their phosphorylation controls CFTR channel activity. Once they are phosphorylated, opening and closing (gating) of the CFTR channel is controlled by ATP binding and hydrolysis at its two NBDs (7). Each NBD contains the Walker A and Walker B nucleotide-binding motifs and the “signature sequence” LSGGQ, which defines the ABC superfamily. It is generally accepted that NBD1 and -2 are dimerized in a head to tail configuration with two ATP molecules sandwiched between the Walker A/B motifs of one NBD and the signature sequences of the counterpart NBD (8). There are lines of convincing evidence that the opening of CFTR chloride channel is associated with this NBD dimerization (9). The detailed dynamics of ATP binding and NBD dimerization in CFTR gating has been intensively investigated (10-12).
Despite a plethora of biochemical and electrophysiological results, structural information of CFTR is limited because of the lack of abundant CFTR protein sources and the difficulties in protein purification and crystallization. So far, only the structure of the NBD1 domain of CFTR has been determined by x-ray crystallography using mouse (13) and human (14) proteins. Ford and co-workers (15, 16) described low resolution structures of wild-type human CFTR by two-dimensional electron crystallography (15) and by the single particle reconstruction technique (16). The two-dimensional crystallography showed two different monomeric structures for CFTR (15). On the other hand, the single particle analysis suggested a structure whose particle size is compatible with a dimeric association by two CFTR molecules (16).
In this study, we purified glycosylated mature CFTR and reconstructed its three-dimensional structure from negatively stained EM images. Our data suggest that two CFTR proteins form an ellipsoidal “tail to tail” dimeric configuration with side orifices in the cytoplasm and that at least part of the R domain is located around the bottom end of the larger cytoplasmic domain.
EXPERIMENTAL PROCEDURES
Expression Constructs, Transfection of HEK293 Cells, and Membrane Preparation—Full-length human CFTR cDNA was subcloned into pcDNA3.1 Zeo (Invitrogen) and tagged with FLAG sequence at either the N (abbreviated N-FLAG CFTR) or C terminus (C-FLAG CFTR). The HEK293 cells were transfected with 4 μg of the DNA construct/1 × 106 cells for 6 h using the calcium phosphate precipitation method (17) to express FLAG-fused CFTR protein. After 48 h, cells were washed twice in TBS (20 mm Tris-HCl, pH 7.4 at 4 °C, and 140 mm NaCl) and harvested using Teflon cell scrapers. After the cells were collected by centrifugation, they were first homogenized with a Teflon homogenizer in 10 volumes (v/w) of TBS and then centrifuged at 1,500 × g for 10 min to remove debris. The supernatant was centrifuged at 100,000 × g for 60 min to obtain the membrane fraction.
Immunofluorescence Experiments—HEK293 cells transfected with CFTR constructs were cultured for 48 h and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS; 136 mm NaCl, 1.4 mm KCl, 10 mm Na2HPO4, and 1.7 mm KH2PO4, pH 7.4) at room temperature for 10 min. For membrane permeabilization, cells were incubated with PBS containing 0.1% saponin and 2 mg/ml bovine serum albumin at room temperature for 30 min. Cells were incubated with Cy3-conjugated anti-FLAG antibodies (Sigma) in PBS containing 2 mg/ml bovine serum albumin and 0.1% saponin at 4 °C for 6 h. They were washed with PBS three times and incubated with PBS containing 1 μg/ml of 4′, 6-diamidino-2-phenylindole at room temperature for 30 min. Samples were washed with PBS and mounted on glass slides with 50% glycerol. Control cells without membrane permeabilization were treated similarly except that saponin treatment was not performed. Samples were observed using an Olympus IX 71 microscope with a DP50 CCD camera (Olympus).
Electrophysiological Measurement—Channel functions of C-FLAG CFTR expressed in Chinese Hamster ovary or HEK293 cells were compared with those reported for wild-type CFTR (10-12, 18, 19) by inside-out patch clamp recording using an EPC10 Patch Clamp Amplifier (HEKA Electronik). The pipette solution contained 140 mm NMDG-Cl, 2 mm MgCl2, 5 mm CaCl2, and 10 mm HEPES, pH 7.4 with NMDG. Cells were perfused with a bath solution containing 145 mm NaCl, 5 mm KCl, 2 mm MgCl2, 1 mm CaCl2, 5 mm glucose, 5 mm HEPES, pH 7.4 with NaOH, and 20 mm sucrose. After the establishment of an inside-out configuration, the patch was perfused with a standard perfusion solution (i.e. intracellular solution) containing 150 mm NMDG-Cl, 2 mm MgCl2, 10 mm EGTA, and 8 mm Tris, pH 7.4 with NMDG. Patches were held at -50 mV in all experiments. CFTR channel currents were recorded at room temperature. The currents were filtered at 100 Hz with an eight-pole Bessel filter (model LPF-8; Warner Instruments) and digitized on line at 500 Hz. CFTR channels were activated by the combined application of 1 mm MgATP and 25 units/ml cAMP-dependent protein kinase. MgATP and cAMP-dependent protein kinase were purchased from Sigma.
Protein Purification—The membrane fraction was homogenized in 5 volumes (v/w) of buffer A (TBS containing 25 mm n-dodecyl β-d-maltoside (DDM) (Sigma), 300 mm MgCl2, 10% glycerol, protease inhibitors, and 0.02% sodium azide). After centrifuging at 100,000 × g for 30 min, the supernatant containing solubilized FLAG-tagged CFTR was loaded onto an anti-FLAG affinity column (Sigma) equilibrated in advance. The column was then washed with 10 column volumes of buffer B (TBS containing 5 mm DDM, 300 mm MgCl2, 10% glycerol, protease inhibitors, and 0.02% sodium azide), and the bound CFTR protein was eluted with buffer B containing 100 μg/ml FLAG peptide (Sigma). The eluates from the FLAG column were analyzed by SDS-PAGE followed by silver staining.
The CFTR-rich fractions were pooled and loaded to the wheat germ agglutinin (WGA)-agarose column (Seikagaku). Unbound protein was washed out by 5 column volumes of buffer B. The bound CFTR protein was eluted by the linear gradient of N-acetylglucosamine (GlcNAc) from 20 to 100 mm. The elution of CFTR protein was monitored by SDS-PAGE analysis. The CFTR-rich fractions were pooled, concentrated with a Microcon filter unit (YM-100; Millipore) and further purified by Superdex 200 size exclusion chromatography (SEC) in a SMART System (GE Healthcare) using buffer C (TBS containing 5 mm DDM, 500 mm MgCl2, 10% glycerol, and 0.02% sodium azide). The elution of protein from the SEC was monitored by its UV absorbance at 280 nm and also by SDS-PAGE analysis.
Estimation of Stokes Radius by SEC—The distribution coefficient, Kav, was calculated from the equation Kav = (Ve - V0)/(Vt - V0) where Ve is the elution volume of high molecular weight standards (GE Healthcare) or CFTR protein. Column void volume (V0) was measured with blue dextran 2000, and Vt represents total bed volume. The Stokes radius (RS) of CFTR was determined using a calibration curve constructed by plotting the RS of each reference protein versus (-log Kav)½ according to the relationship (-log Kav)½ = α(β + RS) (20, 21). All the standards and the CFTR were solubilized in the same buffer used for CFTR purification. The standard proteins used were thyroglobulin (RS ≈ 85.0 Å), ferritin (RS ≈ 61.0 Å), catalase (RS ≈ 52.2 Å), and aldolase (RS ≈ 48.1 Å). The elution of CFTR was repeated five times; the data are presented by average ± S.D.
SDS and Blue-Native PAGE—The standard method of Laemmli (22) was applied for SDS-PAGE. Samples were mixed with an equal volume of sample buffer containing 62.5 mm Tris-HCl, pH 6.8, 2% SDS, 25% glycerol, 0.04 m dithiothreitol, and 0.01% bromphenol blue and then incubated at 60 °C for 15 min. Proteins were separated in a 5-20% polyacrylamide gel and visualized by silver staining. For Western blots, electrophoresed proteins in the gel were transferred to a polyvinylidene difluoride membrane and analyzed with alkaline phosphatase-labeled anti-FLAG antibodies (Sigma). For blue-native PAGE, purified CFTR was mixed with an equal volume of sample buffer (1% Coomassie G-250, 100 mm NaCl, 20% glycerol, and 100 mm bis-Tris, pH 7.2) and electrophoresed in a 4-16% polyacrylamide gel using a cathode buffer containing 50 mm bis-Tris, 50 mm Tricine, pH 6.8, and 0.002% Coomassie G-250 and an anode buffer containing 50 mm bis-Tris and 50 mm Tricine, pH 6.8 (23, 24). Electrophoresis started at 50 V for 30 min and was continued at 150 V for 90 min at room temperature. After destaining the gel with a solution containing 50% methanol and 10% acetic acid, the protein in the gel was visualized by silver staining or analyzed by Western blotting. The molecular mass of intact CFTR was estimated from a calibration curve obtained by plotting the mobility of standard proteins relative to the dye front against the logarithm of their molecular masses. The standards used were thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and bovine serum albumin (67 kDa).
Chemical Cross-linking—The TBS component in the buffer was substituted with PBS by dialysis. Glutaraldehyde or disuccinimidyl suberate was mixed with CFTR-containing solution to the indicated final concentration at room temperature for 30 min. Cross-linking was terminated by incubation with an equal volume of SDS sample buffer at 60 °C for 15 min. The proteins were separated in a 2-15% acrylamide gel and analyzed by Western blotting using anti-FLAG antibody. A protein sample treated similarly but without cross-linker was prepared as a control.
Transmission Electron Microscopy—Purified CFTR proteins of ∼50 μg/ml were adsorbed by thin carbon films supported by copper mesh grids, which were rendered hydrophilic in advance by glow discharge in low air pressure. Samples were washed with five drops of double distilled water, negatively stained with 2% uranyl acetate solution for 30 s twice, blotted, and dried in air. Micrographs of negatively stained particles were recorded in a JEOL 100CX transmission electron microscope at ×52,100 magnification with 100-kV acceleration voltage and an electron dose of ∼30 electrons/Å2 at the specimen level. The magnification calibration was performed by a grating replica (Nissin EM Co.). Images were recorded on SO-163 image films (Eastman Kodak Co.) developed with a D19 developer (Kodak) and digitized with a Scitex Leafscan 45 scanner (Leaf Systems) at a pixel size of 1.92 Å at the specimen level.
Molecular complexes between CFTR and the antibodies were generated by mixing the purified CFTR with an anti-FLAG M2 antibody (Sigma) or an anti-R domain MAB1660 antibody (R&D Systems) at 4 °C for 30 min. After removing excessive antibodies by SEC, the complexes were negatively stained and observed by EM. To obtain clearer images of the CFTR·anti-FLAG antibody complexes, Fab fragments were generated from the antibodies by papain digestion and conjugated with colloidal gold (BBInternational). The conjugate was isolated from non-reacted Fab molecules by 10-30% glycerol gradient centrifugation and then mixed with CFTR at 4 °C for 30 min. Protein G conjugated with colloidal gold (BBInternational) was used to obtain clearer images of MAB1660 binding. The protein G-gold was mixed with CFTR·MAB1660 complexes on the WGA-agarose, and then excessive gold was washed out. The gold-labeled complexes were eluted with 100 mm GlcNAc, negatively stained, and observed by EM.
Automated Particle Selection and Image Analysis—We have developed a single particle image analysis method using neural network (25-27) and simulated annealing (28, 29) named SPINNS (30). The following image analysis was performed using our SPINNS and IMAGIC V (31). CFTR projections were first picked up using the autoaccumulation method with simulated annealing (SA) (28). 291 particles in 160 × 160-pixel sub-frames were selected and used to train a three-layer pyramidal-type neural network (NN) (25, 26). Using the trained NN, 5,039 particles were selected. After background subtraction, the particles selected by the NN were aligned rotationally and translationally (32, 33) by the reference-free method (26). The aligned images were sorted into 150 classes by the modified growing neural gas network method (27). Their class averages were adopted as new references, and this cycle, from alignment to averaging, was repeated 11 times.
The Euler angles of the class averages were automatically determined by the echo-correlated three-dimensional reconstruction method with SA (29). In this step, C2 symmetry was imposed because top and bottom images in averaged projections exhibited 2-fold symmetry. These Euler angles were used to calculate a primary three-dimensional structure by the simultaneous iterative reconstruction technique (34). The reprojections from the volume were used as references for multireference alignment, and raw images in the library were aligned and further clustered, providing improved cluster averages. From these averages, a new three-dimensional map was generated by the reconstruction method using SA without a three-dimensional reference.
The three-dimensional map was further refined by the projection matching method (35) followed by an optimization using echo-correlated reconstruction. This cycle was repeated for seven cycles. Particle images that correlated poorly with the projections from the three-dimensional structure were automatically rejected using the cross-correlation function. The final reconstruction includes 4,206 particles, 83.5% of all the selected images. The resolution of the final three-dimensional map was assessed using the Fourier shell correlation function (36) at the threshold of 0.5.
RESULTS
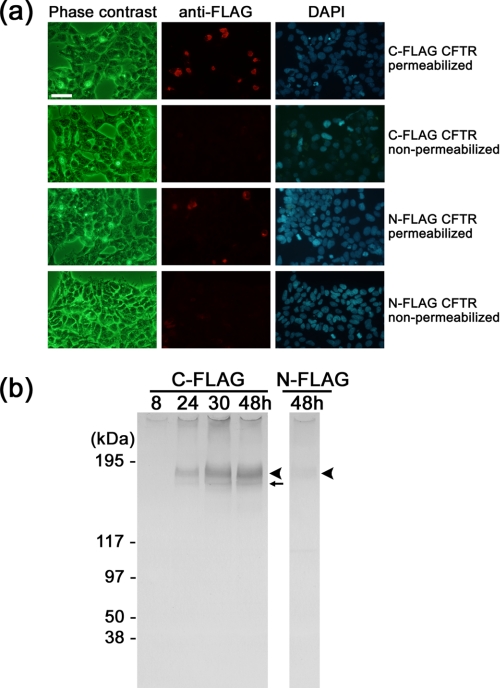
Heterologous Expression of CFTR Protein—We first constructed vectors containing human CFTR cDNA tagged with FLAG sequence at either the N or C terminus and transfected HEK293 cells with the vectors by the calcium phosphate precipitation method. The expressions of CFTR were compared by immunofluorescence using Cy3-conjugated anti-FLAG antibody. After the membrane permeabilization of the cells, the antibody reacted with ∼10% of cells expressing C- or N-FLAG CFTR (Fig. 1a, first and third rows). The level of Cy3 fluorescence was higher in the cells transfected with C-FLAG CFTR than those with N-FLAG CFTR. In contrast, the non-permeabilized control cells were negative to the antibody (Fig. 1a, second and fourth rows), confirming that both N and C termini are located inside the cells. The expressed CFTR proteins were solubilized with DDM and analyzed by Western blotting using anti-FLAG antibody. The antibody detected a broadened band at the expected size for the mature fully glycosylated CFTR (Fig. 1b, black arrowheads; estimated size, 177 kDa (169 kDa for the monomeric CFTR protein plus FLAG tag and associated glycan)) (37). A sharp band beneath the mature CFTR is likely the core-glycosylated immature CFTR (indicated by a small arrow) (37). The expression of C-FLAG CFTR increased during the 30-h period after transfection and reached its plateau at 48 h. The expression of N-FLAG CFTR was faintly detected at the corresponding position. We confirmed that both constructs retain ATP and cAMP-dependent protein kinase phosphorylation-dependent channel functions (see below). However, because the expression level of N-FLAG CFTR was much lower than that of C-FLAG CFTR, we adopted C-FLAG CFTR for further studies.
FIGURE 1.
Expression of recombinant CFTR in HEK cells. a, immunofluorescent images of HEK293 cells expressing FLAG-tagged CFTR. Top row, cells transfected with C-FLAG CFTR and permeabilized with 0.1% saponin. CFTR protein was detected using Cy3-conjugated anti-FLAG antibody (middle panel, red). Cell nuclei were visualized by 4′, 6-diamidino-2-phenylindole (DAPI)(right panel, blue). Second row, cells expressing C-FLAG CFTR were examined without saponin treatment. The cells were negative to the FLAG antibody, suggesting that the C-terminal epitope is inside the cell. Third row, cells transfected with N-FLAG CFTR and permeabilized by saponin. The level of Cy3 immunofluorescence is lower than that of C-FLAG CFTR. Fourth row, the non-permeabilized cells transfected with N-FLAG CFTR were also negative to the antibody. Cells were cultured for 48 h after transfection. The scale bar represents 50 μm. b, Western blot with anti-FLAG antibody. Left panel, cells were transfected with C-FLAG CFTR and cultured for the indicated periods. Proteins were solubilized from the membrane using DDM and precipitated with anti-FLAG gel. Expression of CFTR increased with the culture period up to 48 h. Right panel, the expression of N-FLAG CFTR was very low but was detected at the same size as C-FLAG CFTR. The broad bands at 177 kDa are glycosylated mature CFTR (arrowheads), and the sharp bands that move faster than the mature protein are core-glycosylated but immature CFTR (small arrow).
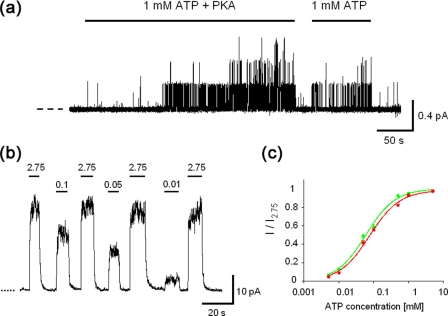
Electrophysiological Measurement of C-FLAG CFTR—The channel function of C-FLAG CFTR was examined using the patch clamp technique. Both cAMP-dependent protein kinase and ATP were applied to the cytoplasmic (bath) side of the inside-out patch obtained from HEK293 cell expressing C-FLAG CFTR (Fig. 2a). Once the C-FLAG CFTR was phosphorylated, the channel was opened by ATP alone (Fig. 2a). Single channel conductance (∼6 picosiemens) and the ATP-dependent gating behavior are similar to those in non-FLAG-tagged wild-type CFTR (10-12, 18, 19). The ATP dependence of the channel activity of C-FLAG CFTR is almost identical between the HEK293 and Chinese Hamster ovary expression systems (Fig. 2, b and c) and also comparable to that of non-FLAG-tagged wild-type CFTR (10-12, 18, 19). These data confirmed that the mechanisms of cAMP-dependent protein kinase phosphorylation-induced ATP-dependent gating and the ion conducting functions were retained in C-FLAG CFTR.
FIGURE 2.
Channel function of C-FLAG CFTR. a, activation of C-FLAG CFTR by cAMP-dependent protein kinase (PKA)-dependent phosphorylation and ATP. Single channel currents were obtained from an inside-out patch from HEK293 cells expressing C-FLAG CFTR. After phosphorylation, ATP alone can gate C-FLAG CFTR. Vm = -50 mV. The dashed line indicates the closed level. b, ATP-dependent macroscopic currents of cAMP-dependent protein kinase-phosphorylated C-FLAG CFTR in the inside-out configuration. The concentrations of ATP in the bath are indicated above. Vm = -50 mV. c, ATP concentration dependence of macroscopic currents of C-FLAG CFTR expressed in HEK293 (red) and Chinese Hamster ovary (green) cells. Vm = -50 mV. These results are also comparable to the channel function of non-FLAG-tagged wild-type CFTR (10-12, 18, 19).
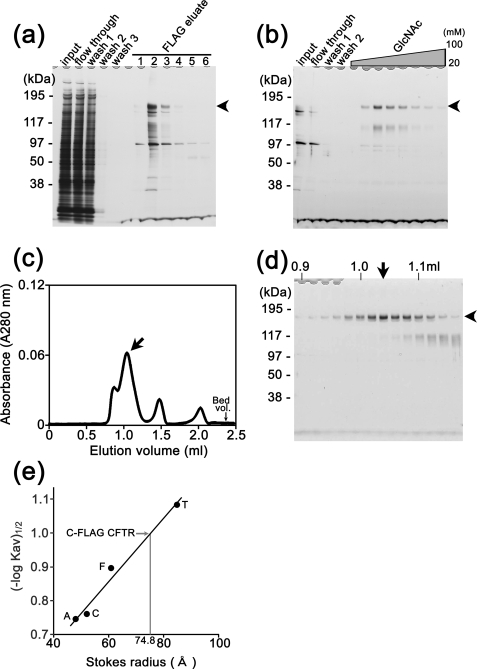
Purification of CFTR Proteins from Heterologously Expressed HEK293 Cells—CFTR proteins were purified from the membrane fraction by a combination of anti-FLAG immunoaffinity chromatography, lectin affinity chromatography, and SEC. The CFTR bound to the FLAG affinity column was competitively eluted with 100 μg/ml FLAG peptides and analyzed by SDS-PAGE. Although CFTR was electrophoresed as a predominant band at 177 kDa, several contaminants including a sharp band at 100 kDa and an immature CFTR moving slightly faster than the mature protein were also observed (Fig. 3a). To isolate mature CFTR, the eluate from the FLAG column was further subjected to WGA chromatography, which has been shown to be effective for CFTR purification (15). The bound CFTR was eluted using a linear gradient of GlcNAc from 20 to 100 mm in which the CFTR was mainly eluted between 40 and 60 mm (Fig. 3b).
FIGURE 3.
Purification of the CFTR protein from transiently expressed HEK cells. Silver staining of aliquots at each purification step from anti-FLAG (a) and from WGA columns (b). Proteins bound to the WGA column were eluted by a linear gradient of GlcNAc from 20 to 100 mm. Bands of fully glycosylated mature CFTR are indicated by arrowheads. c, CFTR-rich eluates from the WGA column were concentrated using the Microcon YM-100 filter unit and analyzed by Superdex 200 SEC. CFTR was eluted in a sharp peak at 1.04-ml elution (indicated by an arrow). d, SDS-PAGE analysis of the SEC fractions. The intensity of the CFTR band in the gel corresponds to absorption in SEC. The fraction at 1.04 ml (arrow) was used for EM image analysis. e, from the elution volumes of CFTR and the standards, the RS of CFTR was calculated as 74.8 ± 1.1 Å (mean ± S.D., n = 5). T, thyroglobulin (RS, 85.0 Å); F, ferritin (RS, 61.0 Å); C, catalase (RS, 52.2 Å); A, aldolase (RS, 48.1 Å).
Although the CFTR was highly concentrated after this purification step, a broad band at 110 kDa still remained (Fig. 3b). It was recognized by anti-FLAG antibody (data not shown) and speculated to be degraded CFTR. To remove this contaminant, the eluate from the WGA column was concentrated using a Microcon YM-100 filter unit and further purified by Superdex 200 SEC. The elution of CFTR from the column was monitored by UV absorption at 280 nm (Fig. 3c), and the aliquot of each fraction was analyzed by SDS-PAGE followed by silver staining (Fig. 3d). A sharp peak at 1.04-ml elution in the SEC was confirmed to be CFTR (Fig. 3d). The 110-kDa contamination was fainter than the band of CFTR and mainly eluted at 1.12-1.16 ml. A rise at 0.87-ml elution and large absorbance at 1.5 ml did not accompany detectable proteins; these are speculated to be absorptions due to micelles of lipids derived from the plasma membrane. Absorbance of FLAG peptides was observed at 2.00-ml elution. An aliquot at 1.04-ml elution (Fig. 3, c and d, arrow) was used for the EM study. This fraction contained highly concentrated mature CFTR with a minimal contamination of the 110-kDa proteins.
Using the purified CFTR, the hydrated size (Stokes radius, RS) of CFTR was calculated from the elution volume in SEC (20, 21) where CFTR was eluted between the thyroglobulin (eluted at 0.991 ml, RS is 85.0 Å) and ferritin (eluted at 1.129 ml, RS is 61.0 Å). The RS of CFTR was determined to be 74.8 ± 1.1 Å (mean ± S.D., n = 5) from the calibration curve (Fig. 3e).
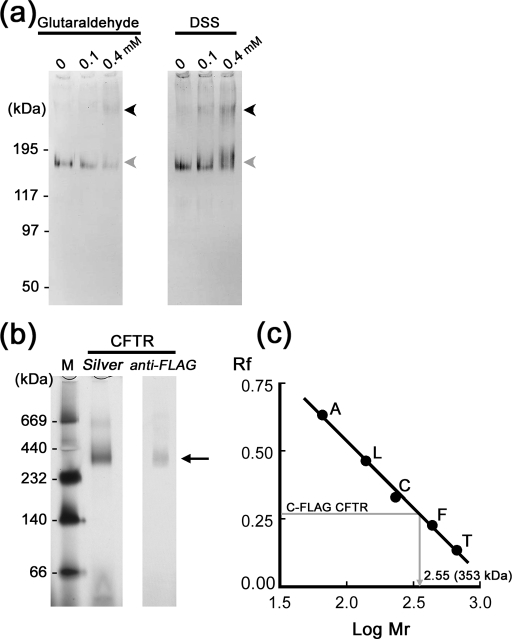
Chemical cross-linking and blue-native PAGE were used to determine the stoichiometry. Purified CFTR was treated with various concentrations of glutaraldehyde or disuccinimidyl suberate and analyzed by SDS-PAGE followed by Western blotting using anti-FLAG antibody (Fig. 4a). The TBS in the buffer composition was replaced with PBS by dialysis in advance. The band of CFTR treated with either chemical was shifted to a higher position with a molecular weight expected for a CFTR dimer. To more accurately estimate the molecular mass of the purified CFTR, we ran the blue-native PAGE and found the band of CFTR between the standards of 232 and 440 kDa (Fig. 4b, indicated by arrow). From the calibration curve obtained from the standard proteins, the molecular mass of the purified CFTR was determined to be 353 kDa (Fig. 4c), nearly double the size of the CFTR monomer (177 kDa including tag and glycan). These results suggest that purified CFTR proteins may assume a dimer configuration.
FIGURE 4.
Chemical cross-linking and native PAGE analysis of CFTR. a, purified CFTR was treated with glutaraldehyde or disuccinimidyl suberate (DSS) at the indicated concentrations and separated by SDS-PAGE followed by Western blotting using anti-FLAG antibody. The band of CFTR (gray arrowheads) was shifted upward at the corresponding position of a dimer (black arrowheads) by treatments with both cross-linkers. b, blue-native PAGE of CFTR. Purified CFTR was separated in a blue-native gel and visualized by silver staining (Silver) or Western blotting using anti-FLAG antibody (anti-FLAG). The CFTR protein was detected between the standards of 232 and 440 kDa (arrow). c, calculation of molecular mass from the blue-native PAGE. From the mobility of proteins relative to the dye front (Rf), the molecular mass of native CFTR was calculated to be 353 kDa, suggesting dimeric assembly (i.e. 177 kDa × 2 = 354 kDa). T, thyroglobulin (669 kDa); F, ferritin (440 kDa); C, catalase (232 kDa); L, lactate dehydrogenase (140 kDa); A, bovine serum albumin (66 kDa).
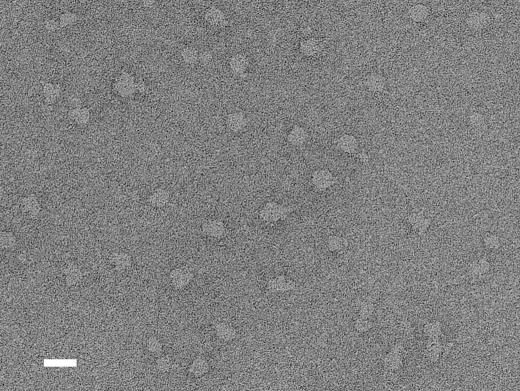
Electron Microscopy and Three-dimensional Reconstruction of CFTR—Purified CFTR proteins were blotted onto a glow-discharged carbon film supported by a copper mesh grid, negatively stained with 2% uranyl acetate, and imaged using an electron microscope at ×52,100 magnification. Variously shaped particles of uniform size were observed (Fig. 5). Most particles were triangular or polygonal with round corners. The variation in shapes is interpreted to reflect different orientations of the same molecule on the grid. The elliptical particles, although rarely observed, are postulated to be top views of the dimeric CFTR, whereas the triangular or polygonal shapes likely represent side views.
FIGURE 5.
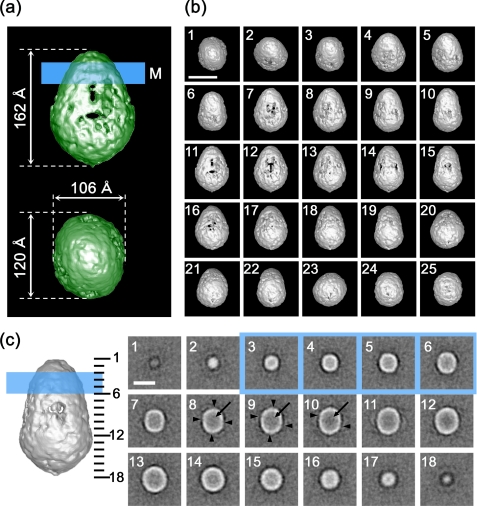
Electron microscopy of negatively stained CFTR. After adsorption to hydrophilic carbon film, negatively stained samples were imaged on a JEOL 100CX electron microscope at a magnification of ×52,100 with 100-kV acceleration voltage. The CFTR particles were observed as uniformly sized projections. For statistical analysis, 291 particles were automatically picked up by the autoaccumulation method and utilized as training data for the three-layer NN autopicking system. Using the trained NN, 5,039 particles were picked up and used for the analysis. The scale bar represents 200 Å.
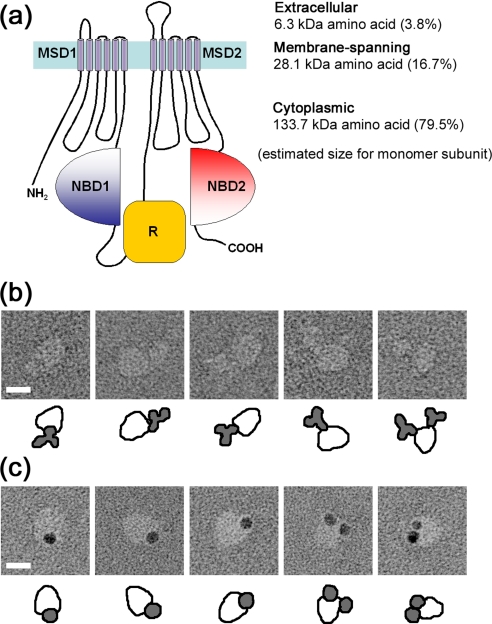
Because the hydropathy plot of CFTR predicted that 79.5% of the CFTR sequence is in the cytoplasm (see Fig. 9a), the larger domain of the protein is speculated to be cytoplasmic. To confirm this hypothesis, the FLAG tag at the cytoplasmic C terminus (38) was decorated with the anti-FLAG antibody, and the complex was negatively stained and observed by EM. The antibodies indeed attached to the larger domain of CFTR, confirming that this side of the molecule represents the cytoplasmic domain (Fig. 6a). It should be noted that we frequently observed CFTR particles bearing two antibodies (Fig. 6a, right end column). This observation further supports the dimeric assembly of CFTR. The bound antibodies were much more clearly observed using the Fab fragments conjugated with colloidal gold. Gold particles were again observed on the larger domain of CFTR (Fig. 6b).
FIGURE 9.
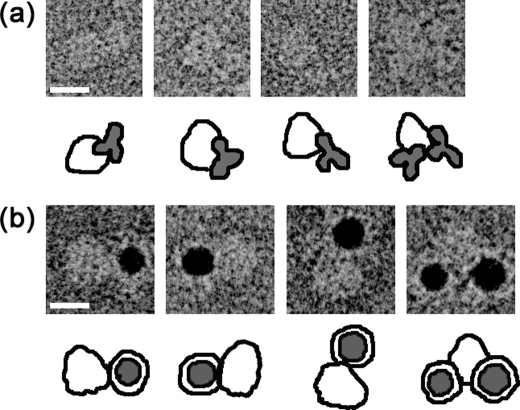
Binding of anti-R domain antibody to CFTR. a, transmembrane topology predicted for CFTR. The subunit is composed of two repeated motifs, each of which comprises an MSD and a cytoplasmic NBD. An R domain is located between NBD1 and MSD2. The molecule is divided into 3.8% extracellular, 16.7% membrane-spanning, and 79.5% cytoplasmic domains, calculated from the amino acid sequence. b, gallery of negatively stained CFTR·anti-R domain antibody (MAB1660) complexes. MAB1660 specifically binds to the larger domain of CFTR. c, gallery of CFTR·MAB1660·protein G-gold complexes. Protein G-gold was mixed with CFTR·MAB1660 complexes on the WGA column, and then excessive gold was washed out. Similar to the observation in b, the protein G-gold again binds to the larger domain. CFTR particles bearing two gold particles were also observed. The scale bars represent 100 Å.
FIGURE 6.
Assignment of the cytoplasmic C terminus of CFTR. a, gallery of negatively stained CFTR·anti-FLAG-antibody complexes. Schematic diagrams of CFTR (open particles) and antibodies (filled in gray) are presented below. The location of the cytoplasmic C terminus was assigned to the larger end of the CFTR molecule. CFTR decorated with two FLAG antibodies was frequently observed (right end column). b, gallery of CFTR·Fab-gold complexes. Fab fragments of anti-FLAG antibodies were conjugated with colloidal gold and then mixed with purified CFTR. Similar to the observation in a, the gold conjugate binds to the periphery of the larger domain. CFTR particles bearing two gold particles were also observed (right end column). The scale bars represent 100 Å.
The projections of CFTR were picked up by a combination of two automatic programs: the autoaccumulation method using SA (28) and the three-layered neural network method (25, 26). The three-dimensional structure was reconstructed with echocorrelated reconstruction methods using SA assuming C2 symmetry in our SPINNS (25-30) and other algorithms in the IMAGIC V software (31) (see “Experimental Procedures” for details). The final reconstruction included 4,206 particles, 83.5% of all the selected images.
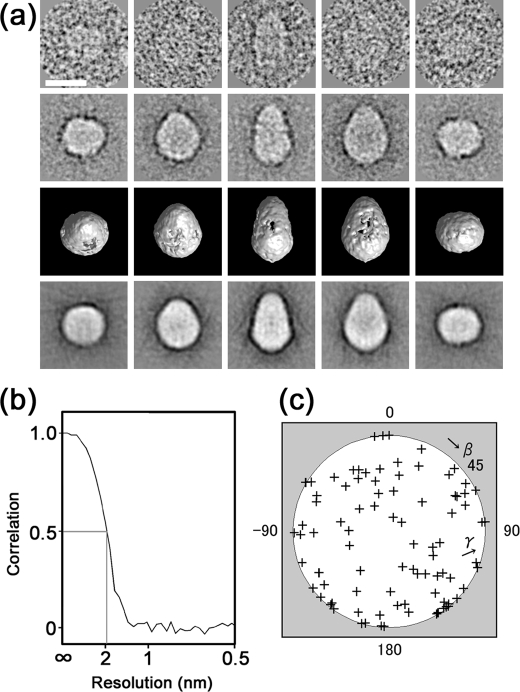
Representative raw images are presented (Fig. 7a, first row) with their corresponding class averages (second row) and with the surface representations and the projections of the reconstructed three-dimensional structure (third and fourth rows). A high level of consistency was observed in size, shape, and inner structure among these data sets (Fig. 7a), indicating successful three-dimensional reconstruction from the original particle images. According to the Fourier shell correlation function (36), the resolution limit is 2 nm by the correlation coefficient >0.5 criterion (Fig. 7b). A plot of the Euler angles of the 94 adopted class averages shows that CFTR is almost randomly oriented on the grid surface (Fig. 7c).
FIGURE 7.
Three-dimensional reconstruction of CFTR. a, raw images of CFTR with different Euler angles (row 1) are compared with the corresponding two-dimensional averages (row 2), the surface views of the three-dimensional reconstruction (row 3), and the reprojections of the three-dimensional reconstruction (row 4) consistent through the reconstruction. Protein is displayed in bright shades. The scale bars represent 100 Å. b, Fourier shell correlation function indicates the resolution limit of 2 nm by the Fourier shell correlation >0.5 criterion. c, plot of the Euler angles (β, γ) of 94 adopted class averages demonstrates almost random orientations of CFTR molecules on the carbon surface.
Structural Features of the CFTR Molecule—The surface representation demonstrates that CFTR is an ellipsoidal molecule of 162 Å in height. The elliptical top view has a major diameter of 120 Å and minor diameter of 106 Å (Fig. 8a). The three-dimensional map is contoured at an isosurface containing a volume corresponding to 276 kDa assuming a protein density of 1.37 g/cm3, which is 81.7% of the dimeric CFTR mass (338 kDa) calculated from the amino acid composition. The putative position of the membrane-spanning region is indicated by a blue line in Fig. 8a (∼30 Å in width) so that the ratio of each domain is close to the prediction from the hydropathy plot (i.e. the volumes of extracellular, membrane-spanning, and cytoplasmic domains are 3.8, 16.7, and 79.5%, respectively; Fig. 9a).
FIGURE 8.
Surface representation of CFTR. a, side and top views of the CFTR protein. CFTR presumably in a closed state is shown to be an ellipsoidal particle with dimensions of 120 × 106 × 162 Å. The molecular mass enclosed by the isosurface is 276 kDa, corresponding to 81.7% of the dimeric CFTR protein. The putative position of the plasma membrane is indicated on the side view by a blue line of 30 Å in thickness (M) so that the ratio of each domain is close to the prediction from the amino acid sequence. b, surface views from 25 different angles. c, sections normal to the symmetric axis at 9.5-Å intervals through the molecule. The number in each panel corresponds to the slice position indicated on the narrower side view (left). Slices included in the transmembrane domain are marked by blue. Rifts in the cytoplasmic surface (arrowheads in panels 8-10) correspond to the orifices in the surface representation. The inside of the molecule is mostly low density (arrows), suggesting that stain permeates into the molecule. The scale bars represent 100 Å.
The surface of the dome-shaped extracellular domain and the MSD is seamless and smooth, whereas the cytoplasmic surface is rough and contains orifices under the putative transmembrane region. Large orifices of ∼50 × 30 Å are prominent in the center of the wider side views (Fig. 8b, panels 11 and 12), and small elliptical orifices of 20 × 25 Å are prominent at the narrower side views (panel 15). These orifices might correspond to the region of intracellular loops between MSDs (1).
Sections normal to the symmetry axis demonstrate the tightly packed internal density at extracellular and transmembrane domains (Fig. 8c, panels 1-6). In contrast, the density inside the cytoplasmic domain is low (arrows, panels 8-10) probably because of the stain permeation through the orifices (arrowheads).
To determine the location of the R domain in the reconstruction, purified CFTR was mixed with an R domain-specific antibody, MAB1660, and the complex was negatively stained and observed by EM. The antibodies attached to the larger domain suggesting that the R domain is located around the bottom end of the cytoplasmic domain (Fig. 9b). CFTR particles bearing two antibodies were frequently observed, and this again supports the dimeric stoichiometry of reconstructed CFTR (Fig. 9b, right end). The MAB1660 bound to the CFTR molecule was clearly demonstrated by the binding of gold-conjugated protein G where one or two gold conjugates bound to the larger domain of CFTR (Fig. 9c). When two gold conjugates were observed for one CFTR particle, these two gold conjugates were closely located next to each other at the cytoplasmic end of the particle, indicating that the CFTR particle assumes a tail to tail configuration. Perhaps because of the steric hindrance and increased ion strength in the buffer composition, not all the epitopes were bound with antibodies or gold conjugates.
DISCUSSION
Purification of CFTR Protein—The insect Sf9 expression system has been shown to express a large quantity of CFTR proteins (39). We adopted human embryonic kidney HEK293 cells because, contrary to the insect cell system, mammalian cells allow production of mature CFTR proteins with full glycosylation, which enables us to use WGA chromatography for protein purification. As the CFTR protein is reported to bind to various chaperones and regulatory proteins in the cell and to form a global protein interaction network (40), we developed a purification protocol aimed at removing tightly bound associated proteins from the CFTR. One successful approach was the careful selection of buffer components. We increased the ionic strength in the buffer to eliminate bound proteins. By adding 300-500 mm magnesium chloride in the buffer, we could obtain highly purified CFTR proteins without disrupting the subunit assembly. Alkaline treatment of the plasma membrane, a method that has been commonly applied to remove associated proteins from the membrane-integral proteins (41), was not adopted in this study. Our experiments demonstrated that CFTR purified using alkaline treatment was eluted at a very early fraction in SEC where aggregated proteins are frequently eluted (data not shown). Another important key was WGA column chromatography, which concentrates fully glycosylated CFTR and excludes immature CFTR and associated proteins (15). To eliminate minor contaminants, we applied SEC as the final step for CFTR purification; this step was not used in the previous structural study (15). At the end of this purification process, fully glycosylated mature CFTR proteins were purified as a sharp single peak in SEC.
Structure of the Reconstructed CFTR—In this study, using the single particle analysis of the negatively stained EM images, we reconstructed the three-dimensional structure of unphosphorylated CFTR under a nucleotide-free condition. We applied 2-fold symmetry in the three-dimensional reconstruction. Because of a small number of symmetry axes, we used our echocorrelated reconstruction method. Because this method does not compress projections into sinograms for a posteriori angular assignment of the projections, it enables accurate three-dimensional reconstruction of the molecules without high point symmetries. The particles are also picked up using our programs with SA and NN to avoid any human intention and preference.
The structure of wild-type human CFTR, reconstructed at 2-nm resolution, was an ellipsoidal molecule with 120 × 106 × 162-Å dimensions. It was larger than the two different conformations of CFTR with 6-7-nm diameter obtained from two-dimensional crystallography (15). Because these reported CFTR structures were suggested to be monomeric (15), the vertical dimension of 120 × 106 Å in our reconstruction is comparable with that expected for a dimer.
The R domain is unique in CFTR among the ABC transporter family and important for the regulation of gating. The R domain contains multiple phosphorylation sites, and the level of their phosphorylation regulates CFTR channel activity (42). Recently Baker et al. (43) suggested that these phosphorylation sites are allocated in the multiple helical segments of the R domain and interact with NBD1 when they are unphosphorylated. However, once they are phosphorylated, the helicity around them is reduced, hence reducing their interaction with NBD1 (43). In our experiments, the R domain should be mostly unphosphorylated and presumably bound to NBD1 (and possibly to NBD2 as well). Both monoclonal antibodies against the C-terminal FLAG and R domain bound to the cytoplasmic end of CFTR, indicating that the R domain/NBD system is organized in the larger cytoplasmic domain (Figs. 6 and 9). The fact that the anti-R domain antibody can bind to the CFTR particle indicates that at least the epitope of the R domain is located at a position accessible from the outside of the molecule under the unphosphorylated condition.
In contrast to the large cytoplasmic domains, the dome-shaped extracellular and membrane-spanning domains are relatively small. This is in good agreement with a homology model of CFTR using the crystal structure of Sav1866 as a template (44). The low resolution of our EM structure does not reveal the extracellular pore entrance of the CFTR channel. Thus, it is unclear whether the ion conduction pathway is formed by one or two CFTR molecules.
Orifices observed under the putative transmembrane region, presumably in the intracellular loops region (44), were prominent (Fig. 8a). Similar side pores are also reported in the voltage-sensitive sodium channel (45) and Kv1.2 potassium channel (46). They are thought to be part of the exit pathway for permeant ions into the cytosol. Whether these side pores have equivalent function for CFTR remains unclear. Further improvement in resolution using cryo-EM single particle analysis may shed light on the functional role of these side pores.
Dimeric Stoichiometry of the CFTR Molecule—Analysis of the quaternary structure of CFTR is essential to understand the mechanisms of ion permeation and gating regulation of CFTR; however, the stoichiometry of functional CFTR has not been clearly determined yet (47). Bear et al. (48) succeeded to reconstitute a cAMP-dependent protein kinase-activated ATP-dependent low conductance chloride channel in lipid bilayer using purified CFTR proteins. This study provided evidence that the CFTR molecule alone is sufficient to form a functional channel. Subsequent biochemical and physiological studies indicated that CFTR behaves as a monomer (49), and the monomer has both channel function and ATPase activity when expressed in Sf9 cells (50). Zhang et al. (51) proposed, from analyses of magnitudes and distributions of subconductance states of CFTR mutations in the sixth transmembrane segment, that a single CFTR polypeptide can form an anion-conducting pore.
On the other hand, many studies suggested that the majority of CFTR molecules in the plasma membrane exist and function as dimers. Freeze-fracture images of CFTR protein expressed in Xenopus oocytes demonstrated that CFTR forms a dimer-like structure at the membrane-spanning region (52). Also atomic force microscopic studies of CFTR expressed in Xenopus oocytes showed that CFTR assumes a 2-fold symmetry to the central pore (53). Tandemly linked heterodimers of wild-type CFTR and a ΔR mutant CFTR showed mixed gating properties between these channels, indicating that the two CFTR molecules interact to form a single ion conductance pore (54). Raghuram et al. (55) revealed that the activity of the CFTR channel is enhanced by intermolecular interactions via bivalent PDZ domains, supporting the hypothesis that bivalent PDZ domains of sodium hydrogen exchange regulatory factor regulate CFTR by cross-linking two CFTR molecules at the C-terminal tails. Ramjeesingh et al. (56) reported that CFTR exists as monomers, dimers, and multimers in mammalian cells. Among them, dimeric CFTR is the predominant form in the plasma membrane.
In the present study, both the chemical cross-linking experiments and blue-native PAGE revealed that CFTR has a molecular mass corresponding to a dimer. A diameter derived from the SEC (estimated at 149.6 Å, assuming it is a hydrated sphere) also agrees well with the dimensions of a CFTR dimer. Furthermore two antibodies simultaneously bound to the large cytoplasmic domain of a single CFTR particle in both experiments using anti-FLAG and anti-R domain antibodies, supporting the dimeric assembly of the purified CFTR.
The dimeric structure in the current study contradicts the previously reported structures of CFTR using two-dimensional crystals (15). However, our results corroborate a more recent report that the EM structure of CFTR has a dimension that is consistent with two copies of the homology model of CFTR (16). It is interesting to note that the thickness of the reconstructed molecule in Awayn et al. (16) was ∼5 nm, approximately half of that in our reconstruction (∼10-nm thickness), and the structure was labeled with only a single (rather than two) nickel-nitrilotriacetic acid-nanogold C-terminal decorator. These discrepancies may be due to differences in methods for protein purification.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK55835-09 and R01 HL53445-11 (to T.-C. H.). This work was also supported by a grant-in-aid for scientific research on priority areas, structure of biological macromolecular assemblies (to K. M. and C. S.); by a grant from the Japan New Energy and Industrial Technology Development Organization (NEDO) (to C. S.); by a grant from PRESTO of the Japan Science and Technology Agency (to T. O.); and by Japan Society for the Promotion of Science (JSPS) Grant 19590215 (to Y. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CFTR, cystic fibrosis transmembrane conductance regulator; ABC, ATP-binding cassette; bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; DDM, n-dodecyl-β-d-maltoside; EM, electron microscopy; HEK, human embryonic kidney; MSD, membrane-spanning domain; NBD, nucleotide-binding domain; NMDG, N-methyl-d-glucamine; NN, neural network; PBS, phosphate-buffered saline; R domain, regulatory domain; RS, Stokes radius; SA, simulated annealing; SEC, size exclusion chromatography; SPINNS, single particle image analysis method using neural network and simulated annealing; TBS, Tris-buffered saline; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)-ethyl]glycine.
References
- 1.Riordan, J. R., Rommens, J. M., Kerem, B., Alon, N., Rozmahel, R., Grzelczak, Z., Zielenski, J., Lok, S., Plavsic, N., Chou, J. L., Drumm, M. L., Iannuzzi, M. C., Collins, F. S., and Tsui, L. C. (1989) Science 245 1066-1073 [DOI] [PubMed] [Google Scholar]
- 2.Welsh, M. J., and Smith, A. E. (1993) Cell 73 1251-1254 [DOI] [PubMed] [Google Scholar]
- 3.Tsui, L. C., and Durie, P. (1997) Hosp. Pract. 32 115-118, 123-129, 134 [DOI] [PubMed] [Google Scholar]
- 4.Gabriel, S. E., Brigman, K. N., Koller, B. H., Boucher, R. C., and Stutts, M. J. (1994) Science 266 107-109 [DOI] [PubMed] [Google Scholar]
- 5.Sears, C. L., and Kaper, J. B. (1996) Microbiol. Rev. 60 167-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya, S. K. (1995) J. Indian Med. Assoc. 93 237-238 [PubMed] [Google Scholar]
- 7.Chen, T. Y., and Hwang, T. C. (2008) Physiol. Rev. 88 351-387 [DOI] [PubMed] [Google Scholar]
- 8.Davidson, A. L., and Chen, J. (2004) Annu. Rev. Biochem. 73 241-268 [DOI] [PubMed] [Google Scholar]
- 9.Vergani, P., Lockless, S. W., Nairn, A. C., and Gadsby, D. C. (2005) Nature 433 876-880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bompadre, S. G., Cho, J. H., Wang, X., Zou, X., Sohma, Y., Li, M., and Hwang, T. C. (2005) J. Gen. Physiol. 125 377-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bompadre, S. G., Ai, T., Cho, J. H., Wang, X., Sohma, Y., Li, M., and Hwang, T. C. (2005) J. Gen. Physiol. 125 361-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou, Z., Wang, X., Li, M., Sohma, Y., Zou, X., and Hwang, T. C. (2005) J. Physiol. 569 447-457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis, H. A., Buchanan, S. G., Burley, S. K., Conners, K., Dickey, M., Dorwart, M., Fowler, R., Gao, X., Guggino, W. B., Hendrickson, W. A., Hunt, J. F., Kearins, M. C., Lorimer, D., Maloney, P. C., Post, K. W., Rajashankar, K. R., Rutter, M. E., Sauder, J. M., Shriver, S., Thibodeau, P. H., Thomas, P. J., Zhang, M., Zhao, X., and Emtage, S. (2004) EMBO J. 23 282-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis, H. A., Zhao, X., Wang, C., Sauder, J. M., Rooney, I., Noland, B. W., Lorimer, D., Kearins, M. C., Conners, K., Condon, B., Maloney, P. C., Guggino, W. B., Hunt, J. F., and Emtage, S. (2005) J. Biol. Chem. 280 1346-1353 [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg, M. F., Kamis, A. B., Aleksandrov, L. A., Ford, R. C., and Riordan, J. R. (2004) J. Biol. Chem. 279 39051-39057 [DOI] [PubMed] [Google Scholar]
- 16.Awayn, N. H., Rosenberg, M. F., Kamis, A. B., Aleksandrov, L. A., Riordan, J. R., and Ford, R. C. (2005) Biochem. Soc. Trans. 33 996-999 [DOI] [PubMed] [Google Scholar]
- 17.Graham, F. L., and van der Eb, A. J. (1973) Virology 52 456-467 [DOI] [PubMed] [Google Scholar]
- 18.Bompadre, S. G., Sohma, Y., Li, M., and Hwang, T. C. (2007) J. Gen. Physiol. 129 285-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou, Z., Wang, X., Liu, H. Y., Zou, X., Li, M., and Hwang, T. C. (2006) J. Gen. Physiol. 128 413-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurent, T., C., and Killander, J. (1964) J. Chromatogr. 14 317-330 [Google Scholar]
- 21.Mio, K., Ogura, T., Hara, Y., Mori, Y., and Sato, C. (2005) Biochem. Biophys. Res. Commun. 333 768-777 [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 23.Schagger, H., and von Jagow, G. (1991) Anal. Biochem. 199 223-231 [DOI] [PubMed] [Google Scholar]
- 24.Schagger, H., Cramer, W. A., and von Jagow, G. (1994) Anal. Biochem. 217 220-230 [DOI] [PubMed] [Google Scholar]
- 25.Ogura, T., and Sato, C. (2001) J. Struct. Biol. 136 227-238 [DOI] [PubMed] [Google Scholar]
- 26.Ogura, T., and Sato, C. (2004) J. Struct. Biol. 145 63-75 [DOI] [PubMed] [Google Scholar]
- 27.Ogura, T., Iwasaki, K., and Sato, C. (2003) J. Struct. Biol. 143 185-200 [DOI] [PubMed] [Google Scholar]
- 28.Ogura, T., and Sato, C. (2004) J. Struct. Biol. 146 344-358 [DOI] [PubMed] [Google Scholar]
- 29.Ogura, T., and Sato, C. (2006) J. Struct. Biol. 156 371-386 [DOI] [PubMed] [Google Scholar]
- 30.Yazawa, M., Ferrante, C., Feng, J., Mio, K., Ogura, T., Zhang, M., Lin, P. H., Pan, Z., Komazaki, S., Kato, K., Nishi, M., Zhao, X., Weisleder, N., Sato, C., Ma, J., and Takeshima, H. (2007) Nature 448 78-82 [DOI] [PubMed] [Google Scholar]
- 31.van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R., and Schatz, M. (1996) J. Struct. Biol. 116 17-24 [DOI] [PubMed] [Google Scholar]
- 32.Frank, J. (2006) Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State, Oxford University Press, New York
- 33.van Heel, M., Gowen, B., Matadeen, R., Orlova, E. V., Finn, R., Pape, T., Cohen, D., Stark, H., Schmidt, R., Schatz, M., and Patwardhan, A. (2000) Q. Rev. Biophys. 33 307-369 [DOI] [PubMed] [Google Scholar]
- 34.Penczek, P., Radermacher, M., and Frank, J. (1992) Ultramicroscopy 40 33-53 [PubMed] [Google Scholar]
- 35.Penczek, P. A., Grassucci, R. A., and Frank, J. (1994) Ultramicroscopy 53 251-270 [DOI] [PubMed] [Google Scholar]
- 36.Harauz, G., and van Heel, M. (1986) Optik 73 146-156 [Google Scholar]
- 37.Cheng, S. H., Gregory, R. J., Marshall, J., Paul, S., Souza, D. W., White, G. A., O'Riordan, C. R., and Smith, A. E. (1990) Cell 63 827-834 [DOI] [PubMed] [Google Scholar]
- 38.Denning, G. M., Ostedgaard, L. S., Cheng, S. H., Smith, A. E., and Welsh, M. J. (1992) J. Clin. Investig. 89 339-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Riordan, C. R., Lachapelle, A. L., Marshall, J., Higgins, E. A., and Cheng, S. H. (2000) Glycobiology 10 1225-1233 [DOI] [PubMed] [Google Scholar]
- 40.Wang, X., Venable, J., LaPointe, P., Hutt, D. M., Koulov, A. V., Coppinger, J., Gurkan, C., Kellner, W., Matteson, J., Plutner, H., Riordan, J. R., Kelly, J. W., Yates, J. R., III, and Balch, W. E. (2006) Cell 127 803-815 [DOI] [PubMed] [Google Scholar]
- 41.Steck, T. L., and Yu, J. (1973) J. Supramol. Struct. 1 220-232 [DOI] [PubMed] [Google Scholar]
- 42.Gadsby, D. C., and Nairn, A. C. (1999) Physiol. Rev. 79 S77-S107 [DOI] [PubMed] [Google Scholar]
- 43.Baker, J. M., Hudson, R. P., Kanelis, V., Choy, W. Y., Thibodeau, P. H., Thomas, P. J., and Forman-Kay, J. D. (2007) Nat. Struct. Mol. Biol. 14 738-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendoza, J. L., and Thomas, P. J. (2007) J. Bioenerg. Biomembr. 39 499-505 [DOI] [PubMed] [Google Scholar]
- 45.Sato, C., Ueno, Y., Asai, K., Takahashi, K., Sato, M., Engel, A., and Fujiyoshi, Y. (2001) Nature 409 1047-1051 [DOI] [PubMed] [Google Scholar]
- 46.Long, S. B., Campbell, E. B., and Mackinnon, R. (2005) Science 309 897-903 [DOI] [PubMed] [Google Scholar]
- 47.Riordan, J. R. (2005) Annu. Rev. Physiol. 67 701-718 [DOI] [PubMed] [Google Scholar]
- 48.Bear, C. E., Li, C. H., Kartner, N., Bridges, R. J., Jensen, T. J., Ramjeesingh, M., and Riordan, J. R. (1992) Cell 68 809-818 [DOI] [PubMed] [Google Scholar]
- 49.Chen, J. H., Chang, X. B., Aleksandrov, A. A., and Riordan, J. R. (2002) J. Membr. Biol. 188 55-71 [DOI] [PubMed] [Google Scholar]
- 50.Ramjeesingh, M., Li, C., Kogan, I., Wang, Y., Huan, L. J., and Bear, C. E. (2001) Biochemistry 40 10700-10706 [DOI] [PubMed] [Google Scholar]
- 51.Zhang, Z. R., Cui, G., Liu, X., Song, B., Dawson, D. C., and McCarty, N. A. (2005) J. Biol. Chem. 280 458-468 [DOI] [PubMed] [Google Scholar]
- 52.Eskandari, S., Wright, E. M., Kreman, M., Starace, D. M., and Zampighi, G. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 11235-11240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schillers, H., Shahin, V., Albermann, L., Schafer, C., and Oberleithner, H. (2004) Cell Physiol. Biochem. 14 1-10 [DOI] [PubMed] [Google Scholar]
- 54.Zerhusen, B., Zhao, J., Xie, J., Davis, P. B., and Ma, J. (1999) J. Biol. Chem. 274 7627-7630 [DOI] [PubMed] [Google Scholar]
- 55.Raghuram, V., Mak, D. O., and Foskett, J. K. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 1300-1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramjeesingh, M., Kidd, J. F., Huan, L. J., Wang, Y., and Bear, C. E. (2003) Biochem. J. 374 793-797 [DOI] [PMC free article] [PubMed] [Google Scholar]