Abstract
The human cytomegalovirus (HCMV) UL82-encoded tegument protein pp71 has recently been shown to activate viral immediate-early (IE) gene expression by neutralizing a cellular intrinsic immune defense instituted by the ND10 protein hDaxx. Pp71 localizes to ND10 upon infection and induces the degradation of hDaxx. Here, we report the successful generation of a recombinant HCMV expressing enhanced yellow fluorescent protein (EYFP) fused to the N terminus of pp71. Intriguingly, insertion of the EYFP-UL82 coding sequence into the HCMV AD169 genome gave rise to a recombinant virus, termed AD169/EYFP-pp71, that replicates to significantly higher titers than wild-type AD169. In particular, we noticed strongly increased protein levels of pp71 after AD169/EYFP-pp71 inoculation. Although the high abundance of pp71 resulted in augmented packaging of the tegument protein into viral particles, no increased hDaxx degradation was detectable upon AD169/EYFP-pp71 infection. In contrast, further investigation revealed a significantly enhanced viral DNA replication compared to wild-type AD169. Thus, we hypothesize that an as-yet-unidentified function of pp71 contributes to the enhanced infectivity of AD169/EYFP-pp71. This assumption is additionally supported by the observation that increased early and late gene expression after AD169/EYFP-pp71 infection occurs independent of elevated IE protein levels. Finally, immunofluorescence analyses confirmed that hDaxx determines the ND10-localization of pp71 upon infection, since pp71 exhibited a nucleolar distribution in the absence of hDaxx. Taken together, we generated a recombinant HCMV that constitutes a useful tool not only to dissect the in vivo dynamics of pp71 subnuclear localization more precisely but also to explore new features of this viral transactivator.
Human cytomegalovirus (HCMV), a member of the beta-subgroup of herpesviruses, generally causes asymptomatic infections in immunocompetent individuals. However, HCMV is of considerable clinical importance for immunocompromised persons such as organ transplant recipients, tumor and AIDS patients, or prenatally infected newborns (31).
Similar to those of other herpesviruses, the HCMV open reading frames (ORFs) are expressed in a temporally regulated cascade consisting of three sequential phases, termed immediate-early (IE), early, and late (10, 29, 45). IE genes are transcribed first and encode critical regulatory proteins that function in part by controlling expression of viral early and late genes. The most abundantly expressed IE genes (UL122/123) are transcribed from the major IE locus. This gene region is controlled by a strong and complex regulatory element known as the major IE enhancer-promoter (MIEP), which directs the production of the IE1 and IE2 protein that are critical for initiating the lytic replication cycle of the virus (14, 27). While the MIEP itself is responsive to many cellular signaling pathways, it additionally is activated by structural protein components of the incoming virion. Specifically, the tegument protein pp71 (the gene product of ORF UL82) is capable of activating IE gene expression from the MIEP in synergism with other tegument proteins such as pUL35 and pUL69 (25, 38, 41, 46, 47). Virus mutants with UL82 deleted enter productive replication inefficiently after infection of cells at a low multiplicity of infection (MOI), demonstrating that, although the MIEP is intrinsically very active, stimulation of IE transcription by pp71 is important for initiation of the viral gene expression program (5, 6). Thus, pp71 is thought to play a significant role in initiating the lytic life cycle of HCMV.
The exact mechanism by which pp71 regulates IE gene expression has been the subject of intensive investigation (5, 25, 28). Recently, a model has been proposed in which pp71 has to inactivate a cellular intrinsic immune defense instituted by the promiscuous transcriptional repressor hDaxx in order to facilitate viral gene expression. The hDaxx protein constitutes a cellular repressor that associates via its SUMO-interacting motif with SUMOylated DNA-binding transcription factors and recruits histone deacetylases (HDACs) to targeted promoters to silence transcription (19, 39). In addition, hDaxx has been identified as a component of a cellular subnuclear structure known as the nuclear domain 10 (ND10; alternatively termed PML-NBs, for promyelocytic leukemia nuclear bodies, or PODs, for PML oncogenic domains). ND10 are defined by the presence of the major ND10 constituents PML, hDaxx, and Sp100, which assemble in distinct foci within the interchromosomal space of the nucleus. Accumulating evidence suggests that ND10 are part of an antiviral defense mechanism of the cell, with individual components of this structure contributing independently to the silencing of viral IE gene expression (32, 34, 43, 44, 48).
Based on the ability of pp71 to directly interact with hDaxx, the HCMV tegument protein has been found to localize at ND10 accumulations immediately upon infection (18, 22). Although initial studies implied that pp71 and hDaxx associate at ND10 to cooperatively activate the MIEP (18), it is now clear from multiple subsequent studies that hDaxx silences the MIEP and that pp71 relieves this repression (32, 34, 43, 44, 48). Consistent with this assumption, overexpression of the cellular restriction factor hDaxx abolishes HCMV infection, while downregulation of hDaxx by usage of small interfering RNA technology, on the contrary, results in increased gene expression and virus replication (7, 32, 44, 48). The hDaxx-mediated repression of viral IE gene expression correlates with changes of the chromatin structure around the MIEP since knockdown (kd) of hDaxx results in the loss of transcriptionally repressive and the gain of transcriptionally active chromatin at the MIEP (48). This regulation appears to involve HDACs, since treatment of infected cells with HDAC inhibitors relieves the repression of viral IE gene expression (34, 48).
In order to successfully antagonize hDaxx-mediated intrinsic immune defense, tegument-delivered pp71 induces the degradation of hDaxx at the start of a lytic infection (34, 44), which has been postulated to occur in a proteasome-dependent (34), ubiquitin-independent manner (20). Consistent with this, HCMV inefficiently enters productive infection in the absence of pp71 (5, 6), unless hDaxx protein levels are depleted prior to infection, thus annihilating the impaired growth phenotype associated with a pp71-deficient mutant (7, 32, 44). Failure of pp71 to overcome hDaxx repression blocks viral IE gene expression and may promote the establishment of latent HCMV infections (35). However, whether hDaxx is a major factor contributing to the control of HCMV latency is still controversially discussed (15, 35).
In order to elucidate the dynamics of pp71-hDaxx/ND10 interaction more precisely, since this represents a critical step in initiating the lytic replication cycle of HCMV, we generated a recombinant virus expressing an enhanced yellow fluorescent protein (EYFP)-tagged version of pp71 to visualize the viral transactivator protein during infection of living cells. Surprisingly, this recombinant virus, designated AD169/EYFP-pp71, exhibited an enhanced-growth phenotype compared to wild-type (wt) HCMV based on strongly elevated pp71 protein levels present during infection, as well as in the tegument of AD169/EYFP-pp71-derived viral particles. Moreover, we provide evidence that an as-yet-unidentified function of pp71 contributes to the augmented replication efficiency of AD169/EYFP-pp71. Finally, by performing immunofluorescence experiments, we could not only confirm the necessity of hDaxx for the dot-like accumulation of pp71 at ND10 directly upon infection but, in addition, demonstrate that pp71 localizes to nucleoli in the absence of the transcriptional repressor. Thus, AD169/EYFP-pp71 represents a promising tool not only to further analyze pp71 subnuclear distribution during infection but also to identify new functions of this important viral transactivator.
MATERIALS AND METHODS
Cells and viruses.
Primary human foreskin fibroblast (HFF) cells were maintained in Eagle minimal essential medium (Gibco/BRL, Eggenstein, Germany) supplemented with 5% fetal calf serum. HFFs with a stable small interfering RNA-mediated kd of PML (siPML2 cells [43]) or hDaxx (siDaxx1 cells [44]) were cultured in Dulbecco minimal essential medium (Gibco/BRL) supplemented with 10% fetal calf serum and 5 μg of puromycin/ml. siPML2-mCh-PML HFFs illustrating PML-kd cells in which PML expression was reconstituted (reintroduction of PML isoform VI fused to the monomeric red autofluorescent protein mCherry) were maintained in Dulbecco minimal essential medium supplemented with 10% fetal calf serum, along with 2.5 μg of puromycin/ml and 1 μg of blasticidin/ml (43).
Viral stocks of wt AD169 and the recombinant AD169/EYFP-pp71 virus were prepared and titrated via IE1p72 fluorescence as described previously (26). Briefly, HFFs were infected with various dilutions of virus stocks. After 24 h of incubation, cells were fixed and stained with monoclonal antibody (MAb) p63-27 directed against IE1p72 (3). Subsequently, the number of positive cells was determined and used to calculate virus titers indicated in IE protein-forming units (IEU). Alternatively, viral stocks were titrated based on genomic equivalents imported into cells upon infection. For this, cells were infected with various amounts of viral inocula, followed by the preparation of genomic DNA at 14 h postinfection (hpi) using a DNeasy tissue kit (Qiagen, Hilden, Germany). Thereafter, viral input DNA was quantified by HCMV-specific real-time PCR exactly as described elsewhere (26, 43). In subsequent experiments, viral inocula were normalized for infection to yield equivalent amounts of viral genomes per cell.
For the detection of IE1-positive cells, HFFs (3.0 × 105 cells per well of a six-well plate) were infected with either wt AD169 or recombinant AD169/EYFP-pp71 with the indicated amount of virus. The cells were fixed 24 hpi, followed by immunostaining with MAb p63-27. The number of positive cells/well was then determined by epifluorescence microscopy. Each experiment was performed in triplicate, and standard deviations were calculated.
Purification of viral particles.
Purification of wt HCMV and recombinant AD169/EYFP-pp71 virus particles from the supernatant of infected HFF cells was performed when the culture displayed a distinctive cytopathic effect. Supernatants were collected and cleared from cell debris by centrifugation for 10 min at 2,000 rpm. After sedimentation of virus particles by ultracentrifugation (23,000 rpm, 10°C, 70 min; Beckman SW27 rotor; Beckman Coulter, Krefeld, Germany) and resuspension in 0.04 M sodium phosphate buffer (pH 7.4), the viral particle pellets were loaded onto a glycerol-tartrate gradient (2) and again subjected to ultracentrifugation (23,000 rpm, 10°C, 70 min; Beckman SW27 rotor). The virion fractions obtained from the gradient were diluted in 0.04 M sodium phosphate buffer (pH 7.4), pelleted by centrifugation, and resuspended using the same buffer.
Oligonucleotides and plasmids.
Oligonucleotides used for cloning or generation of recombinant bacterial artificial chromosomes (BACs) were obtained from Biomers (Ulm, Germany); the sequences are listed in Table 1. The eukaryotic expression plasmid EYFP-pp71 (alternatively termed pHM1532), expressing EYFP in fusion to the N terminus of pp71, was generated by PCR amplification of the pp71 coding sequence (ORF UL82) using the primers pp71-5-korr and pp71-3′nt1680korr-aa559, along with vector pCB6-pp71 (16) as a template. The resulting PCR product was cleaved with BamHI and XhoI and inserted into the BamHI-SalI-digested vector pEYFP-C1 (BD Biosciences Clontech, Heidelberg, Germany). Plasmid pHM2218, which after cleavage with SpeI and AseI gives rise to a linear recombination cassette comprising 500 nucleotides of the 3′ end of ORF UL83, a kanamycin resistance gene, as well as the EYFP-UL82 (EYFP-pp71) fusion sequence, was constructed as follows. In a first step, the indicated UL83 sequence was created by PCR amplification with primers 5′ and 3′ UL83-pHM1638, as well as BAC pHB15 as a template. Thereafter, the UL83 sequence was integrated into plasmid pHM1638 harboring the kanamycin resistance cassette by cleavage with AseI and NotI. The resulting plasmid pHM2215 was further modified by subcloning of the EYFP-pp71 coding sequence from plasmid pHM1532 via PacI and SpeI to finally obtain plasmid pHM2218.
TABLE 1.
Oligonucleotides used for the construction of plasmids and recombinant BACs
| Oligonucleotide | Sequence |
|---|---|
| pp71-5-korr | CATAGGATCCGCATGTCTCAGGCATCGTCCTCGC |
| pp71-3′nt1680korr-aa559 | CATACTCGAGCTAGATGCGGGGTCGACTGC |
| 5′UL83-pHM1638 | CATAGGCGCGCCCTTAAGGCCGAGTCCACCGTCG |
| 3′UL83-pHM1638 | CATAGCGGCCGCAGGGTCAGGGGATGCGCAAAGG |
| 3′Seq-pp71-rev-ATG | GCTTCGGCCTCGCTGATCG |
| 3′kontrBACpp71up | CGGGCCATGCATCGCCTCGACGCC |
BAC mutagenesis.
For construction of the pHB15/EYFP-pp71 BAC (reviewed in reference 1), a linear recombination cassette was utilized to insert the EYFP-pp71 fusion sequence via homologous recombination into the AD169-derived BAC pHB15 (17). The linear recombination fragment necessary for mutagenesis comprising a kanamycin resistance marker, along with 5′ and 3′ genomic sequences (UL83 and UL82 sequences, respectively) required for homologous recombination, was excised from plasmid pHM2218 by AseI and PacI treatment and used for electroporation of competent Escherichia coli strain BW25113 harboring the parental BAC pHB15 (9, 17). The E. coli cells also contained plasmid pKD46, which inducibly expressed the recombination enzymes gam, bet, and exo of bacteriophage λ (9). Bacterial clones harboring recombinant HCMV BACs were selected at 37°C on agar plates containing 30 μg of kanamycin/ml and 30 μg of chloramphenicol/ml. After a first characterization of the recombinant HCMV BACs via restriction enzyme digestion and Southern blotting, the kanamycin resistance marker was removed by site-specific recombination in E. coli (8). For this, the selected bacteria were transformed with Flp recombinase expression plasmid pCP-20 and incubated on chloramphenicol- and ampicillin-positive agar plates at 30°C for 1 to 2 days. Finally, a few colonies were selected for chloramphenicol resistance at 43°C for 1 day, resulting in the loss of plasmid pCP-20, which harbors a temperature-sensitive origin of replication. The integrity of the resulting pHB15/EYFP-pp71 BAC was finally confirmed by restriction enzyme digestion, Southern blot, and PCR analyses with the primers 3′Seq-pp71-rev-ATG and 3′kontrBACpp71up.
Viral nucleic acid isolation and analysis.
Small amounts of BAC DNA were isolated by a standard alkaline lysis procedure from 5-ml cultures grown overnight (36). Large-scale BAC preparations were obtained from 500-ml cultures grown overnight by using the Nucleobond AX 100 kit (Machery Nagel, Düren, Germany) according to the manufacturer's instructions. BAC DNA was characterized by restriction digestion using the enzyme XhoI, followed by a separation of the resulting DNA fragments via electrophoresis at 30 V overnight using a 0.7% agarose gel. For Southern blot analysis, the DNA was transferred onto a nylon membrane (Biodyne B transfer membrane; PALL, Portsmouth, United Kingdom) and probed with biotinylated DNA spanning the complete pp71 coding sequence, according to the instructions of the manufacturer (NEBlot Phototope kit; Biolabs, Frankfurt, Germany).
Reconstitution of recombinant CMVs.
HFF cells seeded into six-well dishes at a density of 3.5 × 105 cells/well were cotransfected with 1 μg of the purified BAC DNA, along with 0.5 μg of the pp71 expression plasmid pCB6-pp71 and 0.5 μg of a vector coding for the Cre recombinase using SuperFECT transfection reagent (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. The HCMV tegument protein pp71 was coexpressed since it is known to enhance the infectivity of viral DNA (4); coexpression of Cre recombinase is necessary in order to remove the BAC cassette and the chloramphenicol resistance gene, which are located between two loxP sites within the HCMV BAC pHB15 (17). One week after transfection, the cells were transferred into 25-cm2 flasks and incubated until plaques appeared.
Antibodies, indirect immunofluorescence, and Western blot analysis.
The polyclonal antisera raised against exon 5 of IE2 (referred to as anti-pHM178), the tegument protein pp71 (SA2932), or pUL84 of HCMV were generated by immunizing rabbits with the respective prokaryotically expressed proteins. MAb-UL44 BS 510 (UL44), MAb-MCP 28-4 (MCP; UL86), MAb-pp65 65-33 (pp65), and MAb-pp71 CMV355 (pp71; UL82) were kindly provided by B. Plachter (University of Mainz, Germany), W. Britt (University of Birmingham, AL), and T. Shenk (Princeton University, Princeton, NJ). MAb p63-27 for detection of IE1 and MAb 41-18 directed against pp28 (UL99) were described elsewhere (3, 37). For detection of endogenous hDaxx either the mouse MAb MCA2143 (Serotec, Oxford, United Kingdom) or a rabbit MAb from Epitomics (Burlingame, CA) were used. Endogenous PML was detected by using the polyclonal antiserum H-238 obtained from Santa Cruz (Santa Cruz, CA). The MAb AC-15, which recognizes β-actin, was purchased from Sigma-Aldrich (Deisenhofen, Germany). Anti-mouse, as well as anti-rabbit, horseradish peroxidase-conjugated secondary antibodies were obtained from Dianova (Hamburg, Germany), while Alexa 488- and Alexa 555-conjugated secondary antibodies were purchased from Molecular Probes.
For indirect immunofluorescence analysis, 3 × 105 HFF cells were grown on coverslips for HCMV infection. The conditions for subsequent fixation and immunodetection of viral and cellular proteins were as described previously (43).
For Western blotting, extracts from infected cells were prepared in sodium dodecyl sulfate (SDS) loading buffer, separated on SDS-containing 8 to 15% polyacrylamide gels, and transferred to nitrocellulose membranes. Western blotting and chemiluminescence detection were performed as previously described (24). Quantification of Western blot results was performed by densitometry using the software program AIDA image analyzer (version 3.10.039; Raytest, Straubenhardt, Germany).
Time-lapse microscopy of live cells.
For live cell imaging, cells were seeded into two-well, chambered coverglass units with coverslip quality glass bottoms (Lab-Tek; Nunc, Karlsruhe, Germany) at a density of 2.0 × 105cells per well for infection with AD169/EYFP-pp71. The next day, time-lapse microscopy was performed by using a Zeiss Axiovert-135 inverted fluorescence microscope (Zeiss, Jena, Germany) with a cooled Spot color digital camera (RT KE/SE, 7.0 monochrome w/o IR; Diagnostic Instruments, Sterling Heights, MI). Camera image acquisition was controlled by MetaVue software (Meta-Imaging series; Universal Imaging Corp., Downington, PA). Single images or timed image sequences obtained with a ×63 objective lens were exported as BMP files from the MetaVue software. Individual frames were prepared for presentation using Adobe Photoshop.
RESULTS
Construction and verification of a recombinant HCMV expressing an EYFP-pp71 fusion protein.
In order to examine the in vivo dynamics of pp71 accumulation with regard to an ND10 association directly upon infection, we generated a recombinant HCMV expressing EYFP fused to the N terminus of pp71. For engineering of this recombinant virus, designated AD169/EYFP-pp71, the BAC technology was used (for a review, see reference (1). To this end, a linear recombination cassette containing the EYFP-pp71 (UL82) coding sequence was electroporated into E. coli harboring the AD169-derived BAC pHB15, along with plasmid pKD46 encoding the recombination functions of phage λ (9) (for a detailed description, see Materials and Methods). The recombination fragment also contained a kanamycin gene as a resistance marker, along with the UL82 and UL83 sequences required for homologous recombination. After integration of the recombination fragment into the HCMV genome, the kanamycin cassette was excised at its flanking frt sites by Flp recombinase (8). Positive recombinant pHB15/EYFP-pp71 BACs were subsequently identified by PCR, using the primers 3′Seq-pp71-rev-ATG and 3′kontrBACpp71up, which were located within ORFs UL82 and UL83, respectively (Fig. 1D). As expected, insertion of the EYFP coding sequence with a size of 0.7 kb resulted in amplification of 1-kb fragments in the case of recombinant pHB15/EYFP-pp71 BACs (Fig. 1D, lanes 2 to 5) compared to only 0.3-kb PCR products of wt pHB15 (Fig. 1D, lane 1). Furthermore, to ascertain that the recombination did not alter the BAC genome apart from the desired EYFP-pp71 introduction, the structural integrity of the obtained pHB15/EYFP-pp71 BACs was investigated by restriction enzyme cleavage with XhoI (Fig. 1B). The digestion of four independent pHB15/EYFP-pp71 genomes yielded complex patterns identical to those of pHB15 wt DNA, with the following predicted exception: consistent with the insertion of the EYFP coding sequence, recombinant pHB15/EYFP-pp71 BACs exhibited a slower migrating fragment (10.2 kb; Fig. 1B, lanes 2 to 5, indicated by a triangle) than wt pHB15 genomes (9.5 kb; Fig. 1B, lane 1, indicated by a circle). The correct integration of the EYFP-pp71 recombination cassette could finally be confirmed by Southern blot hybridization experiments (Fig. 1C) using the XhoI-digested BAC DNAs, together with a biotinylated probe spanning the entire ORF UL82 as schematically indicated in the image of Fig. 1A. Successful recombination again resulted in the detection of the slower-migrating fragment in case of pHB15/EYFP-pp71 DNA (Fig. 1C, lanes 2 to 5) compared to the wt pHB15 BAC (Fig. 1C, lane 1). Thus, we could verify that recombination events took place at the correct site of the HCMV genome.
FIG. 1.
Construction of a recombinant HCMV expressing an EYFP-tagged version of the viral tegument protein pp71. (A) Schematic representation of the HCMV genome region in which the EYFP-pp71 coding sequence was inserted. The scheme shows the XhoI restriction sites along with the probe used for Southern blotting, as well as the primer pair of the PCR in relation to the relevant genomic segment of HCMV. (B) To ensure the integrity of the obtained recombinant BACs, XhoI digestion was performed, followed by the separation of fragments by agarose gel electrophoresis. Integration of the EYFP coding sequence resulted in slower migration of a 10.2-kb fragment (indicated by a triangle) compared to the corresponding 9.5-kb fragment of wt pHB15 (indicated by a circle). (C) Southern blot analysis of XhoI digests of BAC DNAs as indicated, using a biotinylated probe spanning the complete UL82 gene sequence. (D) For identification of positive recombinant HCMV genomes, PCR analysis of bacterial clones harboring the indicated BACs was performed using oligonucleotides specifically binding within ORF UL82 and UL83, respectively. Lanes: 1, wt BAC pHB15; 2 to 5, recombinant pHB15/EYFP-pp71 BACs.
Reconstitution and growth analysis of the AD169/EYFP-pp71 virus.
Next, infectious virus was reconstituted by transfecting HFFs with purified pHB15/EYFP-pp71 BAC DNA. One week after transfection, the cells were transferred into 25-cm2 flasks and cultured for about 2 weeks until plaques appeared. Thereafter, viral progeny virus was recovered and used for infection of fresh HFF cultures to prepare AD169/EYFP-pp71 virus stocks, which were titrated via IE1p72 staining (see Materials and Methods).
To investigate the growth kinetics of the recombinant EYFP-pp71-expressing virus, HFF cells were infected in parallel with wt AD169 and AD169/EYFP-pp71 at an MOI of 1 (Fig. 2A) or 0.1 (Fig. 2B), respectively. The viral inocula used for generating multistep growth curves were standardized for comparable IE1 expression levels. Aliquots of the supernatants of infected cells were collected at various times after infection as indicated (3, 5, 7, 9, and 13 days postinfection) to determine the titer of infectious progeny virus via IE1 fluorescence (3). Surprisingly, infection with the recombinant AD169/EYFP-pp71 virus yielded peak titers that were clearly above those obtained for wt AD169. While in terms of a low MOI (MOI = 0.1; Fig. 2B) an enhanced viral particle release of AD169/EYFP-pp71 compared to wt AD169 was detectable starting at day 9 after addition of the viruses, the effect was even more pronounced at a high MOI (MOI = 1; Fig. 2A). Under the latter conditions, AD169/EYFP-pp71 replicated to titers approximately 1 log beyond that of wt AD169 already at the end of the first round of replication (5 dpi) and thus showed an enhanced replication throughout the entire course of the infection experiment (Fig. 2A). This effect could be observed for four independently reconstituted EYFP-pp71 viruses derived from individual pHB15/EYFP-pp71 BAC clones (data not shown), which argues against the selection of a virus with an unanticipated mutation that enhances infectivity. Consequently, insertion of the EYFP-pp71 coding sequence into the viral genome affects replication of AD169/EYFP-pp71 in an unexpected manner, since it results in a significantly increased production of infectious recombinant viruses compared to wt AD169.
FIG. 2.
Multistep growth curves of HCMV AD169 and AD169/EYFP-pp71. HFFs were infected in parallel with AD169 and AD169/EYFP-pp71 using 1 (A) or 0.1 (B) infectious units/cell. For this, infection settings with viral inocula of AD169 (⧫) and AD169/EYFP-pp71 (▪) identical for IE1 expression were chosen (viral inocula were titrated via IE1p72 fluorescence; see Materials and Methods). At the indicated time points after infection (3, 5, 7, 9, and 13 days postinfection), cell culture supernatants were harvested for quantification of virus titers via IE1 staining. The standard deviations as indicated for each data point on the graphs are derived from three independent experiments.
Analysis of the protein expression kinetics of AD169/EYFP-pp71 reveals strongly enhanced pp71 protein levels especially at late stages of the replication cycle.
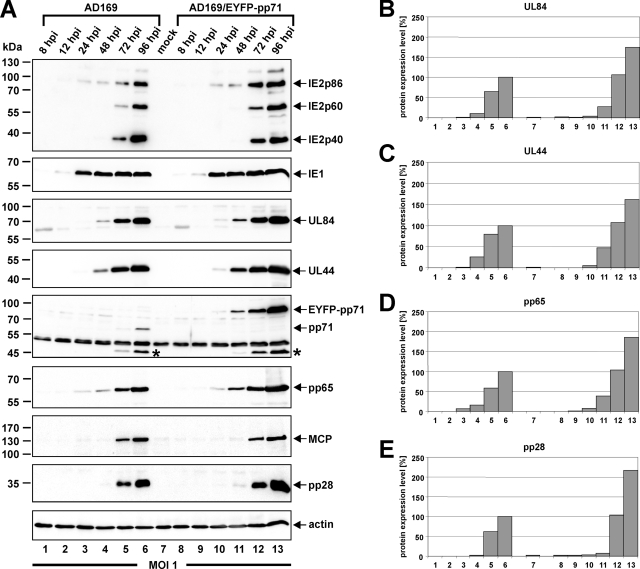
In order to examine the observed augmented replication efficacy of AD169/EYFP-pp71 more precisely and find out possible explanations for this unexpected phenotype, the protein accumulation kinetics during one round of AD169/EYFP-pp71 replication should be monitored. Again, HFF cells were infected in parallel with wt AD169 and the recombinant AD169/EYFP-pp71 virus at an MOI of 1 using infection conditions that were normalized for comparable IE1 expression. Thereafter, total cell lysates were prepared at various times postinfection (8, 12, 24, 48, 72, and 96 hpi) to analyze the expression pattern of viral IE (IE1; IE2 isoforms IE2p86, IE2p60, and IE2p40), early-late (UL44, UL84, pp71, and pp65), and true-late proteins (MCP and pp28) via Western blotting (Fig. 3). Detection of pp71 revealed the presence of a higher-molecular-mass variant in the case of the recombinant AD169/EYFP-pp71 virus compared to pp71 of wt AD169, thus confirming the correct expression of the chimeric EYFP-pp71 fusion protein (Fig. 3A, fifth panel, compare wt pp71 of lane 6 to recombinant EYFP-pp71 of lane 13). Intriguingly, however, EYFP-pp71 was not only expressed in greater abundance than wt pp71 but was also detectable at earlier time points in the replication cycle (Fig. 3A, fifth panel, compare lanes 3 to 6 to lanes 10 to 13). In addition, expression of early and late proteins was similarly affected after infection at a high MOI with AD169/EYFP-pp71 since an increased accumulation of UL84, UL44, pp65, or pp28 was verified (Fig. 3). This could account for the increased release of infectious recombinant viruses, as observed in growth kinetics (Fig. 2). According to the usage of viral inocula adjusted to result in an equivalent IE1 expression, no significant difference in the protein levels of IE1 was noticeable between AD169 and the recombinant AD169/EYFP-pp71 virus throughout the entire replication cycle (Fig. 3, compare upper panels). In contrast, however, while IE2p86 protein levels did not differ between AD169 and AD169/EYP-pp71 at IE and early times postinfection (Fig. 3A, compare lanes 1 to 4 and lanes 8 to 11), increased levels of IE2 isoforms IE2p86, IE2p60, and IE2p40 were detected during the late phase of AD169/EYP-pp71 replication (Fig. 3A, compare lanes 5 and 6 and lanes 12 and 13). Therefore, as a consequence of the experimental setup, only effects of augmented EYFP-pp71 expression that affect stages of viral replication after the onset of IE transcription such as enhanced early-late expression kinetics of AD169/EYFP-pp71 could be identified.
FIG. 3.
Comparison of viral protein accumulation after infection with AD169/EYFP-pp71 and wt AD169. HFFs were infected in parallel with AD169 and AD169/EYFP-pp71 at an MOI of 1 to monitor the protein expression kinetics of both viruses via Western blotting. At different times after virus inoculation (8, 12, 24, 48, 72, and 96 hpi) cell lysates were prepared, subjected to SDS-polyacrylamide gel electrophoresis, and subsequently transferred to a nitrocellulose membrane. (A) Via immunoblotting, the protein levels of the viral IE proteins IE1 and IE2 (isoforms IE2p86, IE2p60, and IE2p40) and the early-late proteins UL84, UL44, pp71 (UL82), and pp65 (UL83), as well as the true late proteins MCP (UL86) and pp28 (UL99), were analyzed. Cellular β-actin levels served as controls for equal protein loading. Arrows indicate the localization of the detected proteins. Asterisks mark the localization of a 45-kDa protein that is detected in infected cells by anti-pp71 antiserum SA2932. (B to E) Densitometric quantification of UL84 (B), UL44 (C), pp65 (D), and pp28 (E) protein amounts standardized on the respective β-actin protein levels. The amount of protein detected at 96 hpi with wt AD169 was set to 100%.
Enhanced initiation of IE gene expression after infection at a low MOI with AD169/EYFP-pp71.
Since pp71 is known to play a crucial role in initiating the replicative cycle of HCMV by stimulating IE transcription (5), we sought to determine whether IE gene expression is also positively influenced after infection with the recombinant AD169/EYFP-pp71 virus. In order to investigate this, infection experiments had to be performed with viral inocula of AD169 and AD169/EYFP-pp71 that were standardized for a comparable uptake of viral DNA. To initially determine the amount of genomic equivalents of AD169 and AD169/EYFP-pp71 that are required to obtain equal numbers of IE1-positive cells (identical IE-forming units), real-time PCR was performed for quantification of the viral DNA content 14 h after infection of HFFs with 104 IEU of each virus. As depicted in Fig. 4A, on average only about half of the viral DNA load was necessary in case of AD169/EYFP-pp71 to initiate IE gene expression as efficiently as the wt AD169 virus.
FIG. 4.
Analysis of IE gene expression after infection with AD169/EYFP-pp71 compared to wt AD169. The recombinant virus AD169/EYFP-pp71 initiates productive infection more efficiently than wt AD169 due to an augmented IE gene expression. (A and B) HCMV-specific real-time PCR was performed to quantify viral genomes after infection of HFF cells with AD169 and AD169/EYFP-pp71 using viral inocula that were either adjusted to comparable IE1 expression levels (A; 104 IEU of AD169 and AD169/EYFP-pp71) or an equivalent viral DNA uptake (B; 0.1 infectious units/cell of AD169). Viral and cellular DNA was extracted from infected cells at 14 hpi. Experiments were performed in triplicate, and the standard deviations are indicated. (C) Dissection of IE gene expression after infection with AD169 and AD169/EYFP-pp71 using viral inocula normalized for identical viral uptake (see Materials and Methods). HFFs (grown on coverslips) were infected with 5, 10, or 20 IEU per well of AD169, as well as equivalent genome copies of AD169/EYFP-pp71. At 24 hpi the number of IE1-positive cells was assessed via indirect immunofluorescence analysis. Error bars illustrate standard deviations derived from three independent experiments.
The outcome of this experiment already suggested an enhanced IE transcription after infection with the recombinant AD169/EYFP-pp71 virus. To finally obtain conclusive evidence for this assumption, the infection experiment was repeated, this time using viral inocula of AD169 and AD169/EYFP-pp71 containing equivalent genome copies of both viruses, as verified by quantitative real-time PCR (Fig. 4B). At 24 hpi, HFFs infected with equal genome copies of AD169 (20 IEU/well) and AD169/EYFP-pp71 were fixed and analyzed for IE expression via IE1 staining. As defined by indirect immunofluorescence analysis, under these infection conditions, indeed, approximately six times more IE1-positive cells were detectable after infection with AD169/EYFP-pp71 compared to wt AD169 (Fig. 4C). Furthermore, for AD169/EYFP-pp71 we noticed a nonlinear correlation between viral input DNA and cells that initiate IE1 gene expression as detected after infection with 5, 10, or 20 IEU/well (see Fig. 4C). In summary, the results indicate that AD169/EYFP-pp71 enters productive replication more effectively than wt AD169 due to more cells initiating IE gene expression, which is observable at least after infection at a low MOI with the recombinant virus.
Increased amounts of pp71 are packaged into AD169/EYFP-pp71 particles.
The existence of strongly augmented EYFP-pp71 protein levels at late stages of AD169/EYFP-pp71 infection, along with the ability of the recombinant virus to initiate IE gene expression more efficiently than wt AD169, gave rise to the idea that more of the tegument protein pp71 could be packaged into AD169/EYFP-pp71 particles. To investigate whether the tegument composition of AD169/EYFP-pp71 was altered in comparison to wt AD169 with regard to an increased incorporation of EYFP-pp71, virions were purified from the supernatant of infected cell cultures by glycerol-tartrate gradient centrifugation (2) and thereafter subjected to Western blotting. In order to evaluate an equal number of viral particles for wt and the recombinant virus, the protein amount of the virion preparations was adjusted to a comparable content of the major capsid protein (MCP) (Fig. 5A, compare lane 1 illustrating wt AD169 with lane 2 constituting AD169/EYFP-pp71). As expected, in the case of purified viral particles of AD169/EYFP-pp71 a markedly increased amount of the regulatory protein pp71 could be detected which, due to the higher molecular mass, was clearly identified as the chimeric EYFP-pp71 fusion protein (Fig. 5D; quantification of the results is shown in Fig. 5E).
FIG. 5.
Evaluation of the tegument protein composition of wt AD169 and AD169/EYFP-pp71 virions. Virus particles from the supernatant of HFFs infected with AD169 or AD169/EYFP-pp71 were separated from cell fragments via low-speed centrifugation. Thereafter, the particles were purified by sedimentation in a glycerol-tartrate gradient, thus resulting in virion fractions, which were used for immunoblot analyses. Virion components were detected using MAbs against MCP (UL86) (A), pp65 (UL83) (B), and pp28 (UL99) (C) or polyclonal antiserum SA2932 directed against pp71 (UL82) (D). Lane 1 contains virion preparations of AD169-infected cells, and lane 2 contains virion preparations of AD169/EYFP-pp71-infected cells. (E) Densitometric quantification of pp71 (UL82), pp65 (UL83), and pp28 (UL99) protein amounts standardized on the respective ΜCP (UL86) protein levels. MCP protein levels were set to 100%.
On the contrary, no significant difference in the abundance of other tegument-associated components such as pp28 (Fig. 5C) could be observed between wt AD169 (Fig. 5, lane 1) and the recombinant EYFP-pp71 virus (Fig. 5, lane 2), except for the integration of slightly reduced amounts of the major structural protein pp65 into virions of AD169/EYFP-pp71 (Fig. 5B; see also Fig. 5E). This is most probably a consequence of spatial limitations resulting from the additional incorporation of EYFP-pp71. Nevertheless, the identification of augmented quantities of pp71 in isolated infectious particles of AD169/EYFP-pp71 correlates with the observation that more cells initiate IE gene expression after infection with the recombinant virus (Fig. 4C).
HDaxx degradation is not significantly increased after AD169/EYFP-pp71 infection.
The experiments described above suggested that elevated amounts of tegument-based pp71 could be responsible for the capability of AD169/EYFP-pp71 to initiate lytic replication more efficiently than wt AD169. Only recently, the molecular mechanism through which pp71 activates HCMV IE gene expression and thereby promotes productive infection has been discovered. As initially identified by Saffert and coworkers, pp71 is able to neutralize the intrinsic immune defense instituted by the cellular hDaxx protein, which normally silences HCMV IE gene expression by recruiting HDACs to the MIEP (34, 48). Directly after infection, the HCMV tegument protein pp71 relieves this cellular repression mechanism by inducing the proteasomal degradation of hDaxx (34, 44). In light of these facts, the question arose as to whether hDaxx degradation could be enhanced upon AD169/EYFP-pp71 inoculation due to an increased tegument concentration of pp71, thus leading to a higher number of infected cells. To resolve this question, HFF cells were infected in parallel with AD169 and AD169/EYFP-pp71 using viral inocula standardized for comparable viral uptake (Fig. 6A). Thereafter, cell lysates were prepared in 60-min intervals and assayed for hDaxx protein levels via immunoblotting. The time course analysis demonstrated a distinct reduction in hDaxx abundance after infection with both viruses, but no complete hDaxx degradation was noticeable (Fig. 6; see also Fig. 8B and 9B). Moreover, the experiment did not reveal any detectable difference in hDaxx protein levels between wt AD169- and AD169/EYFP-pp71-infected cells, thus indicating that hDaxx degradation is not considerably enhanced after infection with the recombinant virus (Fig. 6, compare AD169 lanes to EYFP-pp71 lanes). One possible explanation for this could be the use of high viral loads, which might lead to a saturation of hDaxx degradation. However, treatment of cells with reduced viral doses completely annihilated the detection of hDaxx degradation (data not shown). On the contrary, infection with AD169/EYFP-pp71 resulted in significantly enhanced IE1 protein expression as assessed by Western blot analysis, demonstrating an enhancing effect of EYFP-pp71 on viral replication in the absence of increased hDaxx degradation (Fig. 6B).
FIG. 6.
Examination of hDaxx protein levels in AD169-infected compared to AD169/EYFP-pp71-infected fibroblasts. HFFs were either not infected (mock) or were infected with wt HCMV AD169 and AD169/EYFP-pp71, respectively, using viral inocula that were normalized to an equivalent DNA content as verified by HCMV-specific quantitative real-time PCR (A). (B and C) At the indicated times postinfection, cell lysates were harvested and analyzed by Western blotting for the expression of IE1p72 (B) or hDaxx (C) and β-actin (C), respectively. The latter was included as an internal loading control.
FIG. 8.
Analysis of pp71 subnuclear localization during infection with AD169 and AD169/EYFP-pp71. Pp71 precisely colocalizes with PML and hDaxx in ND10. (A) siPML2/mCh-PML cells live infected with AD169/EYFP-pp71 (17 infectious units/cell) were examined for EYFP-pp71 subnuclear distribution with respect to a mCh-PML association directly after infection via live-cell imaging. Already at 20 min postinoculation pp71 exhibits perfect colocalization with ND10. (B) Time course analysis for monitoring of pp71 subnuclear localization with regard to an hDaxx/ND10 association. HFFs infected with AD169/EYFP-pp71 (17 infectious units/cell) were harvested at the indicated times postinfection for indirect immunofluorescence analysis using the mouse MAb MC2143 for the detection of hDaxx. Pp71 was visible through its EYFP fusion moiety. DAPI served for the staining of cell nuclei. (C) Comparison of pp71 subnuclear localization via antibody detection of the tegument protein upon infection (MOI = 10) with either AD169/EYFP-pp71 (a to h) or wt AD169 (i to q) using the mouse MAb CMV355.
FIG. 9.
Effect of PML or hDaxx ablation on the subnuclear distribution of pp71. (A) Confirmation of the generation of PML- and hDaxx-kd cells via Western blotting. The upper panel shows detection of the various SUMOylated, as well as non-SUMOylated, isoforms of PML in vector (lane 1)-, siC- (lane 2), siDaxx1 (lane 3)-, and siPML2 (lane 4)-transduced cells using the polyclonal antiserum H-238. The major PML isoform is indicated by an arrow. In the middle panel, hDaxx protein levels were detected with an anti-hDaxx rabbit monoclonal antiserum from Epitomics. In the lower panel, β-actin detection served as a loading control. (B) Immunofluorescence analysis of pp71 subnuclear localization in PML-kd cells. In the absence of PML, pp71 still colocalizes with rearranged hDaxx accumulations. PML-negative siPML2 cells grown on coverslips were either mock infected (a to d) or infected with wt AD169 (i to m) or AD169/EYFP-pp71 (e to h and n to q) at an MOI of 10. Mock-infected and infected cells harvested at the indicated times postinfection (2 and 4 hpi) were fixed and costained for the ND10 component hDaxx using the mouse MAb MAC2143. Pp71 was visualized either by staining with the MAb CMV355 in the case of wt AD169 or by the EYFP autofluorescence in the case of AD169/EYFP-pp71. Cell nuclei were counterstained with DAPI. (C) In the absence of hDaxx, pp71 exhibits a mainly nucleolar localization pattern. HDaxx-negative siDaxx1 cells grown on coverslips were infected with wt AD169 (g to i) or AD169/EYFP-pp71 (a to f and h to m) at an MOI of 17. At the indicated time points after infection (1 to 4 hpi) the cells were fixed and analyzed for pp71 subnuclear distribution. DAPI was used for counterstaining of cell nuclei, and pp71 was visible either through antibody detection with the MAb CMV355 (AD169) or its EYFP fluorescent tag (AD169/EYFP-pp71).
Enhanced viral DNA replication after infection with AD169/EYFP-pp71.
Due to the fact that we observed an increased late protein accumulation after infection with AD169/EYFP-pp71 under conditions where no enhanced IE gene expression was present (Fig. 3), we speculated that additional, as-yet-unidentified, intrinsic functions of pp71 could contribute to the augmented replication efficiency of AD169/EYFP-pp71. Since the expression of late genes is intimately linked to viral DNA replication, we decided to analyze whether viral DNA accumulation is affected in the case of AD169/EYFP-pp71 infection. In order to compare the DNA accumulation kinetics of AD169/EYFP-pp71 to that of wt AD169, TaqMan PCR was performed for quantification of viral genome equivalents during one round of HCMV replication. For this, HFFs were infected in parallel with AD169 and AD169/EYFP-pp71 at MOIs of 0.1 and 1 (as determined for AD169; Fig. 7), with viral inocula normalized for identical DNA content. As depicted in Fig. 7, comparable copy numbers of viral genomes were detected for both viruses up to at least 24 hpi due to the adjusted viral inputs used. In parallel to the induction of viral DNA replication starting at approximately 24 to 48 hpi, however, AD169/EYFP-pp71 exhibited a significantly increased accumulation of viral progeny DNA (Fig. 7). As a consequence of this, approximately 3 times (MOI = 1; Fig. 7A) to 10 times (MOI = 0.1; Fig. 7B) more genomic equivalents of AD169/EYFP-pp71 compared to wt AD169 could be measured at the end of one replication round. Hence, increased amounts of EYFP-pp71 as present after infection with AD169/EYFP-pp71 positively influence viral DNA replication, especially under low-MOI conditions. This further strengthens the assumption that, in addition to the well-characterized role of pp71 in initiating lytic replication of HCMV by inducing IE gene expression, pp71 also possesses as-yet-uncharacterized functions during later stages of infection.
FIG. 7.
Characterization of the DNA accumulation kinetics of AD169/EYFP-pp71 compared to wt AD169. (A and B) Real-time PCR for quantification of the accumulation kinetics of viral progeny DNA during one round of wt AD169 and AD169/EYFP-pp71 replication. HFFs were infected in parallel with AD169 (⧫) and AD169/EYFP-pp71 (▪) at an MOI of 1 (as calculated for AD169 [A]) or 0.1 (as calculated for AD169 [B]) using viral inocula that were normalized to yield identical intracellular genome copies at 14 h after infection with both viruses (see Materials and Methods). At the respective time points after infection (8, 12, 24, 48, 72, and 96 hpi), DNA was extracted and subjected to HCMV-specific real-time PCR to monitor viral DNA replication during one replicative cycle of AD169/EYFP-pp71 compared to wt AD169. Standard deviations resulting from three independent experiments are indicated.
Tegument-delivered EYFP-pp71 colocalizes with ND10 immediately upon infection in live cells.
In a next step, we tested whether the expression of fluorescently labeled pp71 from the generated recombinant virus facilitates the monitoring of pp71 subnuclear localization during infection with AD169/EYFP-pp71. Fixed-cell studies have demonstrated that tegument delivered pp71 is transported to the nucleus of infected cells, where it can be found in direct association with the nuclear substructure ND10 at IE times of infection (18, 22). To further examine the ND10 localization of pp71 in live-infected cells, siPML2/mCh-PML HFFs stably expressing the major ND10 constituent PML (isoform VI) in fusion with the red autofluorescent protein mCherry (mCh-PML) were utilized for AD169/EYFP-pp71 infection. In former experiments, these primary human fibroblasts only expressing PML isoform VI have been confirmed as a useful tool to visualize ND10 (43). For live-cell imaging, siPML2/mCh-PML cells seeded onto coverslip glass chamber units were infected with a high infectious dose of AD169/EYFP-pp71 (MOI = 17) in order to gain detectable EYFP-pp71 signals. Subsequent live cell microscopy revealed that already as early as 20 min after addition of the virus, pp71 could be found within the nucleus of infected cells, where it accumulated in dot-like structures that precisely colocalized with PML and thus ND10 domains (Fig. 8A). This finding indicates that identification and targeting of incoming viral components by the antiviral nuclear substructure ND10 is a very fast process that occurs immediately after virus entry. Unfortunately, due to photobleaching of the faint EYFP-pp71 signals we were not able to monitor the dynamics of pp71-ND10 interaction of one single cell over an extended time period using conventional epifluorescence equipment. To investigate the fate of pp71-ND10 accumulations in more detail, indirect immunofluorescence analysis was performed using fixed HFFs similarly infected with AD169/EYFP-pp71 at an MOI of 17. The examination of individual cells in this time course study revealed that pp71 remained associated with ND10 as a perfect colocalization with the transcriptional repressor hDaxx up to 3 hpi (Fig. 8Ba to m). However, in addition to the initial formation of distinct pp71 foci in association with hDaxx (1 hpi; Fig. 8Bb), the viral tegument protein also exhibited an additional diffuse distribution pattern throughout the nucleus as infection progressed (2 and 3 hpi; Fig. 8Bf and k). This observation potentially reflects a saturation of ND10 accumulation capacity resulting from the large number of viral particles used for infection that were still able to load cells with pp71. Starting at about 4 hpi the IE1-induced dispersal of ND10 was detectable (Fig. 8Bp), resulting in the redistribution of pp71 and hDaxx to apparently different subnuclear sites, since no association of both diffusely localized proteins could be identified any longer (Fig. 8Bn to q). Furthermore, a direct comparison after infection with wt AD169 and AD169/EYFP-pp71 using a pp71-specific MAb did not reveal differences in the subcellular localization of pp71 expressed by the two viruses (Fig. 8C). Taken together, since the behavior of pp71 to associate with hDaxx in ND10 could be reproduced with the recombinant virus, it was concluded from these data that fusion to EYFP has no influence on pp71 subcellular localization, thus indicating that AD169/EYFP-pp71 serves as a valuable tool to visualize the dynamics of pp71 nuclear accumulation during infection.
HDaxx, but not PML, is required for ND10 localization of pp71 at the start of a lytic infection.
Taking advantage of the recombinant EYFP-pp71-expressing virus, the determinants of pp71 subnuclear accumulation at ND10 should be clarified in further detail. In previous studies a specific interaction of pp71 with the ND10 factor hDaxx has been identified in yeast two-hybrid and coimmunoprecipitation experiments while, at the same time, no direct binding to other major ND10 components such as PML or Sp100 was noticeable (18, 22). HDaxx positioning at ND10, in turn, has been shown to be dependent on the presence of PML (21). Thus, it was interesting to elucidate the role of the ND10 constituents PML and hDaxx for pp71 subcellular localization during infection, which should be done by infection of PML or hDaxx kd cells (Fig. 9A). As already demonstrated in prior experiments, depletion of PML from primary human fibroblasts results in a dissociation of the remaining ND10 components Sp100 and hDaxx which exhibit a diffuse, microdispersed distribution pattern in the absence of PML (43). However, upon HCMV infection, a reorganization of ND10-like structures consisting of hDaxx and Sp100 was detectable, since both proteins associated again in distinct foci that were found to colocalize with viral IE2 accumulations early after infection of PML-kd cells (43). Such HCMV-induced punctate hDaxx accumulations were likewise detectable after infection of PML-deficient cells with the recombinant AD169/EYFP-pp71 virus (Fig. 9B, compare subpanel c illustrating hDaxx distribution in uninfected PML-kd cells to subpanel g representing hDaxx localization during infection; compare also to subpanels k and l showing pp71 and hDaxx after infection with wt AD169). Interestingly, pp71 perfectly colocalized with these rearranged ND10-like substructures of hDaxx (Fig. 9Be to h and i to m) which, according to prior findings, also contain the ND10 factor Sp100, as well as the viral IE2 protein (43). The latter, in turn, can be found in direct association with viral DNA in HCMV-infected cells (40).
Similarly to the situation in normal fibroblasts (see Fig. 8Bn to q), IE1-mediated disassembly of ND10-like structures, which occurs also in the absence of PML (43), finally resulted in a translocation of hDaxx and pp71 into a diffuse distribution pattern (Fig. 9Bn to q). Taken together, these data led to the conclusion that PML is not required for the formation of dot-like pp71 accumulations at the very start of infection since ablation of the ND10 scaffold protein does not eliminate the potential of hDaxx and Sp100 to associate with viral nucleoprotein complexes consisting of the tegument protein pp71, the IE protein IE2 as well as viral genomes.
Loss of hDaxx, on the contrary, caused substantial changes to the subnuclear localization of pp71 as observed after infection of the hDaxx-kd cells siDaxx1 with AD169/EYFP-pp71 or AD169 (Fig. 9Cb, e, h, and l). Although, beside a diffuse distribution pattern, distinct aggregations of pp71 were still detectable in the absence of hDaxx, the tegument protein accumulated in much larger globular structures compared to hDaxx-positive cells (Fig. 9Cb, e, h, and l). Interestingly, counterstaining of cell nuclei with DAPI (Fig. 9Ca, d, g, and k) revealed that these pp71 concentrations illustrated agglomerations of the viral transactivator protein at the nucleoli of infected cells (Fig. 9C, see merged images in subpanels c, f, i, and m). Consequently, pp71 no longer associates with ND10 after hDaxx ablation, since PML-NBs, which are still present in hDaxx-kd cells, are normally excluded from nucleoli (42). This observation is consistent with data from HCMV-infected murine hDaxx knockout cells in which pp71 likewise localized at sites distinct from ND10 (22). Based on these findings, hDaxx was considered as the major determinant for pp71 subnuclear localization at ND10, as also already suggested from results obtained with an hDaxx-binding deficient mutant of pp71 that could no longer be recruited to ND10 domains (6, 18).
DISCUSSION
The characterization of the recombinant HCMV AD169/EYFP-pp71 virus which should be utilized for localization studies of the viral tegument protein pp71 in live-infected cells led to some unanticipated results. The integration of an EYFP-pp71 coding sequence into the viral genome of HCMV AD169 yielded a recombinant virus that exhibited a significantly increased release of viral particles compared to wt AD169, as detected by multistep growth curve experiments. Analysis of the protein expression kinetics revealed that the phenotype of an enhanced replication efficacy of AD169/EYFP-pp71 is mainly based on significantly increased amounts of pp71 present within cells infected with the recombinant virus. This could be explained either by an increased protein stability of pp71, implying that the EYFP tag positively influences the half-life of the tegument protein or an enhanced protein expression of the EYFP-pp71 fusion protein as a result of the insertion of the EYFP coding sequence upstream of the UL82 ORF, since the genuine UL82 promoter sequence remained unchanged.
The high abundance of EYFP-pp71 present after infection with the recombinant virus had several effects on the viral life cycle of AD169/EYFP-pp71 that could account for the increased release of progeny virus. For instance, analysis of the tegument composition of AD169/EYFP-pp71 revealed that strongly elevated amounts of EYFP-pp71 at late stages of infection resulted in markedly increased packaging of the tegument protein into viral particles of the recombinant virus, while the levels of other tegument components such as pp28 were not altered. At the same time, marginally reduced amounts of the most abundant tegument constituent pp65 were detected in AD169/EYFP-pp71 virion preparations, albeit a slightly increased expression of pp65 in AD169/EYFP-pp71-infected cells. This is most probably due to spatial restrictions resulting from the massive incorporation of EYFP-pp71, since increasing evidence suggests that herpesviral teguments illustrate highly structured and organized rather than unstructured and random protein accumulations (30).
Since pp71 has been shown to be required for efficient initiation of the lytic replication cycle at the very start of HCMV infection (5), the question arose as to whether the augmented tegument concentration of pp71 could account for the enhanced IE gene expression as observed after inoculation with AD169/EYFP-pp71 at a low MOI. The tegument protein pp71 is delivered to the nucleus of infected cells immediately upon infection to inactivate the cellular repressor hDaxx that silences IE transcription by recruitment of HDACs to the MIEP. The pp71-mediated degradation of hDaxx, which occurs through a proteasome-dependent (34), ubiquitin-independent pathway (20), relieves this repression, resulting in loss of transcriptionally repressive and gain of transcriptionally active chromatin structures at the MIEP (48). However, surprisingly, no enhanced degradation of hDaxx as a consequence of increased amounts of tegument-delivered pp71 was noticeable upon infection with AD169/EYFP-pp71 compared to wt AD169. One potential explanation for this could be that infection at a high MOI leads to a saturation of the degradative mechanism, thus precluding the enforcement of hDaxx degradation by higher amounts of pp71. Consequently, since we never observed a total disappearance of hDaxx signals upon HCMV infection, one would have to postulate that only part of the cellular hDaxx pool is amenable to pp71-mediated degradation. This would be consistent with our previous observation that a preferential depletion of hypophosphorylated hDaxx isoforms can be detected during the first hours after HCMV infection (44). On the other hand, however, in contradiction to prior findings of Saffert and colleagues (34), treatment of cells with reduced viral doses in order to circumvent a potential saturation problem completely abrogated the detection of hDaxx degradation. This may indicate that, besides hDaxx degradation, pp71 uses additional, thus-far-undefined mechanisms in order to augment the viral gene expression program. This hypothesis is further supported by our observation that, after high-multiplicity infection with AD169/EYFP-pp71, significantly increased early and late protein levels were detectable in the absence of elevated IE gene expression (see Fig. 3). Moreover, this finding implies that, in addition to the well-defined role of pp71 as a transactivator of IE gene expression, this viral regulatory protein could have a more general effect on transcription from the HCMV genome during later phases of the replication cycle. This is in accordance with previous observations of Baldick et al. (4), demonstrating that pp71 is capable of increasing the infectivity of transfected HCMV DNA through an effect that is separable from increased IE protein production. Consistent with such a notion, an enhanced accumulation of viral progeny DNA was detected after infection with AD169/EYFP-pp71 compared to wt AD169. Presumably, this is a consequence of increased E gene expression after AD169/EYFP-pp71 infection, since many E proteins are necessary for viral DNA synthesis (31). However, a direct involvement of pp71 in viral genome replication cannot be ruled out at the moment. Nonetheless, the recombinant AD169/EYFP-pp71 virus should illustrate a valuable tool to further elucidate these as-yet-uncharacterized functional properties of pp71.
When applied in live cell imaging experiments, the recombinant AD169/EYFP-pp71 virus was a useful tool for monitoring the characteristics of pp71 subnuclear localization in more detail. In accordance with previous results from fixed-cell studies with wt AD169 (18, 22), infection of mCh-PML expressing cells with AD169/EYFP-pp71 exhibited EYFP-pp71 signals that perfectly colocalized with ND10 domains. In addition, the live-cell microscopy experiments revealed that the ND10 association of pp71 occurred very rapidly upon virus entry since it was already detectable as early as 20 min after addition of the virus. Hence, targeting of incoming viral components by ND10 is a very fast antiviral response of the cell, which has been shown to occur through the deposition of ND10 proteins in novel structures that are juxtaposed to the sites of incoming viral nucleoprotein complexes, rather than by movement of preexisting ND10 domains (11, 13). Thereafter, pp71 stays associated with PML-NBs until these structures are dispersed through the action of the IE1 protein. In contrast to IE2, pp71 does not retain its dot-like distribution after disruption of ND10 but illustrates a diffuse localization pattern instead, indicating that the subnuclear determinants for both viral regulatory proteins differ. While in the case of IE2, viral DNA has been identified as the decisive structure for its subnuclear localization (40), a component of ND10 domains obviously is responsible for the formation of pp71 foci during the initial stage of HCMV infection. In previous studies it was proposed that the ND10 constituent hDaxx determines pp71 subnuclear accumulation at ND10 since a direct interaction between both proteins could be identified in yeast two-hybrid and coimmunoprecipitation experiments (18, 22). Consistent with this, the tight spatial association between pp71 and hDaxx, as observed in normal fibroblasts, was still visible in PML-kd cells being devoid of genuine ND10 domains. Distinct pp71 foci, which likewise formed in the absence of PML, perfectly colocalized with the redistributed hDaxx accumulations that are induced upon HCMV infection of PML-kd HFFs. According to results obtained in prior immunofluorescence analyses, these rearranged ND10-like structures of hDaxx additionally contain the ND10 constituent Sp100, as well as the viral IE2 polypeptide in association with viral DNA (40, 43). Hence, ablation of PML does not have an influence on the dot-like distribution of pp71 during infection, nor does it prevent the remaining ND10 factors hDaxx and Sp100 to associate with incoming viral components such as pp71, IE2, or viral genomes. Thus, it appears that detectable levels of PML are not required to coordinate the behavior of other ND10 proteins such as Sp100 and hDaxx in response to HCMV infection since all three major ND10 constituents apparently individually contain determinants that enable their localization to sites associated with viral nucleoprotein complexes (12, 43).
Conclusive evidence for hDaxx being the decisive determinant for the punctate localization of pp71 at ND10 was finally obtained after infection of hDaxx-negative cells with AD169/EYFP-pp71. Loss of hDaxx induced significant changes to the subnuclear distribution of pp71, since the viral tegument protein could no longer be found in tiny foci but accumulated in massive globular structures that turned out to form at the nucleoli of infected cells. Since PML-NBs, which still exist in hDaxx-kd cells, are normally excluded from this subnuclear compartment (42), it can be concluded that depletion of hDaxx indeed abolishes the localization of pp71 at ND10, as was also suggested from infection studies with murine hDaxx knockout cells (22). The observation of a pp71 association with nucleoli in the absence of hDaxx, instead of a diffuse nuclear distribution, suggests that an additional protein interaction mediates the specific targeting of pp71 to this subnuclear structure. In this respect, it should be mentioned that we detected an interaction of pp71 with a nucleolar protein, termed MSP58 (33), by yeast two-hybrid screening (H. Hofmann, H. Sindre, and T. Stamminger, unpublished data). Interestingly, MSP58 has also been described as an hDaxx-binding protein that is able to attenuate the trans-repressive effect of hDaxx on gene expression (23). Thus, it is tempting to speculate that pp71, in addition to inducing a limited degradation of hDaxx, may also be able to neutralize hDaxx-mediated repression in conjunction with MSP58. Experiments to address this hypothesis are in progress.
Acknowledgments
We thank W. Britt (University of Alabama), B. Plachter (Mainz, Germany), T. Shenk (Princeton, NJ), and G. Hahn (Munich, Germany) for providing reagents.
This study was supported by the SFB473, the IZKF Erlangen, the elite graduate school BIGSS, the GRK1071, and the Wilhelm Sander Stiftung.
Footnotes
Published ahead of print on 20 August 2008.
REFERENCES
- 1.Adler, H., M. Messerle, and U. H. Koszinowski. 2003. Cloning of herpesviral genomes as bacterial artificial chromosomes. Rev. Med. Virol. 13111-121. [DOI] [PubMed] [Google Scholar]
- 2.Almeida, J., D. Lang, and P. Talbot. 1978. Herpesvirus morphology: visualization of a structural subunit. Intervirology 10318-320. [DOI] [PubMed] [Google Scholar]
- 3.Andreoni, M., M. Faircloth, L. Vugler, and W. J. Britt. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23157-167. [DOI] [PubMed] [Google Scholar]
- 4.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 714400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate-early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 9714506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantrell, S. R., and W. A. Bresnahan. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 797792-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantrell, S. R., and W. A. Bresnahan. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 806188-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1589-14. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demarchi, J. M. 1981. Human cytomegalovirus DNA: restriction enzyme cleavage maps and map locations for immediate-early, early, and late RNAs. Virology 11423-38. [DOI] [PubMed] [Google Scholar]
- 11.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 795078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., S. Rechter, P. Papior, N. Tavalai, T. Stamminger, and A. Orr. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 807995-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D., G. Sourvinos, C. Leiper, J. B. Clements, and A. Orr. 2004. Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J. Virol. 781903-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groves, I. J., and J. H. Sinclair. 2007. Knockdown of hDaxx in normally non-permissive undifferentiated cells does not permit human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 882935-2940. [DOI] [PubMed] [Google Scholar]
- 16.Hensel, G. M., H. H. Meyer, I. Buchmann, D. Pommerehne, S. Schmolke, B. Plachter, K. Radsak, and H. F. Kern. 1996. Intracellular localization and expression of the human cytomegalovirus matrix phosphoprotein pp71 (ppUL82): evidence for its translocation into the nucleus. J. Gen. Virol. 77(Pt. 12)3087-3097. [DOI] [PubMed] [Google Scholar]
- 17.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 747720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann, H., H. Sindre, and T. Stamminger. 2002. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 765769-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollenbach, A. D., C. J. McPherson, E. J. Mientjes, R. Iyengar, and G. Grosveld. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 1153319-3330. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, J., and R. F. Kalejta. 2007. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367334-338. [DOI] [PubMed] [Google Scholar]
- 21.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. H. Yeh, J. F. Strauss, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147221-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishov, A. M., O. V. Vladimirova, and G. G. Maul. 2002. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 767705-7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, D. Y., and H. M. Shih. 2002. Essential role of the 58-kDa microspherule protein in the modulation of Daxx-dependent transcriptional repression as revealed by nucleolar sequestration. J. Biol. Chem. 27725446-25456. [DOI] [PubMed] [Google Scholar]
- 24.Lischka, P., Z. Toth, M. Thomas, R. Mueller, and T. Stamminger. 2006. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 261631-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 664434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorz, K., H. Hofmann, A. Berndt, N. Tavalai, R. Mueller, U. Schlotzer-Schrehardt, and T. Stamminger. 2006. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 805423-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 751870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall, K. R., K. V. Rowley, A. Rinaldi, I. P. Nicholson, A. M. Ishov, G. G. Maul, and C. M. Preston. 2002. Activity and intracellular localization of the human cytomegalovirus protein pp71. J. Gen. Virol. 831601-1612. [DOI] [PubMed] [Google Scholar]
- 29.McDonough, S. H., and D. H. Spector. 1983. Transcription in human fibroblasts permissively infected by human cytomegalovirus strain AD169. Virology 12531-46. [DOI] [PubMed] [Google Scholar]
- 30.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 32.Preston, C. M., and M. J. Nicholl. 2006. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 871113-1121. [DOI] [PubMed] [Google Scholar]
- 33.Ren, Y., R. K. Busch, L. Perlaky, and H. Busch. 1998. The 58-kDa microspherule protein (MSP58), a nucleolar protein, interacts with nucleolar protein p120. Eur. J. Biochem. 253734-742. [DOI] [PubMed] [Google Scholar]
- 34.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 803863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saffert, R. T., and R. F. Kalejta. 2007. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J. Virol. 819109-9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 743842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schierling, K., T. Stamminger, T. Mertens, and M. Winkler. 2004. Human cytomegalovirus tegument proteins ppUL82 (pp71) and ppUL35 interact and cooperatively activate the major immediate-early enhancer. J. Virol. 789512-9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih, H. M., C. C. Chang, H. Y. Kuo, and D. Y. Lin. 2007. Daxx mediates SUMO-dependent transcriptional control and subnuclear compartmentalization. Biochem. Soc. Trans. 351397-1400. [DOI] [PubMed] [Google Scholar]
- 40.Sourvinos, G., N. Tavalai, A. Berndt, D. A. Spandidos, and T. Stamminger. 2007. Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells. J. Virol. 8110123-10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamminger, T., M. Gstaiger, K. Weinzierl, K. Lorz, M. Winkler, and W. Schaffner. 2002. Open reading frame UL26 of human cytomegalovirus encodes a novel tegument protein that contains a strong transcriptional activation domain. J. Virol. 764836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, J. L. de, and D. R. van. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101(Pt. 4)773-784. [DOI] [PubMed] [Google Scholar]
- 43.Tavalai, N., P. Papior, S. Rechter, M. Leis, and T. Stamminger. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 808006-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavalai, N., P. Papior, S. Rechter, and T. Stamminger. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82126-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wathen, M. W., D. R. Thomsen, and M. F. Stinski. 1981. Temporal regulation of human cytomegalovirus transcription at immediate-early and early times after infection. J. Virol. 38446-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 683943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winkler, M., S. Schmolke, B. Plachter, and T. Stamminger. 1995. UL69 of HCMV, a homolog of the HSV ICP27, is contained within the tegument of virions and activates the major IE enhancer of HCMV in synergy with the tegument protein pp71(UL82). Scand. J. Infect. Dis. Suppl. 998-9. [Google Scholar]
- 48.Woodhall, D. L., I. J. Groves, M. B. Reeves, G. Wilkinson, and J. H. Sinclair. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate-early promoter. J. Biol. Chem. 28137652-37660. [DOI] [PubMed] [Google Scholar]