Abstract
MicroRNAs (miRNAs) play a central role in the regulation of multiple biological processes including the maintenance of stem cell self-renewal and pluripotency. Recently, the miRNA cluster miR302-367 was shown to be differentially expressed in embryonic stem cells (ESCs). Unfortunately, very little is known about the genomic structure of miRNA-encoding genes and their transcriptional units. Here, we have characterized the structure of the gene coding for the human miR302-367 cluster. We identify the transcriptional start and functional core promoter region which specifically drives the expression of this miRNA cluster. The promoter activity depends on the ontogeny and hierarchical cellular stage. It is functional during embryonic development, but it is turned off later in development. From a hierarchical standpoint, its activity decays upon differentiation of ESCs, suggesting that its activity is restricted to the ESC compartment and that the ESC-specific expression of the miR302-367 cluster is fully conferred by its core promoter transcriptional activity. Furthermore, algorithmic prediction of transcription factor binding sites and knockdown studies suggest that ESC-associated transcription factors, including Nanog, Oct3/4, Sox2, and Rex1 may be upstream regulators of miR302-367 promoter. This study represents the first identification, characterization, and functional validation of a human miRNA promoter in stem cells. This study opens up new avenues to further investigate the upstream transcriptional regulation of the miR302-367 cluster and to dissect how these miRNAs integrate in the complex molecular network conferring stem cell properties to ESCs.
MicroRNAs (miRNAs) are endogenous small noncoding RNAs ranging from 19 to 25 nucleotides (nt) in length, encoded by the genome, and transcribed as long primary transcripts (pri-miRNAs) (19). The biogenesis of mature miRNAs involves both nuclear and cytoplasmic processing steps. Within the nucleus, pri-miRNAs are thought to function as substrates for capping, polyadenylation, splicing, and editing reactions (2, 22, 25). Then, they are cropped by the RNase III Drosha (20) to the hairpin-shaped precursor (pre-miRNA) which is exported to cytoplasm through the nuclear transporter exportin 5 (23). Subsequently, the cytoplasmic Dicer (another RNase III-like endonuclease) processes the pre-miRNAs into 20- to 22-nt duplexes (16), with one strand being preferentially loaded onto the RNA-induced silencing complex and acting as the mature miRNA. This ribonucleoprotein complex acts as a negative gene expression regulator promoting the direct inhibition of mRNA translation and/or the destabilization of the target mRNAs (3). The target recognition is mediated by a nonperfect base pairing between the miRNA and a region within the 3′ untranslated region of the messenger.
miRNAs are phylogenetically conserved and have been shown to be instrumental in a wide variety of key biological processes including cell cycle regulation, apoptosis, control of metabolic pathways, imprinting, differentiation, and maintenance of “stemness,” among others (1), suggesting that miRNAs might be as important as transcription factors in controlling gene expression in higher eukaryotes.
To date, abundant information has been gained in the identification and clustering of miRNAs. In fact, more than 500 human miRNAs have been annotated in the MirBase (7). However, a better understanding of their targets and elucidation of their specific biological functions remain to be pursued. Also, our knowledge about the transcription of miRNA-encoding genes, which is the first step of miRNA biogenesis, is very limited. Expanding our knowledge about the transcription of miRNA-encoding genes should offer insights about miRNA regulation. Intergenic miRNA genes are believed to contain their own transcriptional units whereas intronic miRNAs or those associated with Alu mobile elements in humans are more likely transcribed by their host genes (5). Although polymerase III (Pol III)-driven transcription has been reported for miRNAs interspersed with Alu repeats (5), experimental evidence suggests that miRNAs are mostly class II genes (21). Few miRNA promoters have been empirically characterized (6, 11, 13, 14, 21, 25), while some others have just been inferred from biocomputing analysis (28). Importantly, the location and characterization of the core promoter region of miRNA genes are essential to understand the mechanisms and conditions of their activation.
As noted above, one of the biological functions of miRNAs seems to be the regulation of self-renewal versus differentiation in stem cells (4, 8). Accordingly, some miRNAs are differentially expressed in stem cells, suggesting a specialized but incompletely understood role in stem cell regulation (10, 24). In addition, even though the miRNAs are phylogenetically conserved, very little information is available regarding the miRNA expression profile and specificity in hierarchical versus ontogenetically different stages of stem cell development.
The polycistronic cluster of miRNAs miR302-367 was identified to be expressed in human embryonic stem cells (hESCs) and their malignant counterpart, human embryonic carcinoma cells (hECCs) (24). The murine homologue to human miR302a had been previously cloned also from murine ESCs (mESCs) but was not detectable in other adult cell lines, suggesting that this miRNA cluster may be specifically expressed at the embryonic ontogeny stage (10). The human cluster miR302-367 contains eight different miRNAs cotranscribed in a polycistronic way (24): miR302a, miR302a*, miR302b, miR302b*, miR302c, miR302c*, miR302d, and miR367. The first seven constitute the miR302 family with a highly conserved sequence.
Based on the specific expression of this miR302-367 cluster in hESCs and hECCs and its potential underlying regulatory mechanism in tuning stem cell properties, we wanted to expand our knowledge about the biology and transcription of this miRNA cluster. We provide key insights into the structure of the gene coding for the human miRNA cluster miR302-367. It is located within the first intron of the gene. Importantly, we have identified the transcriptional start and functional core promoter region, demonstrating the robustness of this promoter in driving the expression of this miRNA cluster. Functionally, we show that the promoter transcriptional activity depends on the ontogeny and hierarchical cellular stage. This study identifies and experimentally characterizes a new independent miRNA promoter which, to the best of our knowledge, represents the first human miRNA promoter characterized and functionally validated in human stem cells. Importantly, this study suggests that key ESC-specific factors such as Nanog, Oct3/4, Rex1, and Sox2 act as upstream regulators of the miR302-367 cluster. Taken together, this study opens up new avenues to further investigate the upstream transcriptional regulation of the miR302-367 cluster-encoding gene and to dissect how these molecules fit in the complex molecular network conferring stemness properties to ESCs.
MATERIALS AND METHODS
Cell culture.
Human ECC line NTERA-2 was obtained from the ATCC and grown in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 4 mM l-glutamine at 37°C in a 10% CO2 atmosphere. D3 mESCs were cultured on 0.1% gelatin-coated flasks in DMEM supplemented with 10% FBS, 2 mM l-glutamine and 103 units/ml leukemia inhibitory factor ([LIF] ESGRO; Chemicon, Temecula, CA). For differentiation studies, D3 cells were seeded on nonadherent plates and cultured in DMEM supplemented with 10% FBS and 2 mM l-glutamine in the absence or presence of 100 μM of retinoic acid (RA). HS293 (kindly provided by Outi Hovatta, Karolinska Institute, Stockholm, Sweden) and BGV01 hESCs were maintained on mitomycin C-inactivated human foreskin fibroblasts in knockout (KO) DMEM (Gibco, Invitrogen, Grand Island, NY) or DMEM-F12, respectively, supplemented with 20% KO serum replacement (Gibco-Invitrogen), 2 mM nonessential amino acids, 2 mM l-glutamine, 10 nM β-mercaptoethanol, and 4 ng/ml basic fibroblast growth factor (bFGF; Invitrogen). SHEF1 (kindly provided by Peter Andrews, University of Sheffield, United Kingdom) and HS181 (kindly provided by Outi Hovatta) hESC lines were maintained on Matrigel-coated flasks in human foreskin fibroblast-conditioned medium supplemented with 8 ng/ml bFGF. Medium was replaced daily. For differentiation assays, HS181 hESCs were maintained on Matrigel-coated flasks in the presence of differentiation medium (80% KO DMEM, 20% FBS, 2 mM nonessential amino acids, 2 mM l-glutamine, 10 nM β-mercaptoethanol, 10 μM RA, and 50 ng/ml BMP4) that was replaced every 3 days.
Non-stem cell lines NIH 3T3, 293T, REH, and MCF-7 were maintained in DMEM with 10% FBS and 2 mM l-glutamine. Human adipose tissue-derived mesenchymal stem cells (hMSCs; kindly provided by Javier Garcia-Castro, CIBM, Granada, Spain) were derived and expanded as previously described in detail (26).
miRNA profiling.
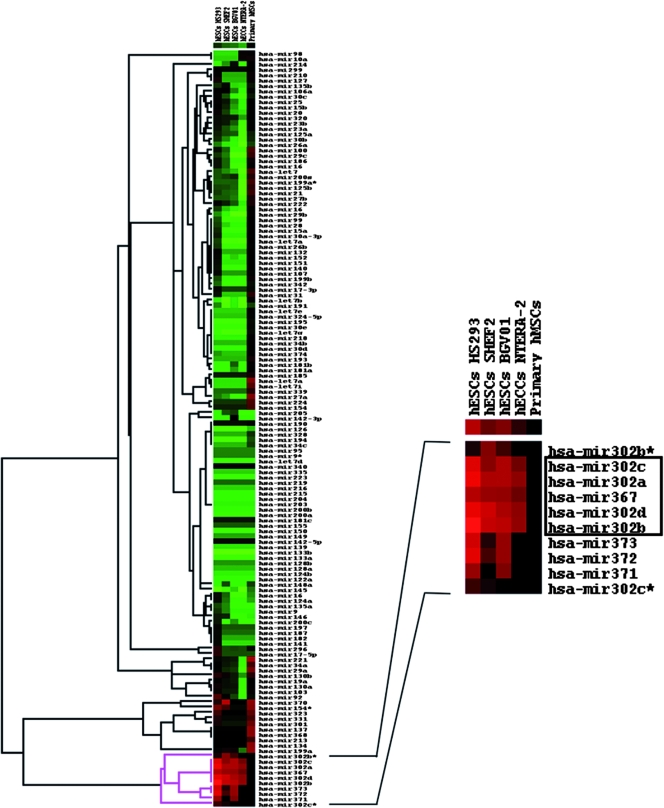
Total RNA was obtained with Trizol reagent (Invitrogen). miRNA profiling was performed by real-time PCR using the TaqMan MicroRNA assay human panel (Applied Biosystems, Foster City, CA) following the manufacturer's instructions. Ten nanograms of total RNA was used for each reaction. PCRs were repeated at least twice, and the mean relative expression level was calculated. The housekeeping glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used for data normalization (Applied Biosystems). An equimolar mixture of commercially available total RNA from 19 (adipose, bladder, brain, cervix, colon, esophagus, heart, kidney, liver, lung, ovary, prostate, skeletal muscle, small intestine, spleen, testes, thymus, thyroid and trachea) human normal adult tissues (Ambion, Austin, TX) was used as the reference sample to establish the relative miRNA expression levels in different hESCs, hMSCs and hECCs. Heat map representations and clustering of data (Fig. 1) were performed using the Cluster and Treeview software programs (Eissen Lab, Lawrence Berkeley National Laboratory and University of California at Berkeley).
FIG. 1.
Heat map representation of relative miRNA expression profiling. Expression levels of a panel of 170 miRNAs were determined by quantitative real-time RT-PCR. Data were normalized against GAPDH mRNA levels, and the expression level is relative to the adult tissue mix reference sample. Pure green spots represent at least 20-fold downregulation of the miRNA in the cell line under study in comparison to the reference sample. Pure red spots indicate at least 20-fold upregulation of the miRNA in the sample under study in comparison to the reference. Black spots indicate no difference in the miRNA expression level in the sample compared to the reference. Heat map representation and data clustering were performed with Cluster and Treeview software.
Northern blotting.
For intracellular localization experiments, total RNA was obtained with Trizol, and nuclear and cytoplasmic RNA fractions were obtained as described by Hwang and coworkers (12). RNA samples were resolved in 12% denaturing polyacrylamide gels and transferred to a nylon membrane using a semidry transfer unit. Membranes were hybridized in PerfectHyb Plus buffer (Sigma, St. Louis, MO) using 5′ end radiolabeled oligonucleotide probes (see Table S1 in the supplemental material). Briefly, 5 pmol of the oligonucleotide was labeled with 70 μCi of [γ32P]ATP and 1 unit of T4 polynucleotide kinase for 45 min at 37°C. The probe was then purified through a Sephadex G25 chromatography column to eliminate the nonincorporated radionucleotide. The whole probe was used for the hybridization step. The Northern blots were visualized, and signals were quantified using a phosphorimaging system (Storm 420; GE Healthcare, Buckinghamshire, United Kingdom). For pri-miRNA detection, poly(A)+ and poly(A)− fractions were obtained from total RNA using a Micro(A) Purist kit (Ambion). Samples were resolved in a 4% denaturing polyacrylamide gel. Transfer and hybridization were performed as described above. The probe was made by mixing equimolar amounts of three oligonucleotides designed to anneal within exons 1, 2, and 3 (see Table S1 in the supplemental material). Probe labeling, hybridization, and imaging steps were performed as mentioned above. The approximate size of the band assigned as putative pri-miRNA was extrapolated from comparison to electrophoretic mobility of the 18S rRNA (1.9 kb) visualized on the gel by ethidium bromide staining.
RACE assays.
5′ Rapid amplification of cDNA ends (RACE) assays were performed as follows. 5′ Triphosphates were eliminated by treatment of 15 μg of total RNA with 30 units of highly concentrated calf intestinal alkaline phosphatase (Promega, Madison, WI) in the presence of 40 units of RNase inhibitor (Applied Biosystems). Thus, only primary capped RNAs would conserve a 5′ phosphate group. Calf intestinal alkaline phosphatase was eliminated by phenol extraction and ethanol precipitation. Then, the cap structure was removed by treatment with tobacco acid pyrophosphatase (TAP; Epicentre Technologies, Madison, WI) at 37°C for 60 min in a total reaction volume of 50 μl containing 50 mM sodium acetate (pH 6.0), 10 mM EDTA, 1% β-mercaptoethanol, and 0.1% Triton X-100. Control RNA was incubated under the same conditions in the absence of the enzyme. Reactions were stopped by phenol-chloroform extraction, followed by ethanol-sodium acetate precipitation. Precipitated RNAs were redissolved in water and mixed with 500 pmol of 5′ RNA adapter (5′-GGC GAC UGG AGC ACG AGG ACA CUG ACA UGG ACU GAA GGA GUA GAA A-3′). The adapter was synthesized by in vitro T7 polymerase transcription using the oligonucleotide 5′-RACET7GGADAPTOR, T7 primer (see Table S1 in the supplemental material), and the RiboMAX large-scale RNA production system-T7 (Promega). A mixture of treated total RNA and RNA adaptor was heat denatured at 65°C for 5 min and then chilled on ice for 15 min. The adapter was ligated at 15°C for 24 h with 20 units of T4 RNA ligase (New England Biolabs, Ipswich, MA) in the presence of 40 units of RNase inhibitor. The RNA was extracted by a conventional phenol-chloroform method, ethanol precipitated, and then reverse transcribed with random hexamers and a High-Capacity cDNA reverse transcription kit (Applied Biosystems), according to the manufacturer's instructions. The products of reverse transcription (RT) were amplified by the use of a 4-μl aliquot of the 20-μl RT reaction mixture, 20 pmol of each gene-specific (5′RACE_GSprimer1 or 5′RACE_GSprimer2) (see Table S1 in the supplemental material) and adapter-specific (5′RACE_primer) (see Table S1 in the supplemental material) primer, a 250 μM concentration of each deoxynucleoside triphosphate, 2 units of Ampli-Taq Gold DNA polymerase (Applied Biosystems), and 1× Taq buffer. Products were separated on 2% agarose gels, and bands of interest were excised, gel eluted, cleaned up, and cloned into pGemT vector (Promega). Independent clones were analyzed and sequenced.
In order to determine the 3′ end of the primary transcript, we took advantage of the poly(A) tail and the known 5′ end to perform a complete cDNA RT-PCR. For retrotranscription, we used 0.5 μg of a poly(A)+ RNA fraction from NTERA-2 cells, and 20 pmol of oligo(dT)15 with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Reaction mixture lacking the enzyme and the RNase inhibitor was heat denatured for 2 min at 70°C and cooled down to 25°C. Then, 7.5 units of enzyme and 40 units of RNase inhibitor were added. The reaction was performed in two sequential steps of 30 min at 25°C and 42°C, respectively, with a final inactivation step of 5 min at 85°C. For the first round of amplification, the oligonucleotide 5′cDNA_hs302 (see Table S1 in the supplemental material) and oligo(dT) were used together with 1 μl of the 20-μl RT reaction mixture. The PCR protocol was adapted to allow specific hybridization of both primers: 95°C for 2 min; 30 cycles of 95°C for 30 s, 55°C for 20 s, 30°C for 20 s, 55°C for 20 s, and 72°C for 30 s; and 72°C for 10 min. Then, a reamplification was carried out with the oligonucleotide 5′cDNA_hs302Nested (see Table S1 in the supplemental material) and oligo(dT), and products were resolved on a 2% agarose gel, eluted, cleaned up, and cloned into pGemT vector. Clones were analyzed and sequenced.
Cell transfection.
All cell lines (293T, NIH 3T3, REH, MCF-7, hMSCs, hECCs, and mESCs) were routinely transfected with Lipofectamine 2000 following the manufacturer's instructions (unless otherwise indicated) except hESCs that were transfected by nucleofection. Nucleofection assays were carried out with the Amaxa Nucleofector (Amaxa, Gaithersburg, MD) using hESC Nucleofection Kit II (Amaxa) and the program A23, as previously described (17).
Luciferase assays.
Firefly and Renilla luciferase measurements were done with a Dual-Luciferase Reporter Assay System (Promega). All experiments were performed in triplicate, and data are shown as normalized means ± standard deviations.
Retrotranscription and real-time PCR.
Total RNA was purified with Trizol reagent. One microgram of total RNA was used for the RT reaction with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and random hexamers, according to the manufacturer's instructions. Real-time PCR was performed with Quatitect Sybr green PCR Master Mix (Qiagen, Valencia, CA) and analyzed with an Mx3005P real-time PCR system (Stratagene, La Jolla, CA). For primer sequences see Table S1 in the supplemental material.
RNA interference-mediated knockdown assays.
Specific small interfering RNAs (siRNAs) and scrambled control siRNAs were purchased from Sigma. siRNA sequences are shown in Table S1 in the supplemental material. NTERA-2 cells were cotransfected with pGL3-PROM_974 (200 ng), Renilla reporter plasmid (30 ng), and the corresponding siRNA (2 μg) using Lipofectamine 2000, following the manufacturer's instructions. Experiments were performed in triplicate. Cells were analyzed 24 h after transfection.
RESULTS
Cell type specificity of the miR302-367 cluster expression.
Large-scale miRNA expression profiling efforts have been hindered due to the cumbersome nature and low sensitivity of conventional Northern blot analysis. Indeed, the information available about the cell type specificity of the human miR302-367 cluster using Northern blotting accounts for only a few cell lines including hESCs (24, 27). Higher-resolution methodologies such as microarray and real-time PCR assays further supported the high-level expression of this cluster in mESCs, hESCs, and hECCs (teratocarcinoma cell lines, e.g., NTERA-2) (9, 15, 18).
We have used a real-time PCR assay to investigate the expression profile of over 170 miRNAs in different primary hESC lines (HS293, SHEF2, and BG01V), an hECC cell line (NTERA-2), and primary adult hMSCs. The aim of this miRNA profiling was to determine the cell type specificity of this cluster expression rather than quantifying its abundance in each cell type. Thus, we compared the expression level of these miRNAs in each stem cell sample with the corresponding level of expression in a mixture of commercially available total RNA from 19 human normal adult tissues (see Materials and Methods). Our results confirm high-level expression of all miRNAs that integrate the cluster miR302-367 in all hESC lines studied and hECCs. Strikingly, these miRNAs are barely detectable in hMSCs and normal adult tissues (data not shown). In fact, when the miRNA expression data were compared to the reference sample, more than 20-fold overexpression was detected (Fig. 1). These data along with the rapid downregulation of the endogenous miR302-367 cluster expression upon differentiation (18, 27; also Fig. S1 in the supplemental material) provide robust experimental support toward a potential role for this miR302-367 cluster in the biology of ESCs.
Intracellular localization of miR302 family.
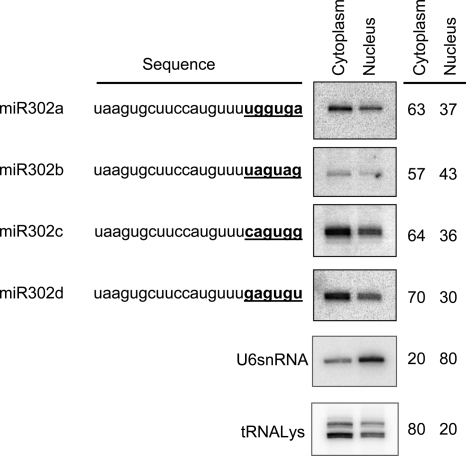
The major members of the miR302 family (302a, 302b, 302c, and 302d) share a high sequence homology, differing only in the 3′ hexanucleotides. The 3′ sequence motifs found are UGGUGA, UAGUAG, CAGUGG, and GAGUGU for miR302a, miR302b, miR302c, and miR302d, respectively (Fig. 2). Interestingly, it has recently been reported that the 3′ hexanucleotide of miR29b (AGUGUU) has a specific role acting as a nuclear localization element that promotes nuclear enrichment of this molecule (12). We wanted to investigate whether the sequence variation in the 3′ end of these miR302 members also determines a similar nuclear localization. Northern blotting was used to determine the ratio of nuclear/cytoplasmic expression levels of miRNAs miR302a, miR302b, miR302c, and miR302d in hECCs (Fig. 2). Membranes were probed with U6 snRNA and lysine-tRNA as nuclear and cytoplasmic markers, respectively, to verify subcellular fractionation (less than 20% reciprocal leaking between fractions was consistently detected). In contrast to the previously reported 3′ hexanucleotide of miR29b but similar to the miR29a, the four miR302 members studied displayed a predominant cytoplasmic localization (range, 57% to 70%) with no relevant differences detected among them (Fig. 2), suggesting that the 3′ hexanucleotide does not promote nuclear localization for any of these miR302 family members.
FIG. 2.
Intracellular distribution of miR302 family members. Northern blot analysis was used to determine the nuclear versus cytoplasmic abundance of miR302a, miR302b, miR303c, and miR302d. All membranes were reprobed with U6 snRNA and tRNA-Lys to verify proper RNA fractionation.
Structure of the genomic and transcriptional units of the cluster miR302-367.
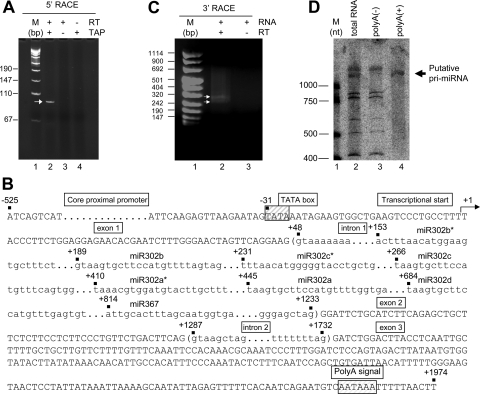
To date, the only available information about the genomic structure and transcription of this miRNA cluster is limited to its genomic localization in human chromosome 4 and the fact that these eight miRNAs are cotranscribed in a polycistronic way (24). Unfortunately, no data are available about the genomic structure of the miR302-367-encoding gene or its transcriptional units. Here, we first aimed at unraveling the nature of the pri-miRNA by means of 5′ and 3′ RACE assays. As described in detail in Materials and Methods, we used an adaptation of the 5′ RACE standard protocol that allowed us to simultaneously determine the transcription start and the capping status of the transcript. Total RNA extracted from NTERA-2 was dephosphorylated so that only cap-protected RNAs retain the 5′ phosphate groups. Upon removing the cap structure with TAP treatment, the modified RNA was ligated to the RACE adaptor RNA. Then, potential ligated molecules were retrotranscribed and amplified using gene-specific and adaptor-specific primers. A clear amplification band was detected only for a TAP-treated RNA sample, with no amplification observed in the absence of either TAP treatment or RT enzyme (Fig. 3A). Cloning and sequencing of the 5′ RACE product confirmed the gene specificity of the assay and identified the first transcribed nucleotide of the miR302-367 pri-miRNA. The transcriptional start of the primary transcript was found to be a thymine localized 153 nucleotides upstream the coding sequence of miR302b*, the first miRNA of the cluster (Fig. 3B).
FIG. 3.
Transcriptional unit of miR302-367 gene. (A) 5′ RACE assay shows specific PCR amplification only in the presence of RT and TAP treatment (lane 2). Identical reactions without TAP treatment (lane 3) or without RT (lane 4) gave no amplification. M, molecular marker. (B) Sequence diagram depicting the gene structure and main genomic components of the human miR302-367 transcriptional unit. The first nucleotide to be transcribed is numbered as +1 (transcription start). Canonical sequence motifs for TATA box and polyadenylation signal are boxed. Intron sequences are shown in brackets and shortened due to their extension. The miR302-367 cluster is located within the first intron. (C) Adapted 3′ RACE assay displaying specific PCR product. No band was observed in the absence of RT. Two bands illustrated by arrows were clearly detectable. (D) Detection of putative polyadenylated unspliced pri-miRNA by Northern blot analysis in NTERA-2. Total RNA (7 μg) was loaded in lane 1. From the same amount of total RNA, poly(A)− (lane 2) and poly(A)+ (lane 3) fractions were loaded.
Once the transcriptional start was determined, we proceeded to find the 3′ end of the transcript by using an adapted 3′ RACE assay. Since the transcript was 5′ capped, it would probably be polyadenylated. Therefore, we used an oligo(dT) RT primer and skipped the ligation step of the second RNA adaptor. The PCR primer was designed to anneal to the identified 5′ end, providing the complete cDNA. Interestingly, two different cDNA molecules were detected after the RT-PCR (Fig. 3C). Cloning and sequencing of the two 3′ RACE PCR products confirmed that they represent different transcripts resulting from the alternative splicing of the primary RNA (Fig. 3B and Fig. 4). These assays suggest that the gene coding for the cluster miR302-367 is a Pol II gene with a capped and polyadenylated transcription product. The miR302-367 coding gene contains three exons and two introns with alternative splicing, which may or may not include exon 2 (Fig. 4). The miRNA cluster is located within the first intron (Fig. 3B), and none of the two mature spliced forms of the mRNA seems to code for any protein (data not shown). The primary transcript 3′ end is positioned 8 to 14 nt downstream of a canonical polyadenylation signal (AATAAA) (Fig. 3B). We did not detect further transcripts generated by premature termination at cryptic polyadenylation signals present at the 5′ region of intron 1 (Fig. 3B and Fig. 4). To confirm the presence of the polyadenylated pri-miRNA, a Northern blot was performed in poly(A)− and poly(A)+ RNA fractions. As expected, a clear band representing the putative pri-miRNA was successfully detected in both total RNA and the poly(A)+ fraction but was absent in the poly(A)− fraction (Fig. 3D).
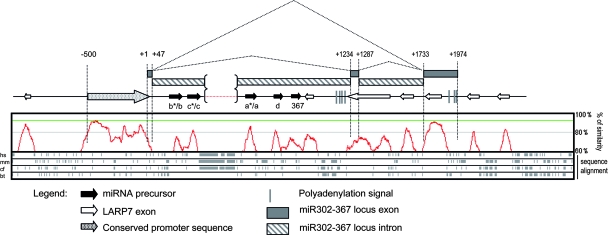
FIG. 4.
Representation displaying the structure homology of the miR302-367 locus. The upper part of the figure depicts a schematic representation of the human miR302-367 locus. The transcriptional start (+1) is shown. Black arrows represent the miRNA precursor. Gray boxes represent the miR302-369 exons. Lined boxes indicate the introns. The polyadenylation signal is represented as gray vertical lines. The conserved promoter sequence is identified upstream of the transcriptional start as a dotted arrow. White arrows represent LARP7 exons. Alternative splicing of the primary RNA is also shown. The lower part of the figure shows an overall profile of sequence similarity (represented in red) obtained from the multiple alignment of H. sapiens (hs), M. musculus (mm), C. familiaris (cf), and B. taurus (bt) homologues. Multiple sequence alignment was performed using the Lagan program of Vista tools.
In order to identify important conserved functional motifs within the miR302-367 gene, we performed a multiple sequence alignment among distinct eutherian species: Homo sapiens, Mus musculus, Bos taurus, and Cannis familiaris (Fig. 4). The miR302-367 gene was found to overlap with the LA-related protein 7 (LARP7) gene, coded by the opposite genomic DNA strand. Figure 4 depicts the conserved regions corresponding with LARP7 exons. In addition to these LARP7 exons, stretches with a high degree of similarity and containing the miRNAs can be identified. Intriguingly, a highly conserved region, not related to the LARP7 coding sequence, was located just upstream of the transcriptional start of the miR302-367 gene. We therefore hypothesized that this conserved region upstream of the transcriptional start of the miR302-367 gene would constitute a candidate proximal promoter of the miR302-367 gene.
Identification and functional characterization of the miR302-367 proximal core promoter.
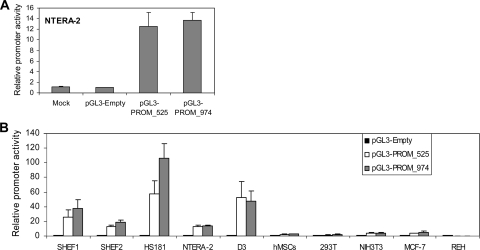
The sequence alignment profile among all the species compared identified an approximately 525-bp region upstream of the transcriptional start of the miR302-367 as the highest conserved sequence (Fig. 4), making it a strong candidate promoter for the miR302-367 gene. In an attempt to characterize whether this motif truly represents the potential promoter of the miR302-367 gene, two different regions were PCR amplified from the genomic DNA of the hECC line NTERA-2 using specific primers (see Table S1 in the supplemental material): a 525-bp fragment (PROM_525) and a larger 974-bp fragment including additional upstream sequence (PROM_974). Amplified PCR products were subcloned upstream of the luciferase reporter gene in the pGL3Basic vector (Promega). The potential transcriptional activity of both constructs (pGL3-PROM_525 and PGL3-PROM_974), compared to the empty vector, was initially assessed by transient transfection of the hECC line NTERA-2. All measurements were normalized to Renilla activity. Dual luciferase assays clearly demonstrated that both regions drove the transcription of the reporter gene, resulting in a 15-fold increase in luciferase activity (Fig. 5A). Interestingly, there were no significant differences in transcriptional activity between the PROM_525 and PROM_974 regions, suggesting that this 525-bp motif may represent the putative core promoter for the miR302-367 gene.
FIG. 5.
Transcriptional activity of genomic regions PROM_525 and PROM_974. (A) Both genomic regions were PCR amplified from the hECC line NTERA-2 and subcloned into the promoterless luciferase reporter vector pGL3Basic (Promega). Either the pGL3-PROM_525 or pGL3-PROM_974 construct was cotransfected along with a Renilla reporter plasmid into the hECC line NTERA-2. Luciferase activity was measured 24 h after transfection. Luciferase activity was normalized against Renilla activity and expressed as promoter activity relative to the empty vector (pGL3-Empty). Data from three independent experiments are represented as means ± standard deviations. (B) The transcriptional activity of both the 525-bp and the 974-bp motifs was determined in multiple cell types at different ontogenetic stages of development: genetically stable hESC lines SHEF1, SHEF2, and HS181; the transformed hECC line NTERA-2; and mESC line D3 are ESC lines. hMSCs are adult MSCs. In addition to these primary cells with stem cell potential, four transformed cell lines with no stem cell capacity were studied: the human embryonic kidney cell line 293T, the murine embryonic fibroblastic cell line NIH 3T3, and the human adult breast line MCF-7 and B-cell leukemic line REH. Data are presented as described for panel A.
The transcriptional activity of the miR302-367 promoter is restricted to the ESC compartment.
Our miRNA profiling (Fig. 1) confirmed cell type specificity for miR302-367 expression: it is highly expressed in hESCs and hECCs but not in adult hMSCs and normal tissues. Accordingly, upon identification and validation of the proximal core promoter of the miR302-367 gene, we wanted to characterize whether its transcriptional activity is also cell type specific and developmentally controlled. We assayed the promoter transcriptional activity of both the 525-bp and the 974-bp motifs in different cell types comprising different ontogeny stages (embryonic versus adult) and displaying distinct developmental potential (stem cell versus non-stem cell lines).
Embryonic cells (hESCs lines SHEF1, SHEF2, and HS181; hECC line NTERA-2; mESC line D3; and the non-stem cell lines 293T and NIH 3T3) were compared to adult cells (primary hMSCs and the non-stem cell lines REH and MCF-7). Transfections and dual luciferase assays were carried out as shown in Fig. 5A. In line with the miRNA profiling data, this miR302-367 promoter uniquely displayed robust transcriptional activity at an embryonic stage (hESCs, hECCs, and mESCs) but was switched off later in development (hMSCs and adult cell lines) (Fig. 5B). Similar transcriptional activity between the PROM_525 and PROM_974 regions was observed (Fig. 5B). Only the hESC line HS181 showed a slightly higher transcriptional activity with the PROM_974 region. This further confirms that the highly conserved 525-bp region may represent the putative core promoter driving the basal expression of the miR302-367 gene, though additional sequences present in the PROM_974 construct may contain enhancing elements. These data also highlight that promoter transcriptional activity is ontogenetically regulated since it is restricted to an embryonic stage of development and turned off later in development, which supports the idea that the embryonic-stage-specific expression of this miR302-367 cluster (Fig. 1) is conferred by its promoter transcriptional activity.
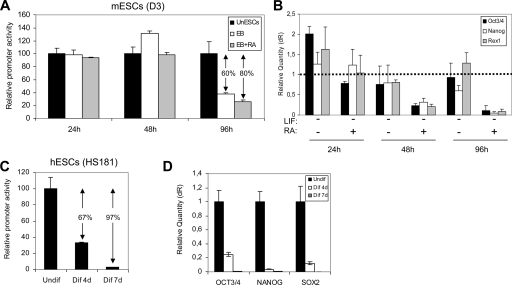
Even though the promoter transcriptional activity is restricted to an embryonic stage, we were made curious by the fact that the promoter transcriptional activity was off in not only adult stem cells (hMSCs) but also embryonic cell lines with no stem cell potential (293T and NIH 3T3) (Fig. 5B). This raises a question of whether cellular differentiation has an impact on promoter activity. We therefore studied whether the differentiation of ESCs induces changes in the promoter transcriptional activity. The mESC line D3 and the hESC line HS181 were cotransfected with a Renilla reporter and the pGL3-PROM_974 construct. The construct containing the larger 974-bp motif promoter was used in order to account for potential cis-acting elements upstream of the 525-bp core promoter, which might be cooperating with the core promoter in controlling the expression of the miR302-367 gene. For mESCs, the transcriptional activity was analyzed 24, 48, and 96 h after differentiation was induced. Differentiation was induced through embryoid body formation in nonadherent culture substrate in the absence of LIF, with or without RA supplement. Differentiation of hESCs was induced by removal of bFGF and addition of serum supplemented with RA and BMP4 (see Materials and Methods). Promoter activity was analyzed at day 0 (undifferentiated hESCs), day 4, and day 7 after induction of differentiation. As shown in Fig. 6A and C, respectively, differentiation of both mESCs and hESCs parallels the loss of transcriptional activity for the miR302-367 gene promoter. Loss of promoter activity becomes evident for mESCs 96 h after transfection under differentiating conditions. Promoter activity decays 80% and 60% upon differentiation with or without RA, respectively (Fig. 6A). In hESCs, the silencing of PROM_974 is very pronounced (67%) by day 4 of differentiation, being almost total (97%) by day 7. To verify the loss of promoter transcriptional activity in differentiating ESCs, the differentiation was traced by assessing with real-time RT-PCR the RNA expression of Oct3/4, Nanog, Rex1, and Sox2, four key transcription factors associated with pluripotency (Fig. 6B and D). In line with the decay of promoter activity in D3 cells 96 h after induction of differentiation (Fig. 6A), significant decreased expression of Oct3/4, Nanog, and Rex1 was observed by real-time RT-PCR (Fig. 6B). As expected, the lower level of promoter activity in the presence of RA was accompanied by a more pronounced downregulation of the ESC transcription factors. Similarly, the decreased activity of PROM_974 in differentiating hESCs (Fig. 6C) is accompanied by a drastic reduction in transcription factor levels (Fig. 6D). Taken together, our data indicate that miR302-367 promoter activity depends on the ontogeny and hierarchical cellular stage. The promoter is functional during embryonic development (hESCs, mESCs, and hECCs), but it is turned off later in development (hMSCs and transformed cell lines). From a hierarchical standpoint, promoter activity decays upon differentiation of ESCs, demonstrating that it is restricted to the ESC compartment and that the ESC-specific expression of the miR302-367 cluster is fully conferred by its core promoter transcriptional activity.
FIG. 6.
Loss of promoter transcriptional activity upon ESC differentiation. (A) The construct pGL3-PROM_974 and the Renilla reporter were cotransfected in mESCs. Undifferentiated mESCs (UnESC) were maintained on gelatin-coated plates in the presence of LIF (solid black bars). mESCs were differentiated through embryoid body formation in the absence (open bars) or presence (gray bars) of RA. The promoter activity was assessed at 24, 48, and 96 h after transfection. Luciferase activity was normalized against Renilla. The promoter activity is expressed relative to the undifferentiated mESCs. (B) Real-time RT-PCR analysis for the ESC-specific transcription factors Oct3/4, Nanog, and Rex1. Data acquisition and analysis were performed using the Mx3500P device (Stratagene). RNA expression is normalized against β-actin and relative to undifferentiated ESCs, which were used as the calibrator. dR, baseline-corrected raw fluorescence data. The dotted line defines the relative RNA expression of the calibrator sample. (C) hESCs were differentiated by prolonged culture on Matrigel-coated plates in the presence of differentiation medium (80% KO DMEM, 20% FBS, 2 mM l-glutamine, 2 mM nonessential amino acids, 50 μg/ml penicillin-streptomycin, and 10 nM β-mercaptoethanol, supplemented with 10 μM RA and 50 ng/ml of BMP4). Cells were assayed for PROM_974 activity at days 0, 4, and 7 after induction of differentiation (Undif, Dif 4d, and Dif 7d, respectively). Cells were cotransfected with construct pGL3-PROM_974 and the Renilla reporter 24 h before promoter activity measurements. Promoter activity is expressed relative to the undifferentiated hESCs (day 0). ↓, potential transcriptional activators; *, potential transcriptional repressor. (D) Real-time RT-PCR analysis for the ESC-specific transcription factors. Data acquisition and analysis were performed using the Mx3500P device (Stratagene). RNA expression is normalized against GAPDH and relative to undifferentiated hESCs (Undif), which were used as the calibrator.
Transcriptional regulation of the miR302-367 gene.
We have confirmed that the ESC-specific expression of the miR302-367 cluster is conferred by its core promoter region, which was consequently characterized. Then, we wanted to investigate the potential transcription factors involved in the upstream regulation of this gene. First, an algorithmic prediction of potential transfactor binding sites within the PROM_974 was performed using MatInspector software (Genomatix Software GmbH) and the Transfac, version 7.0, database (BIOBASE, GmbH). The most relevant results are summarized in Table 1. Interestingly, despite the relative simplicity of this promoter, putative binding sites for Oct3/4, Nanog, Rex1, and Sox family members were identified.
TABLE 1.
Algorithmic prediction of transcription factor putative binding sites within the PROM_974 region
| Transcription factor | Transfactor position (5′→3′)
|
DNA stranda | Matrix similarityb | P value | Sequencec | |
|---|---|---|---|---|---|---|
| Start (nt) | End (nt) | |||||
| Sox2 | 30 | 46 | − | 0.966 (0.94) | 0.003 | AAAAAACAATATCCCAA |
| Rex1 | 52 | 64 | + | 0.763 (0.78) | 0.008 | GAAGCCATGCTAG |
| Nanog | 175 | 191 | − | 0.947 (0.94) | 0.064 | TTGTGAATGTATATATT |
| Cdx2 | 364 | 382 | − | 0.854 (0.84) | 0.043 | TTTTACTTTTATAAATTAA |
| Oct3/4 | 380 | 394 | − | 0.809 (0.81) | 0.034 | TTAATGCTAAAATTT |
| Sox2 | 644 | 660 | − | 0.950 (0.94) | 0.003 | GGGTAACAATGAGTGCT |
| Nanog | 738 | 754 | + | 0.944 (0.94) | 0.064 | GTGGTAATGGTTTTAGC |
| Rex1 | 920 | 933 | − | 0.764 (0.78) | 0.008 | TCAGCCACTTCTA |
+, positive strand; −, negative strand.
Optimal matrix similarity as defined by the MatInspector software is given in parentheses. Minimum value, 0; maximum value, 1.
Boldface letters represent core sequences.
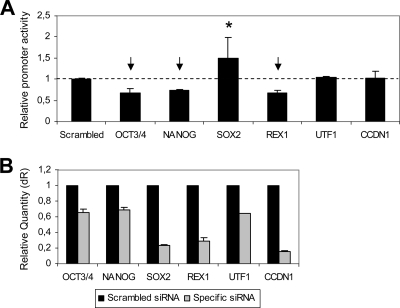
Based on these encouraging in silico results, we used a loss-of-function approach in order to experimentally demonstrate the potential role of these key ESC transfactors in the transcriptional control of the miR302-367 gene. The PROM_974 activity was evaluated 24 h after siRNA-mediated knockdown of candidate transfactors: Oct3/4, Nanog, Rex1, and Sox2. Two additional ESC-related transcription factors were tested (UTF-1 and CCDN1). hECCs were cotransfected with the pGL3-PROM_974 construct, the Renilla reporter plasmid, and the corresponding siRNA. A scrambled siRNA with no homology to human or mouse sequences was used as a control and reference sample. The promoter activity measured 24 h posttransfection yielded the results shown in Fig. 7A. Oct3/4, Nanog, and Rex1 inhibition had a negative effect on PROM_974 activity, suggesting that they may act as transcriptional activators of the miR302-367 gene. In contrast, repression of Sox2 was consistently associated with a significant increase in PROM_974 activity, indicating its potential role as a negative regulator of the miR302-367 promoter. Conversely, efficient downregulation of CCDN1 and UTF1 (Fig. 7B) had no effect on promoter activity. Efficiency of the siRNAs was assessed by real-time RT-PCR. The siRNAs reduced their specific mRNA targets by 30 to 80% at 24 h after transfection (Fig. 7B). These in silico and siRNA-mediated knockdown results support the proof of principle that the miR302-367 gene is a downstream factor of the Nanog, Oct3/4, Rex1, and Sox2 regulation network.
FIG. 7.
Regulation of the miR302-367 promoter activity by ESC transcription factors. (A) The activity of the miR302-367 promoter (PROM_974 region) was evaluated after siRNA-mediated knockdown of potential regulating transcription factors: Oct3/4, Nanog, Rex1, and Sox2. Two additional ESC-associated transcription factors were tested: UTF-1 and CCDN1. Cells were cotransfected with the pGL3-PROM_974 construct, the Renilla reporter plasmid, and the corresponding siRNA. A scrambled siRNA with no homology to mouse or human sequences was used as a control. Measurements were performed 24 h posttransfection. Normalized promoter activity is expressed relative to scrambled siRNA-transfected cells (dotted line). The mean ± standard deviation from three independent experiments is shown. (B) siRNA-mediated inhibition of the transcription factors studied was confirmed by real-time PCR 24 h posttransfection. Data were normalized to GAPDH. Scrambled siRNA-transfected cells were used as a calibrator sample. dR, baseline-corrected raw fluorescence data.
DISCUSSION
miRNAs are emerging as key regulators of multiple key biological processes including differentiation and self-renewal of stem cells (1), suggesting that they might be as important as transcription factors in controlling gene expression. Recently, the miR302-367 cluster has been described to be differentially expressed in mESCs and hESCs as well as in their malignant counterparts, hECCs (24). Importantly, however, very little is known about the genomic structure of the miR302-367 cluster-encoding gene and its transcriptional units, which would represent the first step of miRNA biogenesis. Improved knowledge about the transcription of the miR302-367-encoding gene should offer insights about miRNA regulation and should clarify how these small molecules integrate in the complex network of transcription factors and signaling pathways underlying pluripotency in stem cells.
Here, we first performed a comparative expression profile of more than 170 miRNAs in different ESC lines and primary human adult stem cells (hMSCs). Our results further confirmed that all miRNAs that comprise the cluster miR302-367 are expressed at high levels in all hESC lines and hECCs while being barely detectable in hMSCs and multiple normal adult tissues. These data provide robust experimental support toward a potential role for this miR302-367 cluster in the biology of ESCs. Interestingly, all the members within the miR302-367 cluster displayed similar expression profiles in normal genetically stable hESCs compared to genetically unstable hESCs (BGV01) and hECCs (NTERA-2), indicating that the miR302-367 members seem to be equally expressed in ESCs regardless of genome stability and cellular genetic background. A closer analysis of our miRNA profiling reveals further potential relevant information. First, miRNA profiling indicates that undifferentiated hESCs display, in general, much lower levels of miRNA expression than adult tissues and adult primary hMSCs, which suggests that miRNA downregulation may be necessary for proper lineage specification and differentiation. Second, in sharp contrast to the ESC-specific miR302-367 cluster, there are some miRNA molecules whose expression may be influenced by the genetic stability of the ESCs. For instance, miR370 and miR154* are highly expressed in genetically stable hESCs but not in the karyotypically abnormal hESC line BGV01 and hECC NTERA-2. Finally, our miRNA profiling revealed various miRNAs upregulated in adult primary stem cells compared to ESCs (miR221, miR34a, miR29a, miR137, miR368, miR213, miR134, miR199a, miR339, miR27a, miR224, and miR154). The biological role of this set of miRNAs in differentiation, proliferation, and self-renewal of adult MSCs needs further investigation.
We then analyzed in depth the genomic and transcriptional units of this miR302-367 cluster. We have characterized the structure of the gene coding for the human miR302-367 cluster. We show that this miRNA cluster is located within the first intron of the gene. The transcriptional start of the primary transcript was found to be a thymine located 153 nt upstream of the coding sequence of miR302b*, the first miRNA of the cluster. The pri-miRNA is a Pol II-capped and polyadenylated transcript. The two spliced transcription products identified by the 3′ RACE assays support the idea that the miR302-367 coding gene contains three exons and two introns with alternative splicing, which may or may not include exon 2. The primary unspliced RNA has the expected strong secondary structure derived from the presence of miRNA hairpin precursors. Thus, low efficiency during retrotranscription may be the reason that this molecule is not detected by 3′ RACE assays.
The major members of the miR302 family (miR302a, -302b, -302c, and -302d) share high sequence homology, differing only in the 3′ hexanucleotide. It has been recently reported that miR29a and miR29b have different intracellular localizations owing to their specific 3′ hexanucleotides (12). In sharp contrast to miR29a, the miR29b 3′ hexanucleotide (AGUGUU) acts as a nuclear localization motif, promoting nuclear enrichment of this molecule (12). The study by Hwang et al. suggested that miRNAs sharing common 5′ sequences, considered to be largely redundant, might display distinct functions because of the influence of cis-acting regulatory motifs. Accordingly, the miR302 members displayed identical 5′ sequences. Moreover, the miR302d hexanucleotide (GAGUGU) shows high sequence similarity to the 3′ hexanucleotide of miR29b (AGUGUU) (conserved residues are in boldface) and with the relaxed consensus of this motif, established by mutagenesis (residues in boldface in AGNGUN) (12). However, our results show that all four miR302 family members display a predominant cytoplasmic localization with no significant differences detected among them, suggesting that the 3′ hexanucleotide does not promote nuclear localization for any of these miRNAs. On the other hand, because the 3′ ends are conserved across species, we cannot exclude an alternative functional role, for example, providing an additional degree of target specificity or specific localization to the cytoplasm. The seed sequence (nt 2 to 7 at the miRNA 5′ end) of the miRNAs is identical, and so they probably exert redundant regulation of common targets. This seed sequence is also partially shared by some members of the mESC-specific cluster miR290-295. However, our results from in silico comparative genomics further support the hypothesis that both clusters have evolved independently (11). In contrast to the miR290-295 locus, apparently specific for eutherian mammals and highly variable among them, the locus for the miR302-367 cluster is extremely conserved from chick to human (data not shown). This argues for a more ancient origin of the miR302-367 cluster and a possible evolutionary convergence of both clusters, resulting in regulation of common targets.
The core message of our work is the characterization and functional validation of the proximal core promoter driving the expression of the miR302-367-encoding gene. Both upstream genomic regions tested for transcriptional activity, PROM_525 and PROM_974, drove the transcription of the reporter gene, resulting in a 15- to 110-fold increase. The absence of relevant differences between both constructs suggests that the 525-bp motif may represent the putative core promoter for the miR302-367 gene. However, based on the results from the HS181 cell line, we cannot rule out the existence of upstream cis-acting elements with a role in the in vivo transcriptional regulation of the miRNA cluster. Our cell biology data indicate that the miR302-367 promoter activity depends on the ontogeny and hierarchical cellular stage. The promoter activity is functional during embryonic development (hESCs, mESCs, and hECCs), but it is turned off later in development (hMSCs and multiple transformed cell lines). From a hierarchical standpoint, miR302-367 promoter activity decays upon differentiation of both mouse and human ESCs, demonstrating that its activity is restricted to the ESC compartment and that the ESC-specific expression of the miR302-367 cluster is fully conferred by its core promoter transcriptional activity. Importantly, our data are further supported by the fact that endogenous miR302-367 expression is also downregulated during hESC differentiation (see Fig. S1 in the supplemental material). (The tight regulation of the promoter activity in transiently transfected cells suggests that the silencing of the endogenous miRNA cluster in somatic and non-stem embryonic cells does not occur via heterochromatinization of the locus, as has been proposed for the mESC-specific cluster miR290-295 (11)). Alternatively, the expression of the miR302-367 cluster may be controlled by ESC-specific factor(s) probably common between human and mouse. In mESC differentiation assays, where the activity of the promoter is evaluated at very short times, a lag period between a decrease in key transfactor levels and reduction of the reporter activity is observed. This might position the miR302-367 gene as a downstream target of one or more of these ESC-associated transcription factors rather than an upstream determinant of ESC phenotype. Alternatively, the lag period may be explained by a slow turnover of miR302-367-inducing transfactors. The first hypothesis is supported by the presence in the promoter sequence of potential binding sites for Oct3/4, Nanog, Rex1, and Sox2, among others. Furthermore, the siRNA-mediated downregulation of these factors clearly affects the promoter activity. To the best of our knowledge, the results of this study represent the first identification, characterization, and functional validation of a human miRNA promoter in human stem cells. This study also reveals important clues about miR302-367 promoter regulation and should open up new avenues to further investigate the upstream transcriptional regulation of the miR302-367 cluster and to dissect how these miRNAs integrate in the complex molecular network conferring stem cell properties to ESCs.
Supplementary Material
Acknowledgments
This work was funded by the Consejería de Salud, Junta de Andalucía, Spain (grants reference 0004/2005 to A.B.-D.J. and 0030/2006 to P.M.). This work was partially supported by a grant from The José Carreras International Foundation (FIJC-05/ED-Thomas 2006) to P.M.
We thank all members of the Andalusian Stem Cell Bank for their insights. We are especially indebted to Javier García-Castro (Andalusian Stem Cell Bank, Granada, Spain), Peter Andrews (Sheffield University, United Kingdom), and Outi Hovatta (Karolinska Institute, Stockholm, Sweden) for providing key reagents and to Ruth Rubio and Vicente Agustin for their outstanding technical assistance.
Footnotes
Published ahead of print on 25 August 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ambros, V. 2004. The functions of animal miRNAs. Nature 435350-355. [DOI] [PubMed] [Google Scholar]
- 2.Aukerman, M. J., and H. Sakai. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 152730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, E., S. Y. Kim, M. A. Carmell, E. P. Murchison, H. Alcorn, M. Z. Li, A. A. Mills, S. J. Elledge, K. V. Anderson, and G. J. Hannon. 2003. Dicer is essential for mouse development. Nat. Genet. 35215-217. [DOI] [PubMed] [Google Scholar]
- 5.Borchert, G., W. Lanier, and B. Davidson. 2006. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 131097-1101. [DOI] [PubMed] [Google Scholar]
- 6.Cai, X., C. H. Hagedorn, and B. R. Cullen. 2004. Human miRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 101957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths-Jones, S. 2004. The microRNA registry. Nucleic Acids Res. 32D109-D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatfield, S. D., H. R. Shcherbata, K. A. Fischer, K. Nakahara, R. W. Carthew, and H. Ruohola-Baker. 2005. Stem cell division is regulated by the microRNA pathway. Nature 435974-978. [DOI] [PubMed] [Google Scholar]
- 9.Hohjoh, H., and T. Fukushima. 2007. Marked change in microRNA expression during neuronal differentiation of human teratocarcinoma NTera2D1 and mouse embryonal carcinoma P19 cells. Biochem. Biophys. Res. Commun. 362360-367. [DOI] [PubMed] [Google Scholar]
- 10.Houbaviy, H. B., M. Murray, and P. Sharp. 2003. Embryonic stem cell-specific microRNAs. Dev. Cell 5351-358. [DOI] [PubMed] [Google Scholar]
- 11.Houbaviy, H. B., L. Dennis, R. Jaenisch, and P. Sharp. 2005. Characterization of a highly variable eutherian microRNA gene. RNA 111245-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang, H. W., E. A. Wentzel, and J. T. Mendell. 2007. A hexanucleotide element directs microRNA nuclear import. Science 31597-100. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, S. M., L. Shin-Yi, and F. J. Slak. 2003. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 259364-379. [DOI] [PubMed] [Google Scholar]
- 14.Johnston, R. J., and O. Hobert. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426845-849. [DOI] [PubMed] [Google Scholar]
- 15.Josephson, R., C. J. Ording, Y. Liu, S. Shin, U. Lakshmipathy, A. Toumadje, B. Love, J. D. Chesnut, P. W. Andrews, M. S. Rao, and J. M. Auerbach. 2007. Qualification of embryonal carcinoma 2102Ep as a reference for human embryonic stem cell research. Stem Cells 25437-446. [DOI] [PubMed] [Google Scholar]
- 16.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 152654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakshmipathy, U., B. Pelacho, K. Sudo, J. L. Linehan, E. Coucouvanis, D. S. Kaufman, and C. M. Verfaillie. 2004. Efficient transfection of embryonic and adult stem cells. Stem Cells 22531-543. [DOI] [PubMed] [Google Scholar]
- 18.Lakshmipathy, U., B. Love, L. A. Goff, R. Jörnsten, R. Graichen, R. P. Hart, and J. D. Chesnut. 2007. microRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 161003-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, Y., K. Jeon, J. T. Lee, S. Kim, and V. N. Kim. 2002. microRNA maturation: stepwise processing and subcellular localization. EMBO J. 214663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. P. LeeProvost, O. Rådmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425415-419. [DOI] [PubMed] [Google Scholar]
- 21.Lee, Y., M. Kim, J. Han, K. H. Yeom, S. Lee, S. H. Baek, and V. N. Kim. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 234051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luciano, D. J., H. Mirsky, J. Nicholas, N. J. Vendetti, and S. Maas. 2004. RNA editing of a miRNA precursor. RNA 101174-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund, E., S. Güttinger, A. Calado, J. E. Dahlberg, and U. Kutay. 2004. Nuclear export of microRNA precursors. Science 30395-98. [DOI] [PubMed] [Google Scholar]
- 24.Suh, M. R., Y. Lee, J. Y. Kim, S. K. Kim, S. H. Moon, J. Y. Lee, K. Y. Cha, H. M. Chung, H. S. Yoon, S. Y. Moon, V. N. Kim, and K. S. Kim. 2004. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270488-498. [DOI] [PubMed] [Google Scholar]
- 25.Tam, W. 2001. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene 274157-167. [DOI] [PubMed] [Google Scholar]
- 26.Yañez, R., M. L. Lamana, J. García-Castro, I. Colmenero, M. Ramírez, and J. A. Bueren. 2006. Adipose tissue-derived mesenchymal stem cells have in vivo immunosuppressive properties applicable for the control of the graft-versus-host disease. Stem Cells 242582-2591. [DOI] [PubMed] [Google Scholar]
- 27.Yu, J., F. Wang, G. H. Yang, F. L. Wang, Y. N. Ma, Z. W. Du, and J. W. Zhang. 2006. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem. Biophys. Res. Commun. 34959-68. [DOI] [PubMed] [Google Scholar]
- 28.Zhou, X., J. Ruan, G. Wang, and W. Zhang. 2007. Characterization and identification of microRNA core promoters in four model species. PLoS Comput. Biol. 3412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.