Abstract
Replication of the double-stranded, circular human papillomavirus (HPV) genomes requires the viral DNA replicase E1. Here, we report an initial characterization of the E1 cistron of HPV type 16 (HPV-16), the most common oncogenic mucosal HPV type found in cervical and some head and neck cancers. The first step in HPV DNA replication is an initial burst of plasmid viral DNA amplification. Complementation assays between HPV-16 genomes carrying mutations in the early genes confirmed that the expression of E1 was necessary for initial HPV-16 plasmid synthesis. The major early HPV-16 promoter, P97, was dispensable for E1 production in the initial amplification because cis mutations inactivating P97 did not affect the trans complementation of E1− mutants. In contrast, E1 expression was abolished by cis mutations in the splice donor site at nucleotide (nt) 226, the splice acceptor site at nt 409, or a TATAA box at nt 7890. The mapping of 5′ mRNA ends using rapid amplification of cDNA ends defined a promoter with a transcription start site at HPV-16 nt 14, P14. P14-initiated mRNA levels were low and required intact TATAA (7890). E1 expression required the HPV-16 keratinocyte-dependent enhancer, since cis mutations in its AP-2 and TEF-1 motifs abolished the ability of the mutant genomes to complement E1− genomes, and it was further modulated by origin-proximal and -distal binding sites for the viral E2 gene products. We conclude that P14-initiated E1 expression is critical for and limiting in the initial amplification of the HPV-16 genome.
High-risk (HR) oncogenic mucosal human papillomavirus (HPV) types are the major cause of most, if not all, carcinomas of the uterine cervix and many other anogenital tumors, and they are found in 20 to 30% of cancers of the head and neck (HNC). HPV type 16 (HPV-16) is the most prevalent HR HPV: it is present in nearly one-half of cervical carcinomas and more than 90% of HPV-associated HNC (reviewed in reference 32). Recently, preventive vaccines against HPV-16 and HPV-18, the second most common HR HPV, have become available. While the vaccines are highly efficacious, they cannot eliminate existing infection. The development of therapies targeted at HPV infection requires a profound understanding of the viral genes involved in replication and their regulation.
Upon introduction into the basal layer cell of skin or stratified squamous epithelia, the double-stranded, closed circular HPV plasmid genome undergoes an initial amplification (11, 18, 45). HPV plasmid amplification in the initial stages of infection appears to be tightly restricted, potentially in part to limit triggering intracellular defense mechanisms by unchecked viral replication and/or to minimize host immune responses to high levels of viral polypeptides. A low steady-state viral copy number in established HPV infections suggests limited levels of cellular factors or the regulated synthesis of viral factors responsible for supporting HPV replication.
Papillomaviral DNA replication requires an origin of replication (ori) and two viral proteins: the DNA helicase E1 and the sequence-specific transcriptional activator product of the full-length E2 open reading frame (ORF) (reviewed in reference 15). First described for the model bovine papillomavirus (BPV) type 1 (4, 23, 39), the findings have been rapidly extended to mucosal HPV types (2, 5, 53). The DNA sequence of mucosal HPV origins appears to be well conserved, with an A/T-rich core flanked by E2 binding sites. The E1 molecules assemble at the core to form a hexamer under the steric guidance of the full-length E2 protein in the form of dimers bound to conserved E2 binding sites which flank the ori sequence (33, 43). The E1 protein serves as a replication initiator with ATP-dependent helicase activity that promotes ori unwinding (3, 20) and may also displace histone H1, further influencing ori function (46). The E1 hexamer also recruits cellular replication factors, including replication factor A, DNA polymerase α, and topoisomerase I, to assemble a functional replicating complex or “replisome” (6).
Although E1-dependent HPV ori replication has been studied in great detail, relatively little is known about the structure and regulation of the HPV E1 gene. Expression of the early viral gene products, such as E1, may be modulated at the level of transcription initiation by cellular and viral transcription factors, by posttranscriptional control, such as alternative splicing or polyadenylation, or by selective translation from polycistronic RNA messages (reviewed in reference 55). It has been assumed that E1 mRNA(s) initiates at the major early P97 promoter and arises through differential splicing in a fashion similar to that of the E7 mRNA (9, 36), yet candidate E1 mRNAs have not been unequivocally identified, possibly due to their very low levels.
In this study, we have characterized the E1 cistron of HPV-16 as a genetic entity that encodes the E1 polypeptide and controls its expression during initial HPV genome amplification. Mutations within a dense, complex genome such as that of HPV can have pleiotropic effects and thus yield ambiguous phenotypes. We have developed a complementation assay in which HPV replication depends on the expression of HPV genes from mutant HPV plasmid genomes that fail to amplify individually. Our results show that, during the initial amplification of the HPV-16 genome, E1 expression requires a polycistronic, spliced message that initiates at a novel TATAA-dependent promoter upstream of P97 with a major start site at HPV-16 nucleotide (nt) 14.
MATERIALS AND METHODS
Plasmid construction.
The replication-competent HPV-16 W12E plasmid (GenBank accession number AF125673) was a gift from Paul Lambert (22). All HPV-16 mutants were PCR generated, and the PCR fragments were digested and cloned into the HPV-16 W12E parent. The HPV-16 E1− precursor plasmid (a gift from the zur Hausen laboratory), which contains a frameshift mutation by a deletion at position 1087 in the 3′ portion of the E1 ORF (10), was excised with NcoI and BbsI and then cloned into the HPV-16 W12E plasmid. The E1 5′ ORF− construct, however, contains an amber mutation of amino acid 3 within the E1 ORF. The E8− construct was constructed by introducing an amber mutation into the E8 ORF at position 1281. Amber mutations were also introduced into the E6− construct at position 211, the E7− construct at position 714, and the E2 DNA binding domain (DBD−) construct at position 3717; all three mutations introduced unique XbaI sites. All PCR-generated plasmids were verified by automated sequencing (DNA Core Facility, University of Iowa).
The following synthetic oligonucleotide primers were used to introduce mutations into the HPV-16 W12E plasmid (as illustrated in Fig. 1). The wild-type (wt) sequence is capitalized, while nucleotide substitutions are in lowercase letters and oligonucleotide positions in the HPV-16 genome are in parentheses: E1 5′ ORF- (nt 865 to 889), 5′-CCATGGCTtagCCTGCAGGTACCAATG-3′; E8- (nt 1274 to 1293), 5′-CTGAAGTaGAAACTCAGCAG-3′; E2 DBD- (nt 3539 to 3559), 5′-GCGCtcTAgAACCATGGTGGACAGTGCTCCAATCCTC-3′; E6- (nt 208 to 234), 5′-gccctctagaGACGTGAGGTATATGAC-3′; E7- (nt 706 to 729), 5′-AGAGCCCtcTAgAATATTGTAACC-3′; SD226- (nt 217 to 235), 5′-GCGACGTGAGGcATATGAC-3′; SA409- (nt 399 to 422), 5′-GTTAATTcGaTGTATTAACTGTC-3′; SA526- (nt 514 to 539), 5′-GTCTTGTTGCcGATCgTCAAGAACAC-3′; SD880- (nt 863 to 891), 5′-CCATGGCTGATCCTGCAGGcACCAATGG-3′; E2#1 mut (nt 30 to 66), 5′-GCGTAACCGAAATCGGTTGAgttGAAACCGGTTAGTA-3′; E2#2 mut (nt 17 to 50), 5′-TATAAAACTAAGGGCGTAACCGAAATCtGTTGAA-3′; E2#3 mut (nt 7850 to 7879), 5′-TGTGTGCAAAggGTTTT GGGTTACACATTT-3′; E2#4 mut (nt 7850 to 7879), 5′-TGTGTGCAAAggGTTTTGGGTTACACATTT-3′; Sp1#1 mut (nt 17 to 50), 5′-TATAAAcagccGGGCGTAACCGAAATCGGTTGAA-3′; TATAA 65- (nt 45 to 75), 5′-GTTGAACCGAAACCGGTTAGTAgAgAcGCAG-3′; TATAA 7890- (nt 7870 to 7895), 5′-agcttcAAcACTAAACTACAATAATTCATG-3′; and Enhancer- (nt 7670 to 7715), 5′-CTATGatCCAAgtCCTTAacaACCGCTGTTctGttgcgATTTTTGG-3′.
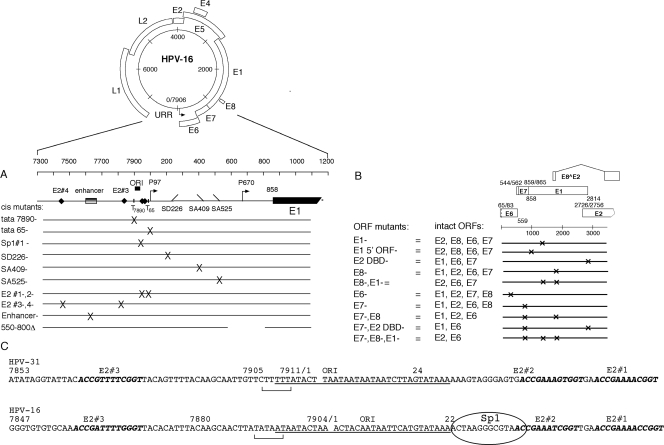
FIG. 1.
HPV-16 constructs used in this study. (A) Genomic map of the 7.9-kb HPV-16 plasmid and cis mutants used in this study. ORI, origin of replication; T, TATAA box. The P97 early and putative P670 late gene promoters are indicated by arrows. (B) Schematic of mutants introduced into the ORFs of viral genes (i.e., E1, E2, E6, E7, and E8) within the intact genome where “DBD−” refers to a mutation in the E2 DBD and “E1 5′ ORF−” to a mutation of the beginning of the E1 ORF. The remaining intact ORFs are indicated for each plasmid construct. (C) Comparison of defined origin of replication (ORI) in the related HPV-31 genome to the putative HPV-16 origin of replication, where E2 binding sites are indicated in boldface type, TATAA boxes have brackets, and the HPV-16 Sp1#1 binding site is circled.
Cell culture.
Stock cultures of SCC13 cells (an HPV-negative squamous cell carcinoma line) (35) were grown on irradiated J2 fibroblast feeder cells in E medium containing 0.5 μg/ml hydrocortisone, 0.1 nM cholera toxin, 5 μg/ml transferrin, 5 μg/ml insulin, 2 nM 3,3′-5-triodo-l-thyronine, and 5 ng/ml epidermal growth factor (18). For transient assays, SCC13 cultures were plated at a density of 2.4 × 106 cells per 60-mm plate in the absence of irradiated feeder cells.
Transcription assays.
Total RNA was harvested from transiently transfected SCC13 cell cultures 17 h posttransfection by using RNAqueous kits (Ambion, Austin, TX) prior to significant detectable plasmid replication as measured by Southern blotting. HPV-16 transcripts were detected by 5′ rapid amplification of cDNA ends (RACE) using a FirstChoice RLM-RACE kit (Ambion, Austin TX), using 10 μg of total RNA per sample and nested HPV-16 primers (see list in Fig. 4). All results were confirmed in two independent experiments, and the specific PCR products were excised, purified, and sequenced following resolution on 1.0% agarose to confirm their identity.
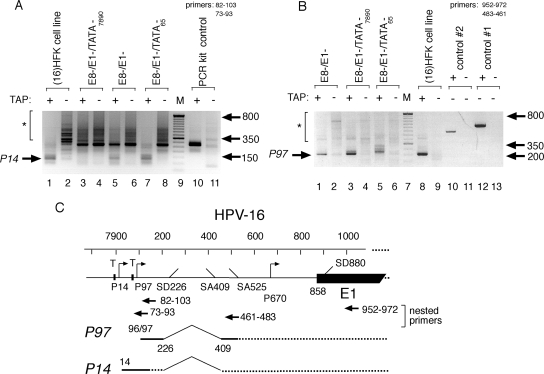
FIG. 4.
HPV-16 E1 transcripts originate from a TATAA-dependent promoter upstream of P97. (A) Specific transcripts were amplified by 5′ RACE, using nested primers as indicated, which detected early gene transcripts in tobacco acid pyrophosphatase (TAP)-treated total RNA isolated from SCC13 cells transiently transfected with HPV-16 plasmids. Mutations in HPV-16 ORFs and TATAA boxes (as illustrated in Fig. 1) are indicated. T, TATAA box; M, 50-bp molecular marker ladder (Invitrogen). P14/E1-specific amplification products are indicated by an arrow. The clonal (15) HFK cell line harbors stably replicating HPV-16 W12E plasmids. “PCR kit control” refers to a murine PCR template/primer set provided in the 5′ RACE kit (lane 10), while the asterisk indicates nonspecific products. (B) Reciprocal 5′RACE analysis for P97 transcription in the same RNA samples analyzed in panel A. Controls #1 (primer nt 952 to 972) and #2 (primer nt 483 to 461) also contain primer nt 22 to 38 and HPV-16 template (lanes 10 and 12) or no template (lanes 11 and 13). The asterisk indicates nonspecific products in lanes 1 to 8. (C) Map of early transcripts associated with the HPV-16 P14 and P97 transcription start sites and related splice donor and acceptor sites.
Plasmid amplification assays.
For transient replication assays, HPV-16-W12E DNA constructs were first cleaved from pUC vector sequences with BamHI and then religated at 5 μg/ml for 16 h. Ligated DNAs, reproducibly containing 30 to 50% of HPV sequences as single circles, were purified over plasmid purification columns (MaxiKit; Qiagen, Valencia, CA), and 3 μg was transfected into SCC13 cells with Effectene (Qiagen, Valencia, CA). Beta-galactosidase staining of control cultures transfected with pCMV-β-gal indicated transfection efficiencies of >50%. Cultures were split 1:2 at 24 h posttransfection and cultured for up to 5 days before total DNA harvesting (QIAamp DNA blood kit; Qiagen, Valencia, CA). Total DNA concentrations were determined by the optical density at 260 nm. A total of 4 μg of each DNA sample was then digested with DpnI and linearized with BamHI and XbaI before Southern blotting. Digestion of DNA samples was confirmed by visualization of ethidium bromide-stained agarose gels following electrophoresis. Replication-defective HPV-16 plasmids included as negative blot controls also served as DpnI digestion controls. DpnI-digested, whole-cell DNAs from transfected SCC13 cultures were resolved on 1.0% agarose gels, depurinated in 0.25 M HCl, and blotted directly onto positively charged nylon membranes (Hybond-XL; Amersham Biosciences Corp., Piscataway, NJ) by alkaline transfer with 0.4 N NaOH. Aliquots of linearized HPV-16 DNA (1 to 30 pg) were included as positive Southern blotting controls and to normalize band intensities (quantified by scanning densitometry) between autoradiograms. Blots were hybridized at 65°C with probes (1.5 × 106 cpm/ml of hybridization buffer) containing a representative equimolar cocktail of PCR-amplified segments of the HPV-16 W12E (nt 6226 to 3873/4471 to 6000) genome and [α-32P]dATP/dCTP labeled by random priming (HotPrime kit; GenHunter Corp., Nashville, TN).
RESULTS
HPV-16 E1 and E2 gene products are required for initial plasmid amplification.
To define the viral gene products required for initial HPV-16 replication, we first generated a series of mutations within cis elements of the transcription- and replication-competent HPV-16 W12E genome that have been shown to be required for efficient expression of the E6 and E7 early transforming genes (Fig. 1A and C). Mutations were also engineered within the ORFs encoding several of the principal early viral gene products (i.e., the E1, E2, E6, E7, and E8 ORFs) thought to be critical for the establishment and immortalization phases of the viral life cycle (Fig. 1B). For example, HPV-16 genomes containing amber mutations within the E1 and E2 ORFs are replication defective and incapable of immortalizing primary keratinocytes in long-term transfection assays (data not shown). In this study, we focused on the effect of these ORF mutations, as well as mutations in cis regulatory sequences, on initial plasmid amplification in complementation assays performed by transient transfections using an HPV-negative, immortalized squamous cell carcinoma cell line (SCC13).
As has been previously demonstrated for BPV (52) and HPV replication (5), the ORFs of the E1 and E2 genes were necessary for initial HPV-16 plasmid amplification in our assays (Fig. 2, lanes 2 and 3). The E6 product, however, appeared to have no significant effect in parallel (Fig. 2, lane 4), as has been shown for HPV-31 (50). However, mutation of the HPV-16 E8 ORF resulted in a >30-fold increase in replication (Fig. 2, lane 6), indicating that E8 is a potent inhibitor of transient replication of HVP-16, as has also been demonstrated for transient replication of HPV-31 (57). Interestingly, and in contrast to observations for a similar disruption of E7 in HPV-31 (50), mutation of the HPV-16 E7 ORF (Fig. 2, lane 5) also relieved the inhibition of plasmid amplification, resulting in a >15-fold increase in plasmid levels.
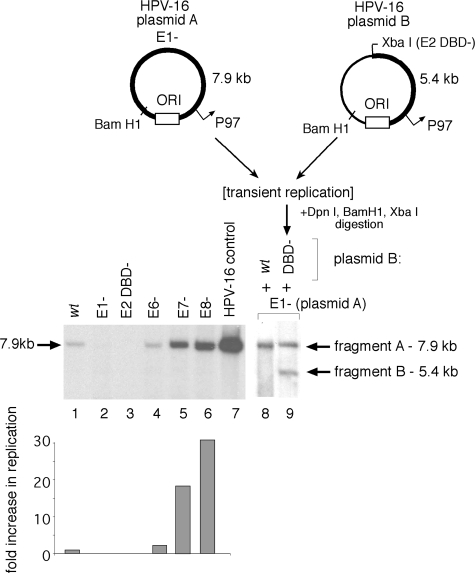
FIG. 2.
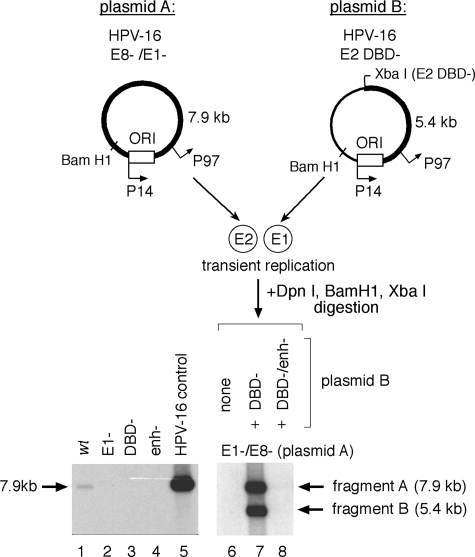
Products of the E1 and E2 viral ORFs are required for initial HPV-16 plasmid amplification. Replication of mutant HPV plasmids and complementation of HPV-16 E1 function in SCC13 cell transfections, using equimolar cotransfection of HPV-16 genomes as measured by Southern blotting, where ORI refers to the viral origin of replication. Relative replication was expressed as the increase over the normalized wt activity and quantified by scanning densitometry of detected DpnI-resistant HPV genomic fragments; shown are averages from two to three independent experiments. Fragments “A” and “B” were derived from their respective plasmid sources. Linearized HPV-16 genome (30 pg) was included as a positive blotting control.
Replication of an otherwise replication-defective (E1−) plasmid can be rescued by the addition of an E1-expressing plasmid (as shown in Fig. 2, lane 9). Cotransfection of an HPV-16 plasmid defective for E1 expression (Fig. 2, plasmid “A” or “E1−”) with one defective in E2 expression (Fig. 2, plasmid “B” or “E2 DBD−”) resulted in comparable amplifications for the two plasmid species, as a complementation model would predict (Fig. 2, compare lanes 2 and 3 to lane 9). Cotransfection of a wt plasmid with a plasmid defective for E1 expression demonstrated that the E1− construct did not inhibit plasmid amplification (Fig. 2, compare lanes 1 and 8). Since the termination linker inserted into plasmid “B” (the E2 DBD− construct) also introduces a novel XbaI site, digestion of extracted DNA with both BamHI and XbaI results in discrete fragments generated from the two input plasmids that can be resolved by electrophoresis and independently tracked by Southern blotting. This revealed equivalent amplifications of both species, as demonstrated by the comparable intensities of the 7.9-kb (fragment A) and 5.4-kb (fragment B) digestion products derived from plasmids A and B, respectively (Fig. 2, lane 9). Both plasmid constructs used in this complementation assay, however, express additional viral products capable of modulating amplification of both viral plasmid species. The E8 gene product, for example, which has been shown to be a robust inhibitor of replication and early gene expression in HPV-31 (44) and HPV-16 (Fig. 2), potentially competes with the full-length E2 during assembly of an efficient replication complex at the ori, and it also downregulates the activity of viral gene promoters via E8 binding to promoter-proximal E2 binding sites.
Major early HPV-16 promoter P97 is not required for E1-dependent plasmid amplification.
To further elucidate the individual contribution of endogenously expressed E1 and E2 gene products to initial plasmid amplification, we refined our complementation assay by applying a reductionist approach in which each plasmid species provides a critical component to viral replication. One plasmid provides E2 (but not E1), while the other provides E1 (but not E2) in the absence of the potent modulator E8, which could otherwise mask the contribution of E1 and E2 to HPV replication (Fig. 3). Furthermore, complementation in the absence of the E8 product offered an additional practical advantage by reproducibly providing a severalfold-higher baseline replication activity with which we could quantify subtle negative phenotypes from our mutant plasmid constructs that might otherwise fall below the level of detection. We mutated the E1 gene by inserting a termination linker into the ORF and the E8 gene by inserting a termination codon into the ORF which abrogated E8 expression but did not interrupt translation of E1. We transfected a construct containing an intact E2 ORF (Fig. 3, plasmid “A”) with equimolar quantities of a second construct, containing a linker insertion in the E2 DBD which disrupts both E2 and E8 expression but does not impact the E1 ORF (Fig. 3, plasmid “B”); both the E2 and the E1 expressor plasmid species must be present in the cell for plasmid amplification to take place (Fig. 3, lanes 6, 10, and 15).
FIG. 3.
Functional HPV-16 E1 can be expressed from a promoter other than P97. Replication in SCC13 cells transiently transfected with equimolar quantities of complementary HPV-16 W12 constructs was assayed by Southern blotting as previously described. Mutations in the HPV-16 ORFs and cis elements are illustrated in Fig. 1. E1−, E1 ORF; E8−, E8 ORF; T(65) and T(7890), TATAA boxes at nucleotide positions 65 and 7890; Sp1#1−, mutation of Sp1#1 site. Linearized HPV-16 genome (30 pg) was included as a positive blotting control.
Previous studies with homologous HPV-18 suggested that cis sequences flanking the E1 ORF were critical for potential E1 mRNA production, but the precise origin of these putative E1 transcripts was not precisely defined (34). Similar studies examining other mucosal HPV types suggested that E1 expression was dependent on the activity of the E6/E7 early promoter and its splicing signals (19). We introduced mutations into cis segments of the HPV-16 genomes (as depicted in Fig. 1A) which were thought to influence early gene RNA message structure and regulation to assay their effect on E1-dependent activity in our complementation system (Fig. 3). In vitro footprinting assays (T. Ishiji, T. H. Haugen, and L. P. Turek, unpublished data) had revealed multiple TATAA boxes in the HPV-16 upstream regulatory region (URR) capable of binding the TATAA binding protein (TBP) as well as two binding sites for the cellular trans-activator, Sp1, located proximal to and upstream of the P97 E6/E7 promoter (51) (Fig. 1A and C).
While mutation of the TATAA box at nt 65 (Fig. 3, lane 2) (which is required for P97 transcription), or deletion of the putative late gene promoter P670 (Fig. 3, lane 8) (13), had no effect on E1-dependent replication, disruption of the TATAA box at nt 7890 (Fig. 3, lane 16) appeared to be critical for efficient expression of functional E1. Mutation of Sp1 binding site 1 (the Sp1#1 site) (Fig. 3, lane 9), which has been shown to drive the P97 E6/E7 promoter (48), exhibited no significant effect on E1-dependent amplification. Expression of functional E1 appears to require the splice donor site at position 226 (SD226) (Fig. 3, lane 11), as well as the splice acceptor site at position 409 (SA409) (Fig. 3, lane 12), both of which are shared by transcripts originating from P97 that are critical for E7 expression. Mutation of SA526, however, had no negative effect on E1-dependent amplification (Fig. 3, lane 13). We also confirmed that mutation of the 5′ ATG of the E1 ORF (Fig. 3, lane 7) was sufficient to abrogate E1-dependent activity in these assays. The replication-defective plasmid “B” species were tested individually as negative controls (Fig. 3, lanes 18 to 25). These results show that HPV-16 E1-dependent plasmid amplification is modulated by splicing of mRNA encoding the E1 gene product which originates from a novel promoter.
HPV-16 E1 transcripts originate from a promoter other than P97.
Since it appeared that E1-dependent activity in our system is derived from transcripts that may originate from a promoter upstream of P97, i.e., functional E1 levels are still expressed from mutants lacking HPV-16 early (P97) and late (P670) gene expression (Fig. 3), we sought potential upstream start sites of E1 messages via 5′ RACE. We used nested primers capable of annealing upstream of the P97 start site (Fig. 4C), utilizing RNA isolated from cells transfected with E1 expression plasmids, and mutant constructs, serving as RNA templates (Fig. 4A).
In RNA isolated from an HPV-16-positive keratinocyte cell line, which harbors stably replicating extrachromosomal HPV plasmids (M. J. Lace, J. R. Anson, A. J. Klingelhutz, J. H. Lee, A. D. Bossler, T. H. Haugen, and L. P. Turek, submitted for publication), and in RNA from SCC13 cells transiently transfected with the E1−/E8− plasmid, a transcript with a 5′ end initiating at nt +14 was detected (Fig. 4A, lanes 1 and 5, respectively). Transcripts originating at nt +14 were not affected by a mutation of the TATAA box at nt +65, which drives P97 transcription (Fig. 4A, lane 7) but were eliminated by mutation of the TATAA box at nt 7890, located immediately upstream of the putative nt +14 start site (Fig. 4A, lane 3). Conversely, the use of 5′ RACE primers which anneal downstream of the P97 start site resulted in only P97 transcripts being detected in RNA from an HPV-immortalized, clonal human foreskin keratinocyte (HFK) cell line (Fig. 4B, lane 8). Similar results were observed for SCC13 cells transiently transfected with either wt HPV-16 or a construct harboring a mutation in the TATAA box at nt 7890 (Fig. 4B, lanes 1 and 3, respectively). However, mutation of the TATAA box at nt 65 resulted in the detection of a longer transcript, with a start site at nt 14 (Fig. 4B, lane 5). By using an RNase protection assay, a P97 transcript was detected at abundant levels compared to the essentially undetectable transcript levels originating from P14 in the same RNA sample (data not shown), indicating limited E1 transcription from P14 relative to transcription from the major early P97 E6/E7 promoter.
cis elements within the HPV-16 URR modulate E1 expression and origin function.
Mutations were then introduced into the defined binding sites for cellular and viral trans-acting factors, which were previously shown to be critical for E6/E7 gene expression from the HPV-16 P97 promoter, to determine their specific contribution to regulating the expression of functional E1 in the context of our complementation assays. Mutations introduced into the enhancer abolished replication activity in an otherwise wt plasmid (Fig. 5, lane 4) as well as E1-dependent plasmid amplification in a complementation assay (Fig. 5, compare lane 7 to lane 8), indicating that the keratinocyte-dependent, AP-2/TEF-1 enhancer, which is a critical cis element supporting P97 E6/E7 promoter activity (7, 8, 21), also appears to drive E1 expression.
FIG. 5.
The HPV-16 keratinocyte-dependent enhancer is required for E1 expression. Complementation of E1 function in transient replications in SCC13 cells as measured by Southern blotting. enh−, enhancer mutant; E1−, E1 ORF mutant; E8−, E8 ORF mutant; DBD−, mutation of the E2 DBD. Linearized HPV-16 genome (30 pg) was included as a positive blotting control.
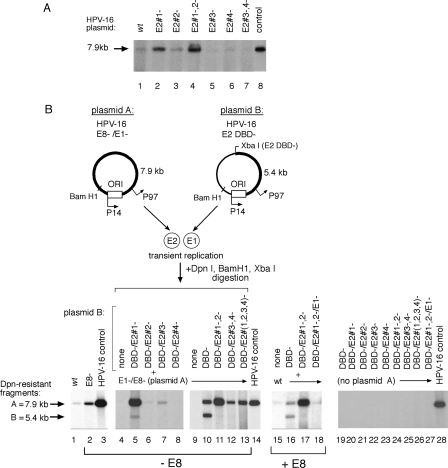
We also mutated the four defined E2 binding sites both upstream and downstream of the HPV-16 ori, with nucleotide substitutions abrogating E2 binding (data not shown), to determine their effect on viral plasmid amplification. Alteration of the proximal E2 binding site 1 (E2#1 site) resulted in a 2.5-fold increase in plasmid amplification (Fig. 6A, lane 2), while alteration of the E2#2 site had little effect (Fig. 6A, lane 3) compared to alteration of the upstream E2#3 or E2#4 site, which reduced replication levels >2-fold (Fig. 6A, lanes 5 to 7) compared to those in the wt genome (Fig. 6A, lane 1). The simultaneous alteration of all four E2 sites produced a composite effect with residual replication of this construct below wt levels (data not shown). These results, however, offered limited insight into the complex mechanism of HPV replication modulated by these cis sequences, since the experiment could not distinguish among their influences on the expression of E1, E2, and/or other viral gene products or their effects on the assembly of a replication complex at the ori in this context.
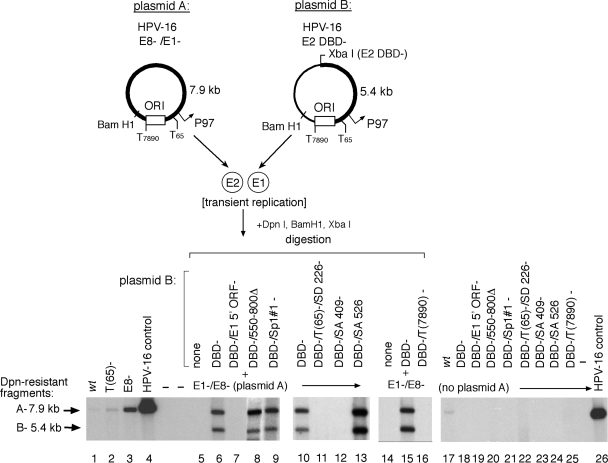
FIG. 6.
E2 binding sites in the HPV-16 promoter positively and negatively modulate E1 expression and ORI function. (A) Mutation of E2 sites both positively and negatively influence HPV-16 initial amplification in transiently transfected SCC13 cells as measured by Southern blotting. (B) Complementation of E1 function in transient replication assays using mutated HPV-16 constructs (as illustrated in Fig. 1), where E2#1−,#2− and E2#3−,#4− indicate mutations of the P97 promoter-proximal and -distal E2 binding sites, respectively. Linearized HPV-16 genome (30 pg) was included as a positive blotting control.
We then incorporated the E2 site mutations listed above into plasmid constructs that could delineate the effect of these cis sequence alterations on E1 expression or ori function in complementation assays (Fig. 6B). Since the termination linker inserted into the E2 DBD− constructs also inserts a novel XbaI site, BamHI and XbaI digestion generates discrete fragments that can be resolved by electrophoresis and tracked by Southern blotting (as demonstrated in Fig. 2). Cotransfection of the two input plasmids containing wt E2 binding sites resulted in equivalent replication of both species (Fig. 6B, lane 10) as demonstrated by the comparable intensities of the 7.9-kb (fragment A) and 5.4-kb (fragment B) digestion products, derived from plasmids A and B, respectively. In contrast, mutation of the E2#1 site in the E1-expressing plasmid resulted in a >4-fold increase in overall replication but also produced a reduction in replication of the E2#1 mutant plasmid “B” relative to levels for the E2#1 wt plasmid “A” (Fig. 6B, lane 5). This result implies that mutation of the E2#1 site resulted in increased E1 expression but that the ori function of this plasmid is impaired. Plasmid amplification in this system is not required for E1 expression, since a plasmid that can express functional E1 can support replication of other HPV plasmid species even when it cannot amplify effectively due to an impaired ori.
In contrast, mutation of either the E2#2 or the E2#3 site reduced E1 expression two- to threefold, as measured by the ability of this construct to support replication, and disrupted ori function in the plasmids containing the E2#2 or E2#3 mutation (Fig. 6B, lanes 6 to 7). Mutation of the E2#4 site, however, reduced E1 expression >8-fold but exhibited no apparent effect on ori function, since the two plasmid species (containing a wt or mutant E2#4 site) amplified at comparable, reduced levels (Fig. 6B, compare lane 10 to lane 8). The simultaneous mutation of two or more E2 binding sites resulted in a predicted composite phenotype, indicating competing positive and negative effects of the E2 sites on ori function and/or E1 expression (Fig. 6B, lanes 11 to 13). Replication-defective plasmids were run as negative controls (Fig. 6B, lanes 19 to 27).
To verify that the increase in replication observed with the “derepressed,” proximal E2 mutant constructs (Fig. 6B, lane 5, for example) was due to a concomitant increase in E1 expression alone, we introduced a mutation into the E1 ORF of the DBD−/E2#1−,#2− construct (Fig. 6B, lane 18) in appropriate complementation assays (Fig. 6B, lanes 15 to 18) where all viral gene products were expressed, including the E8 repressor. While mutation of proximal E2 binding sites (plasmid B) produced a >6-fold increase in overall replication compared to that of the wt plasmid (Fig. 6B, compare lane 16 to lane 17), the addition of a mutation in the E1 ORF returned replication to wt levels (Fig. 6B, compare lanes 15 and 18). These results demonstrated that the observed increase in replication activity was due primarily to increased E1 expression in these assays and not overexpression of another viral gene product.
Disruption of the E7 ORF enhances E1 expression.
The presence of an intact E7 ORF appears to have an inhibitory effect on HPV-16 plasmid amplification. We noted that the insertion of a termination linker in the 3′ portion of the E7 ORF resulted in increased initial plasmid amplification (Fig. 2, lane 5). To elucidate the mechanism responsible for the observed phenotype of this E7 ORF mutant, we used our complementation assay to determine if this E7 mutation indeed influenced E1 and/or E2 expression and whether it functioned in cis or in trans (Fig. 7A).
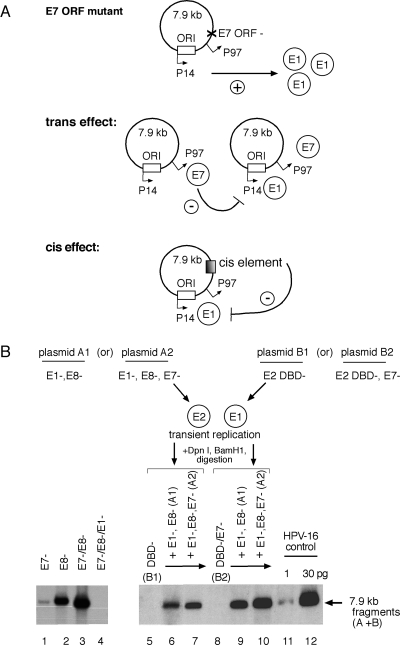
FIG. 7.
Disruption of the E7 ORF results in a cis-dependent increase in E1 expression. (A) Model illustrating E7 ORF-dependent modulation of E1 expression via a trans or cis mechanism. (B) Complementation assays monitored E1-dependent amplification in transiently transfected SCC13 cells, using HPV-16 plasmids containing E7 ORF mutants (E7−) in cis or trans by examining composite digest patterns of linearized, DpnI-resistant fragments from plasmids expressing E2 (plasmid A1 or A2) or expressing E1 (plasmid B1 or B2) via Southern blotting. Aliquots of linearized HPV-16 plasmid genomes (1 and 30 pg) were included as positive blotting controls.
Simultaneous disruption of the E8 and E7 ORFs resulted in an additive, severalfold increase in replication activity (Fig. 7B, lanes 1 to 3), indicating that the respective mutations in the E8 and E7 ORFs likely targeted different mechanisms; the E8− ORF abrogated repression of gene expression and replication by an E8 gene product, while the E7− ORF disrupted an as-yet-undefined mechanism that also inhibited plasmid amplification. We engineered E2 or E1 expressor constructs (Fig. 7B, plasmids A1 and A2 and plasmids B1 and B2, respectively), which harbored the E7− mutation to see if either influenced E1-dependent plasmid amplification. Total replication in these complementation assays was measured by linearizing both plasmid species prior to Southern blotting. Parallel experiments were also performed in which DpnI-resistant fragments derived from plasmid A or B were tracked independently by appropriate digestion to confirm that the two plasmid species were amplifying at similar levels (data not shown). Disruption of the E7 ORF in the E2-expressing plasmid A2 (i.e., the E1−,E8−,E7− construct) resulted in a <2-fold increase in amplification over that observed with the E1−,E8− construct (Fig. 7B, lanes 6 and 7), indicating that the E7 ORF mutation may influence E2 expression in cis. However, complementation assays using the E7 ORF mutation within the E1-expressing plasmid B2 (i.e., the DBD−/E7− construct) resulted in a >4-fold increase in activity (Fig. 7B, compare lanes 6 and 7 to lane 9), indicating that (in the presence of E7 expression) modulation of E1 expression in cis is sufficient to account for the bulk of the phenotype observed in these assays. When E7 expression is removed from the complementation system (Fig. 7B, lane 10), a <2-fold further increase in activity was detected, indicating a potential minor contribution of E7 to either E2 or E1 expression in trans.
These results demonstrate that mutation of the E7 ORF influences E1-dependent initial plasmid amplification primarily in cis, presumably via an increase in E1 expression. There also appears to be a minor effect on E2 expression when the E7 ORF mutation is introduced into the E2-expressing plasmid, but whether this is due to a cis effect on E2 expression or a lack of E2-dependent repression of E1 expression in trans cannot be determined.
DISCUSSION
In this study, we have characterized critical features of the E1 cistron of HPV-16 during initial genome amplification (also referred to as transient replication), a critical first step that precedes and determines the establishment of persistent infection. We have developed a complementation assay in which HPV replication depends on the expression of HPV genes from mutant HPV plasmid genomes that fail to amplify individually. This approach readily distinguishes between cis and trans effects of different mutations that could not be discerned by mutagenesis of a single genome. Furthermore, because both plasmids replicate and can be distinguished by restriction patterns on Southern blots, the assay identifies any cis mutations that alter viral ori function of the modified HPV genome. Finally, in contrast to complementation assays based on the overexpression of viral proteins from vector plasmids or retroviruses that have been used in previous studies, the HPV gene products are expressed in response to specific cellular and viral factors at native, potentially limiting levels, thus permitting the study of their regulation.
HPV-16 E1 is expressed from a spliced mRNA initiating at a previously undetected promoter, P14.
High-level transcripts of the papillomavirus early gene region originate at a major early gene promoter located just 5′ of the E6 ORF, designated P97, in HPV-16 (7, 21). Surprisingly, our mutational analysis first excluded the P97 promoter as necessary for E1 production during the initial amplification. Mutations in the TATAA box at nt 65 or in Sp1#1 at nt 28 immediately 5′ of the P97 promoter inactivated P97-initiated transcripts yet had no effect on E1 production in the complementation assay. Similarly, the deletion of the P670 late promoter had no effect on E1 expression. In contrast, the mutation of a putative TATAA box at nt 7890 profoundly reduced E1-dependent plasmid amplification. The TATAA(7890) sequence and the TATAA(65) motif were found to bind the TBP subunit of TFIID equally well in DNase I footprinting assays (Ishiji et al., unpublished). The 5′ RACE results identified a transcription start site at nt 14, located 24 nt downstream of the TATAA(7890), that was inactivated by the TATAA(7890) mutation. We designated this novel TATAA-dependent promoter P14.
P14 appears to be the principal source of E1 transcripts during the initial amplification of HPV-16 DNA. We cannot exclude the possibility that it serves to initiate mRNAs for additional viral gene products at this or later stages of infection. We also do not know whether E1 transcripts originate from P14, P97, or possibly other promoters described for other mucosal HPVs (30, 42) or for HPV-16 (37, 47) during the plasmid maintenance or vegetative amplification stages of infection.
The products of the HPV-16 E1, E2, E6, and E7 early genes are thought to be transcribed from alternatively spliced RNA transcripts detected in HPV-immortalized human keratinocytes that harbor extrachromosomal plasmid genomes as shown by PCR-based methodologies (see reference 55 for a review). We found that a mutation in either the RNA splice donor site SD226 or the splice acceptor site SA409, which are localized within the E6 ORF and used for efficient E7 expression, also effectively abolished the E1 function in HPV-16. A previous study had noted that the disruption of analogous splicing sites in HPV-31 disrupted transient replication yet did not identify which of the viral gene(s), E6, E7, E1 or E2, was affected (19). Taken together, our results indicate that the principal source of E1 expression during the initial amplification of viral DNA is an mRNA initiated at P14 and spliced from SD226 to SA409.
Regulation of HPV-16 E1 expression by cellular and viral factors.
Papillomavirus particles do not contain transcriptionally active proteins; viral transcription is initially activated by cellular transcription factors. The control regions of mucosal HPVs encompass, in addition to the ori sequences, keratinocyte-dependent enhancers that contain large numbers of repeated binding sites for cellular factors (7, 8, 12, 18, 21). Mutations or variations in many of these motifs also affect viral genome replication (17, 19, 40, 54). We found that enhancer function was required for E1 production. Mutations in HPV-16 genomes that abolish the binding of the cellular transcription factors TEF-1 (21) and AP-2 to enhancer motifs abrogated transcription activation function of the enhancer and failed to express E1 in complementation assays. It will be of interest to test whether other transcription factors implicated by mutation analysis in the regulation of replication of an extrachromosomal HPV ori, for example, YY1 (24), AP-1 (18), TBP (14), and CDP (28), contribute to E1 expression, ori replication, or both, since the complementation assay can distinguish between these effects.
We found that E1 expression also is regulated by viral factors. The conserved binding sites in the HPV early promoter for the viral E2 protein also modulate initial amplification of HPV-16, as has been shown for HPV-31 (19, 45). In contrast, we were able to distinguish the effect of abrogating E2 binding to these sequences on E1 expression from the contribution these sites make to ori function in initial plasmid amplification. The distal E2 site (E2#4), for example, supported E1 expression but had no effect on ori function, while the E2 sites flanking the ori influenced both ori function (as one would predict for critical components of a minimal ori structure) and E1 expression from the overlapping P14 promoter. However, mutation of the E2#1 site, which is immediately upstream of the major early promoter P97, resulted in both increased E1 expression and impairment of ori function. E2 binding to the E2#1 site could simply support recruitment of E1 and proper assembly of a functional replisome, as shown for the E2#2 and E2#3 sites flanking the ori (this study). Loss of E2 binding to the E2#1 site has also been shown to derepress transcription from the major early promoter P97 (T. H. Haugen, unpublished data), which could serve as an additional source of E1 mRNA in its derepressed state. Mutation of the E2#1 site could derepress the P14 promoter as well, resulting in increased E1 levels.
It is perplexing that the steady-state levels of P14 transcripts are low compared to those of P97 despite the fact that P14 and P97 are modulated by the same trans-acting cellular and viral factors. It also is unclear why the highly active P97 promoter does not generate E1-expressing mRNA(s) during initial amplification, because precursor RNAs originating at P14 and P97 undergo similar splicing. Differences in translation initiation of spliced or unspliced transcripts capable of expressing E1 could contribute to this effect. Posttranscriptional regulation of HPV early gene expression by leaky ribosomal scanning of polycistronic messages has been proposed for HPV-16 E6 and E7 expression (41), by distance-dependent translation reinitiation, as suggested for the synthesis of HPV-18 E7 (49), or by ribosomal shunting, as recently suggested for modulating the expression of HPV-18 E1 polycistronic messages (34). The cis components of these polycistronic RNA messages which preferentially direct efficient E1 translation, however, remain to be defined.
While we cannot completely rule out subtle contributions to transient viral replication from other viral gene products, such as E5 and E4, it appears that E1 and E2 expression, in the context of the intact genome, is necessary and sufficient to drive initial HPV amplification. The presence of an intact HPV-16 E7 ORF, however, appeared to inhibit transient replication in our assays. E7 expression has been shown to support differentiation-dependent replication of HPV-18 (27) and HPV-31 (25) by altering the interaction of E7 with members of the E2F family of regulatory factors, which have been shown to modulate plasmid replication (26). However, this positive effect may take place only in persistently infected keratinocytes that undergo differentiation in the suprabasal layers to support DNA replication. Alternatively, the loss of E7 expression could also influence E1 expression by disrupting the interaction of E7 with other cellular transcription factors, such as IRF-1 (1, 31), or TBP (26). These models could support the role of E7-dependent modulation of E1 expression and/or replication in trans.
However, since in our assays we observed a significant increase in E1-dependent replication only when the E7 ORF is mutated in cis, we favor a mechanism where E1 expression is modulated primarily in cis rather than in trans by an E7 gene product. Our observations support a mechanism of alternate initiation of E1 versus E7 translation from polycistronic HPV mRNA, similar to a model previously proposed (49, 56). The authors demonstrated that alternative translation of E7 versus E6 from bicistronic mRNA is tightly regulated not only by mRNA splicing but also by its 5′ cap structure in relation to the translation initiation site. Disruption of the E6 ORF in cis, by linker insertion, enhanced the expression of the downstream E7 gene via enhanced translation initiation. Similarly, our data support such a mechanism, where translation initiation of the downstream E1 gene could be enhanced when the upstream E7 ORF is disrupted, as in our E7− construct. The E7 ORF mutant acting predominantly in cis explains the increase in E1-dependent plasmid amplification in our complementation assays, as opposed to the E7 protein acting as an inhibitor of E1 expression or replication in trans (Fig. 7).
Complex mechanisms control E1-dependent HPV-16 plasmid amplification.
Upon HPV infection, the production of mRNA encoding viral replication factors precedes the expression of other early gene products (29). HPV infection progresses from an establishment phase to a maintenance phase where a stable, low copy number viral load is supported by differential modes of replication which are thought to be tightly linked to E1 levels (reviewed in reference 16). Since a stable viral load is a feature of HPV persistence, tightly regulated expression of limiting levels of E1 is critical in the maintenance phase of the viral life cycle.
This study demonstrates that modulation of E1 expression results in altered initial plasmid amplification. Several potential checkpoints for negative regulation of E1 expression are apparent. The proximal E2 binding sites mediate E1 repression, presumably via the E2 gene product(s). Our results suggest that, similar to HPV-31 (44, 57), HPV-16 also may encode an E8^E2 repressor product, since a translation termination mutation in the short E8 ORF acted in trans to increase the amplification of both complementing plasmids. Furthermore, the overlap between the P14 promoter and the replication origin represents a setting where E1 overexpression could form a negative feedback loop via competition between replication and transcription factors. Under these conditions, binding to the ori by increased E1 levels could sterically inhibit further E1 expression from the overlapping P14 promoter, as previously suggested for E1-dependent inhibition of BPV early gene transcription (38). Furthermore, at the posttranscriptional level, the expression of E7 may lead to inefficient initiation of E1 translation.
Differential regulation and temporal expression of E1 would contribute to an effective viral strategy of limited initial amplification of the viral genome, allowing immortalization of the host cell to progress while evading cellular immune defenses potentially triggered by elevated viral plasmid replication or high levels of viral gene products. Furthermore, coordinate regulation of early viral gene products during early immortalization events would maintain a critical stoichiometry of limiting concentrations of these viral gene products. Such a strategy would be predicted to contribute to maintaining a stable viral copy number as viral infection is established.
In summary, this study has used a novel approach to analyze the structure and regulation of the HPV-16 E1 cistron in E1-dependent initial viral amplification. Both E1 expression and ori function have been shown to be tightly regulated by conserved cis elements within the HPV URR interacting with both cellular and viral factors. Similar complementation assays will permit further insight not only into the structure of E1 mRNA transcripts and modulation of E1 expression but into defining the role of other viral gene products in the initial amplification of the extrachromosomal viral genome.
Acknowledgments
We thank Aloysius Klingelhutz and Alice Fulton for critical reviews of the manuscript.
This work was supported by the Department of Veterans Affairs.
Footnotes
Published ahead of print on 2 August 2008.
REFERENCES
- 1.Arany, I., K. J. Grattendick, W. E. Whitehead, I. A. Ember, and S. K. Tyring. 2003. A functional interferon regulatory factor-1 (IRF-1)-binding site in the upstream regulatory region (URR) of human papillomavirus type 16. Virology 310280-286. [DOI] [PubMed] [Google Scholar]
- 2.Auster, A. S., and L. Joshua-Tor. 2004. The DNA-binding domain of human papillomavirus type 18 E1: crystal structure, dimerization, and DNA binding. J. Biol. Chem. 2793733-3742. [DOI] [PubMed] [Google Scholar]
- 3.Castella, S., G. Bingham, and C. M. Sanders. 2006. Common determinants in DNA melting and helicase-catalysed DNA unwinding by papillomavirus replication protein E1. Nucleic Acids Res. 343008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, G., and A. Stenlund. 2002. Sequential and ordered assembly of E1 initiator complexes on the papillomavirus origin of DNA replication generates progressive structural changes related to melting. Mol. Cell. Biol. 227712-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang, C. M., M. Ustav, A. Stenlund, T. F. Ho, T. R. Broker, and L. T. Chow. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. USA 895799-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clower, R. V., J. C. Fisk, and T. Melendy. 2006. Papillomavirus E1 protein binds to and stimulates human topoisomerase I. J. Virol. 801584-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cripe, T. P., A. Alderborn, R. D. Anderson, S. Parkkinen, P. Bergman, T. H. Haugen, U. Pettersson, and L. P. Turek. 1990. Transcriptional activation of the human papillomavirus-16 P97 promoter by an 88-nucleotide enhancer containing distinct cell-dependent and AP-1-responsive modules. New Biol. 2450-463. [PubMed] [Google Scholar]
- 8.Cripe, T. P., T. H. Haugen, J. P. Turk, F. Tabatabai, P. G. Schmid, M. Dürst, L. Gissmann, A. Roman, and L. P. Turek. 1987. Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 63745-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doorbar, J., A. Parton, K. Hartley, L. Banks, T. Crook, M. Stanley, and L. Crawford. 1990. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology 178254-262. [DOI] [PubMed] [Google Scholar]
- 10.Dürst, M., L. Gissmann, H. Ikenberg, and H. zur Hausen. 1983. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl. Acad. Sci. USA 803812-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frattini, M. G., and L. A. Laimins. 1994. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc. Natl. Acad. Sci. USA 9112398-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gloss, B., H. U. Bernard, K. Seedorf, and G. Klock. 1987. The upstream regulatory region of the human papilloma virus-16 contains an E2 protein-independent enhancer which is specific for cervical carcinoma cells and regulated by glucocorticoid hormones. EMBO J. 63735-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grassmann, K., B. Rapp, H. Maschek, K. U. Petry, and T. Iftner. 1996. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J. Virol. 702339-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartley, K. A., and K. A. Alexander. 2002. Human TATA binding protein inhibits human papillomavirus type 11 DNA replication by antagonizing E1-E2 protein complex formation on the viral origin of replication. J. Virol. 765014-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hebner, C. M., and L. A. Laimins. 2006. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev. Med. Virol. 1683-97. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann, R., B. Hirt, V. Bechtold, P. Beard, and K. Raj. 2006. Different modes of human papillomavirus DNA replication during maintenance. J. Virol. 804431-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubert, W. G. 2005. Variant upstream regulatory region sequences differentially regulate human papillomavirus type 16 DNA replication throughout the viral life cycle. J. Virol. 795914-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubert, W. G., T. Kanaya, and L. A. Laimins. 1999. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5′ of the minimal origin. J. Virol. 731835-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubert, W. G., and L. A. Laimins. 2002. Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression. J. Virol. 762263-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes, F. J., and M. A. Romanos. 1993. E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res. 215817-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishiji, T., M. J. Lace, S. Parkkinen, R. D. Anderson, T. H. Haugen, T. P. Cripe, J. H. Xiao, I. Davidson, P. Chambon, and L. P. Turek. 1992. Transcriptional enhancer factor (TEF)-1 and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in human keratinocytes and cervical carcinoma cells. EMBO J. 112271-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeon, S., B. L. Allen-Hoffmann, and P. F. Lambert. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 692989-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert, P. F. 1991. Papillomavirus DNA replication. J. Virol. 653417-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, K. Y., T. R. Broker, and L. T. Chow. 1998. Transcription factor YY1 represses cell-free replication from human papillomavirus origins. J. Virol. 724911-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longworth, M. S., R. Wilson, and L. A. Laimins. 2005. HPV31 E7 facilitates replication by activating E2F2 transcription through its interaction with HDACs. EMBO J. 241821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maldonado, E., M. E. Cabrejos, L. Banks, and J. E. Allende. 2002. Human papillomavirus-16 E7 protein inhibits the DNA interaction of the TATA binding transcription factor. J. Cell Biochem. 85663-669. [DOI] [PubMed] [Google Scholar]
- 27.McLaughlin-Drubin, M. E., J. L. Bromberg-White, and C. Meyers. 2005. The role of the human papillomavirus type 18 E7 oncoprotein during the complete viral life cycle. Virology 33861-68. [DOI] [PubMed] [Google Scholar]
- 28.Narahari, J., J. C. Fisk, T. Melendy, and A. Roman. 2006. Interactions of the cellular CCAAT displacement protein and human papillomavirus E2 protein with the viral origin of replication can regulate DNA replication. Virology 350302-311. [DOI] [PubMed] [Google Scholar]
- 29.Ozbun, M. A. 2002. Human papillomavirus type 31b infection of human keratinocytes and the onset of early transcription. J. Virol. 7611291-11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozbun, M. A., and C. Meyers. 1998. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J. Virol. 722715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park, J.-S., E.-J. Kim, H.-J. Kwon, E.-S. Hwang, S.-E. Namkoong, and S.-J. Um. 2000. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein: implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J. Biol. Chem. 2756764-6769. [DOI] [PubMed] [Google Scholar]
- 32.Psyrri, A., and D. DiMaio. 2008. Human papillomavirus in cervical and head-and-neck cancer. Nat. Clin. Pract. Oncol. 524-31. [DOI] [PubMed] [Google Scholar]
- 33.Remm, M., R. Brain, and J. R. Jenkins. 1992. The E2 binding sites determine the efficiency of replication for the origin of human papillomavirus type 18. Nucleic Acids Res. 206015-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Remm, M., A. Remm, and M. Ustav. 1999. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J. Virol. 733062-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rheinwald, J. G., and M. A. Beckett. 1981. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 411657-1663. [PubMed] [Google Scholar]
- 36.Rohlfs, M., S. Winkenbach, S. Meyer, T. Rupp, and M. Dürst. 1991. Viral transcription in human keratinocyte cell lines immortalized by human papillomavirus type-16. Virology 183331-342. [DOI] [PubMed] [Google Scholar]
- 37.Rosenstierne, M. W., J. Vinther, C. N. Hansen, M. Prydsoe, and B. Norrild. 2003. Identification and characterization of a cluster of transcription start sites located in the E6 ORF of human papillomavirus type 16. J. Gen. Virol. 842909-2920. [DOI] [PubMed] [Google Scholar]
- 38.Sandler, A. B., S. B. Vande Pol, and B. A. Spalholz. 1993. Repression of bovine papillomavirus type 1 transcription by the E1 replication protein. J. Virol. 675079-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sedman, J., and A. Stenlund. 1995. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 146218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sen, E., S. Alam, and C. Meyers. 2004. Genetic and biochemical analysis of cis regulatory elements within the keratinocyte enhancer region of the human papillomavirus type 31 upstream regulatory region during different stages of the viral life cycle. J. Virol. 78612-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stacey, S. N., D. Jordan, A. J. Williamson, M. Brown, J. H. Coote, and J. R. Arrand. 2000. Leaky scanning is the predominant mechanism for translation of human papillomavirus type 16 E7 oncoprotein from E6/E7 bicistronic mRNA. J. Virol. 747284-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steger, G., M. Rehtanz, and C. Schnabel. 2001. Identification of a promoter in position 56 within the long control region of human papillomavirus type 18. Arch. Virol. 1462069-2084. [DOI] [PubMed] [Google Scholar]
- 43.Stenlund, A. 2003. E1 initiator DNA binding specificity is unmasked by selective inhibition of non-specific DNA binding. EMBO J. 22954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stubenrauch, F., M. Hummel, T. Iftner, and L. A. Laimins. 2000. The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 741178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stubenrauch, F., H. B. Lim, and L. A. Laimins. 1998. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J. Virol. 721071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swindle, C. S., and J. A. Engler. 1998. Association of the human papillomavirus type 11 E1 protein with histone H1. J. Virol. 721994-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, S. H., C. C. Baker, W. Stunkel, and H. U. Bernard. 2003. A transcriptional initiator overlaps with a conserved YY1 binding site in the long control region of human papillomavirus type 16. Virology 305486-501. [DOI] [PubMed] [Google Scholar]
- 48.Tan, S. H., B. Gloss, and H. U. Bernard. 1992. During negative regulation of the human papillomavirus-16 E6 promoter, the viral E2 protein can displace Sp1 from a proximal promoter element. Nucleic Acids Res. 20251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang, S., M. Tao, J. P. McCoy, Jr., and Z.-M. Zheng. 2006. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 804249-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas, J. T., W. G. Hubert, M. N. Ruesch, and L. A. Laimins. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. USA 968449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ushikai, M., M. J. Lace, Y. Yamakawa, M. Kono, J. Anson, T. Ishiji, S. Parkkinen, N. Wicker, M.-E. Valentine, I. Davidson, L. P. Turek, and T. H. Haugen. 1994. trans activation by the full-length E2 proteins of human papillomavirus type 16 and bovine papillomavirus type 1 in vitro and in vivo: cooperation with activation domains of cellular transcription factors. J. Virol. 686655-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, V. G., M. West, K. Woytek, and D. Rangasamy. 2002. Papillomavirus E1 proteins: form, function, and features. Virus Genes 24275-290. [DOI] [PubMed] [Google Scholar]
- 54.Wooldridge, T. R., and L. A. Laimins. 2008. Regulation of human papillomavirus type 31 gene expression during the differentiation-dependent life cycle through histone modifications and transcription factor binding. Virology 374371-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng, Z. M., and C. C. Baker. 2006. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front. Biosci. 112286-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, Z. M., M. Tao, K. Yamanegi, S. Bodaghi, and W. Xiao. 2004. Splicing of a cap-proximal human papillomavirus 16 E6E7 intron promotes E7 expression, but can be restrained by distance of the intron from its RNA 5′ cap. J. Mol. Biol. 3371091-1108. [DOI] [PubMed] [Google Scholar]
- 57.Zobel, T., T. Iftner, and F. Stubenrauch. 2003. The papillomavirus E8-E2C protein represses DNA replication from extrachromosomal origins. Mol. Cell. Biol. 238352-8362. [DOI] [PMC free article] [PubMed] [Google Scholar]