Abstract
Lytic reactivation from latency is critical for the pathogenesis of Kaposi's sarcoma-associated herpesvirus (KSHV). We previously demonstrated that the 691-amino-acid (aa) KSHV Rta transcriptional transactivator is necessary and sufficient to reactivate the virus from latency. Viral lytic cycle genes, including those expressing additional transactivators and putative oncogenes, are induced in a cascade fashion following Rta expression. In this study, we sought to define Rta's direct targets during reactivation by generating a conditionally nuclear variant of Rta. Wild-type Rta protein is constitutively localized to cell nuclei and contains two putative nuclear localization signals (NLSs). Only one NLS (NLS2; aa 516 to 530) was required for the nuclear localization of Rta, and it relocalized enhanced green fluorescent protein exclusively to cell nuclei. The results of analyses of Rta NLS mutants demonstrated that proper nuclear localization of Rta was required for transactivation and the stimulation of viral reactivation. RTA with NLS1 and NLS2 deleted was fused to the hormone-binding domain of the murine estrogen receptor to generate an Rta variant whose nuclear localization and ability to transactivate and induce reactivation were tightly controlled posttranslationally by the synthetic hormone tamoxifen. We used this strategy in KSHV-infected cells treated with protein synthesis inhibitors to identify direct transcriptional targets of Rta. Rta activated only eight KSHV genes in the absence of de novo protein synthesis. These direct transcriptional targets of Rta were transactivated to different levels and included the genes nut-1/PAN, ORF57/Mta, ORF56/Primase, K2/viral interleukin-6 (vIL-6), ORF37/SOX, K14/vOX, K9/vIRF1, and ORF52. Our data suggest that the induction of most of the KSHV lytic cycle genes requires additional protein expression after the expression of Rta.
Kaposi's sarcoma-associated herpesvirus (KSHV; also known as human herpesvirus-8) is the etiologic agent of the human cancers primary effusion lymphoma (PEL) and Kaposi's sarcoma (KS) (8, 22). Since B-cell infection predicts future KS development and latent KSHV infection is established before KS onset, the reactivation of productive (lytic) KSHV infection from the latently infected B-cell reservoir is a necessary step in KS development (2, 51, 54, 78). Deciphering the mechanisms that function at the molecular level to control KSHV reactivation is therefore essential for understanding the pathogenesis of the virus.
During KSHV reactivation in PEL tissue culture models, de novo expression of viral genes unfolds sequentially in a cascade fashion (31, 44). A fraction of the cells in which the virus initiates reactivation support completion of the gene expression program, resulting in the production of progeny virus and lysis of the host cells (46, 47, 60). The reactivation program has thus been termed the KSHV lytic cycle. Similarly to other DNA viruses, the KSHV lytic cycle genes have been classified as immediate early (IE), delayed early (DE), or late (L) genes. Genes whose expression is resistant to protein synthesis inhibitors have been formally classified as IE genes (62, 67, 81). Genes whose transcription requires viral DNA replication have been classified as L genes (44). By default, lytic genes that have not been classified as IE or L fall into the DE class. Twelve of the 17 viral genes that are candidates for contributing directly to KSHV pathogenesis are expressed during lytic reactivation, primarily with DE kinetics (13, 17, 44, 53, 57). Those observations suggest that the expression of the candidate DE pathogenic genes is one mechanism by which KSHV lytic reactivation contributes to KS development.
We and others have demonstrated that the KSHV Rta protein, expressed by the open reading frame (ORF50), is necessary and sufficient for lytic viral reactivation in tissue culture models of latency (24, 46, 47, 68, 79). The viral gene expression program induced by ectopic Rta in latently infected PEL cells is similar to that induced by chemicals that stimulate complete viral reactivation (14, 56). Rta is expressed with IE kinetics (67, 81) and encodes a 691-amino-acid (aa) protein that transactivates the putative promoters of many DE genes when they are cloned upstream of reporters (e.g., firefly luciferase [10, 12, 18, 32, 45-47, 63, 72, 73, 82]). These promoters include those for the candidate viral pathogenic genes. Rta functions as a tetramer to transactivate viral transcription and reactivate the lytic cycle (3). Rta specifies genes for transactivation by binding directly to promoter DNA and interacting with the cellular DNA binding proteins recombination signal binding protein Jk (RBP-Jk), Octamer 1 (Oct-1), CAAT-enhancer binding protein α (C/EBPα), and c-Jun (6, 7, 37, 39, 74, 76). The requirement for each of these molecular interactions in Rta function appears to be cell and promoter specific (59).
Studies of Rta-dependent reactivation at the single-cell level suggest that the expression of Rta alone does not ensure completion of the lytic cycle and the production of mature virions from every infected cell (46, 47). To understand the factors that determine the progression of the lytic cycle during Rta-dependent reactivation demands the identification of the authentic, direct-transcriptional targets of Rta in infected cells. However, the strategies described in the current literature have been insufficient for this purpose. Genome-wide analyses of the kinetic order of KSHV gene expression during reactivation fail to reveal Rta's direct transcriptional targets because additional transactivators are expressed downstream of Rta (23, 26, 30, 34, 59, 75). Other lytic cycle proteins that are induced by Rta inhibit viral transcription and reactivation (5, 29, 36, 40). Mechanistic studies of Rta-dependent reactivation are also complicated by reports that ectopic Rta can transactivate the transcription of Rta from the endogenous KSHV genome (11, 14, 24, 56). The results of transient assays of the Rta-dependent transactivation of promoter-reporter plasmids in transfected cells can be difficult to interpret since they rely on testing viral promoters removed from their authentic contexts in the viral genome.
In this study, our goal was to establish a strategy to overcome the shortcomings described above. We generated a conditionally nuclear variant of Rta by fusing it to a modified hormone-binding domain (HBD) of the murine estrogen receptor (ER) (Rta-ER). We employed Rta-ER to identify Rta's direct transcriptional targets in the viral genome in infected cells. Addition of the protein synthesis inhibitor hygromycin permitted the identification of the Rta-dependent transcriptome in the absence of translation of the other viral transcriptional regulatory proteins. We demonstrate that a C-terminal (aa 516 to 530) lysine-rich sequence is necessary for nuclear localization and transactivation by Rta. The deletion of this nuclear localization sequence (NLS) was critical, permitting tight control of Rta's nuclear localization by the addition or omission of the synthetic hormone 4-hydroxytamoxifen (4-OHT) to the cell growth medium.
MATERIALS AND METHODS
Plasmids.
All plasmids were propagated and purified as described previously (59). Plasmids constructed by PCR amplification were verified by DNA sequencing.
For pcDNA3.1-FLc50-RFP-Timer (expresses WT Rta), full-length (FL) ORF50 was PCR amplified from the template plasmid pGem3-FLc50 (46); the 3′ primer removed the translation termination codon. The product was digested with EcoRI and EcoRV (introduced by primers) and ligated with the plasmid pcDNA3.1/V5-HisB (Invitrogen) that had been digested with the same enzymes. The resultant clone was named pcDNA3.1-V5-FLc50 and expressed FL Rta fused C terminally to the V5 epitope tag and a six-His epitope. Next, the V5 and His tags were replaced, in frame, by the mutant red fluorescent protein (RFP) DsRed1-E5: the insert encoding this RFP was excised from the plasmid pTimer-1 (Clontech) by digestion with SacII and HpaI and ligated into pcDNA3.1-V5-FLc50 that had been digested with SacII and PmeI.
For pcDNA3.1-ORF50ΔNLS1-RFP-Timer (expresses Rta with NLS1 deleted from the first AUG of ORF50 exon 2, at nucleotide [nt] 181 [RtaΔNLS1]), pcDNA3.1-FLc50-RFP-Timer was digested with BsiWI (ORF50 nt 544) and EcoRI (in the polylinker 5′ of the insert) and ligated to the insert of pBS1.3 (described in reference 46) that was digested with the same enzymes.
For pcDNA3.1-ORF50ΔNLS2-RFP-Timer (expresses RtaΔNLS2; see Fig. 1A), two PCR products, spanning nt 1 to 1545 (PCR I) and 1591 to 2076 (PCR II), respectively, were sequentially cloned. PCR I was digested with EcoRV/NheI (introduced by the primers) and ligated to pcDNA3.1/Zeo (Invitrogen) that had been digested with the same enzymes. PCR II was digested with NheI/EcoRV and ligated to the plasmid constructed from PCR I that had been digested with NheI and NruI. This created the plasmid named pcDNA3.1Zeo-ORF50ΔNLS2. This plasmid then was used as a template to PCR amplify the FL insert (PCR product PCR III), digested with BsiWI (internal)/EcoRV (introduced by the 3′ primer), and ligated to pcDNA3.1-FLc50-RFP-Timer that had been digested with the same enzymes.
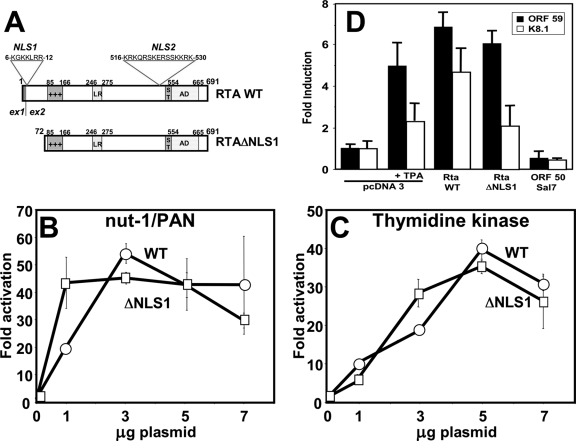
FIG. 1.
Putative NLS1 is dispensable for Rta function. (A) Schematics of WT Rta (top) and RtaΔNLS1 (bottom). Numbers refer to amino acid positions. ex, exon; +++, basic-amino acid rich; LR, leucine repeat; ST, serine-threonine amino acid rich; AD, transcriptional activation domain. (B) BL-41 cells were coelectroporated with the indicated amounts of plasmids expressing WT Rta (WT) or RtaΔNLS1 (ΔNLS1), the pGL3-nut-1 promoter/reporter (expressing luciferase), and pcDNA3.1-His-lacZ (expressing β-galactosidase) as an internal control. The level of activation was calculated by comparison to that of the reporter coelectroporated with empty expression vector. (C) BL-41 cells were electroporated similarly to the description for panel B, but the pGL3-TK promoter/reporter (expressing luciferase) was tested. (D) BCBL-1 cells were coelectroporated with the indicated plasmids and a plasmid expressing hepatitis delta virus large delta antigen and treated with tetradecanoyl phorbol acetate (TPA), as indicated. At 72 h postelectroporation, the percentage of transfected cells expressing large delta antigen and coexpressing ORF59 or K8.1 was calculated for each transfection. Levels of induction were calculated by comparison to that in cells transfected with the empty vector, which was normalized as onefold. Error bars show standard deviations for each transfection.
For pcDNA3.1-ORF50ΔNLS1,2-RFP-Timer (expresses Rta with NLS1 and NLS2 deleted [RtaΔNLS1,2]), PCR III was amplified and digested as described in the paragraph above and then ligated to pcDNA3.1-ORF50ΔNLS1-RFP-Timer that had been digested with the same enzymes.
ORF50 Sal 7 is a plasmid clone of the 7-kb SalI fragment containing the genomic ORF50/Rta locus and its natural promoter (47).
For pEGFP-C1-NLS1 and pEGFP-C1-NLS2 (express Rta NLS1 and NLS2, respectively, fused to enhanced green fluorescent protein [eGFP]), double-stranded oligonucleotides encoding NLS1 and NLS2, respectively, of ORF50 were cloned in frame with eGFP at the BamHI/SalI sites of pEGFP (Clontech).
For pcDNA3.1-ZeoFL50-ER-Tm (expresses WT Rta ER protein), pBS-SK-ER-Tm (containing the tamoxifen [TM]-responsive HBD of the murine ER; a gift of Gerard Evan [42]) was digested with BamHI and EcoRI to release the ER-TM fragment, which was ligated into pcDNA3.1/Zeo (Invitrogen) that had been digested with the same enzymes. This created the plasmid pcDNA3.1ZeoER-Tm. Rta was then constructed as a C-terminal fusion to the ER-TM domain by using a two-step procedure. First, Rta was PCR amplified by using the plasmid pGem3-FLc50 (46) as a template and a 3′ primer that changed the Rta termination codon from TGA to TGG (Trp). The 3′ primer also introduced a BamHI restriction site to that end; following amplification, the PCR product was digested with BamHI (which also cuts internally in Rta, at nt 335). The resulting fragment was ligated into pcDNA3.1ZeoER-Tm, which had been digested with BamHI and treated with shrimp alkaline phosphatase (Promega) to inhibit self-ligation. The resultant plasmid was named pcDNA3.1-50Bam-ER-TmZeo. The N terminus of Rta was added in the second step by PCR amplifying Rta nt 1 to 815, using pGem3-FLc50 as template and a 5′ primer that introduced an NheI restriction site 5′ to the Rta start codon. This PCR product was digested with NheI and NarI (which cuts at Rta nt 630) and ligated with pcDNA3.1-50BamER-TmZeo that had been digested with the same enzymes.
For pcDNA3.1-ORF50ΔNLS1-ER-Tm (expresses ER and Rta with NLS1 deleted[RtaΔNLS1-ER]), pcDNA3.1-50BamER-TmZeo was digested with Nar I (ORF50 nt 630) and NheI (which cuts in the polylinker), and ligated to the Nar I/SpeI fragment of pBS-1.3 (46).
For pcDNA3.1-ORF50ΔNLS1,2-ER-Tm (expresses RtaΔNLS1,2-ER), the BstEII fragment from pcDNA3.1-ORF50ΔNLS2-GFP-Timer was ligated to pcDNA3.1-ORF50ΔNLS1-ER-Tm.
The pGL3 promoter expresses firefly luciferase under the control of the simian virus 40 early promoter (Promega). pcDNA3-His-lacZ expresses β-galactosidase under the control of the human cytomegalovirus major IE enhancer (Invitrogen).
Cell culture, transfections, and inductions.
BL-41 and BCBL-1 cells were propagated, transfected, and induced as previously described (3, 6). Luciferase/β-galactosidase reporter assays were performed exactly as previously described (3). At least two transfections were performed in triplicate for each experiment.
HeLa cells were propagated in complete DME as previously described (7). For transfections, 5 × 104 cells were seeded on coverslips in each well of six-well plates and grown overnight (ON). Approximately 4 h before transfection, the medium was replaced with fresh complete medium. DNA cocktails were prepared, consisting of 10 μg of total DNA, 10 μl 10× NTE (10 mM Tris-Cl, pH 7.4, 100 mM NaCl, 1 mM EDTA), 12.5 μl 2 M CaCl2, and H20 to a final volume of 100 μl. The DNA cocktails were added dropwise to an equal volume of 2× transfection buffer (100 μl 0.5 M HEPES, 810 μl H20, 90 μl 2 M NaCl, 2 μl 1 M Na2HPO4) in a 1.5-ml centrifuge tube, and streams of bubbles were blown through the mixture 5 to 10 times using a pipette. The mixture was left at room temperature for 15 to 30 min and then added dropwise to the wells containing medium and cells. DNA precipitates were distributed by rotating plates in a figure-eight motion, and the plates were returned to the incubator for ON growth. The medium/precipitates were aspirated, cells were washed with fresh medium, and then 3 ml of medium was added to each well. The plates were returned to the incubator for additional ON growth of cells.
Vero cells infected with the recombinant KSHV construct rKSHV.219 (Vero-rKSHV.219) were grown in complete Dulbecco's modified Eagle's medium supplemented with 6 μg/ml puromycin as previously described (71). For transfection, cells were seeded at 1 × 106 per 10-cm plate to obtain approximately 50 to 70% confluence the following day and transfected with TransIT-LT1 transfection reagent with 10 μg DNA according to the manufacturer's (Mirus) instructions. For gene expression studies, 4-OHT (0.1 mM; Sigma), cycloheximide (CHX) (10 or 25 μg/ml; Sigma), hygromycin B (300 μg/ml; Invitrogen), or neomycin (5 μl/ml; Sigma) was added to the cultures at the times indicated in each figure.
Immunofluorescence.
The levels of reactivation in BCBL-1 cells were quantitated by measuring the KSHV DE marker ORF59 or the L marker K8.1, as described in reference 47. The results of at least two experiments performed in duplicate were quantitated. The levels of reactivation in Vero-rKSHV.219 cells were quantitated by measuring the K8.1 marker at 48 h after the addition of 4-OHT, using the formulas described in reference 3.
For adherent Vero-rKSHV.219, HeLa, or 293 cells, coverslips were washed three times with 1× phosphate-buffered saline (PBS), and liquid was aspirated completely. Fresh 4% paraformaldehyde (wt/vol in 1× PBS) was added to fix the cells at room temperature for 30 min. The coverslips were washed three times with 1× PBS and then incubated in permeabilization buffer (1× PBS, 0.1% Triton X-100, 0.1% sodium citrate) at 4°C for 10 min. Cells were washed twice with 1× PBS and incubated in blocking buffer (1× PBS, 1.0% Triton X-100, 0.5% Tween-20, 3% bovine serum albumin) at room temperature for 30 min. The primary antisera (anti-ORF50 or anti-ORF57 diluted 1:1,000 in blocking buffer [47, 59]) were added to cover the cells (∼250 μl) and incubated at room temperature for 1 h. Coverslips were washed three times with 1× PBS and then incubated with secondary antibodies (goat anti-rabbit conjugated to tetramethyl rhodamine iso-thiocyanate [TRITC; MP Biomedical] or Alexa Fluor 350 [Invitrogen molecular probe] diluted 1:1,000 in blocking buffer) at room temperature for 1 h in the dark. Coverslips were washed three times with 1× PBS, and all liquid at the edges of the coverslips was removed by aspiration. A drop of DNA mounting solution (standard or containing the DNA dye 4′,6-diamidino-2-phenylindole [DAPI]; Vector Laboratories) was placed on the slide, and coverslips were mounted inverted and sealed to the slides with nail polish. Slides were viewed immediately or stored at 4°C in the dark. Nuclear/cytoplasmic ratios were determined with the software program ImageJ (1).
All immunofluorescence procedures were performed under conditions in which autofluorescence from GFP or RFP fused to ORF50 was not visible.
KSHV infections.
Vero-rKSHV.219 cells were transfected with the pcDNA3.1-ORF50ΔNLS1,2-ER-Tm plasmid. At 16 h posttransfection, cells were left untreated or treated with 0.1 mM 4-OHT, as indicated in Fig. 7. The supernatant medium was collected 7 days after the addition of 4-OHT, and cells and debris were removed by centrifugation and passage of supernatants through 0.4-μm filters. Filtered virus was transferred to 80%-confluent 293 cells plated on coverslips in 35-mm plates. Infected 293 cells were detected by GFP expression from rKSHV.219 at 2 days postinfection.
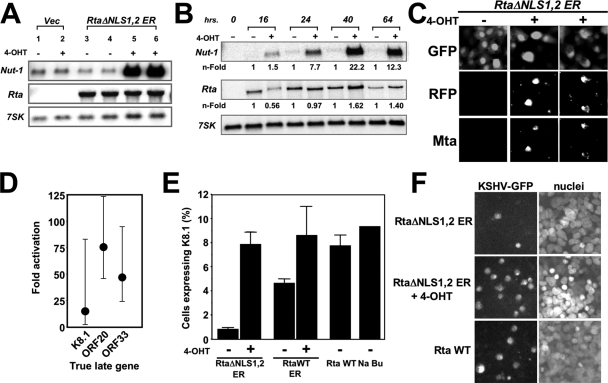
FIG. 7.
Nuclear relocalization of Rta reactivates the complete KSHV lytic cycle. (A) Vero-rKSHV.219 cells were transfected with the RtaΔNLS1,2-ER expression vector. Three hours following transfection, half of the cells were left untreated or treated with 4-OHT, as indicated. At 40 h following 4-OHT addition, total cellular RNA was purified and analyzed by Northern blotting; probes are indicated at left. Vec, vector. (B) Vero-rKSHV.219 cells were transfected with RtaΔNLS1,2-ER plasmid and treated with 4-OHT as described for panel A. At the indicated times following 4-OHT addition, total cellular RNA was purified and analyzed by Northern blotting; probes are indicated at left. The blot was quantitated by phosphorimager; each signal for Nut-1/PAN and ORF50 was normalized to the corresponding signal for 7SK. The level of induction was calculated by comparison to the signal for untreated cells at each time point. (C) Vero-rKSHV.219 cells were transfected and treated with 4-OHT as described for panel B. At 24 h after the addition of 4-OHT, cells were analyzed by indirect immunofluorescence using ORF50/Mta-specific primary antiserum and Alexa Fluor 350-conjugated secondary antibody. GFP and RFP fluorescence and immunofluorescent images of representative cells were captured digitally. (D) Vero-rKSHV.219 cells were transfected and treated with 4-OHT as described for panel B. At 72 h following 4-OHT addition, total cellular RNA was purified and analyzed by real-time qRT-PCR with primers that detect the indicated true L transcripts. (E) Vero-rKSHV.219 cells were transfected with the indicated plasmids, and indicated cells were treated with 4-OHT as described for panel A. At 48 h following 4-OHT addition, viral reactivation was quantitated by measuring the percentage of cells expressing the true L marker K8.1. Positive controls were transfection of WT Rta plasmid or treatment with sodium butyrate (Na Bu). Results are from two experiments performed in triplicate, in each of which 500 K8.1 positive cells were counted. (F) Vero-rKSHV.219 cells were transfected with the indicated plasmids, and the indicated cells were treated with 4-OHT as described for panel A. Seven days after the addition of 4-OHT, cell supernatants were transferred to naïve 293 cell monolayers. KSHV-infected 293 cells were detected by GFP expression, and nuclei were detected with DAPI. Infection was quantitated by counting 500 GFP-positive cells in triplicate experiments. Error bars show standard deviations. +, treated; −, not treated.
RNA isolation.
RNA isolation was performed as described in reference 59.
Northern blotting.
Northern blotting was performed as described in reference 46. The double-stranded probes were the EcoRI fragment of pCR2.1-nut-1 (+) (for nut-1/PAN), the BamHI/HindIII fragment of pRSET-0.8 (for ORF50/Rta) (47), and the AccI/KpnI fragment of pGem-7SK (for 7SK) (59). The probes were labeled with [α32P]dCTP by using a HexaLabel DNA labeling kit (Fermentas Life Sciences) and purified by chromatography (Spin-25; USA Scientific). The signals on the Northern membranes were quantitated by using a phosphorimager.
Western blotting.
Western blotting was performed as described in reference 59. Antitubulin antibody was purchased from Sigma.
Microarray printing, cDNA labeling, hybridization, and data analysis.
The KSHV microarray consisted of 60- to 70-mer oligonucleotides designed to detect 96 predicted KSHV ORFs and splice variants identical to those published by Lu et al. (44) and updated to include newer annotated transcripts. Detector oligonucleotides for human housekeeping genes (Operon Biotechnologies) were also synthesized. Oligonucleotides were spotted in triplicate onto poly-l-lysine-coated glass microscope slides by using a GeneMachines Omnigrid 100 arrayer (Genomic Solutions) and SMP3 pins (Telechem).
Six pairs of total-RNA samples from five independent transfections were analyzed. The RNA quality was assessed by visual inspection by formaldehyde-agarose gel electrophoresis and Northern blotting. Three or 6 μgs of each RNA sample (equal amounts per pair) were adjusted to 11 μl with distilled water. A 2.2-μl amount of random hexamers (2 μg/μl; Sigma-Genosys) was added and the mixture was heated at 98°C for 2 min and then snap-cooled on ice. An 11.1-μl amount of master mix (5.0 μl first-strand buffer, 2.5 μl 100 mM dithiothreitol, 2.3 μl low-dT deoxynucleotide triphosphate mix [5 mM A, G, and C and 0.2 mM T stock], containing either 1.5 μl Cy3 [in mixture without 4-OHT] or Cy5 [in mixture with 4-OHT] [PerkinElmer]) was added. The mixture was subsequently supplemented with 1.2 μl SuperScript (Invitrogen) and then incubated for 10 min at 25°C, followed by 90 min at 42°C. Microcon 10 columns (YM 10; Millipore) were primed with 400 μl 1× TE buffer (Tris-EDTA, pH 8.0) by centrifugation at room temperature for 10 min. Paired Cy3-labeled and Cy5-labeled cDNAs were combined, added to the primed column, and purified by centrifugation. When approximately 25 μl of liquid remained above the membrane, an extra 200 μl 1× TE buffer was added to wash the column. Centrifugation was continued until approximately 4.5 μl of liquid remained above the membrane. The purified, labeled cDNAs were transferred to a new tube and combined with an equal volume of hybridization mixture (0.5 μl 10 mg/ml tRNA [Sigma], 0.5 μl 10 mg/ml salmon sperm DNA [Sigma], 1.0 μl 20× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 2.5 μl formamide [Fisher], and 1.0 μl 1% sodium dodecyl sulfate [SDS]). The mixture was heated for 2 min at 98°C and cooled for 5 min at room temperature immediately before being loaded on the microarray slides.
Prior to hybridization, the slides were blocked in prehybridization solution (3.0 g bovine serum albumin [fraction V; Roche], 1.2 ml 10% SDS, 106.0 ml Milli-Q water) at 42°C for 1 h. The prehybridized slides were washed with nuclease-free water for 2 min and isopropanol for 2 min and subsequently dried by centrifugation. The labeled cDNAs were applied to slides, covered by a 22- by 22-mm coverslip (Corning), sealed into a hybridization chamber (GeneMachines), and hybridized at 50°C overnight. After hybridization, coverslips were removed by submerging the slides in solution I (2× SSC, 1% SDS), and slides were rinsed in solution II (1× SSC, 0.05% SDS) for 1 min and solution III (0.06× SSC) for 2 min. Slides were dried by centrifugation and scanned immediately with a GenePix 4000B scanner (Molecular Devices). The images were processed by using GenePix 5.1. The data were filtered by removing all spots that were below the background noise or flagged as “bad.” Spots were considered to be below the background noise if the sum of the median intensities of the two channels was less than twice the highest mean background of the chip. The chips were normalized by the print-tip loess method (80). The ratio of the mean median intensity of Cy5 over the mean median intensity of Cy3 was determined for each spot (dye ratios).
Replicate dye ratios from identical RNA samples hybridized to independent arrays were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Replicate dye ratios from independent RNA samples were then normalized to the averaged dye ratios of the KSHV true L ORF25/major capsid protein transcript (as quantitated by Northern blotting, the major capsid protein transcript was unaffected by Rta WT-ER and mutants in the presence of protein synthesis inhibitors).
The five resulting data sets were tested for significant changes in levels of gene expression by using two methods. For method 1 (to find the z-score), the dye ratios of each spot for RtaΔNLS1,2-ER were divided by the dye ratios for the ER vector to determine the spot-specific effects of 4-OHT addition. The means of the triplicate dye ratios were calculated to determine the gene-specific effects of 4-OHT addition. Each data set was log transformed and fit to a normal (Gaussian) distribution, and the means of the normalized gene-specific values were calculated. The resulting set of gene-specific means was fit to a normal distribution, outliers were excluded by using Peirce's criterion (hypothesis: one outlier per gene measurement) (35), and the average number of standard deviations from the mean was calculated for each gene (z-score). For method 2 (to find q values), gene-specific t tests and q values (minimum false-positive rate that can be attained when calling a change in gene expression significant) were calculated by comparing the dye ratios of each spot for RtaΔNLS1,2-ER to the dye ratios for the ER vector by using the software program Significance Analysis of Microarrays (SAM) (70).
The levels of change in expression for genes whose means were the greatest number of standard deviations from the population mean and with the lowest q values were quantitated by using real-time reverse transcriptase PCR (RT-PCR).
qRT-PCR.
Primers for quantitative PCR (qPCR) were designed by using Primer3 software (http://frodo.wi.mit.edu) as described in reference 16; sequences are available from the authors by request. Five or 100 ng of total RNA (per experiment) were quantitated by using an iScript one-step RT-PCR kit with Sybr green (Bio-Rad) according to the manufacturer's recommendations, with the following modifications: the total reaction mixture volume was 25 μl, containing 12.5 μl 2× Sybr green RT-PCR mix, 0.5 μl iScript reverse transcriptase, 0.75 μl forward primer, 0.75 μl reverse primer, and nuclease-free water. The mixtures were amplified by incubation at 50°C for 10 min and 95°C for 5 min, followed by 50 cycles of 95°C for 10 s, 55°C for 30 s, and 72°C for 1 min, in a Corbett RotorGene instrument. The reaction products were examined by melting curve analysis and 2.5% agarose gel electrophoresis to ensure that a single amplicon was generated, without primer dimers. In each experiment, one control lacking RT was included to ensure that amplicons were not generated from contaminating viral genomic DNA in the extracts. The respective take-off cycles for each reaction were determined by using the comparative quantitation function of the Corbett Research software package and used as the threshold cycles (CT) to calculate the levels of change in expression by the ΔΔCT method (43). The control reactions for ΔΔCT used GAPDH primers for internal controls or matched primer pairs to compare viral genes in matched transfections in the presence or absence of 4-OHT. The means and standard deviations of the results for each gene were calculated from the results of at least two reactions from at least two different transfections.
Microarray data accession number.
Raw and normalized microarray data are available at the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession number GSE 11432.
RESULTS
Rta does not require the putative NLS1.
We and others have previously performed structure-function analyses of the KSHV ORF50/Rta protein, whose results are summarized in Fig. 1A. Rta contains an N-terminal, basic DNA binding domain (45, 65), a proline-rich leucine repeat that is required for the tetramerization of Rta (3), and a C-terminal transcriptional activation domain (46, 72). Rta is constitutively localized to the nuclei in transfected and infected cells (47).
Further analysis of the amino acid sequence of Rta revealed two distinct lysine/arginine-rich domains, which we hypothesized might function as NLSs. NLS1 (aa 6 to 12) and NLS2 (aa 516 to 530) are indicated in Fig. 1A. We deleted exon 1 from a genomic clone of Rta, generating an insert that expresses Rta protein initiating at methionine 72 (see Fig. 1A). We call this mutant RtaΔNLS1.
We first tested whether or not RtaΔNLS1 could activate transcription from two promoters that we previously showed were activated by the WT Rta protein (46, 47). We cotransfected reporter vectors for the Nut-1/PAN or thymidine kinase (TK) promoters together with increasing amounts of the WT or RtaΔNLS1 expression vectors into BL-41 cells. We calculated the change in the level of activation by comparison to the basal activity of each promoter transfected with empty vector alone. The transactivation of both promoters by either WT Rta or RtaΔNLS1 was indistinguishable. The nut-1 promoter was activated by both proteins to a maximum of 50- to 60-fold (Fig. 1B), while the TK promoter was activated by both proteins to about 40-fold (Fig. 1C). These data suggest that aa 1 to 71 of Rta, including the putative NLS1, are dispensable for the transactivation of KSHV DE promoters.
We also tested RtaΔNLS1 for its ability to reactivate KSHV from latency in BCBL-1 cells. We coelectroporated BCBL-1 cells with the Rta expression plasmids and a plasmid expressing the hepatitis delta antigen to permit the quantitation of transfected cells. At 48 h after electroporation, we counted the percentage of transfected cells scoring positive for either the DE protein ORF59 or the true L protein K8.1 by indirect immunofluorescence. Each value was compared to that for BCBL-1 cells transfected with the empty expression vector to calculate the change in the level of reactivation.
The results presented in Fig. 1D show that the treatment of vector-transfected cells with tetradecanoyl phorbol acetate leads to about a fivefold and twofold induction of ORF59 and K8.1, respectively. The ectopic expression of either WT Rta or RtaΔNLS1 results in similar induction of the lytic markers (Fig. 1D); these data suggest that aa 1 to 71 of Rta, including NLS1, are also dispensable for stimulating the reactivation of latent KSHV. The control DNA for the effects of transfection, the ORF50 genomic locus controlled by its native promoter (ORF50 Sal 7), was unable to reactivate the virus since Rta is not expressed from this clone in untreated PEL cells (Fig. 1D). The results presented in Fig. 1 therefore show that aa 1 to 71, including NLS1, are not required for Rta transactivation or the stimulation of KSHV reactivation.
Putative NLS2 is required for transactivation by Rta.
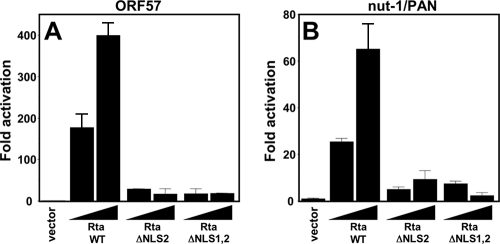
We also deleted aa 516 to 530, designated NLS2 (Fig. 1A), in the context of the FL WT Rta or RtaΔNLS1. This strategy generated the mutants RtaΔNLS2 and RtaΔNLS1,2, respectively. We compared these two mutants for their abilities to activate reporters for the ORF57 and nut-1/PAN promoters in transfections of BL-41 cells. As we have previously shown (3, 6, 7, 46, 47, 59), WT Rta transactivates the ORF57 promoter by at least 400-fold (Fig. 2A) and the nut-1 promoter by 60- to 80-fold (Fig. 2B). However, neither RtaΔNLS2 alone nor the double-mutant RtaΔNLS1,2 transactivated either promoter. We concluded that NLS2 was required for Rta to transcriptionally transactivate both promoters.
FIG. 2.
Putative NLS2 is required for Rta function. BL-41 cells were coelectroporated with plasmids expressing the indicated proteins and the levels of activation calculated as described in the legend to Fig. 1B. Reporter plasmids were pGL3-ORF57 (A) or pGL3-Nut-1 (B), both expressing luciferase. Black triangles indicate relative amounts of transfected expression vectors. Error bars show standard deviations.
NLS2, but not the putative NLS1, is required for nuclear localization of Rta.
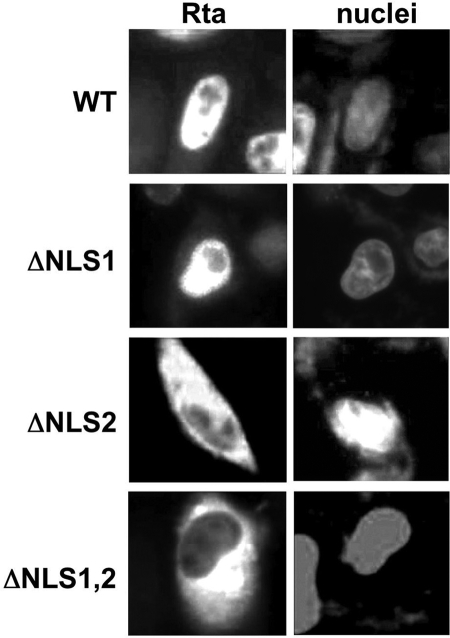
To determine whether nuclear localization corresponded with the transactivation phenotypes of the NLS mutants, we transfected HeLa cells with the vectors expressing WT Rta or each of the mutants and determined the subcellular localization of each by indirect immunofluorescence. Using the nuclear stain DAPI as a reference, RtaΔNLS1 was found to localize completely to the nuclei of HeLa cells, indistinguishably from WT Rta (Fig. 3).
FIG. 3.
NLS2, but not putative NLS1, is required for nuclear localization of Rta. HeLa cells were transfected with plasmids expressing the indicated proteins and analyzed by indirect immunofluorescence using Rta-specific antiserum. Nuclei were visualized by DAPI stain. Images of representative cells were captured digitally and converted to grayscale.
However, the deletion of putative NLS2 resulted in a strikingly different subcellular localization for Rta. Both RtaΔNLS2 and RtaΔNLS1,2 were well expressed but were confined to the cytoplasm of transfected HeLa cells. (Fig. 3). For both mutants, we observed nearly 100% discordance between the nuclear stain (DAPI) and the Rta stain in all transfected cells. The deletion of NLS2 resulted in a similar inability to enter the nucleus in CV-1 and SLK cells (not shown). Together with the results shown in Fig. 1 and 2, these data suggested that aa 1 to 71, including the putative NLS1, were not required for (i) nuclear localization of Rta, (ii) transactivation by Rta, or (iii) reactivation of the virus from latency. These results also suggested that the failure of the RtaΔNLS2 and RtaΔNLS1,2 proteins to transactivate transcription was due to their failure to access the nuclei of transfected cells.
NLS2, but not putative NLS1, is sufficient to relocalize a heterologous protein to the nucleus.
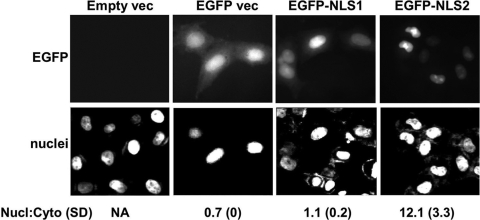
To determine whether each of the putative NLS motifs could redirect a heterologous protein to the cell nucleus, we fused each independently to the ORF of eGFP. As shown in Fig. 4, GFP alone is distributed nonspecifically between the nuclei and cytoplasm of transfected HeLa cells. Fusion of the putative NLS1 to eGFP had little effect on its subcellular localization (Fig. 4). However, fusion of NLS2 to eGFP resulted in dramatic and exclusive relocalization of eGFP to HeLa cell nuclei (Fig. 4). Considered together with the data presented above, Rta NLS2 is necessary and sufficient to localize proteins to cell nuclei.
FIG. 4.
NLS2 is sufficient to completely relocalize eGFP to the cell nuclei. HeLa cells were transfected with plasmids expressing the indicated proteins and analyzed by DAPI staining of nuclei. GFP and DAPI images of representative cells were captured digitally and converted to grayscale. Nuclear/cytoplasmic localization ratios (Nucl:Cyto) are shown under the images; SD, standard deviations; NA, not applicable.
Nuclear/cytoplasmic localization of Rta can be regulated by fusion to a heterologous HBD.
Deletion of the putative C-terminal NLS2 of Rta resulted in constitutive relocalization of the mutant protein to the cytoplasm of transfected cells and inhibition of its ability to transactivate transcription. It has been reported that a protein's nuclear localization can be controlled in a hormone-dependent fashion if fused to the HBD of the murine ER gene (42). Since the ER HBD lacks the ER DNA binding domain (DBD), it lacks the ability to bind to its cognate DNA elements but can be directed to DNA by the DNA binding specificity of the protein to which it is fused. This approach has been used successfully to generate conditionally active, hormone-dependent variants of c-Myc, v-Myb, MyoD, Epstein-Barr virus EBNA2, and adenovirus E1a, among others (19, 20, 28, 33, 66).
To determine whether relocalization of the NLS2 mutant of ORF50 to the cell nucleus could rescue its activity, we fused WT Rta and RtaΔNLS1,2, at their respective C-termini, to a modified form of the ER HBD. This modified HBD is called ER-TM and contains a point mutation, G525R, that eliminates drawbacks of the original ER HBD (42). In this way, ER-TM no longer binds 17β-estradiol that is frequently present in cell culture media and it lacks endogenous transcriptional activity; however, it remains responsive to activation by the synthetic steroid 4-OHT.
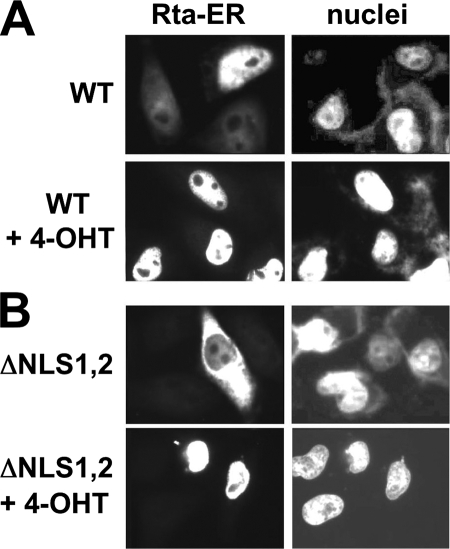
In HeLa cells, most but not all Rta WT-ER was redistributed to the cytoplasm in the absence of 4-OHT but still retained observable nuclear localization (Fig. 5A, top panel). In contrast, RtaΔNLS1,2-ER was found constitutively in the cytoplasm in the absence of inducing hormone 4-OHT (Fig. 5B, top panel), i.e., we observed complete discordance between the nuclear (DAPI) stain and the ORF50/Rta stain in all transfected cells. These data suggest that ER-TM is incompletely dominant to the endogenous Rta NLS2 in controlling the distribution of Rta WT-ER in transfected cells. In WT Rta, NLS2 seems to partially overcome the cytoplasmic retention of the ER fusion by heat shock proteins in the absence of inducing hormone. We saw a similar incomplete dominance of the ER domain when it was fused to RtaΔNLS1 (not shown).
FIG. 5.
4-OHT-regulated nuclear localization of Rta. HeLa cells were transfected with plasmids expressing WT Rta-ER (A) or RtaΔNLS1,2-ER (B), and half of each set of transfected cells was treated with 4-OHT. Cells were analyzed and visualized as described in the legend to Fig. 3.
When we added the synthetic hormone 4-OHT to the cell medium, all of the WT Rta (Fig. 5A, bottom panel) and mutant (Fig. 5B, bottom panel, and data not shown) fusion proteins were dramatically relocalized to the nuclei of transfected cells. Therefore, we concluded that the nuclear/cytoplasmic localization of RtaΔNLS1,2-ER, but not Rta WT-ER, was very tightly regulated by the addition or omission of 4-OHT to tissue culture media.
Rta must be nuclear to transactivate transcription.
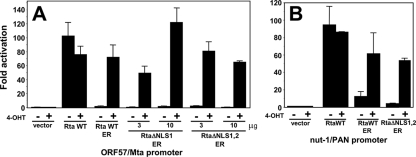
To test the functional significance of the nuclear/cytoplasmic localizations of the Rta-ER fusion proteins, each was cotransfected with the ORF57/Mta promoter-reporter vector into BL-41 cells. At 3 h postelectroporation, 4-OHT was added to half of the transfection cultures and the other half remained untreated. Twenty-four hours later, the cells were harvested, and the luciferase activities of the reporter gene were quantitated.
In the absence of the inducing hormone, Rta WT-ER, RtaΔNLS1-ER, and RtaΔNLS1,2-ER were all unable to transactivate the ORF57/Mta promoter (Fig. 6A). The small amount of nuclear localization of Rta WT-ER and RtaΔNLS1-ER in the absence of hormone (Fig. 5A and data not shown) seems to be inconsequential for transactivation of the ORF57 promoter by either fusion protein. However, the addition of 4-OHT resulted in robust transactivation by all three proteins (50- to 120-fold), suggesting that each protein is rendered active by translocation to the nucleus. We speculate that transactivation by RtaΔNLS1,2-ER at 10 μg did not increase relative to the level at 3 μg due to nonspecific transcriptional squelching over the range of input plasmids tested. The negative control transfections of the empty expression vector (pcDNA3) showed no transactivation in the presence or absence of 4-OHT. The positive control transfections of cognate WT Rta without the ER-TM fusion transactivated the ORF57 promoter regardless of 4-OHT addition, as expected.
FIG. 6.
Nuclear localization of Rta is required for transactivation. BL-41 cells were coelectroporated with plasmids expressing the indicated proteins, and half of each set of transfected cells was treated with 4-OHT. The level of activation was calculated as described in the legend to Fig. 1B. Reporter plasmids were pGL3-ORF57 (A) or pGL3-Nut-1 (B), both expressing luciferase. Error bars show standard deviations. +, treated; −, not treated.
To confirm these data for a second KSHV DE promoter, we repeated the above approach using the nut-1/PAN promoter-reporter vector. Identically to results with the ORF57/Mta promoter, RtaΔNLS1,2-ER only activated nut-1/PAN in cells treated with 4-OHT (Fig. 6B). Interestingly, Rta WT-ER activated the nut-1/PAN promoter even in the absence of hormone (Fig. 6B), a result agreeing with the partial nuclear localization of the WT fusion protein under those conditions (Fig. 5A). Regardless of this effect, when we added the hormone, transactivation by the Rta-ER fusion protein was stimulated by 60- to 80-fold. The positive control transfections of cognate WT Rta (without the ER-TM fusion) transactivated the nut-1 promoter regardless of 4-OHT addition, as expected (Fig. 6B).
Taken together, these data demonstrate a tight correspondence between nuclear localization of WT and mutant Rta proteins and their abilities to transactivate transcription. The transcriptional defect in RtaΔNLS1,2 was the result of its incorrect localization to the cytoplasm, but it transactivated promoters at a level quantitatively similar to that of cognate Rta when it was relocalized to the nucleus by the addition of 4-OHT. The cognate WT Rta protein, not fused to the ER, was constitutively localized to the nucleus of BL-41 cells and robustly transactivated the Mta and nut-1 promoters in the presence or absence of tamoxifen. For both promoters, there was no background stimulation by 4-OHT, as control cells transfected with the empty vector (Fig. 6) showed only basal activity in the absence or presence of the hormone.
Nuclear localization is required for Rta to reactivate KSHV from latency.
To determine whether the entire KSHV lytic cycle could be reactivated by conditionally relocalizing Rta to the cell nucleus, we focused our analyses on the effects of RtaΔNLS1,2-ER since it offered the most-stringent control among the ER HBD fusion proteins. As our infected model cell line, we chose easily transfectable Vero-rKSHV.219 cells (71). This is a subclone of the Vero cell line that is infected with an rKSHV clone derived from the JSC-1 PEL cell line (“rKSHV.219”). The rKSHV.219 virus constitutively expresses eGFP and puromycin resistance and produces infectious virus following various methods of inducing reactivation (71).
Among KSHV genes, the nut-1/PAN promoter is strongly activated by Rta in transient transfections, and the nut-1/PAN transcript is induced by Rta in latently infected PEL cells (47, 68). To test whether regulated nuclear localization of RtaΔNLS1,2-ER would function similarly in latently infected Vero cells, we transfected multiple dishes of Vero-rKSHV.219 cells with the RtaΔNLS1,2-ER expression vector or the empty vector. Half of each set of transfectants was treated with the inducer 4-OHT 3 h later. Analyses of total RNA by Northern blotting showed that background levels of the nut-1/PAN transcript were expressed in untreated cells that were transfected with either vector (Fig. 7A, lanes 1, 3, and 4). The small amount of nut-1 expression in the absence of 4-OHT (Fig. 7A) suggests that a low level of spontaneous reactivation is occurring; presumably, spontaneously expressed endogenous Rta transactivates nut-1. The addition of 4-OHT to the growth medium resulted in dramatic induction of nut-1/PAN in cells expressing RtaΔNLS1,2-ER, but not in vector-transfected cells (Fig. 7A, compare lane 2 with lanes 5 and 6). This proved that the induction of nut-1/PAN expression by 4-OHT required ectopic expression of the Rta fusion protein and was not mediated indirectly by effects of 4-OHT on the cells alone. The middle panel of Fig. 7A shows that the Rta transcript was only detectable in cells expressing RtaΔNLS1,2-ER, regardless of 4-OHT addition; the Northern probe detects both the endogenous ORF50 message and the ectopically expressed RtaΔNLS1,2-ER message, and the two transcripts are indistinguishable in size. These data suggested that the relocalization of Rta to the cell nucleus was capable of reactivating the KSHV lytic cycle in a fashion similar to that in PEL cells.
Similar to other DNA viruses, individual KSHV genes are expressed in an ordered cascade during reactivation, in kinetically classifiable groups (31, 44). To determine the kinetics of the expression of nut-1/PAN in response to nuclear relocalization of RtaΔNLS1,2-ER, we analyzed nut-1/PAN expression over a 64-h period following 4-OHT addition to transfected cells. The Nut-1/PAN transcript was induced at all time points in transfected cells that had been treated with 4-OHT (Fig. 7B, top panel). Nut-1/PAN was easily detected at 16 h and peaked in expression at 40 h posttransfection (22.2-fold). The lack of Nut-1/PAN expression in untreated cells demonstrates tight control of RtaΔNLS1,2-ER function at all time points. Untransfected cells were the control (Fig. 7B, first lane). Rta expression remained relatively constant in transfected cells (Fig. 7B, center panel). Finally, the cellular 7SK RNA showed strong expression regardless of transfection or the addition of 4-OHT (Fig. 7B, bottom panel). These data demonstrate that the RtaΔNLS1,2-ER protein induces Nut-1/PAN expression from the latent KSHV genome in Vero cells in a 4-OHT-dependent manner. The kinetic pattern of nut-1/PAN expression is similar to that of virus reactivated with chemicals (67), with a slightly delayed peak of expression.
The reactivation of rKSHV.219 can also be monitored by the expression of the RFP dsRed, which was cloned into the rKSHV.219 genome under the control of an ectopic copy of the lytic nut-1/PAN promoter (71). We combined the detection of RFP with indirect immunofluorescence by using our ORF57/Mta-specific antiserum (59) 24 h after adding 4-OHT to half of the cells. The results in Fig. 7C show that GFP, expressed constitutively from the rKSHV.219 virus, was easily and uniformly detectable in all Vero-rKSHV.219 cells, regardless of 4-OHT treatment. However, both RFP and Mta were expressed only in the cells that were treated with 4-OHT, and not in cells that were not treated with 4-OHT (Fig. 7C). As a negative control, Vero-rKSHV.219 transfected with empty vector and treated with 4-OHT did not express either marker of reactivation (not shown). The data shown in Fig. 7 therefore confirmed that the relocalization of Rta to infected-cell nuclei induced the expression of both RNA and protein markers of KSHV lytic reactivation.
Nuclear relocalization of Rta reactivates the entire KSHV lytic cycle.
True L genes in KSHV are those whose transcription strictly requires the prior initiation of viral DNA replication (44). In PEL cells, ectopic expression of Rta induced the expression of the true L gene K8.1 in the absence, but not the presence, of ganciclovir, an inhibitor of KSHV replication (47). Those data proved that ectopic Rta protein induced authentic KSHV reactivation in PEL cells.
To determine whether nuclear relocalization of Rta could induce true L gene expression in Vero-rKSHV.219 cells, we asked whether transcription from the true L genes K8.1, ORF20, and ORF33 (44) was induced by the addition of 4-OHT to cells transfected with the RtaΔNLS1,2-ER vector. K8.1, ORF20, and ORF33 were all transactivated following the relocalization of Rta to the nuclei of the infected Vero cells (Fig. 7D). The average range of expression for the three genes was 13- to 76-fold, as determined by real-time qRT-PCR. Negative control reactions in which RT was omitted did not yield a PCR product. Therefore, the relocalization of Rta to the nuclei of KSHV-infected Vero cells induces true L gene expression.
We sought confirmation of the real-time RT-PCR data by quantitating K8.1 protein expression in the KSHV-infected Vero cells. As shown in Fig. 7E, RtaΔNLS1,2-ER only stimulated K8.1 expression in cells treated with 4-OHT. Interestingly, Rta WT-ER induced K8.1 expression even in the absence of hormone (Fig. 7E), agreeing with the partial nuclear localization of the WT fusion protein under those conditions (Fig. 5A). As expected, the positive control transfection of the cognate WT Rta vector (without the ER-TM fusion) and treatment of the cells with the histone deacetylase inhibitor sodium butyrate induced K8.1 expression without further addition of 4-OHT (Fig. 7E).
The ultimate proof of complete, productive viral reactivation is the release of mature, infectious virus to the tissue culture medium of infected cells. At 7 days after the addition of OHT to cells transfected with the RtaΔNLS1,2-ER vector, we transferred clarified supernatants to monolayers of naïve 293 cells and detected rKSHV.219 by observing the 293 cells for GFP expression. Figure 7F shows representative images from these 293 cell monolayers, which were performed in triplicate. In the top panel of Fig. 7F, supernatant from cells transfected in the absence of 4-OHT shows a low level of spontaneously reactivated KSHV that is transmissible. The middle panel of Fig. 7F shows a reproducible, approximately sevenfold increase in the amount of infectious virus produced by 4-OHT-mediated nuclear relocalization of RtaΔNLS1,2-ER protein. Finally, this induction is equivalent to the amount of infectious virus produced by transfecting Vero-rKSHV.219 with cognate, WT Rta expression vector (Fig. 7F, bottom panels). The increase in viral titers observed in all panels of Fig. 7F was similar to the induction of latent virus shown in Fig. 7E.
In summary, our data demonstrate that the entire productive KSHV lytic cycle is induced and viral promoters are transactivated in direct concordance with the nuclear localization of the Rta protein.
Nut-1/PAN is a direct transcriptional target of Rta in infected cells.
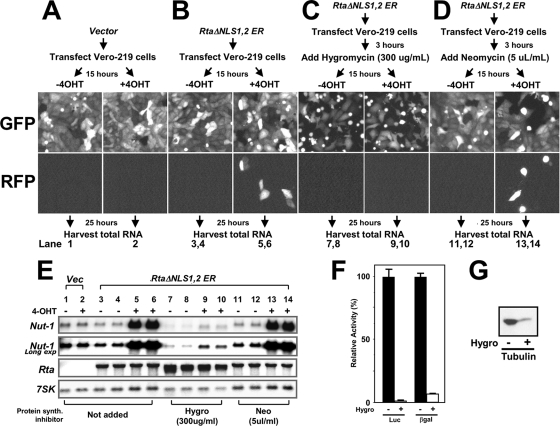
The data above showed that the fusion of RtaΔNLS1,2 to the modified HBD of the ER generated a variant of Rta (RtaΔNLS1,2-ER) whose nuclear localization, transactivation, and stimulation of the complete KSHV lytic cycle could be regulated by the addition of 4-OHT to the tissue culture growth medium. The transfection of RtaΔNLS1,2-ER into Vero-rKSHV.219 cells constituted an ideal system for identifying Rta's direct transcriptional targets in infected cells. Rta could be constitutively expressed in an inactive, cytoplasmic form and then induced to relocalize to the cell nuclei in the presence of protein synthesis inhibitors to prohibit de novo expression of any other viral or cellular transactivators. Furthermore, the shutoff of de novo protein synthesis could be confirmed by monitoring RFP expression from the recombinant virus (rKSHV.219).
Since CHX alone induced the expression of nut-1/PAN and other viral transcripts in these cells (not shown), we tested two other protein synthesis inhibitors, hygromycin and neomycin (Fig. 8A to D) (52). As expected, nut-1/PAN was not induced by 4-OHT treatment (alone) in cells transfected with the negative control empty expression vector (Fig. 8E, lanes 1 and 2, top panels). The positive controls show 4-OHT-dependent induction of nut-1/PAN in Vero-rKSHV.219 cells transfected with RtaΔNLS1,2-ER (Fig. 8E, lanes 3 to 6, top panels). In agreement, Fig. 8B shows that RFP expression, driven by the ectopic nut-1/PAN promoter, is also dependent on the addition of 4-OHT to cells transfected with RtaΔNLS1,2-ER. In cells transfected with RtaΔNLS1,2-ER and treated with either hygromycin or neomycin, nut-1/PAN induction is 4-OHT dependent (Fig. 8E, lanes 7 to 14). The results presented in Fig. 8E also show that the induction of nut-1/PAN in the presence of neomycin (lanes 13 and 14) is much more robust than in the presence of hygromycin (lanes 9 and 10). In fact, the induction of nut-1/PAN in the presence of neomycin is indistinguishable from induction without the addition of a protein synthesis inhibitor (Fig. 8E, compare lanes 11 to 14 with lanes 3 to 6). In contrast, in the presence of hygromycin, 4-OHT-dependent induction of nut-1/PAN is only detectable upon long exposure of the Northern blot (Fig. 8E, second panel down, lanes 7 to 10). Spontaneous nut-1/PAN expression (i.e., in the absence of 4-OHT) is also extinguished in the presence of hygromycin (Fig. 8E, lanes 7 and 8).
FIG. 8.
Nut-1/PAN is a direct transcriptional target of Rta in infected cells. (A, B, C, D) Strategies for testing the protein synthesis inhibitors hygromycin and neomycin in Vero-rKSHV.219 cells are given above the panels. In the experiments whose results are shown in panels B, C, and D, each half of the experimental strategy (untreated or treated with 4-OHT) was performed in duplicate; panels labeled GFP and RFP show images representative of the results of each experiment (converted to grayscale). (E) Total cellular RNAs corresponding to cells in the experiments diagrammed in panels A, B, C, and D were purified and analyzed by Northern blotting; probes are indicated at left. (F) Vero-rKSHV.219 cells were transfected with the reporter vectors pGL3-promoter (expressing firefly luciferase; Promega) and pcDNA3.1-His-lacZ (expressing β-galactosidase; Invitrogen). Cells were left untreated or treated with hygromycin as described forpanel C, and reporter expression was analyzed 40 h after the addition of hygromycin. Error bars show standard deviations. (G) Vero-rKSHV.219 cells were left untreated or treated with hygromycin as described for panel C, and endogenous tubulin expression was analyzed by Western blot 40 h after the addition of hygromycin. −, untreated; +, treated with 4-OHT; hygro, hygromycin; neo, neomycin; Luc, luciferase; βgal, β-galactosidase.
The difference in nut-1/PAN induction shown in Fig. 8E, lanes 7 to 13, might be attributable to different potencies of neomycin and hygromycin. As shown in Fig. 8D, neomycin fails to eliminate de novo expression of RFP induced by 4-OHT treatment, suggesting that translation is not inhibited by neomycin under these conditions. However, Fig. 8C shows that hygromycin extinguishes RFP expression. Therefore, we hypothesize that the more-robust nut-1/PAN expression in cells treated with neomycin compared to that in cells treated with hygromycin results from the expression of additional lytic cycle transactivators only in the neomycin-treated cells. Thus, hygromycin is the ideal protein synthesis inhibitor for the detection of direct transcriptional targets of Rta.
Two additional approaches confirmed that hygromycin extinguished de novo protein synthesis in these cells. Both firefly luciferase and β-galactosidase expression were reduced by more than 95% in Vero-rKSHV.219 cells transfected with reporter vectors (Fig. 8F). As detected by Western blotting, endogenous tubulin expression was reduced by a similar magnitude (Fig. 8G).
Identification of direct transcriptional targets of Rta in infected cells.
To identify all of Rta's direct transcriptional targets, we tested the entire KSHV transcriptome using the strategies outlined in Fig. 8A (vector transfected with or without 4-OHT) and Fig. 8C (RtaΔNLS1,2-ER transfected with or without 4-OHT). As hygromycin treatment for 40 h was cytotoxic (observe rounding in the top panels of Fig. 8C), we harvested total cellular RNA 15 h after 4-OHT addition (30 h after hygromycin addition). The KSHV transcriptome was queried by hybridizing labeled cDNAs to a KSHV oligonucleotide microarray. The microarray contains probes for every KSHV ORF and some of the alternatively spliced transcripts (as originally designed by Lu and colleagues [44]), with updated probes for newly annotated transcripts.
We analyzed the statistical significance of the gene responses using two methods and quantitated individual gene responses by real-time qRT-PCR. The statistical methods, described in detail in Materials and Methods, were (i) generating a normal (Gaussian) curve of the data and determining the number of standard deviations from the mean of each gene's response (z-score) and (ii) performing gene-specific t tests using Significance Analysis of Microarrays (SAM) software (70). The output of the SAM software is a q value, which represents the minimum false-positive rate that can be attained when calling a change in gene expression significant.
Overall, we identified eight genes that were induced at a level at least 1.1 standard deviations from the mean level of induction by relocalizing RtaΔNLS1,2-ER to the nuclei of infected cells while inhibiting de novo protein synthesis (Table 1). These genes were nut-1/PAN, ORF57/Mta, ORF56/Primase, K2/vIL-6, ORF37/SOX, K14/vOX, K9/vIRF1, and ORF52. All had q values of <0.01 (i.e., <1% chance they are false positives). As determined by real-time qRT-PCR, Rta activated six of these genes directly, over a range from 3.5- to 49.2-fold (ORF56 and nut-1/PAN, respectively). K14 and K9 induction were not quantitated by qRT-PCR since three individual sets of primers failed to specifically amplify the corresponding mRNAs.
TABLE 1.
Identification of direct transcriptional targets of the Rta protein
| Gene | No. of deviations from mean level of activationa | q valueb | Level of activation (n-fold) (SD)c |
|---|---|---|---|
| Activated directly by Rtad | |||
| Nut-1/PAN | 3.9 | <0.01 | 49.2 (2.6) |
| ORF57/Mta | 2.7 | <0.01 | 13.4 (3.0) |
| ORF56/Primase | 1.5 | <0.01 | 3.5 (1.2) |
| K2/vIL-6 | 1.4 | <0.01 | 4.1 (0.7) |
| ORF37/SOX | 1.3 | <0.01 | 3.8 (0.8) |
| K14/vOX | 1.2 | <0.01 | ND |
| K9/vIRF1 | 1.2 | <0.01 | ND |
| ORF52 | 1.1 | <0.01 | 12.7 (4.4) |
| Not activated directly by Rtae | |||
| K11.1/vIRF2 | 0.6 | <0.14 | 1.1 (0.2) |
| ORF41 | −0.1 | <0.14 | 1.0 (0.0) |
| ORF36/CHPK | −0.1 | <0.05 | 1.2 (0.7) |
| K8.1 | −0.2 | <0.09 | 1.1 (0.4) |
| ORF34 | −0.4 | <0.14 | 1.1 (0.4) |
| K10.5/vIRF3 | −0.7 | <0.14 | 0.8 (0.1) |
| ORF73/LANA-1 | −1.6 | NSC | 1.1 (0.1) |
| ORF38 | −1.6 | NSC | 1.4 (0.9) |
Values are the average number of standard deviations from the mean of the results for each microarray experiment.
Values were estimated by using Significance Analysis of Microarrays (SAM) software (70). NSC, not significantly changed.
Values were determined by real-time qPCR. ND, not determined.
Includes all viral genes activated by nuclear relocalized RtaΔNLS1,2-ER in the absence of de novo protein synthesis in Vero-rKSHV.219 cells compared to the level of activation by empty vector.
Includes a selection of viral genes not activated by nuclear relocalized. RtaΔNLS1,2-ER in the absence of de novo protein synthesis in Vero-rKSHV.219 cells compared to the level of activation by empty vector.
We also selected eight genes that were not significantly activated by relocalizing RtaΔNLS1,2-ER to the nuclei of infected cells while inhibiting de novo protein synthesis (Table 1). These genes were K11.1/vIRF2, ORF41, ORF36/CHPK, K8.1, ORF34, K10.5/vIRF3, ORF73/LANA-1, and ORF38. q values for these genes ranged from 0.05 to 0.14 (four genes), and two were not significantly changed (Table 1). As quantitated by real-time qRT-PCR, the expression level of these genes only changed from 0.8- to 1.4-fold, confirming their insignificance as direct transcriptional targets of Rta.
DISCUSSION
Reactivation of KSHV from latency in B cells is essential for KS development (4, 21, 50, 55, 61, 78). KSHV Rta is a nuclear, transcriptional transactivating protein that is necessary and sufficient for viral reactivation (24, 46, 47, 68, 79). During reactivation, transcripts encoding Rta are expressed with IE kinetics prior to the detection of KSHV DE transcripts (67, 81). Rta transactivates many KSHV promoters, but none of the corresponding genes had been formally demonstrated to be direct targets of Rta in infected cells. Since progression through the lytic cycle during reactivation is critical for KSHV pathogenesis, delineating the regulation of lytic-gene expression circuits requires the identification of the genes that are regulated directly or indirectly by Rta, independently or in combination with other viral or cellular transcriptional proteins.
To begin to answer these questions, we developed a system in which the Rta protein is the only viral transactivator expressed in KSHV-infected cells so that we could identify Rta's direct transcriptional targets. By fusing Rta to a modified HBD of the murine ER, we generated a variant of Rta whose nuclear localization (Fig. 5) and transactivation (Fig. 6) were strictly controlled by the addition of 4-OHT to the cell culture medium.
To optimize the regulation of the nuclear/cytoplasmic localization of the Rta fusion protein, we characterized two putative lysine/arginine-rich NLSs in ORF50/Rta (Fig. 1A). We showed that the putative NLS1 (aa 6 to 12) was dispensable (Fig. 1 and 3), while NLS2 (aa 516 to 530) was essential for nuclear localization (Fig. 3), transactivation (Fig. 2), and stimulation of reactivation by Rta (Fig. 7). NLS2 was also sufficient to restrict GFP to the nuclei of transfected cells (Fig. 4). The deletion of both NLSs in the ORF50-ER fusion protein generated a variant of Rta (called RtaΔNLS1,2-ER) that was expressed exclusively in the cytoplasm in the absence of 4-OHT (Fig. 5) and failed to transactivate (Fig. 6). Following the addition of 4-OHT to the medium of transfected cells, the fusion protein was redirected completely to the nucleus (Fig. 5), regained its ability to transactivate transcription (Fig. 6), and reactivated the entire KSHV lytic cycle (Fig. 7). These data suggested that the transcriptional defect in RtaΔNLS1,2 was due largely to its inability to access the nucleus and that targeting of RtaΔNLS1,2 to the nucleus in a heterologous fashion can rescue its function. Point mutations in NLS2 have been shown to eliminate Rta transactivation (9, 25), but the nuclear localization of those mutant proteins was not examined.
Our strategy for constructing the NLS1 deletion mutant of Rta, which effectively deleted aa 1 to 72 (Fig. 1A), extends our insight into the structure-function relationships of the cognate protein. We have previously shown that DNA binding and the oligomerization of WT Rta are mediated by aa 1 to 272 and 1 to 414, respectively (3, 45, 46). RtaΔNLS1 was indistinguishable from WT ORF50 in nuclear localization (Fig. 3), transactivation of the nut-1 and TK promoters (Fig. 1B and C), and viral reactivation (Fig. 1D). As both DNA binding and tetramerization of Rta are required for KSHV reactivation (3, 7), our data suggest that aa 1 to 72 are dispensable for both essential molecular interactions of Rta. Chen et al. (12) have reported that an NLS1 deletion mutant of Rta similar to ours was unable to transactivate the K9 and DNA polymerase promoters in transient assays in uninfected cells. That mutant Rta protein was partially localized to cell nuclei, so it is not clear why its transactivation phenotype differed from ours. We found that fusion with NLS1 could partially relocalize GFP to the nuclei of transfected cells, but not as dramatically as the similar GFP fusion of NLS2 (Fig. 4).
The Vero-rKSHV.219 cell line was ideal for identifying the direct transcriptional targets of Rta. The cells were easily transfectable and permitted monitoring of RFP to ensure that de novo protein synthesis was extinguished by treatment with translational inhibitors (Fig. 8). However, our observation that CHX treatment alone increased the expression of nut-1 and other viral transcripts (not shown) suggested that CHX can induce viral lytic cycle gene expression outside the context of full KSHV reactivation in these cells. This effect of CHX was probably due to its established pleiotropy in animal cells (41, 69). As hygromycin did not cause similar pleiotropic effects on these cells, we used it to extinguish protein synthesis (Fig. 8C).
The genes that were significantly activated by RtaΔNLS1,2 in the absence of de novo protein synthesis were determined by statistical analyses of the microarray data and quantitated by real-time qRT-PCR (Table 1). In our model system, the eight direct transcriptional targets of Rta were nut-1/PAN, ORF57/Mta, ORF56/Primase, K2/vIL-6, ORF37/SOX, K14/vOX, K9/vIRF1, and ORF52 (Table 1). The results of previous studies showed that sequences upstream of five of these genes, nut-1/PAN, ORF57, K2, K14, and K9, were sufficient for directing Rta-mediated, transient transactivation of reporter molecules in uninfected cells (12, 15, 34, 39, 46, 47, 63). Our data establish that in the context of the intact viral genome in infected cells (i) those six genes are indeed authentic, direct transcriptional targets of Rta and (ii) their proximal upstream sequences are authentic promoters. The mechanisms used by Rta to transactivate some of these promoters have been well defined by using transient assays. Among the transcriptional mechanisms employed by Rta, direct DNA binding and interactions with the cellular protein RBP-Jk are essential for KSHV reactivation (6, 7, 38).
However, our data provide further support for the idea that Rta-mediated transactivation is very complex. Our data demonstrate that in the context of a viral infection, Rta's target genes cannot be easily predicted only by identifying an Rta or RBP-Jk binding site in the putative promoter of a gene. For example, the nut-1/PAN, K12/kaposin, and ori-Lyt promoters all contain high-affinity Rta binding sites that are required for Rta transactivation in transient assays (10, 64). However, only nut-1/PAN was activated by Rta when we inhibited de novo protein synthesis in infected cells (Fig. 8E and Table 1). Likewise, the presence or absence of particular cis promoter elements did not distinguish between the magnitudes of Rta transactivation of particular genes. For example, the binding of RBP-Jk to the promoters of ORF57/Mta, K8/K-bZIP, K14, and ORF6 is required for Rta-mediated transactivation of those promoters in transient assays (6, 7, 37, 39). However, only ORF57/Mta was activated by Rta when we inhibited de novo protein synthesis.
In fact, the KSHV genome contains 177 consensus RBP-Jk elements (our unpublished data), yet nuclear-relocalized Rta induced only eight genes when we inhibited de novo protein synthesis (Table 1). Our data suggest that regardless of the autonomous mechanism used by Rta to transactivate promoters during reactivation, the transcription of the majority of the lytic genes require de novo expression of additional proteins. There is ample evidence for both viral and cellular proteins that could satisfy this proposed role as Rta-cooperating factors. The most apparent is the Mta transactivator protein encoded by KSHV ORF57, an essential protein whose expression at the single-cell level appears to correspond to viruses that are “committed” to full reactivation (26, 49, 59). Other lytic cycle transactivators are K9/vIRF-1, K-bZIP, and ORF49 (23, 30, 75). Therefore, the induction of some lytic genes by Rta is probably indirectly mediated through the other lytic cycle transactivators.
The requirement for de novo expression of cellular transcriptional regulatory proteins for KSHV reactivation can be interpreted by considering the results of studies of Rta transactivation in transient promoter-reporter assays in uninfected cells. These transient assays have shown that Rta transactivates many promoters from KSHV lytic cycle genes. Since the majority of these genes were not identified in the present study as direct transcriptional targets of Rta in infected cells, we hypothesize that de novo expression of cellular proteins by Rta also contributes to Rta-mediated transactivation of these promoters. For example, Rta transactivates the K-bZIP promoter in uninfected cells, yet we did not detect induction of K-bZIP by Rta-ER in infected cells when de novo protein synthesis was extinguished. Transactivation of the KSHV K-bZIP promoter by Rta requires the cellular protein RBP-Jk (7, 77). While the expression level of RBP-Jk does not change when KSHV reactivates (6), the cellular proteins Oct-1 and C/EBPα are induced during reactivation and contribute to the Rta-mediated transactivation of K-bZIP (6, 76). In fact, C/EBPα is induced by the ectopic expression of Rta. We therefore propose that in infected cells, K-bZIP transactivation by Rta requires the induction of cellular proteins that participate not just as accessory proteins in transactivation but as promoter-binding factors that are essential for reactivation.
The limited number of viral genes autonomously activated by Rta also has significant implications for KSHV pathogenesis and the progression of reactivation. Rta directly activated only one of the nine viral core replication proteins (48), ORF56/Primase (Table 1). Similarly, we have previously demonstrated that less than 10% of PEL cells ectopically expressing Rta coexpress the ORF59 (DNA polymerase processivity) protein during reactivation (46, 47). Therefore, our data provide formal proof that Rta requires additional viral and/or cellular proteins to commit the lytic program to initiate viral DNA replication. On a single-cell level, the expression of Rta is insufficient to commit every cell in an infected population to support productive lytic viral replication. Instead, Rta is sufficient to initiate the lytic reactivation program, but cellular and viral proteins expressed subsequent to Rta are key regulators of successful progression down the lytic cycle cascade.
Indeed, one direct transcriptional target of Rta identified in this study, the ORF57/Mta transactivator, does not autonomously reactivate the KSHV lytic cycle but cooperates with Rta to stimulate reactivation and is required for producing mature KSHV virions (6, 26, 47, 49, 59). “Mosaic” patterns of expression of Mta (or other proteins expressed de novo subsequent to Rta) in a population of Rta-expressing cells could determine which cells progress to complete reactivation and which ones might allow the expression of pathogenic lytic cycle genes in the absence of productive replication and cell lysis. In this regard, Rta also directly transactivated K2/vIL-6, K14/vOX, and K9/vIRF1 (Table 1), each of which encodes a protein with a potential direct effect on the abnormal cell growth phenotypes associated with KSHV infection.
The results of analyses of genes that are expressed as single units of multicistronic transcripts when reactivation is induced with chemicals suggest qualitative differences in transactivation when Rta is the only transactivator expressed. In response to chemical inducers, ORF37 is expressed in two different multicistronic messages that also contain ORF34, -35, and -36 or ORF35 and -36 (27). We detected only ORF37 as a direct target of Rta (Table 1), suggesting that transcriptional start site selection might be altered when Rta is the only transactivator expressed.
Finally, it is likely that we have not detected every direct transcriptional target of Rta using the present experimental conditions. One significant influence on our data is the temporal relationship of treatment with the two chemicals hygromycin and 4-OHT and the RNA harvest. Although we optimized the concentration and length of hygromycin treatment to inhibit de novo protein synthesis (Fig. 8), we could not avoid significant cytotoxicity of the drug under the present conditions (Fig. 8C). The cytotoxicity might have globally lowered intracellular transcript levels. The effective intracellular concentration of the Rta-ER fusion protein might also influence the activation of different genes in a promoter-specific fashion. We optimized our experimental conditions for detection of the nut-1/PAN transcript (Fig. 7 and 8). Two alterations of our protocol might allow for higher intracellular concentrations of the Rta-ER fusion protein: delaying the time prior to hygromycin treatment or altering the time period between 4-OHT addition and RNA harvest. Increased concentrations of the Rta-ER fusion protein might reveal additional targets of Rta that require a higher threshold of the activator. Of course, increased intracellular concentrations of Rta might globally result in decreased overall RNA expression, as Rta is proapoptotic in heterologous systems (58). By blocking de novo protein synthesis, we are likely inhibiting the expression of antiapoptotic proteins that counteract Rta's putative stimulation of programmed cell death in our system. Finally, since CHX induced viral transcription independent of Rta expression (not shown), it is conceivable that hygromycin had the same effect on a subset of Rta's targets and that subsequent relocalization of Rta to cell nuclei resulted in undetectable changes in the level of expression of those transcripts.
Acknowledgments
We gratefully thank Jeff Vieira for the gift of Vero-rKSHV.219 cells and Gerard Evan for the gift of the ER clone.
The research was supported by the American Cancer Society, the American Heart Association, the UMDNJ Foundation, and the NJ Medical School/University Hospital Cancer Center.
Footnotes
Published ahead of print on 20 August 2008.
REFERENCES
- 1.Abramoff, M., P. Magelhaes, and S. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 1136-42. [Google Scholar]
- 2.Ambroziak, J., D. Blackbourn, B. Herndier, R. Glogan, J. Gullet, A. McDonald, E. Lennette, and J. Levy. 1995. Herpesvirus-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268582-583. [DOI] [PubMed] [Google Scholar]
- 3.Bu, W., K. D. Carroll, D. Palmeri, and D. M. Lukac. 2007. The Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50/Rta lytic switch protein functions as a tetramer. J. Virol. 815788-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, T. B., M. Borok, L. Gwanzura, S. MaWhinney, I. E. White, B. Ndemera, I. Gudza, L. Fitzpatrick, and R. T. Schooley. 2000. Relationship of human herpesvirus 8 peripheral blood virus load and Kaposi's sarcoma clinical stage. AIDS 142109-2116. [DOI] [PubMed] [Google Scholar]
- 5.Cannon, M., E. Cesarman, and C. Boshoff. 2006. KSHV G protein-coupled receptor inhibits lytic gene transcription in primary-effusion lymphoma cells via p21-mediated inhibition of Cdk2. Blood 107277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, K. D., W. Bu, D. Palmeri, S. Spadavecchia, S. J. Lynch, S. A. Marras, S. Tyagi, and D. M. Lukac. 2006. Kaposi's sarcoma-associated herpesvirus lytic switch protein stimulates DNA binding of RBP-Jk/CSL to activate the Notch pathway. J. Virol. 809697-9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, K. D., F. Khadim, S. Spadavecchia, D. Palmeri, and D. M. Lukac. 2007. Direct interactions of KSHV/HHV-8 ORF50/Rta protein with the cellular protein Octamer-1 and DNA are critical for specifying transactivation of a delayed-early promoter and stimulating viral reactivation. J. Virol. 818451-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesarman, E., and E. A. Mesri. 2007. Kaposi sarcoma-associated herpesvirus and other viruses in human lymphomagenesis. Curr. Top. Microbiol. Immunol. 312263-287. [DOI] [PubMed] [Google Scholar]
- 9.Chang, P. J., and G. Miller. 2004. Autoregulation of DNA binding and protein stability of Kaposi's sarcoma-associated herpesvirus ORF50 protein. J. Virol. 7810657-10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, P. J., D. Shedd, L. Gradoville, M. S. Cho, L. W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 763168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, P. J., D. Shedd, and G. Miller. 2005. Two subclasses of Kaposi's sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J. Virol. 798750-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J., K. Ueka, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 748623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coscoy, L. 2007. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat. Rev. Immunol. 7391-401. [DOI] [PubMed] [Google Scholar]
- 14.Damania, B., J. H. Jeong, B. S. Bowser, S. M. DeWire, M. R. Staudt, and D. P. Dittmer. 2004. Comparison of the Rta/Orf50 transactivator proteins of gamma-2-herpesviruses. J. Virol. 785491-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, H., M. J. Song, J. T. Chu, and R. Sun. 2002. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 768252-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dittmer, D. P., C. M. Gonzalez, W. Vahrson, S. M. DeWire, R. Hines-Boykin, and B. Damania. 2005. Whole-genome transcription profiling of rhesus monkey rhadinovirus. J. Virol. 798637-8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146403-413. [DOI] [PubMed] [Google Scholar]
- 19.Eilers, M., D. Picard, K. R. Yamamoto, and J. M. Bishop. 1989. Chimaeras of Myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature 34066-68. [DOI] [PubMed] [Google Scholar]
- 20.Engelke, U., D. M. Wang, and J. S. Lipsick. 1997. Cells transformed by a v-Myb-estrogen receptor fusion differentiate into multinucleated giant cells. J. Virol. 713760-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engels, E. A., R. J. Biggar, V. A. Marshall, M. A. Walters, C. J. Gamache, D. Whitby, and J. J. Goedert. 2003. Detection and quantification of Kaposi's sarcoma-associated herpesvirus to predict AIDS-associated Kaposi's sarcoma. AIDS 171847-1851. [DOI] [PubMed] [Google Scholar]
- 22.Ganem, D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1273-296. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez, C. M., E. L. Wong, B. S. Bowser, G. K. Hong, S. Kenney, and B. Damania. 2006. Identification and characterization of the Orf49 protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 803062-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 746207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gwack, Y., H. Nakamura, S. H. Lee, J. Souvlis, J. T. Yustein, S. Gygi, H. J. Kung, and J. U. Jung. 2003. Poly(ADP-ribose) polymerase 1 and Ste20-like kinase hKFC act as transcriptional repressors for gamma-2 herpesvirus lytic replication. Mol. Cell. Biol. 238282-8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han, Z., and S. Swaminathan. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 805251-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haque, M., V. Wang, D. A. Davis, Z. M. Zheng, and R. Yarchoan. 2006. Genetic organization and hypoxic activation of the Kaposi's sarcoma-associated herpesvirus ORF34-37 gene cluster. J. Virol. 807037-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollenberg, S. M., P. F. Cheng, and H. Weintraub. 1993. Use of a conditional MyoD transcription factor in studies of MyoD transactivation and muscle determination. Proc. Natl. Acad. Sci. USA 908028-8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izumiya, Y., S. F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H. J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 771441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayachandra, S., K. G. Low, A. E. Thlick, J. Yu, P. D. Ling, Y. Chang, and P. S. Moore. 1999. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc. Natl. Acad. Sci. USA 9611566-11571. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Jenner, R., M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 751798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1488-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 743586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotz, S., and N. Johnson (ed.). 1985. Encyclopedia of Statistical Sciences, vol. 6, p. 333. John Wiley & Sons, New York, NY. [Google Scholar]
- 36.Lee, B. S., M. Paulose-Murphy, Y. H. Chung, M. Connlole, S. Zeichner, and J. U. Jung. 2002. Suppression of tetradecanoyl phorbol acetate-induced lytic reactivation of Kaposi's sarcoma-associated herpesvirus by K1 signal transduction. J. Virol. 7612185-12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang, Y., J. Chang, S. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jk, the target of the Notch signaling pathway. Genes Dev. 161977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang, Y., and D. Ganem. 2003. Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. USA 1008490-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang, Y., and D. Ganem. 2004. RBP-J (CSL) is essential for activation of the K14/vGPCR promoter of Kaposi's sarcoma-associated herpesvirus by the lytic switch protein RTA. J. Virol. 786818-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao, W., Y. Tang, S. F. Lin, H. J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 773809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin, W. W., and Y. W. Hsu. 2000. Cycloheximide-induced cPLA(2) activation is via the MKP-1 down-regulation and ERK activation. Cell. Signal. 12457-461. [DOI] [PubMed] [Google Scholar]
- 42.Littlewood, T., D. Hancock, P. Danielian, M. Parker, and G. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 231686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 44.Lu, M., J. Suen, C. Frias, R. Pfeiffer, M. H. Tsai, E. Chuang, and S. L. Zeichner. 2004. Dissection of the Kaposi's sarcoma-associated herpesvirus gene expression program by using the viral DNA replication inhibitor cidofovir. J. Virol. 7813637-13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukac, D., L. Garibyan, J. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by the Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 756786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 739348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252304-312. [DOI] [PubMed] [Google Scholar]
- 48.Lukac, D. M., and Y. Yuan. 2007. Reactivation and lytic replication of KSHV, p. 434-460. In A. Arvin, G. Campadelli-Fiume, E. Mocarski, P. Moore, B. Roizman, R. Whitley, and K. Yamanishi (ed.), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 49.Majerciak, V., N. Pripuzova, J. P. McCoy, S. J. Gao, and Z. M. Zheng. 2007. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8α, and K8.1 and the production of infectious virus. J. Virol. 811062-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin, D. F., B. D. Kuppermann, R. A. Wolitz, A. G. Palestine, H. Li, C. A. Robinson, et al. 1999. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. N. Engl. J. Med. 3401063-1070. [DOI] [PubMed] [Google Scholar]
- 51.Mesri, E. A., E. Cesarman, L. Arvanitakis, S. Rafii, M. A. Moore, D. N. Posnett, D. M. Knowles, and A. S. Asch. 1996. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J. Exp. Med. 1832385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327389-394. [DOI] [PubMed] [Google Scholar]
- 53.Montaner, S., A. Sodhi, A. Molinolo, T. H. Bugge, E. T. Sawai, Y. He, Y. Li, P. E. Ray, and J. S. Gutkind. 2003. Endothelial infection with KSHV genes in vivo reveals that vGPCR initiates Kaposi's sarcomagenesis and can promote the tumorigenic potential of viral latent genes. Cancer Cell 323-36. [DOI] [PubMed] [Google Scholar]
- 54.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 3321181-1185. [DOI] [PubMed] [Google Scholar]
- 55.Moore, P. S., L. A. Kingsley, S. D. Holmberg, T. Spira, P. Gupta, D. R. Hoover, J. P. Parry, L. J. Conley, H. W. Jaffe, and Y. Chang. 1996. Kaposi's sarcoma-associated herpesvirus infection prior to onset of Kaposi's sarcoma. AIDS 10175-180. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 774205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicholas, J. 2007. Human herpesvirus 8-encoded proteins with potential roles in virus-associated neoplasia. Front. Biosci. 12265-281. [DOI] [PubMed] [Google Scholar]
- 58.Nishimura, K., K. Ueda, S. Sakakibara, E. Do, E. Ohsaki, T. Okuno, and K. Yamanishi. 2003. A viral transcriptional activator of Kaposi's sarcoma-associated herpesvirus (KSHV) induces apoptosis, which is blocked in KSHV-infected cells. Virology 31664-74. [DOI] [PubMed] [Google Scholar]
- 59.Palmeri, D., S. Spadavecchia, K. Carroll, and D. M. Lukac. 2007. Promoter and cell-specific transcriptional activation by the Kaposi's sarcoma-associated herpesvirus ORF57/Mta protein. J. Virol. 8113299-13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2342-346. [DOI] [PubMed] [Google Scholar]
- 61.Robles, R., D. Lugo, L. Gee, and M. A. Jacobson. 1999. Effect of antiviral drugs used to treat cytomegalovirus end-organ disease on subsequent course of previously diagnosed Kaposi's sarcoma in patients with AIDS. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 2034-38. [DOI] [PubMed] [Google Scholar]
- 62.Saveliev, A., F. Zhu, and Y. Yuan. 2002. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi's sarcoma-associated herpesvirus. Virology 299301-314. [DOI] [PubMed] [Google Scholar]
- 63.Song, M., H. Brown, T.-T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 753129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song, M. J., H. Deng, and R. Sun. 2003. Comparative study of regulation of RTA-responsive genes in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 779451-9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 765000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spitkovsky, D., P. Steiner, J. Lukas, E. Lees, M. Pagano, A. Schulze, S. Joswig, D. Picard, M. Tommasino, M. Eilers, et al. 1994. Modulation of cyclin gene expression by adenovirus E1A in a cell line with E1A-dependent conditional proliferation. J. Virol. 682206-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun, R., S.-F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 732232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 9510866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang, D., J. M. Lahti, J. Grenet, and V. J. Kidd. 1999. Cycloheximide-induced T-cell death is mediated by a Fas-associated death domain-dependent mechanism. J. Biol. Chem. 2747245-7252. [DOI] [PubMed] [Google Scholar]
- 70.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vieira, J., and P. M. O'Hearn. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325225-240. [DOI] [PubMed] [Google Scholar]
- 72.Wang, S., S. Liu, M. Wu, Y. Geng, and C. Wood. 2001. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch. Virol. 1461415-1426. [DOI] [PubMed] [Google Scholar]
- 73.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 7511961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, S. E., F. Y. Wu, H. Chen, M. Shamay, Q. Zheng, and G. S. Hayward. 2004. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J. Virol. 784248-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, S. E., F. Y. Wu, M. Fujimuro, J. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of CCAAT/enhancer-binding protein alpha (C/EBPα) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 779590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, Y., and Y. Yuan. 2007. Essential role of RBP-Jkappa in activation of the K8 delayed-early promoter of Kaposi's sarcoma-associated herpesvirus by ORF50/RTA. Virology 35919-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, F. E. Suggett, D. M. Aldam, A. S. Denton, et al. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346799-802. [DOI] [PubMed] [Google Scholar]
- 79.Xu, Y., D. P. AuCoin, A. R. Huete, S. A. Cei, L. J. Hanson, and G. S. Pari. 2005. A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 793479-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu, F., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 735556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ziegelbauer, J., A. Grundhoff, and D. Ganem. 2006. Exploring the DNA binding interactions of the Kaposi's sarcoma-associated herpesvirus lytic switch protein by selective amplification of bound sequences in vitro. J. Virol. 802958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]