Abstract
Spinal muscular atrophy (SMA) is a recessive neuromuscular disorder caused by the homozygous loss of the SMN1 gene. The human SMN2 gene has a C-to-T transition at position +6 of exon 7 and thus produces exon 7-skipping mRNAs. However, we observed an unexpectedly high level of exon 7-containing SMN2 transcripts as well as SMN protein in testis of smn−/− SMN2 transgenic mice. Using affinity chromatography, we identified several SMN RNA-associating proteins in mouse testis and human HeLa cells, including hnRNP Q. The major hnRNP Q isoform, Q1, directly bound SMN exon 7 in the vicinity of nucleotide +6. Overexpression of hnRNP Q1 promoted the inclusion of exon 7 in SMN2, probably by activating the use of its upstream 3′ splice site. However, the minor isoforms Q2/Q3 could antagonize the activity of hnRNP Q1 and induced exon 7 exclusion. Intriguingly, enhanced exon 7 inclusion was also observed upon concomitant depletion of three hnRNP Q isoforms. Thus, differential expression of hnRNP Q isoforms may result in intricate control of SMN precursor mRNA splicing. Here, we demonstrate that hnRNP Q is a splicing modulator of SMN, further underscoring the potential of hnRNP Q as a therapeutic target for SMA.
Spinal muscular atrophy (SMA) is an autosomal recessive disorder that results in degeneration of motor neurons in the spinal cord, and it is one of the most common genetic diseases causing infant mortality (6, 33). SMA is primarily caused by homologous deletion or mutations of the survival motor neuron 1 (SMN1) gene (22). The duplicate gene, SMN2, in the human genome differs from SMN1 by a few nucleotides. The only difference in the coding region is the C-to-T transition at position +6 of exon 7. This difference is translationally silent but results in exon 7 skipping in the SMN2 mRNA (27, 31). Thus, SMN2 transcripts lacking exon 7, which are abundant in cells, encode a C-terminally truncated SMN protein. The SMN protein is primarily involved in assembly of snRNPs (small nuclear ribonucleoproteins) and may also participate in precursor mRNA (pre-mRNA) splicing and in neuronal mRNA transport (35, 37). The SMN protein with a deletion of exon 7 is less stable and has lower self-oligomerization activity (26, 28). Moreover, this protein is unable to interact with the snRNP Sm proteins and fails to promote splicing in vitro (34, 35). Therefore, SMN2 is insufficient to fully compensate for SMN1 loss in SMA.
Regulation of the SMN pre-mRNA splicing involves several cis elements around exon 7 that either serve as recognition sites for splicing regulatory factors or form secondary structures (26, 39-41). At least three splicing enhancers (SE) are located within exon 7. The C-to-T change at nucleotide +6 of SMN2 likely disrupts the enhancer activity of SE1 and may also create a splicing silencer (3, 16). ASF/SF2 has been predicted to bind a sequence encompassing nucleotides +6 to +11 of exon 7 according to a sequence motif matrices analysis (3). An in vitro splicing assay has shown that excess ASF/SF2 could activate exon 7 inclusion in SMN1, suggesting a positive role of ASF/SF2 in this process (3). The C-to-T transition in SMN2 attenuates the binding of ASF/SF2 and results in inefficient utilization of exon 7. On the other hand, a splicing repressor, hnRNP A1, binds this site in SMN2, perhaps as well as an element within intron 6, and induces exon 7 skipping (16, 17). Therefore, inefficient use of exon 7 in SMN2 may result from the loss of an ASF/SF2-binding enhancer and the gain of an hnRNP A1 responsive silencer.
In addition, several other splicing factors have also been implicated in the regulation of exon 7 inclusion. Human Tra2-β1 interacts with the AG-rich exonic enhancer SE2 that is located downstream of SE1 (11). Indeed, overexpression of Tra2-β1 promotes exon 7 utilization in SMN2 (11, 45). Other regulatory proteins, such as SRp30c, hnRNP G, and RBMY, also can enhance SMN2 exon 7 inclusion through their interaction with human Tra2-β1 (11, 12, 45). None of these splicing effectors was identified through a systematic biochemical approach. Because SMN2 acts as a modifying gene that can influence the severity of SMA (43, 44), a comprehensive search for proteins that regulate SMN2 exon 7 utilization is thus necessary.
In this report, we show for the first time that hnRNP Q may serve as a splicing regulator for alternative splicing of SMN. The hnRNP Q family comprises three isoforms generated by alternative splicing (32). The major isoform, hnRNP Q1, contains an acidic residue-rich domain at the N terminus, three RNA recognition motifs (RRMs) in the central region, and an arginine/glycine (RG)-rich domain at the C terminus. The Q2 isoform has a truncated RRM2 and contains an extended C-terminal region. The Q3 isoform, the longest one, differs from Q1 only by its C-terminal extension which is the same as Q2. hnRNP Q has been reported as a spliceosomal component and is required for efficient in vitro pre-mRNA splicing (32). Moreover, hnRNP Q has been identified in a complex that controls c-fos mRNA stability (9). Of particular interest is that hnRNP Q is a component of cytoplasmic mRNA granules and may act together with the SMN protein in mRNA trafficking in neural cells (37, 38). More importantly, a clinical study has shown that hnRNP Q expression is elevated in SMA patients who have milder symptoms and in their unaffected siblings (10). Therefore, hnRNP Q may have a trans-dominant effect in SMA and thus modulate the severity of SMA. Our present report that hnRNP Q can modulate exon 7 inclusion in SMN2 transcripts further emphasizes its importance in SMA.
MATERIALS AND METHODS
Plasmid construction.
The cDNAs encoding the hnRNP Q isoforms (Q1, Q2, and Q3) and SRp30c were obtained by reverse transcription-PCR (RT-PCR) from HeLa cell poly(A)+ RNAs. The vectors for expressing FLAG-tagged hnRNP Q isoforms (Q1, Q2, and Q3) and truncated Q1 proteins (lacking either the N-terminal acidic domain [ΔN] or C-terminal RG-rich domain [ΔC] or both domains [ΔNC]) were constructed by inserting each coding sequence in frame with the pEF-FLAG, a generous from Ming-Ji Fann (Yang-Ming University, Taipei). The green fluorescent protein (GFP) coding sequence from pEGFP (BD Biosciences) was subcloned into a pCDNA3.1 (Invitrogen) plasmid, in which the hnRNP Q1 cDNA had been inserted; the resulting plasmid produced a GFP-hnRNP Q1 fusion. The hnRNP Q1 coding DNA fragment was subcloned into pET21 (Novagen) to generate the vector for overproduction of recombinant His-tagged hnRNP Q1 in Escherichia coli. The coding sequences of SRp30c, ASF, and hnRNP A1 were PCR amplified and subcloned into pCDNA3.1-FLAG (24). The expression vector of myc-PSF was a kind gift of B. J. Blencowe (University of Toronto, Canada). To knock down the expression of hnRNP Q, the following short hairpin RNA (shRNA)-coding cDNAs were each inserted into a modified pRS vector (OriGene) containing the histone H1 promoter. The shQT shRNA, corresponding to nucleotides 211 to 229 of the human hnRNP Q1 coding sequence (GenBank accession no. AY034483) was targeted to all three hnRNP Q isoforms (shQ1, shQ2, and shQ3). The shQ1 and shQ2/3 were targeted to nucleotides 1855 to 1873 of hnRNP Q1 and to nucleotides 1587 to 1605 of Q2, respectively. A small interfering RNA (siRNA) mixture (siQT) targeting to three isoforms was purchased from Dharmacon. The pCI-SMN2 plasmid was kindly given by Brunhilde Wirth (University of Cologne, Germany) (27). The pCI-SMN2-ug (40) and pCI-SMN2-r20 mutant reporters (see Fig. 4) were generated using a PCR-based mutagenesis method, and their sequences have been verified.
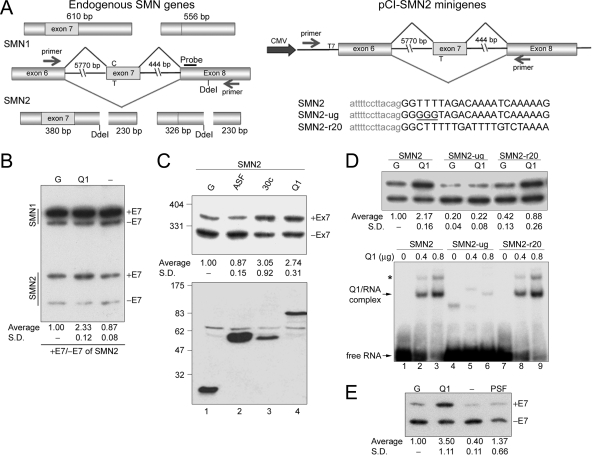
FIG. 4.
hnRNP Q1 promotes inclusion of exon 7 in SMN2. (A) The left diagram shows endogenous SMN1 and SMN2 transcripts and their RT-PCR products amplified using primers as indicated by arrows. Note that only SMN2 contains a DdeI restriction site. Southern blotting was performed using the probe as indicated. The right diagram shows the pCI-SMN2 reporter used in the in vivo splicing assay. Expression of the SMN transcript was driven by the cytomegalovirus (CMV) promoter. (B) HEK293 cells were transfected with GFP (G), GFP-hnRNP Q1 (Q1), or empty (−) expression vector. RT-PCR and following analysis are as described in panel A. E7, exon 7. (C) The pCI-SMN2 reporter was cotransfected into HEK293 cells along with expression vector encoding GFP (G) or GFP-ASF/SF2, SRp30c, or hnRNP Q1. RT-PCR was performed to detect SMN transcripts produced from the reporters described in panel A. Ex7, exon 7. (D) Similar to panel C, the wild-type pCI-SMN2 and its ug and r20 mutants were each cotransfected with expression vector encoding GFP or GFP-hnRNP Q1 (upper panel). His-tagged hnRNP Q1 was incubated with 32P-labeled wild-type or mutant SMN RNA oligonucleotide, and the resulting complexes were fractionated on a nondenaturing polyacrylamide gel (lower panel). The asterisk may represent a dimerized hnRNP Q1/RNA complex The SMN-ug RNA may form a secondary structure or weakly associate with hnRNP Q1 (lanes 4 to 6). (E) The pCI-SMN2 reporter was cotransfected with GFP (G), GFP-hnRNP Q1 (Q1), empty (−) or myc-PSF (PSF) expression vector into human SMA fibroblasts. Calculation of the relative change in exon 7 (E7) inclusion, shown in panels B to D, is described in Materials and Methods, and immunoblotting was performed using anti-GFP, as described for panel C. SD, standard deviation.
Cell culture and transfection.
Human HeLa and HEK293 cells and fibroblasts derived from SMA patients (23) were grown at 37°C in Dulbecco's modified Eagle medium containing 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Transfection was performed using Lipofectamine 2000 (Invitrogen).
RT-PCR.
To analyze the expression of SMN in type III SMA-like mice (smn−/− SMN2), total RNA was extracted from different tissues using a standard protocol (4). RT-PCR was performed using rTth DNA polymerase and the following primers corresponding to the coding region of human SMN gene: nucleotides 179 to 199 (forward) and 528 to 509 (reverse) for exon 2 to 4, nucleotides 483 to 505 (forward) and 770 to 750 (reverse) for exon 4 to 6, and nucleotides 740 to 763 (forward) and 1158 to 1138 (reverse) for exon 6 to 8.
Mouse tissue extracts.
For RNA affinity selection, subcellular extracts were prepared from mouse tissues according to the recommended protocols (Pierce). In brief, mouse tissues were homogenized with ice-cold Cytoplasmic Extraction Reagent I (Pierce). After incubation for 10 min on ice, Cytoplasmic Extraction Reagent II was added and thoroughly mixed with the samples. Nuclei were subsequently separated from the cytoplasmic fraction by centrifugation at 16,000 × g for 5 min. The pellet containing nuclei was resuspended with ice-cold Nuclear Extraction Reagent. After centrifugation as above, the supernatant was collected as the nuclear extract. The extracts were stored at −80°C. Total cell lysate prepared from the brain, liver, and testis of SMA-like mice (smn−/− SMN2) was used for immunoblotting. Antibodies used were anti-hnRNP Q (Sigma), anti-SMN (Abnova), and anti-α-tubulin (NeoMarkers).
RNA affinity chromatography.
Biotinylated RNA oligonucleotides corresponding to SMN1 and SMN2 and containing the 3′ 13 residues of intron 6 and the 5′ 22 residues of exon 7 were synthesized by Dharmacon. Mouse testis and liver nuclear extracts were individually incubated with an equal volume of empty streptavidin beads at 4°C for 1 h to remove proteins that nonspecifically bound to the beads. Subsequently, a 70-μl reaction mixture containing 50 μl of precleaned nuclear extract and 5 μg of biotinylated RNA was incubated under splicing conditions (0.5 mM ATP, 20 mM creatine phosphate, 2.4 mM MgCl, and 20 units of RNasin; Promega) at 30°C for 30 min, followed by a further 3-h incubation with 50 μl of streptavidin beads at 4°C. After extensive washing with NET-2 buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl) containing 0.25% (wt/vol) NP-40, resin-bound samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, and individual protein bands were analyzed by nanoelectrospray mass spectrometry (MS) (see below).
To identify HeLa cell proteins that associate with SMN exon 7, 1.2 μg of biotinylated RNA oligonucleotide was conjugated with 20 μl of streptavidin-agarose resin (Sigma). After a preclearing step, HeLa cell nuclear extracts containing ∼200 μg of proteins were incubated with SMN RNA-conjugated resin under splicing conditions as described above at 30°C for 30 min and then at 4°C for another 2 h. After samples were washed as above, resin-bound proteins were collected for immunoblot analysis. For MS analysis, a 10-fold scale of RNA affinity selection was performed. To compare the SMN RNA binding affinities of hnRNP Q isoforms and fragments, lysates prepared from ∼2 × 106 HEK293 cells containing overexpressed FLAG-tagged protein were subjected to affinity selection as above. Antibodies used for immunoblotting were anti-hnRNP Q, anti-hnRNA1 (Abcam), anti-ASF (Zymed), anti-CA150 (25), and anti-FLAG (M2; Sigma).
MS.
Nanoelectrospray MS was used to identify mouse proteins that associated with SMN exon 7. In-gel digestion of isolated proteins was performed essentially as described previously (8, 42). Gel slices containing the protein of interest were excised and washed with a 1:1 solution of acetonitrile and 200 mM ammonium bicarbonate. The gels were dehydrated with acetonitrile and then rehydrated with 50 mM ammonium bicarbonate. Proteins were digested with 20 ng/ml trypsin in 50 mM ammonium bicarbonate at 37°C for 4 h. Nanoscale capillary liquid chromatography tandem MS (LC-MS/MS) analysis was subsequently performed using an Ultimate capillary LC system (LC Packings) coupled to a QSTARXL quadrupole time-of-flight (TOF) mass spectrometer (Applied Biosystems/MDS Sciex). The nanoscale capillary LC separation was performed on a reverse-phase C18 column. Subsequent ionization (2.0-kV ionization potential) was performed with a PicoTip (FS360-20-10-D-20; New Objective, Cambridge, MA) for online LC-MS; a nanoelectrospray interface was used for LC-MS/MS analysis. Data were obtained using automatic information-dependent acquisition (Applied Biosystems/MDS Sciex). Product ion spectra were generated by nano-LC-MS/MS and searched against NCBI databases for exact matches using the ProID program (Applied Biosystems/MDS Sciex) and the MASCOT search program. A mouse taxonomy restriction was used, and the mass tolerance of both precursor ion and fragment ions was set to 0.3 Da.
HeLa cell proteins that were obtained from RNA affinity selection were fractionated by SDS-PAGE. Samples were stained with Sypro-Ruby (Bio-Rad) and visualized using Typhoon 9410 (Amersham Biosciences). The bands of interest were excised and subjected to in-gel trypsinization followed by matrix-assisted laser desorption ionization-TOF MS analysis using a Voyager-DE STR biospectrometry workstation (Applied Biosystems).
Expression and purification of recombinant hnRNP Q1 protein.
Plasmid pET21-hnRNP Q1 was transformed into E. coli strain BL21(DE3) for overproduction of recombinant His-tagged hnRNP Q1. E. coli lysates were prepared by sonication on ice and subsequently loaded onto a nickel agarose column (Novagen), essentially according to the recommendation of the manufacturer. Bound proteins were eluted with 1 M imidazole and were dialyzed against buffer D, which contained 20 mM HEPES (pH 7.9), 50 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, and 20% glycerol.
Gel mobility shift assay.
SMN RNA oligonucleotides were 5′ end labeled with 32P by T4 polynucleotide kinases (New England Biolabs). His-tagged hnRNP Q1 was incubated with ∼25 fmol (∼5 × 104 cpm) of 5′ end-labeled SMN RNA in a 20-μl reaction containing 10 mM HEPES (pH 7.9), 0.05 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 5 mM MaCl2 1 ng/μl bovine serum albumin, 25 ng/μl tRNA, and 10 U RNasin at 25°C for 15 min. The samples were fractionated on a 6% nondenaturing polyacrylamide gel, followed by autoradiography.
Site-specific RNA labeling and UV cross-linking.
RNA oligonucleotides corresponding to exon 7 nucleotides +6 to +22 of SMN1 and SMN2 were 5′ end labeled. Each 20 pmol of the 32P-labeled RNA was mixed with 60 pmol of unlabeled upstream RNA fragment (corresponding to the last 13 nucleotides of intron 6 and exon 7 nucleotides +1 to +5 of SMN1/2 pre-mRNA) and 40 pmol of bridge DNA, complementary to the above oligonucleotides, in a 10-μl reaction mixture. Ligation was carried out by T4 DNA ligase (Fermentas) at 37°C for 1.5 h (3). The ligated product was then treated with RQ1 DNase (Promega) and gel purified. Cell lysates were incubated with 60,000 cpm of site-specifically labeled RNA in a 10-μl reaction volume under splicing conditions as above at 30°C for 20 min. The reaction mixtures were irradiated in a UV cross-linker (Stratalinker; Stratagene) for 10 min. The reaction mixtures were then treated with 0.1 μg of RNase A1 at 30°C for 15 min, followed by immunoprecipitation.
Immunoprecipitation.
Cell lysates were incubated with antibody-conjugated protein A-Sepharose (GE Healthcare) or anti-FLAG M2 beads (Sigma) at 4°C for 2 h. The beads were extensively washed with 0.5% NP-40-containing NET-2 buffer. Bound proteins were subjected to autoradiography or immunoblotting.
Indirect immunofluorescence.
Indirect immunofluorescence was performed essentially as described previously (21), except that cells were fixed with ice-cold methanol for 10 min for endogenous hnRNP Q detection. Primary antibodies used were polyclonal anti-FLAG (2 μg/ml) and anti-hnRNP Q (2 μg/ml).
In vivo splicing assays.
In vivo splicing assays were carried out as described previously (27). Briefly, 0.7 μg of the SMN1 or SMN2 minigene was cotransfected with 4 μg of the plasmid that encodes a trans-acting factor into ∼5 × 105 HEK293 cells. Total RNA was isolated using Trizol reagent (Invitrogen) 48 h posttransfection and treated with RNase-free RQ1 DNase (Promega). RT was performed by using oligo(dT) and SuperScript III, followed by a 30-cycle PCR using primers specific to the vector sequence GGTGTCCACTCCCAGTTCAA (forward) and SMN coding region (nucleotides 940 to 903; reverse) and GeneTaq (Nippon Gene). A 5% volume of each PCR mixture was analyzed by Southern blotting using 32P-labeled forward oligonucleotide as a probe. Quantitative analysis was performed using a PhosphorImager (Molecular Dynamics) or Scion Image (Beta 4.0.2, PC format of NIH Image; Wayne Rasband, National Institutes of Health). To examine endogenous SMN expression in HEK293 cells, RT-PCR was performed using primers corresponding to SMN nucleotides 740 to 762 (forward) and nucleotides 1349 to 1332 (reverse). The PCR products were digested with DdeI, followed by Southern blotting using the primer corresponding to SMN nucleotides 1097 to 1060 as a probe. For individual transfection, the efficiency of exon 7 inclusion was measured by the following formula: level of exon 7 inclusion/(level of exon 7 inclusion + level of exon 7 exclusion). Since the exon 7-containing product was dominant in the results shown in Fig. 4B, the formula level of exon inclusion/level of exon 7 exclusion was used to represent exon 7 inclusion efficiency.
RESULTS
Enhanced exon 7 inclusion in testis of smn−/− SMN2 mice.
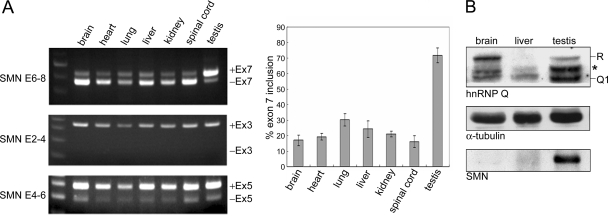
Homozygous loss or mutation of SMN1 is common in SMA patients, and thus a therapeutic strategy is to increase the expression of functional SMN2. Transgenic smn−/− SMN2 mice lacking the mouse smn gene but carrying a copy of human SMN2 provide an animal model of human SMA (13). We had expected that a major portion of the SMN transcripts would lack exon 7 in such SMA-like mice due to the C-to-U change at position +6 of exon 7 in human SMN2. While analyzing SMN2 expression in different tissues of these mice, we found that their testis, unlike other tissues, had an unexpectedly high level of exon 7-containing SMN2 transcripts (Fig. 1A). Such enhanced exon inclusion appeared to be specific to exon 7 because it was not detected with two other alternatively spliced exons, exons 3 and 5 (15) (Fig. 1A). Accordingly, a significant amount of SMN protein was detected in testis but not in brain or liver (Fig. 1B). This finding suggested that a regulatory mechanism in testis could activate exon 7 inclusion in SMN2 mRNAs and thereby produce SMN protein.
FIG. 1.
SMN expression in different tissues of smn−/− SMN2 mice. (A) Total RNA was prepared from smn−/− SMN2 mouse tissues and analyzed by RT-PCR using specific primers (see Materials and Methods). The reaction products were then subjected to agarose gel electrophoresis. Representative data are shown at left. PCR product sizes in SMN exons 6 to 8 (E6-8), exons 2 to 4 (E2-4), and exons 4 to 6 (E4-6) were as follows: exon 7 inclusion (+Ex7), 419 bp; exon 7 exclusion (−Ex7), 367 bp; +Ex3, 350 bp; −Ex3, 149 bp; +Ex5, 288 bp; −Ex5, 192 bp. The bar graph shows exon 7 inclusion efficiency (see Materials and Methods). (B) Immunoblotting shows SMN, hnRNP Q/R, and α-tubulin protein levels in the indicated tissue cell lysates of smn−/− SMN2 mice. Anti-hnRNP Q cross-reacted with hnRNP R proteins. The asterisk may represent hnRNP Q2/3 or hnRNP R isoforms or degraded fragments.
Affinity selection of testis-specific proteins associated with exon 7 of SMN2 mRNA.
The above observation suggested the presence of a testis-specific splicing activator(s) that promotes exon 7 inclusion. To search for such a factor(s), we exploited affinity selection using a biotinylated RNA oligonucleotide spanning the region of SMN2 corresponding to the last 13 nucleotides of intron 6 and the first 22 nucleotides of exon 7. Nuclear extracts prepared from the testis of smn−/− SMN2 mice were subjected to RNA affinity chromatography. Mouse liver extracts were used for comparison because exon 7 inclusion was not particularly induced in this tissue (Fig. 1). Affinity-selected proteins were fractionated by SDS-PAGE, and those with tissue specificity were subsequently analyzed by nanoelectrospray MS. Table 1 profiles the identified RNA-binding proteins that potentially associated with the SMN2 exon 7 RNA. Candidate proteins of potential interest included splicing factors SAP155 and U2AF65 and several RNA binding proteins, such as hnRNP L-like, Q, and M and PSF (Table 1). Identification of SAP155 and U2AF might be due to their interaction with the 3′ end sequence of intron 6 (36). Perhaps some of the identified RNA binding proteins could function as regulators of SMN pre-mRNA splicing or participate in other steps of SMN mRNA metabolism.
TABLE 1.
Identification of RNA binding proteins that potentially associated with the SMN exon 7 RNA oligonucleotide in testis and liver of smn−/− SMN2 transgenic micea
| Tissue type and band no. | Protein description and/or name | Accession no. | Mass (Da)b | MASCOT score | Sequence coverage (%) |
|---|---|---|---|---|---|
| Liver-specific bands | |||||
| 1 | Splicing factor 3B subunit 1, SF3b1; SAP155 | Q99NB9 | 145,724 | 32 | 11 |
| 2 | Proline- and glutamine-rich splicing factor, PSF | Q8VIJ6 | 75,394 | 298 | 17 |
| 3 | Proline- and glutamine-rich splicing factor, PSF | Q8VIJ6 | 75,394 | 49 | 6 |
| 4 | hnRNP D0 | Q60668 | 38,330 | 147 | 16 |
| TIA-1-related protein, TIAR | P70318 | 43,361 | 112 | 7 | |
| Testis-specific bands | |||||
| 1 | Splicing factor U2AF65 | P26369 | 53,483 | 564 | 35 |
| Y box-binding protein 2; FRGY2 homolog | Q9Z2C8 | 38,248 | 305 | 20 | |
| Y box-binding protein 1 | P62960 | 35,709 | 204 | 11 | |
| hnRNP M | Q9D0E1 | 77,597 | 113 | 7 | |
| 2 | Splicing factor 3B subunit 1, SF3b1; SAP155 | Q99NB9 | 145,724 | 611 | 29 |
| 3 | Splicing factor 3B subunit 1, SF3b1; SAP155 | Q99NB9 | 145,724 | 814 | 33 |
| 4 | hnRNP L-like | Q921F4 | 64,084 | 298 | 31 |
| Y box-binding protein 2 | Q9Z2C8 | 38,248 | 197 | 20 | |
| hnRNP Q | Q7TMK9 | 69,590 | 94 | 10 | |
| hnRNP U-like 1 | Q8VDM6 | 95,942 | 79 | 7 |
The testis and liver nuclear extracts of transgenic smn−/− SMN2 micer were subjected to RNA affinity chromatography using the SMN2 exon 7 olgoribonucleotide as bait. For each testis and liver sample, four representative protein bands that appeared in only one of the two tissues were subjected to nanoelectrospray MS analysis (see Materials and Methods).
Theoretical mass.
hnRNP Q binds nearby nucleotide +6 of SMN exon 7 in HeLa cells.
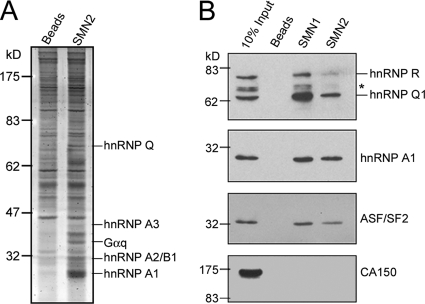
To isolate human SMN exon 7-interacting proteins, we performed RNA affinity chromatography using HeLa cell nuclear extracts. Proteins that bound to the SMN2 oligoribonucleotide were fractionated by SDS-PAGE, and those of high abundance were subjected to a matrix-assisted laser desorption ionization-TOF MS analysis. The hnRNP proteins A1/A2/A3 and Q as well as the G protein subunit Gαq were identified (Fig. 2A). Among these proteins, hnRNP A1 is a known negative regulator of SMN splicing (16), and hnRNP Q was detected in mouse testis (Table 1). Indeed, immunoblotting showed that hnRNP Q was expressed in mouse testis at a relatively higher level than even in brain and liver (Fig. 1B).
FIG. 2.
hnRNP Q association with SMN exon 7 RNA. (A) Affinity selection of SMN2 exon 7-associated proteins was performed by incubation of the HeLa cell nuclear extract with SMN2 oligoribonucleotide-conjugated resin or empty beads. Bound samples were detected by Sypro-Ruby staining on an SDS-polyacrylamide gel. Proteins identified by MS are indicated at right. (B) HEK293 cell proteins were selected by SMN1 or SMN2 exon 7 RNA resin and detected by immunoblotting with antibodies against hnRNP Q, hnRNP A1, ASF/SF2, and CA150. Anti-hnRNP Q cross-reacted with hnRNP R. The asterisk represents hnRNP Q2/3 or potential degraded fragments or isoforms of hnRNP R.
We next used RNA affinity selection followed by immunoblotting to verify the binding of hnRNP Q to SMN exon 7. The result showed that hnRNP Q and its highly related protein hnRNP R associated with both SMN1 and SMN2 exon 7 RNA (Fig. 2B). Both positive controls, hnRNP A1 and ASF, were also detected (Fig. 2B). This in vitro selection apparently revealed that hnRNP Q/R had a relatively higher preference for association with SMN1 than SMN2, whereas hnRNP A1 and ASF/SF2 bound these two SMN RNAs equally; this finding was somewhat different from that of the UV cross-linking assay (see below for the detail). Nevertheless, splicing factor CA150 was not pulled down by either RNA oligonucleotide (Fig. 2B), and the binding of Gαq to the SMN RNA was not verified in this study.
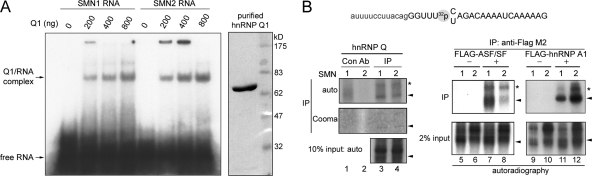
We next prepared recombinant His-tagged hnRNP Q1, the major Q isoform, to examine whether it binds directly to SMN RNA. A gel mobility shift assay showed that hnRNP Q1 by itself bound both radioisotope-labeled SMN1 and SMN2 exon 7 RNA oligonucleotides in a dose-dependent manner (Fig. 3A). Previous reports have indicated that ASF/SF2 and hnRNP A1 contact a cis element nearby nucleotide +6 of exon 7 (3, 16). Therefore, we prepared SMN exon 7 oligonucleotides containing a single 32P label immediately prior to nucleotide +6 (Fig. 3B) for use in UV cross-linking experiments. UV cross-linking was performed in HEK293 cell lysates, followed by immunoprecipitation. Transiently expressed FLAG-tagged ASF and hnRNP A1 were radiolabeled and exhibited considerable preference for SMN1 and SMN2, respectively (Fig. 3B, lanes 7, 8, 11, and 12), which was thus consistent with previous reports (3, 16). Endogenous hnRNP Q was cross-linked to both SMN oligonucleotides, however, with similar efficiency (lanes 1 to 4). This result indicated that hnRNP Q directly contacts SMN exon 7 near nucleotide +6. Perhaps hnRNP Q was present in both SMN1 and SMN2 complexes, but its association with the former was stabilized by other components. Therefore, biotinylated SMN1 RNA pulled down somewhat more hnRNP Q (Fig. 2B). Because hnRNP Q has been implicated as an SMA modifier (10), the finding of its association with SMN exon 7 RNA and direct contact with nucleotide +6 was particularly interesting. Therefore, in this study, we characterized the role of hnRNP Q in the splicing of SMN pre-mRNA.
FIG. 3.
hnRNP Q1 directly binds SMN exon 7 nearby nucleotide +6. (A) Coomassie blue staining shows the purity of recombinant His-tagged hnRNP Q1 (right panel). His-tagged hnRNP Q1 was incubated with 5′ end-labeled SMN1 or SMN2 RNA oligonucleotide. The reaction were analyzed by electrophoresis on a nondenaturing polyacrylamide gel. (B) SMN RNA oligonucleotides containing a single [32P]phosphate prior to nucleotide +6 were used for cross-linking. Lysates were prepared from HEK293 cells that were nontransfected (lanes 1 to 4) or transfected with an empty vector (lanes 5, 6, 9, and 10), or the vector expressing FLAG-tagged ASF/SF2 (lanes 7 and 8) or hnRNP A1 (lanes 11 and 12). These lysates were then incubated with SMN RNA for UV cross-linking. After RNase digestion, immunoprecipitation was performed using anti-mouse immunoglobulin (lanes 1 and 2, control antibody [Con Ab]), anti-hnRNP Q (lanes 3 and 4), or anti-FLAG (lanes 5 to 12). Input and precipitated proteins were visualized by autoradiography. Immunoprecipitated (IP) hnRNP Q is also shown by Coomassie (Cooma) blue staining. Arrowheads indicate hnRNP Q, FLAG-ASF/SF2, and FLAG-hnRNP A1. Radiolabeled signals (asterisks) might result from incomplete RNase digestion.
hnRNP Q1 promotes inclusion of exon 7 in SMN2 mRNA.
Initially, we examined whether overexpression of hnRNP Q1 has any effect on splicing of endogenous SMN1 and SMN2 transcripts in HEK293 cells. hnRNP Q1 fused with GFP was transiently expressed in HEK293 cells. The SMN splicing products were generated by primers specific to exon 6 and exon 8, followed by DdeI restriction digestion, which could distinguish SMN1 from SMN2 RNA (Fig. 4A). Figure 4B shows that overexpression of GFP-hnRNP Q1 enhanced exon 7 inclusion in SMN2. No apparent effect was detected with SMN1 splicing, perhaps due to high levels of exon 7 inclusion even in mock-transfected cells (Fig. 4B).
Next, we used the SMN2 minigene as a reporter, which contained the human SMN2 gene sequence spanning from exon 6 to exon 8, including all of introns 6 and 7 (27) (Fig. 4A). GFP-hnRNP Q1 promoted exon 7 inclusion in the SMN2 transcripts (Fig. 4C, lane 4), consistent with the observation with endogenous SMN2 (Fig. 4B). Overexpression of GFP-SRp30c also induced exon 7 inclusion in SMN2, but GFP-ASF had no detectable effect (Fig. 4C, lanes 2 and 3). Similar results were obtained in HeLa cells (data not shown). The SMN1 minigene was also examined. Since only the full-length transcript was produced from this minigene, whether hnRNP Q1 has any effect on exon 7 inclusion in SMN1 was not determined (data not shown).
Since hnRNP Q1 bound SMN exon 7, we tested its activity on mutated SMN2 minigenes. The SMN2-ug mutant contained three consecutive Gs at positions +3 to +5 of exon 7 whereas the SMN2-r20 mutant was extensively mutated by reversing the sequence of positions +3 to +22 of exon 7 (Fig. 4A). Both mutants had reduced activities to produce the full-length SMN compared to the wild-type reporter (Fig. 4D, upper panel). However, overexpression of GFP-hnRNP Q1 could promote exon 7 inclusion only in SMN2-r20 (Fig. 4D, upper panel). Accordingly, the gel mobility shift assay showed that recombinant His-hnRNP Q1 bound to both the wild-type and SMN2-r20 mutant RNA oligonucleotides (Fig. 4D, lower panel). We apparently reasoned that the SMN2-r20 mutant was responsive to hnRNP Q1-mediated regulation due to its pyrimidine-rich context around position +6 of exon 7. Nevertheless, the binding of hnRNP Q1 to exon 7 is likely essential for its activity on SMN splicing modulation.
Finally, we tested whether hnRNP Q could modulate SMN splicing in cells derived from SMA patients (23). Using the SMN2 minigene, we observed that GFP-hnRNP Q1 promoted exon 7 inclusion in this fibroblast line (Fig. 4E). Such an exon 7 inclusion effect was also detected with the endogenous SMN2 transcripts (see Fig. S1 in the supplemental material), albeit relatively low levels due to the low transfection efficiency of human fibroblasts. Therefore, hnRNP Q1 can function in the context of human SMA cells. Taken together, the above in vivo splicing assays indicated that hnRNP Q likely binds closely to position +6 of SMN exon 7 and promotes its inclusion in various cell lines.
We also performed in vitro splicing of an SMN2-derived pre-mRNA in HeLa nuclear extracts. Splicing of this pre-mRNA would yield exon 7-containing and -skipping mRNAs as well as other intron-retained products (3), but our system was insufficient to generate completely spliced products (see Fig. S2 in the supplemental material). Nevertheless, we observed that recombinant hnRNP Q1 promoted SMN2 spicing to the 3′ splice site of intron 6 instead of intron 7, which may result in exon 7 inclusion (see Fig. S2 in the supplemental material). Therefore, both in vivo and in vitro results indicated the role of hnRNP Q1 in activation of exon 7 utilization.
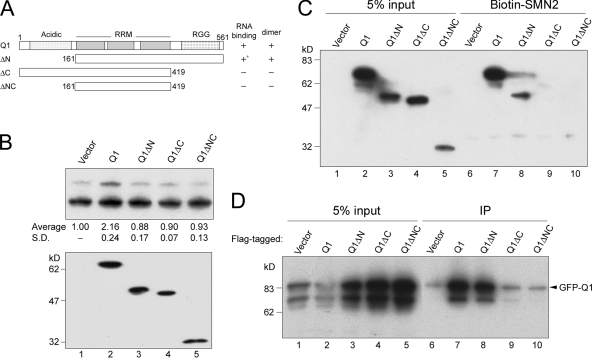
The C-terminal domain of hnRNP Q mediates RNA binding and self-association and is required for exon 7 inclusion.
Next, to determine which domain of hnRNP Q1 contributes to its splicing activity, we examined its truncated versions for SMN splicing. The truncated hnRNP Q1 proteins (ΔN, C, or ΔNC) and each truncated and full-length protein carried a FLAG epitope tag (Fig. 5A). Cotransfection of the hnRNP Q1 expression vector with the SMN2 reporter showed that none of the truncated versions could activate exon 7 inclusion in the SMN2 transcripts (Fig. 5B, lanes 3 to 5), indicating that both domains are required.
FIG. 5.
The C-terminal domain of hnRNP Q1 is essential for SMN2 exon 7 inclusion perhaps by its ability to bind SMN2 exon 7 and form homodimers. (A) Diagram shows hnRNP Q1 and truncated versions used in the splicing assay. The RNA binding and dimerization abilities of hnRNP Q isoforms are summarized; the asterisk indicates weak binding. (B) The pCI-SMN2 reporter was cotransfected with vector expressing FLAG-tagged full-length or truncated hnRNP Q1 proteins (Q1ΔN, Q1ΔC, or Q1ΔNC) into HEK293 cells. Splicing assay, quantitative analysis, and immunoblotting were as described in the legend of Fig. 4. SD, standard deviation. (C) HEK293 cells containing overexpressed FLAG-tagged full-length or truncated hnRNP Q1 were incubated with SMN2 oligoribonucleotide-conjugated resin. SMN RNA-selected proteins were analyzed by immunoblotting with anti-FLAG. Cells transfected with an empty vector served as a control (Vector). (D) GFP-hnRNP Q1 was overexpressed alone (Vector) or coexpressed with FLAG-tagged full-length or truncated hnRNP Q1 in HEK293 cells. Immunoprecipitation (IP) was performed in the presence of RNase using anti-FLAG, and immunoblotting used anti-GFP.
To explore how the N- and C-terminal domains of hnRNP Q function, we first examined the ability of truncated hnRNP Q1 proteins to bind SMN exon 7 RNA using RNA affinity chromatography. HEK293 cell lysates containing full-length, ΔN, ΔC, or ΔNC hnRNP Q1 were incubated with biotinylated SMN exon 7 oligoribonucleotides. The C-terminal truncation severely impaired the SMN RNA binding activity of hnRNP Q1 (Fig. 5C, lane 9), whereas N-terminally truncated Q1 partially retained this activity (∼30% that of full-length hnRNP Q1) (Fig. 5C, lane 8). Perhaps both the N- and C-terminal domains contributed to SMN RNA binding, but the latter, which contains the RG-rich sequence, was more critical for this activity.
Because certain hnRNP-type proteins, such as hnRNP A1 and PTB, may form multimers for their splicing regulatory function (2), we examined whether hnRNP Q1 is also able to self-interact. A GFP-hnRNP Q1 fusion protein was coexpressed with FLAG-tagged full-length or truncated Q1 in HEK293 cells. Using anti-FLAG, we observed that GFP-hnRNP Q1 coprecipitated with FLAG-Q1 (Fig. 5D, lane 7), suggesting that hnRNP Q1 forms a homodimer or even a multimer in vivo. Dimerization of hnRNP Q1 was also demonstrated by in vitro chemical cross-linking using disuccinimidyl suberate (data not shown). Moreover, we observed that the C-terminal, but not the N-terminal, truncation abolished this self-interaction (Fig. 5D, lanes 8 and 9). Therefore, the C-terminal domain of hnRNP Q1 participates in both RNA binding and dimerization, which perhaps are required for SMN exon 7 inclusion. However, how the N-terminal domain of hnRNP Q1 is involved in SMN splicing still remains to be characterized.
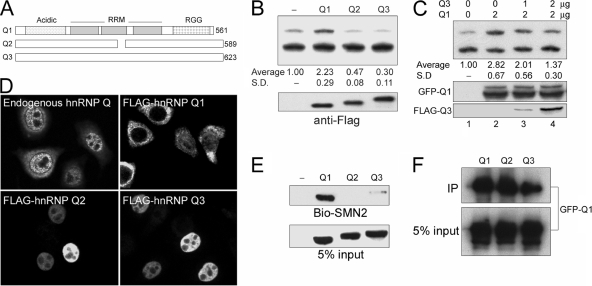
The hnRNP Q isoforms have differential activity in alterative splicing of SMN2.
Since three hnRNP Q protein isoforms (Fig. 6A) are often coexpressed in cells (5, 7, 32), we therefore examined whether they behave similarly in splicing of SMN pre-mRNAs. Surprisingly, in sharp contrast to hnRNP Q1, overexpression of FLAG-tagged Q2 or Q3 protein suppressed exon 7 inclusion in SMN2 (Fig. 6B). Overexpression of hnRNP Q3 reduced the level of Q1-induced exon 7 inclusion (Fig. 6C), suggesting that Q3 acts as a negative regulator and could antagonize the activity of Q1.
FIG. 6.
The hnRNP Q isoforms have opposing activity in alterative splicing of SMN2. (A) Diagram shows three hnRNP Q isoforms. (B) The pCI-SMN2 reporter was cotransfected with an empty vector or vector encoding FLAG-hnRNP Q1, Q2, or Q3. Splicing assay, quantitative analysis, and immunoblotting were performed as in described in the legend of Fig. 4. Ex7 represents exon 7. (C) The pCI-SMN2 reporter was cotransfected with the GFP-hnRNP Q1 expression vector alone or together with the vector encoding FLAG-hnRNP Q3 (lanes 3 and 4). Lane 1 shows a mock transfection. Immunoblotting was performed using anti-GFP and anti-FLAG to detect hnRNP Q1 and Q3, respectively. (D) Indirect immunofluorescence was performed using anti-hnRNP Q in nontransfected HeLa cells or using anti-FLAG in cells that transiently expressed FLAG-hnRNP Q1, Q2, or Q3. (E) HEK293 cell lysates containing overexpressed FLAG-hnRNP Q isoform were subjected to biotinylated-SMN2 (Bio-SMN2) RNA affinity chromatography as in described in the legend of Fig. 5C. (F) GFP-hnRNP Q1 was coexpressed with each of the FLAG-hnRNP Q isoforms in HEK293 cells. Subsequent immunoprecipitation and immunoblotting were performed as in described in the legend of Fig. 5D. SD, standard deviation.
To explore why hnRNP Q1 and Q2/3 isoforms functioned oppositely in SMN splicing, we first examined their subcellular localization. Indirect immunofluorescence showed that overexpressed FLAG-hnRNP Q1 was distributed throughout the cells but was located predominantly in the cytoplasm (Fig. 6D). By contrast, both Q2 and Q3 isoforms were primarily localized in the nucleus (Fig. 6D).
The different subcellular distributions of hnRNP Q isoforms were somewhat intriguing and could not give a straightforward explanation of why overexpression of hnRNP Q2/3 resulted in exon 7 exclusion. Therefore, we examined the ability of the Q2/3 isoforms to bind SMN RNA. Using a biotinylated SMN2 exon 7 RNA oligonucleotide as ligand, we observed that Q2 or Q3 had no or minimal SMN2 RNA binding activity, respectively (Fig. 6E). The same result was obtained with SMN1 RNA (data not shown). We reasoned that the truncated RRM2 of hnRNP Q2 might account for the loss of RNA binding activity, and the extended C-terminal regions of both hnRNP Q2 and Q3 might also hamper RNA binding. Nevertheless, inefficient binding of the hnRNP Q2/3 isoforms to exon 7 RNA was still not sufficient to explain their dominant-negative effect on exon 7 inclusion unless they could sequester an endogenous positive effector(s). We therefore examined whether the Q2 and Q3 isoforms could heterodimerize with Q1 by coprecipitation of overexpressed GFP-hnRNP Q1 with FLAG-tagged hnRNP Q2/3. Figure 6F shows that both hnRNP Q2 and Q3 efficiently interacted with Q1. Therefore, overexpressed hnRNP Q2/3 might sequester endogenous Q1 by forming heteromeric complexes and thereby lead to a suppressive effect on SMN exon 7 inclusion.
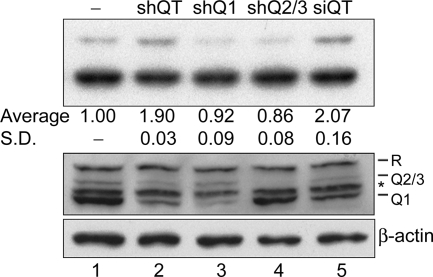
Downregulation of hnRNP Q proteins enhances SMN2 exon 7 inclusion.
The above data revealed differential activities of hnRNP Q isoforms in SMN splicing. Therefore, we next examined the effect(s) of downregulation of hnRNP Q proteins. We transfected either the expression vector encoding shRNAs or siRNA oligonucleotides targeted to all three hnRNP Q isoforms into HEK293 cells (Fig. 7, lanes 2 and 5). Immunoblotting revealed that the levels of hnRNP Q proteins were reduced by shRNA or siRNA to 30 to 40% of that of the mock transfections (Fig. 7, bottom, lanes 2 and 5). Surprisingly, the in vivo splicing assay showed that SMN2 exon 7 inclusion was increased under the conditions of reduced hnRNP Q protein levels (Fig. 7, top, lanes 2 and 5). Therefore, another two shRNAs, specific to hnRNP Q1 and Q2/3, respectively, were tested. Intriguingly, splicing of the SMN2 minigene transcript was not significantly affected by specific depletion of hnRNP Q1 or Q2/3 (Fig. 7, bottom, lanes 3 and 4). All of above observations were reproducible in independent experiments, and the plausible explanations are discussed below.
FIG. 7.
Downregulation of hnRNP Q proteins enhances exon 7 inclusion. HEK293 cells were transfected with empty pRS vector (−) or pRS-shQT, -shQ1, or -shQ2/3 or siRNA mixture siQT. Immunoblotting (bottom) was performed to show the expression level of hnRNP Q/R isoforms and β-actin. The asterisk may represent hnRNP R isoforms or degraded fragments. The splicing assay (top) and quantitative analysis were performed as described in the legend of in Fig. 4. SD, standard deviation.
DISCUSSION
A critical C-to-T transition at position +6 of exon 7 in SMN2 interferes with exon 7 inclusion in mRNA and thus yields a functionally defective exon 7-truncated SMN protein (27, 31). Splicing factors or antisense oligonucleotides that can modify SMN2 exon 7 splicing have shown their potential for therapeutic application (12, 14, 29, 39). We initially observed an unexpectedly high level of full-length SMN2 mRNAs as well as SMN protein in the testis of transgenic smn−/− SMN2 mice (Fig. 1), suggesting a testis-specific regulation of SMN2 pre-mRNA splicing. Using SMN RNA affinity selection, we identified hnRNP Q in both mouse testis and human cell lines. We subsequently demonstrated that hnRNP Q could modulate splicing of SMN2 exon 7. Since hnRNP Q may function together with the SMN protein in axonal growth and act as a genetic modifer of SMA (10, 37), we focused primarily on hnRNP Q in this study. Nevertheless, we also examined PSF, another splicing factor that potentially associated with SMN exon 7 RNA (Table 1) and found that overexpression of PSF also enhanced exon 7 inclusion (Fig. 4E, lanes 3 and 4). Therefore, future experiments are needed to investigate whether any other SMN exon 7-associated factors could also modulate SMN splicing.
Positive role of hnRNP Q1 in SMN pre-mRNA splicing.
We identified hnRNP Q as a potential factor involved in SMN mRNA metabolism by its association with SMN RNA fragments containing the 5′ part of exon 7. Recombinant hnRNP Q1 by itself was able to bind both SMN1 and SMN2 exon 7 RNAs (Fig. 3). Indeed, hnRNP Q associates with SMN RNA in vivo at a site proximal to nucleotide +6 of exon 7 (Fig. 3). Overexpression of hnRNP Q1 can enhance SMN2 exon 7 inclusion (Fig. 4). Activation of SMN2 exon 7 by hnRNP Q1 was at least partially cis element dependent. Perhaps hnRNP Q1 binds exon 7 and subsequently promotes the use of its upstream 3′ splice site (see Fig. S2 in the supplemental material). The C-terminal RG-rich domain of hnRNP Q1 contributed substantially to RNA binding (Fig. 5), as observed with many RNA binding proteins containing a similar domain (1, 18, 30, 46). Moreover, hnRNP Q, also similar to several other hnRNP proteins as well as its C. elegans homolog HRP-2, formed homomers (19, 20). This self-interaction activity was independent of RNA and also involved the C-terminal domain (Fig. 5). C-terminal domain truncation impaired hnRNP Q1-mediated exon 7 inclusion, suggesting a correlation between the RNA binding and self-interaction activities of Q1 and its function in SMN exon 7 selection. However, N-terminally truncated hnRNP Q1 retained partial RNA binding activity and was able to form a dimer, but it failed to activate SMN splicing. Therefore, the N-terminal domain of hnRNP Q1 may have an as yet unidentified activity critical for SMN splicing.
hnRNP Q1 predominantly localized in the cytoplasm (Fig. 6), which somewhat contradicts its function in splicing regulation. We had introduced a nuclear localization signal derived from the simian virus 40 large T antigen into hnRNP Q1, which forced nuclear retention of hnRNP Q1 but, intriguingly, impaired its ability to promote exon 7 inclusion (data not shown). We reasoned that continuous shuttling of hnRNP Q1 (see Fig. S3 in the supplemental material), which may allow its posttranslational modification, is probably required for SMN exon 7 inclusion. Nevertheless, we apparently conclude that hnRNP Q1, when overexpressed, can act as a splicing activator for SMN2.
Negative role of hnRNP Q2/3 in SMN pre-mRNA splicing.
In sharp contrast to hnRNP Q1, overexpression of the minor hnRNP Q isoforms (Q2 or Q3) suppressed exon 7 inclusion in SMN2 and abrogated the positive effect of hnRNP Q1 (Fig. 6). We explain this result by the incapability of hnRNP Q2/3 binding to SMN exon 7 RNA. The C-terminal extension of hnRNP Q2/3 probably impairs exon 7 RNA binding and inclusion activity. Indeed, truncation of this extension from hnRNP Q3, which contains an intact RRM2, restored its activity in exon 7 inclusion (data not shown). How this extended region interferes with the splicing activation activity of hnRNP Q still remains to be investigated. Moreover, because hnRNP Q2 and Q3 were able to interact with Q1 (Fig. 6), their overexpression may prevent endogenous hnRNP Q1 from accessing SMN2 mRNA. The hnRNP Q2/3 isoforms, though expressed at a much lower level than Q1, localized predominantly in the nucleus (Fig. 6). Perhaps these two minor isoforms, when overexpressed, antagonize the function of nuclear hnRNP Q1, thus acting as a negative regulator for SMN2 exon 7 inclusion.
Coordinated function of hnRNP Q isoforms in SMN pre-mRNA splicing.
Our results clearly demonstrated that overexpressed hnRNP Q1 and Q2/3 isoforms had opposing effects on SMN2 exon 7 inclusion (Fig. 6). When overexpressed, hnRNP Q1 acted as a positive regulator for SMN2 exon 7 inclusion. However, specific depletion of Q1, leaving only Q2/3 in cells, had no considerable effect on SMN splicing (Fig. 7). We therefore infer that the Q2/3 isoforms are not deleterious for exon 7 inclusion unless they are overexpressed. Moreover, a physiological level of Q2/3 is probably essential for Q1 function; for example, Q2/3 might facilitate nuclear import of Q1 via heterodimerization. Therefore, specific and extensive depletion of Q2/3 might impair the activity of Q1 and therefore could not elevate the level of exon 7 inclusion (Fig. 7). However, when the expression of all isoforms was concomitantly attenuated, Q1, even at a reduced level, could still activate SMN2 exon 7 with the aid of residual Q2/3. Nevertheless, many possibilities remain to be examined in the future. Together, different isoforms of hnRNP Q, at different expression levels, may fine-tune SMN exon 7 inclusion.
hnRNP Q1 binds SMN exon 7 and thereby activates its inclusion during splicing. hnRNP Q1 might also act in coordination with ASF/SF2 or hnRNP A1, each of which also binds exon 7 near this site, to regulate SMN exon 7 utilization. Indeed, we observed that overexpression of hnRNP A1 abrogated the SMN exon 7 RNA binding and inclusion activities of hnRNP Q1 (data not shown). It is possible that hnRNP A1, due to its high affinity toward SMN2 exon 7, outcompetes Q1 and subsequently induces exon 7 skipping. As to ASF, it had no interaction with hnRNP Q and did not have any effect on hnRNP Q-mediated SMN2 splicing regulation (data not shown). The coordinate function of hnRNP Q with other splicing regulators also remains to be characterized in more detail.
hnRNP Q as a therapeutic tool.
A clinical analysis has shown that SMA patients had reduced levels of both SMN and its interacting proteins including hnRNP Q compared with their unaffected siblings with homologous SMN1 deletions (10). The inverse correlation between hnRNP Q levels and the severity of SMA suggests that hnRNP Q is an important modifier of SMA. We report here that different expression levels and isoforms of hnRNP Q may intricately control alternative splicing of SMN2. Although the detailed mechanisms underlying hnRNP Q-mediated SMN splicing regulation remain to be elucidated, our study indicates that augmenting the expression level of hnRNP Q1 may be a therapeutic strategy for SMA.
Supplementary Material
Acknowledgments
We thank B. J. Blencowe, M.-J. Fann, A. Krainer, and B. Wirth for their plasmids and C.-C. Lai and C.-G. Jou for mass spectrometric analysis. We also thank Y.-S. Lue for HeLa cell nuclear extracts. Finally, we appreciate Tim C. Taylor for editing the manuscript.
This work was primarily supported by intramural funds from Academia Sinica and also by grant NHRI-EX97-9737NI from the National Health Research Institutes of Taiwan.
Footnotes
Published ahead of print on 15 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdul-Manan, N., S. M. O'Malley, and K. R. Williams. 1996. Origins of binding specificity of the A1 heterogeneous nuclear ribonucleoprotein. Biochemistry 353545-3554. [DOI] [PubMed] [Google Scholar]
- 2.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72291-336. [DOI] [PubMed] [Google Scholar]
- 3.Cartegni, L., and A. R. Krainer. 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 30377-384. [DOI] [PubMed] [Google Scholar]
- 4.Chang, J. G., H. M. Hsieh-Li, Y. J. Jong, N. M. Wang, C. H. Tsai, and H. Li. 2001. Treatment of spinal muscular atrophy by sodium butyrate. Proc. Natl. Acad. Sci. USA 989808-9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elvira, G., S. Wasiak, V. Blandford, X. K. Tong, A. Serrano, X. Fan, M. del Rayo Sanchez-Carbente, F. Servant, A. W. Bell, D. Boismenu, J. C. Lacaille, P. S. McPherson, L. DesGroseillers, and W. S. Sossin. 2006. Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics 5635-651. [DOI] [PubMed] [Google Scholar]
- 6.Feldkotter, M., V. Schwarzer, R. Wirth, T. F. Wienker, and B. Wirth. 2002. Quantitative analyses of SMN1 and SMN2 based on real-time LightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 70358-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabanella, F., C. Carissimi, A. Usiello, and L. Pellizzoni. 2005. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum. Mol. Genet. 143629-3642. [DOI] [PubMed] [Google Scholar]
- 8.Gharahdaghi, F., C. R. Weinberg, D. A. Meagher, B. S. Imai, and S. M. Mische. 1999. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20601-605. [DOI] [PubMed] [Google Scholar]
- 9.Grosset, C., C. Y. Chen, N. Xu, N. Sonenberg, H. Jacquemin-Sablon, and A. B. Shyu. 2000. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 10329-40. [DOI] [PubMed] [Google Scholar]
- 10.Helmken, C., Y. Hofmann, F. Schoenen, G. Oprea, H. Raschke, S. Rudnik-Schoneborn, K. Zerres, and B. Wirth. 2003. Evidence for a modifying pathway in SMA discordant families: reduced SMN level decreases the amount of its interacting partners and Htra2-β1. Hum. Genet. 11411-21. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann, Y., C. L. Lorson, S. Stamm, E. J. Androphy, and B. Wirth. 2000. Htra2-β1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc. Natl. Acad. Sci. USA 979618-9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann, Y., and B. Wirth. 2002. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-β1. Hum. Mol. Genet. 112037-2049. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh-Li, H. M., J. G. Chang, Y. J. Jong, M. H. Wu, N. M. Wang, C. H. Tsai, and H. Li. 2000. A mouse model for spinal muscular atrophy. Nat. Genet. 2466-70. [DOI] [PubMed] [Google Scholar]
- 14.Hua, Y., T. A. Vickers, B. F. Baker, C. F. Bennett, and A. R. Krainer. 2007. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 5e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jong, Y. J., J. G. Chang, S. P. Lin, T. Y. Yang, J. C. Wang, C. P. Chang, C. C. Lee, H. Li, H. M. Hsieh-Li, and C. H. Tsai. 2000. Analysis of the mRNA transcripts of the survival motor neuron (SMN) gene in the tissue of an SMA fetus and the peripheral blood mononuclear cells of normals, carriers and SMA patients. J. Neurol. Sci. 173147-153. [DOI] [PubMed] [Google Scholar]
- 16.Kashima, T., and J. L. Manley. 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 34460-463. [DOI] [PubMed] [Google Scholar]
- 17.Kashima, T., N. Rao, and J. L. Manley. 2007. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 1043426-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiledjian, M., and G. Dreyfuss. 1992. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 112655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, J. H., B. Hahm, Y. K. Kim, M. Choi, and S. K. Jang. 2000. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298395-405. [DOI] [PubMed] [Google Scholar]
- 20.Kinnaird, J. H., K. Maitland, G. A. Walker, I. Wheatley, F. J. Thompson, and E. Devaney. 2004. HRP-2, a heterogeneous nuclear ribonucleoprotein, is essential for embryogenesis and oogenesis in Caenorhabditis elegans. Exp. Cell Res. 298418-430. [DOI] [PubMed] [Google Scholar]
- 21.Lai, M. C., R. I. Lin, and W. Y. Tarn. 2001. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. USA 9810154-10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefebvre, S., L. Burglen, S. Reboullet, O. Clermont, P. Burlet, L. Viollet, B. Benichou, C. Cruaud, P. Millasseau, M. Zeviani, et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80155-165. [DOI] [PubMed] [Google Scholar]
- 23.Liang, W. C., C. Y. Yuo, J. G. Chang, Y. C. Chen, Y. F. Chang, H. Y. Wang, Y. H. Ju, S. S. Chiou, and Y. J. Jong. 2008. The effect of hydroxyurea in spinal muscular atrophy cells and patients. J. Neurol. Sci. 26887-94. [DOI] [PubMed] [Google Scholar]
- 24.Lin, J. C., and W. Y. Tarn. 2005. Exon selection in α-tropomyosin mRNA is regulated by the antagonistic action of RBM4 and PTB. Mol. Cell. Biol. 2510111-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, K. T., R. M. Lu, and W. Y. Tarn. 2004. The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol. Cell. Biol. 249176-9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorson, C. L., and E. J. Androphy. 2000. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 9259-265. [DOI] [PubMed] [Google Scholar]
- 27.Lorson, C. L., E. Hahnen, E. J. Androphy, and B. Wirth. 1999. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 966307-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorson, C. L., J. Strasswimmer, J. M. Yao, J. D. Baleja, E. Hahnen, B. Wirth, T. Le, A. H. Burghes, and E. J. Androphy. 1998. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 1963-66. [DOI] [PubMed] [Google Scholar]
- 29.Madocsai, C., S. R. Lim, T. Geib, B. J. Lam, and K. J. Hertel. 2005. Correction of SMN2 Pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol. Ther. 121013-1022. [DOI] [PubMed] [Google Scholar]
- 30.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 707445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monani, U. R., C. L. Lorson, D. W. Parsons, T. W. Prior, E. J. Androphy, A. H. Burghes, and J. D. McPherson. 1999. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 81177-1183. [DOI] [PubMed] [Google Scholar]
- 32.Mourelatos, Z., L. Abel, J. Yong, N. Kataoka, and G. Dreyfuss. 2001. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 205443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearn, J. 1978. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J. Med. Genet. 15409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellizzoni, L., B. Charroux, and G. Dreyfuss. 1999. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl. Acad. Sci. USA 9611167-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pellizzoni, L., N. Kataoka, B. Charroux, and G. Dreyfuss. 1998. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 95615-624. [DOI] [PubMed] [Google Scholar]
- 36.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12340-345. [DOI] [PubMed] [Google Scholar]
- 37.Rossoll, W., S. Jablonka, C. Andreassi, A. K. Kroning, K. Karle, U. R. Monani, and M. Sendtner. 2003. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of β-actin mRNA in growth cones of motoneurons. J. Cell Biol. 163801-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossoll, W., A. K. Kroning, U. M. Ohndorf, C. Steegborn, S. Jablonka, and M. Sendtner. 2002. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum. Mol. Genet. 1193-105. [DOI] [PubMed] [Google Scholar]
- 39.Singh, N. K., N. N. Singh, E. J. Androphy, and R. N. Singh. 2006. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 261333-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh, N. N., E. J. Androphy, and R. N. Singh. 2004. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 315381-388. [DOI] [PubMed] [Google Scholar]
- 41.Singh, N. N., R. N. Singh, and E. J. Androphy. 2007. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 35371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terry, D. E., E. Umstot, and D. M. Desiderio. 2004. Optimized sample-processing time and peptide recovery for the mass spectrometric analysis of protein digests. J. Am. Soc. Mass Spectrom. 15784-794. [DOI] [PubMed] [Google Scholar]
- 43.Wirth, B., L. Brichta, and E. Hahnen. 2006. Spinal muscular atrophy and therapeutic prospects. Prog. Mol. Subcell. Biol. 44109-132. [DOI] [PubMed] [Google Scholar]
- 44.Wirth, B., L. Brichta, B. Schrank, H. Lochmuller, S. Blick, A. Baasner, and R. Heller. 2006. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum. Genet. 119422-428. [DOI] [PubMed] [Google Scholar]
- 45.Young, P. J., C. J. DiDonato, D. Hu, R. Kothary, E. J. Androphy, and C. L. Lorson. 2002. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2 β1. Hum. Mol. Genet. 11577-587. [DOI] [PubMed] [Google Scholar]
- 46.Zanotti, K. J., P. E. Lackey, G. L. Evans, and M. R. Mihailescu. 2006. Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA. Biochemistry 458319-8330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.