Abstract
Design of a partially cysteine-depleted C98S/C239S/C377S/C468A cytochrome P450 3A4 mutant designated CYP3A4(C58,C64) allowed site-directed incorporation of thiol-reactive fluorescent probes into α-helix A‥ The site of modification was identified as Cys-64 with the help of CYP3A4(C58) and CYP3A4(C64), each bearing only one accessible cysteine. Changes in the fluorescence of CYP3A4(C58,C64) labeled with 6-bromoacetyl-2-dimethylaminonaphthalene (BADAN), 7-diethylamino-3-(4’-maleimidylphenyl)-4-methylcoumarin (CPM), or monobromobimane (mBBr) were used to study the interactions with bromocriptine (BCT), 1-pyrenebutanol (1-PB), testosterone (TST), and α-naphthoflavone (ANF). Of these substrates only ANF has a specific effect, causing a considerable decrease in fluorescence intensity of BADAN and CPM and increasing the fluorescence of mBBr. This ANF-binding event in the case of BADAN-modified enzyme is characterized by an S50 of 18.2 ± 0.7, compared with the value of 2.2 ± 0.3 for the ANF-induced spin transition, thus revealing an additional low affinity binding site. Studies of the effect of TST, 1-PB, and BCT on the interactions of ANF monitored by changes in fluorescence of CYP3A4(C58,C64)-BADAN or by the ANF-induced spin transition revealed no competition by these substrates. Investigation of the kinetics of fluorescence increase upon H2O2-dependent heme depletion suggests that labeled CYP3A4(C58,C64) is represented by two conformers, one of which has the fluorescence of the BADAN and CPM labels completely quenched, presumably by photoinduced electron transfer from the neighboring Trp-72 and/or Tyr-68 residues. The binding of ANF to the newly discovered binding site appears to affect the interactions of the label with the above residue(s), thus modulating the fraction of the fluorescent conformer.
Keywords: Cytochrome P450 3A4, α-naphthoflavone, cooperativity, substrate binding site, BADAN, conformational heterogeneity
A number of recent studies on the mechanisms of function and regulation of microsomal monooxygenase systems have focused on the importance of homo- and heterotropic cooperativity exhibited by various mammalian drug-metabolizing cytochromes P450 (1–8). Cooperative behavior has been reported for such enzymes as P450 1A2 (9), 2C9 (10–12), 2C5 (13), 2B6 (14), and 2B4 (15). However, the most prominent cases of P450 cooperativity are found in cytochrome P450 3A4 (CYP3A4), the principal drug-metabolizing enzyme in humans (16). In addition to the homotropic cooperativity observed with a wide variety of substrates (1, 4, 16–18), CYP3A4 also provides important examples of heterotropic activation (19–22), of which the most known instances are observed with α-naphthoflavone (ANF). This flavonoid typically attenuates homotropic cooperativity observed with other substrates (18, 23), while stimulating their metabolism (23, 24). Another representative heterotropic activator and inhibitor of CYP3A4 is quinidine (22, 25–27).
During the last decade our understanding of the mechanisms of CYP3A4 cooperativity has progressed from a static model with multiple binding sites (8, 18, 20, 23, 28) to more complex dynamic models suggesting effector-induced conformational rearrangements of the enzyme along with multiple ligand binding (18, 28–32). The recent advances in X-ray crystallography of mammalian cytochromes P450 provide us with important evidence of a vast conformational mobility of these proteins. The structures of substrate-free and ligand-bound cytochrome P450 2B4 (CYP2B4) recently resolved in this laboratory (33–35) depict an impressive large-scale substrate-induced conformational transition, which changes considerably the openness of the heme pocket. Recently resolved structures of the complexes of CYP3A4 with ketoconazole and erythromycin (36) demonstrate that this heme protein also exhibits substantial conformational plasticity. At the same time, our recent studies on CYP3A4 emphasize an important role of persistent conformational heterogeneity in the mechanisms of interactions with substrates and allosteric ligands (30, 37). Recently we proposed that this heterogeneity, which was first reported by Koley et al. (21, 38, 39), may originate from differences in conformation and/or orientation of the subunits in P450 oligomers (37). We also suggested that the mechanism of action of heterotropic activators, such as ANF, may result from modulation of the architecture of the oligomer, which changes the partitioning between different conformers of the enzyme (30, 37). It seems likely that the conformational difference reflected in the heterogeneity of CYP3A4 is closely related to the conformational dynamics observed in CYP2B4 and now in CYP3A4 itself. Therefore, we in the present study we sought to probe a substrate-induced large-scale rearrangement in CYP3A4, which may be related to the mechanisms of cooperativity.
The main premise of this study was to utilize the fluorescence of a covalently attached, environment-sensitive probe capable of FRET to the heme of CYP3A4 to monitor possible structural rearrangement upon substrate binding. For this purpose we elected to utilize a thiol-reactive fluorescent probe attached to an appropriate cysteine residue in the enzyme. The wild type CYP3A4 possesses six cysteines that are potentially accessible for modification, namely Cys-58, Cys-64, Cys-98, Cys-239, Cys-377, and Cys-468.
In order to incorporate thiol-reactive labels into desired regions of the enzyme we initially sought to design a completely cysteine-depleted variant of the enzyme. This report represents a key interim step in this direction. Here we employ the mutant with four cysteine-depleting replacements, where residues 98, 239 and 377 are replaced with serine and residue 468 with alanine. The resulting quadruple mutant C98S/C239S/C377S/C468A designated here as CYP3A4(C58,C64), possesses only two closely-located potentially reactive cysteines in □-helix A. This location appears to be favorable for probing substrate-induced conformational rearrangements in P450. For instance, the transition between the closed structure of CPI-bound CYP2B4 (34) to the more open conformation of the BIF-bound enzyme (35) changes the distance between Ser-54 at the N-terminal part of helix A and the heme iron from 27.1 to 24.3 Å. This change is large enough to be detected by the modulation of the efficiency of FRET from an appropriate fluorophore to the heme. At the same time, an important conformational change in the loop preceding helix A detected upon the binding of bifonazole to CYP2B4 (35) shows that substrate-induced local conformational changes in this region are also possible. Thus, we decided to probe the effect of substrates on the fluorescence of the thiol-reactive fluorescent probe 6-bromoacetyl-2-dimethylaminonaphthalene (BADAN) incorporated into this partially cysteine-depleted mutant. The chosen dye is known to exhibit an ample solvatochromic shift in the position of its emission band (40–42), which is useful in probing conformational transitions in proteins (40, 43). Moreover, a broad emission band of the probe overlaps considerably with the heme absorbance bands of P450.
In this study we explore the changes in the fluorescence of BADAN-labeled CYP3A4(C58,C64) caused by its interactions with bromocriptine (BCT), ANF, 1-pyrenebutanol (1-PB), and testosterone (TST). While the interactions of BCT with CYP3A4 exhibit no cooperativity (44), prominent homotropic cooperativity is observed with 1-PB, TST, and ANF (17, 18, 45–47), and ANF represents heterotropic cooperativity (vide supra). The studies of substrate-dependent changes in the fluorescence of the probe were complemented by monitoring of the changes in the spin state of the heme iron, which reflect the formation of a functional enzyme-substrate complex. Our results indicate that addition of ANF results in a substantial decrease in BADAN fluorescence, in contrast to 1-PB, BCT and TST. A similar effect of ANF was observed in the protein labeled with another probe, 7-diethylamino-3-(4'-maleimidylphenyl)-4-methylcoumarin (CPM). Our observations suggest the presence of an additional binding site for ANF that is distinct from those revealed by the substrate-induced spin shift.
Experimental Procedures
Materials
1-PB, BADAN, CPM, and monobromobimane (mBBr) were from Invitrogen/Molecular Probes (Eugene, OR). ANF was from Indofine Chemical Company (Hillsbrough, NJ), and bromocriptine (BCT) mesylate was from Sigma Chemicals (St. Louis, MO). The QuikChange site-directed mutagenesis kit was from Stratagene (La Jolla, CA). All other chemicals were of ACS grade and were used without further purification.
Site-Directed Mutagenesis
C98S was generated using the QuikChange site-directed mutagenesis kit and a template consisting of the N-terminally truncated (Δ3–12) and tetrahistidine-tagged CYP3A4 wild type cDNA. CYP3A4(C58,C64) was generated by consecutive single-cysteine replacements of C98S, C98S/C239S, and C98S/C239S/C377S. The following PCR primers were used:
| C98S | 5`- CAGTGCTAGTGAAAGAATCTTATTCTGTCTTCAC-3` |
| 5`- GTGAAGACAGAATAAGATTCTTTCACTAGCACTG-3` | |
| C239S: | 5`- AATATCTCTGTGTTTCCAAGAGAAG-3` |
| 5`- CTTCTCTTGGAAACACAGAGATATT-3` | |
| C377S: | 5`- GAGAGGGTCAGCAAAAAAGATG-3` |
| 5`- CATCTTTTTTGCTGACCCTCTC-3` | |
| C468A: | 5`- CCTTCAAACCTGCTAAAGAAACACAGATCCCCC-3` |
| 5`- GGGGGATCTGTGTTTCTTTAGCAGGTTTGAAGG-3` |
CYP3A4(C58) and CYP3A4(C64) quintuple mutants were obtained using the above CYP3A4(C58,C64) as a template. To generate these constructs we used the following primers
| C58T: | 5`- CCATAAGGGCTTTACTATGTTTGACATGG -3` |
| 5`- CCATGTCAAACATAGTAAAGCCCTTATGG -3` | |
| C64A: | 5`- GTTTGACATGGAAGCTCATAAAAAGTATGG -3` |
| 5`-CCATACTTTTTATGAGCTTCCATGTCAAAC -3` |
All constructs were sequenced to verify that only the desired mutations were present (Protein Chemistry Core Laboratory, The University of Texas Medical Branch, Galveston, TX).
Protein expression, and purification
CYP3A4 and CYP3A4(C58,C64) were expressed as the His-tagged protein in Escherichia coli TOPP3 cells and purified by a combination of metal-affinity and ion-exchange chromatography. Cell growth and protein extraction were performed as previously described (17, 48), except for replacement of the 100 mM MOPS buffer with 100 mM Na-Hepes. The supernatant was diluted to a P450 concentration of 1 µM with 100 mM Na-Hepes buffer, pH 7.4, containing 10% glycerol (v/v) (Buffer A) containing 0.5% CHAPS, and 0.5 M KCl. Starting at this step all buffers contained 2 mM mercaptoethanol unless indicated. The protein was loaded onto a column of Ni-NTA Agarose (Qualigen Inc., Valencia, CA) using 1 mL gel per 60 nmol P450. The column was subsequently washed with 3 bed volumes of buffer A containing 20 mM imidazole and 0.5 M KCl, 30 bed volumes of buffer A containing 0.5 % Igepal CO-630 and 0.5 M KCl, and 10 bed volumes of Buffer A containing 0.2% Igepal CO-630 (Buffer B). Protein was eluted with 200 mM imidazole in Buffer B. The pooled fractions were diluted with 20 volumes of Buffer B and applied to a column of Macro-Prep High S Support resin (Bio-Rad Laboratories, Hercules, CA) equilibrated with buffer B (1 mL of resin per 100 nmol P450). The column was washed with 30 volumes of the same buffer, and the protein was eluted with 150 mM KCl in buffer B. In some instances we used CM Sepharose CL-6B (GE Biosciences, Piscataway, NJ) in place of Macro-Prep High S resin, in which case the concentration of KCl in the elution buffer was increased to 180 mM. The pooled fractions eluted from the ion-exchange column were applied onto the second column of Ni-NTA agarose (1 mL per 100 nmol of protein) to wash out the detergent. The column was washed with 30–50 bed volumes of buffer A containing 3 mM TCEP, which replaces the mercaptoethanol used in prior steps. The protein was eluted with the same buffer containing 150 mM histidine-HCl. The fractions containing P450 were pooled, concentrated to 80 – 120 µM, and dialyzed against two changes of Buffer A containing 1 mM EDTA and 3 mM TCEP. In all cases the protein preparations were characterized by a ratio of optical densities at 418 and 280 nm of 1.4 or higher. The protein was stored at −80 °C as a 80–120 µM solution.
Modification by thiol-reactive probes
An appropriate amount (40 – 60 nmol) of protein was diluted to a concentration of 10 µM with argon-saturated buffer A. TCEP was removed from the sample by dialysis under constant bubbling with argon or by two repetitive dilution/concentration cycles with a Sartocon-1 (MWCO 100,000) concentrator (Sartorius AG, Goetingen, Germany). A 5% stock solution of Igepal CO-630 was added to a final concentration of 0.2%. An appropriate thiol-reactive probe (BADAN, CPM or mBBr) was added at 1.2-fold molar excess to the protein as a 2–3 mM stock solution in 1:1 acetone:methanol. After incubation at 25 °C under argon saturation and constant stirring for 30–60 min (depending on the probe) the reaction was stopped by addition of a 1 M stock solution of DTT to a final concentration of 2 mM. The time course of modification was monitored by an increase in the fluorescence of the probe. The detergent (Igepal CO-630) and the unbound probe were removed from the sample by washing on a column of CM Sepharose CL-6B. The degree of protein modification was determined from a series of spectra of absorbance (340–700 nm wavelength region) obtained in the process of H2O2-dependent heme depletion (49) of the labeled protein. The spectrum of the P450 absorbance bands derived from this series was used together with the standard absorbance spectrum of a glutathione adduct of the probe for the least-squares fit deconvolution of the spectrum of modified protein. In our calculations of the degree of modification we assumed the extinction coefficients of BADAN, CPM and mBBr at their respective absorbance maxima to be of 21, 33 and 5.0 mM−1cm−1 respectively (50).
Absorbance and fluorescence measnurements
Absorbance spectra were recorded with a S2000 CCD rapid scanning spectrometer (Ocean Optics, Inc., Dunedin, FL, USA) using an L7893 UV/VIS fiber-optics light source (Hammamatsu Photonics K.K., Shizuoka, Japan). Fluorescence measurements were performed with an Edinburgh Instruments F900 spectrofluorometer (Edinburgh Instruments Ltd., Edinburgh, UK) equipped with a thermostated cell holder and a magnetic stirrer. In all our fluorometric titration and H2O2-dependent heme depletion experiments the excitation of fluorescence of the probes was performed at 402 nm with a bandwidth of 5 nm, if not otherwise indicated. This wavelength of excitation was selected to minimize excitation of ANF. The emission spectra in the 420 nm – 650 nm wavelength region were recorded with a bandwidth of 2 nm.
The experiments on the effect of heme depletion on the fluorescence of the probes were performed with simultaneous registration of the absorbance and fluorescence spectra. This was achieved using a custom-designed remote cell holder connected to both a F900 fluorometer and S2000 spectrometer with flexible liquid light guides. Change of the light sources was achieved with a computer-controlled beam switch. This design was similar to that described earlier for our variable pathlength titration method (44).
All experiments were performed at 25 °C in 100 mM HEPES buffer (pH 7.4), containing 1 mM DTT and 1 mM EDTA. The series of absorbance and fluorescence spectra obtained in titration experiments were analyzed using principal component analysis method (PCA) as described previously (44, 51, 52). All data treatment procedures and curve fitting were performed using our SPECTRALAB software package (51).
Results
Construction, expression, purification and labeling of a cysteine-depleted quadruple mutant of CYP3A4
Our preliminary studies showed that the wild type CYP3A4 may be labeled by such thiol-reactive probes as CPM or N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-yl)methyl) iodoacetamide (BODYPY-FL iodacetamide) at a molar ratio of label:enzyme up to 2 without any protein precipitation or decrease in the heme protein content, which would be indicative of protein denaturation. Incorporation of a third probe molecule causes some loss of the heme protein, while further labeling results in massive precipitation of the enzyme (data not shown). The kinetics of labeling of the wild type CYP3A4 by CPM is unusually rapid. At 10 µM protein and 12 µM dye the modification of the wild type enzyme at 25 °C takes less then 10 minutes to complete, suggesting the presence of a highly reactive and easily accessible cysteine residue(s) in the enzyme.
A recent study (53) identified residues 96 and 468 as the sites of modification of CYP3A4 by the photoreactive label lapachenole. Analysis of X-ray structures of the enzyme (54, 55) suggest that Cys-239, and, to a lesser extent, Cys-377 are also readily accessible to modification by SH-reactive probes. Therefore, in order to direct the labeling to Cys-58 and Cys-64, as desired, it appeared necessary to eliminate all the above cysteine residues. Appropriate amino acids for these substitutions were chosen based on the sequence alignment of a series of 25 P450 enzymes representing families 1, 2, 3, 4, 11 and 52. This analysis suggested serine or threonine as adequate substitutes at positions 98, 239 and 377, while alanine, glycine or proline residues were expected to be more appropriate at position 468. Based on this analysis we constructed and generated the C98S/C239S/C377S/C468A quadruple mutant (CYP3A4(C58,C64)). The expression level and the yield upon purification of this protein were similar to those of the wild type enzyme, indicating that the substitutions implemented in this construct do not affect protein folding, stability, or incorporation into the membrane.
Incubation of CYP3A4(C58,C64) with BADAN or CPM showed that the reaction of both labels with the protein is quite rapid. The modification of CYP3A4(C58,C64) by these probes at 10 µM protein and a 1.2 label:protein molar ratio requires 20–30 minutes to complete. Our procedure of modification (see Materials and Methods) allowed us to recover >70% of the initial amount of P450 taken into the experiment. The recovered protein was completely labeled in a 1:1 molar ratio. Attempts to incorporate a second molecule of the probe into the protein resulted in its precipitation.
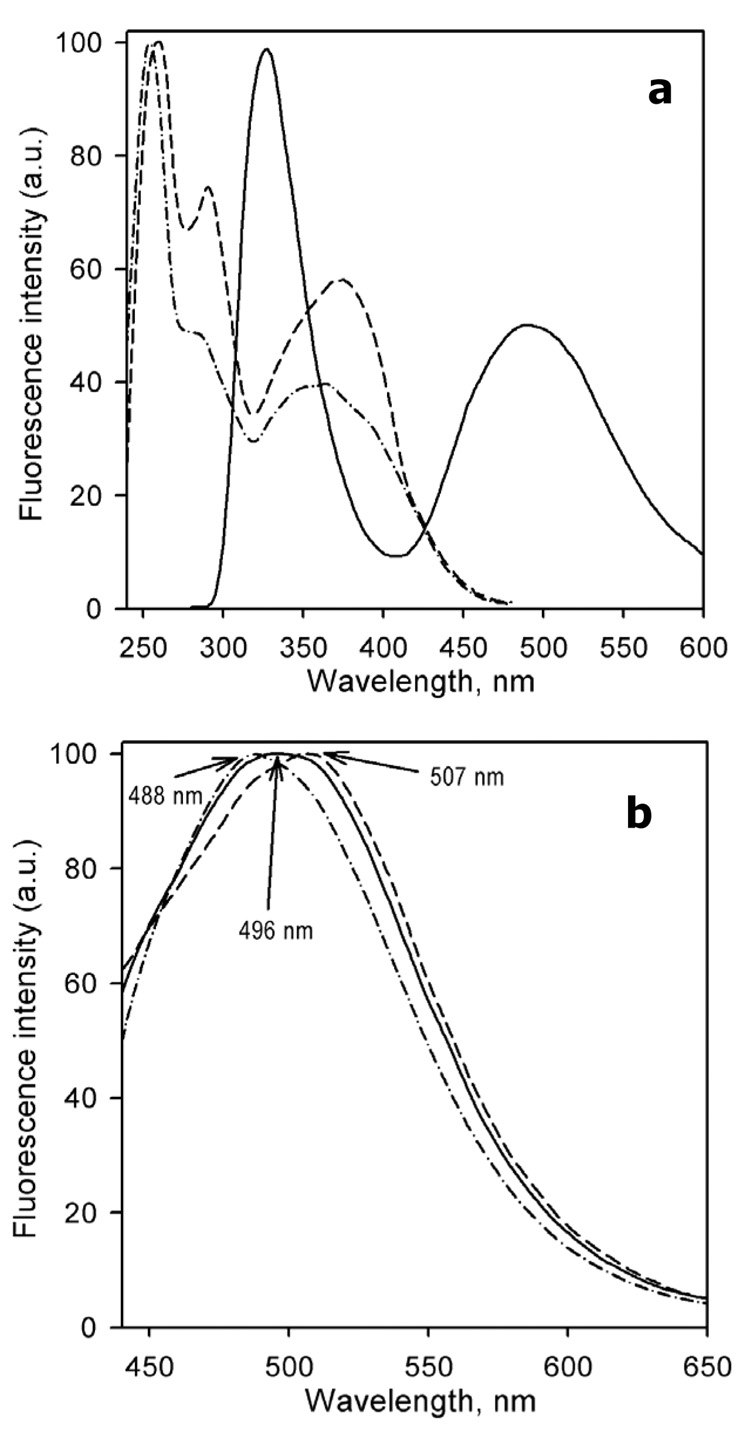
The spectra of emission and excitation of CYP3A4(C58,C64) labeled with BADAN are shown in Fig. 1. The emission spectrum shown in Fig. 1a (solid line) was obtained upon excitation at 260 nm. As the tryptophan residues of the protein are also fluorescent upon excitation at this wavelength, an extensive tryptophan emission band around 330 nm is also present here. The excitation spectrum of CYP3A4(C58,C64)-BADAN (Fig 1a, dashed line) is shown in comparison with the excitation of the glutathione adduct of BADAN (Fig. 1a, dashed-and-dotted line). In addition to the main excitation bands centered at 250–260 and 380–385 nm, which are represented in both spectra, the spectrum of excitation of CYP3A4(C58,C64)-BADAN has an additional band positioned around 290 nm, which may reflect the excitation of the label through FRET from tryptophan. Interestingly, the exact position of the emission maximum of CYP3A4(C58,C64)-BADAN was dependent on the wavelength of excitation (fig. 1b). Upon excitation at 385 nm the emission maximum is positioned at 488 nm, which is indicative of an apolar environment of the probe (40, 41). However, upon excitation at 280 nm, when extensive FRET from tryptophan may complement direct excitation of the probe, the emission band becomes broader and is displaced to 496 nm. Upon excitation at 315 nm, which corresponds to the minimum of the excitation spectrum of BADAN itself, the emission band exhibits a further red-shift – in this case to around 507 nm. This observation may suggest conformational heterogeneity of the protein so that a conformer with more efficient FRET from tryptophan to BADAN has the probe exposed to a more polar environment.
Figure 1.
Spectra of fluorescence emission and excitation of CYP3A4(C58,C64)-BADAN (10 µM solution in 100 mM Na-Hepes puffer, pH 7.4, 1 mM EDTA, 1mM DTT). Panel a shows the spectra of emission (excitation at 260 nm, solid line) and excitation (emission at 490 nm, dashed line) The excitation spectrum of a gluthatione adduct of BADAN (emission at 490 nm) is shown for comparison by a dashed-and-dotted line. Panel b shows the spectra of emission of CYP3A4(C58,C64)-BADAN recorded with excitation at 280 nm (solid line), 315 nm (dashed line) and 385 nm (dashed-and-dotted line). All spectra were recorded at 25 °C in a 5×5 mm quartz cell with emission and excitation bandwidths of 2 nm. The spectra were corrected for the spectral response of the detector. The amplitudes of all spectra were normalized to fit into 0 – 100 units scale.
Effect of cysteine substitutions and labeling on the interactions of CYP3A4 with substrates
To assess the effect of cysteine depletion and labeling on CYP3A4 interactions with substrates we studied the formation of the complexes of CYP3A4, CYP3A4(C58,C64) and their BADAN-labeled derivatives with BCT, 1-PB, and ANF by monitoring the substrate-induced spin shift. In these experiments a series of absorbance spectra in the 340–700 nm wavelength region recorded at increasing concentrations of substrate was interpreted in terms of the changes in concentrations of P450 low-spin, P450 high-spin, and apparent P420 states of the protein (44, 51). These experiments are exemplified in Fig. 2, which shows the results obtained with wild type CYP3A4 and ANF. Consistent with earlier results (30, 44, 47), all three substrates tested in this study displace the spin equilibrium toward the high-spin state, which is indicative of Type-I binding.
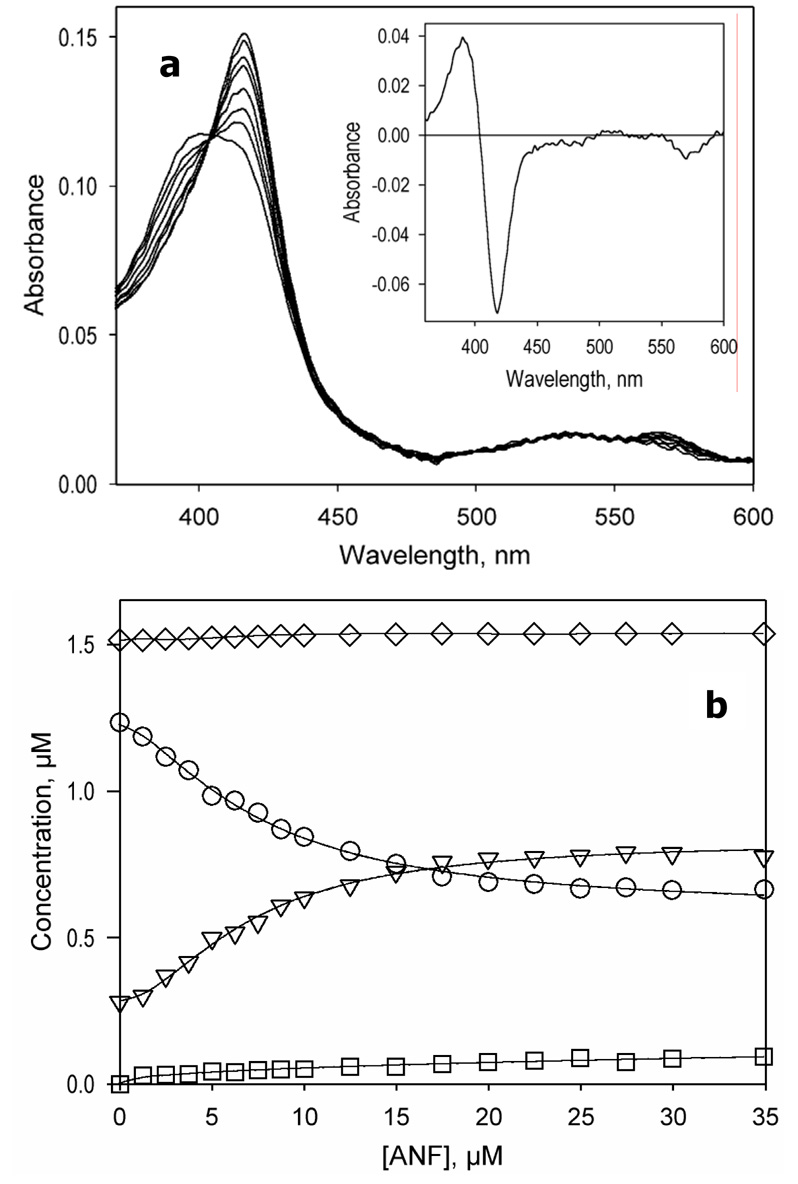
Figure 2.
ANF-induced changes in the absorbance spectra of CYP3A4. (a) A series of absorbance spectra of 1.5 µM enzyme recorded at 0, 1.2, 2.5, 3.7, 6.2, 8.7, 12, and 30 µM ANF. The inset shows the spectrum of the first principal component. (b) ANF-induced changes in the concentration of the high-spin (triangles), low-spin (circles), P420 (squares), and total enzyme (diamonds). Experimental conditions: 100 mM Hepes buffer, pH 7.4, 1 mM DTT, 0.2 mM EDTA, 25 °C.
Plots of the high-spin content in CYP3A4, CYP3A4(C58,C64), and their BADAN-labeled derivatives upon ANF binding are shown in Fig. 3. For all four enzyme variants the titration curves obey the Hill equation with the values of the Hill coefficient (n) in the range of 1.5 – 2.0, which is indicative of positive homotropic cooperativity (Table 1) and consistent with earlier findings (46, 47). The S50 values obtained in these experiments were in the range of 2.2 – 4.8 µM (Table 1), which is consistent with the value of 5.7 ± 1.9 µM obtained by Hosea and co-authors (47). The difference from the S50 value of 17 µM (46) may reflect different experimental conditions, as Domanski and co-authors examined ANF binding in the presence of 0.1 mg/mL dioleylphosphatidylcholine and 0.05% CHAPS.
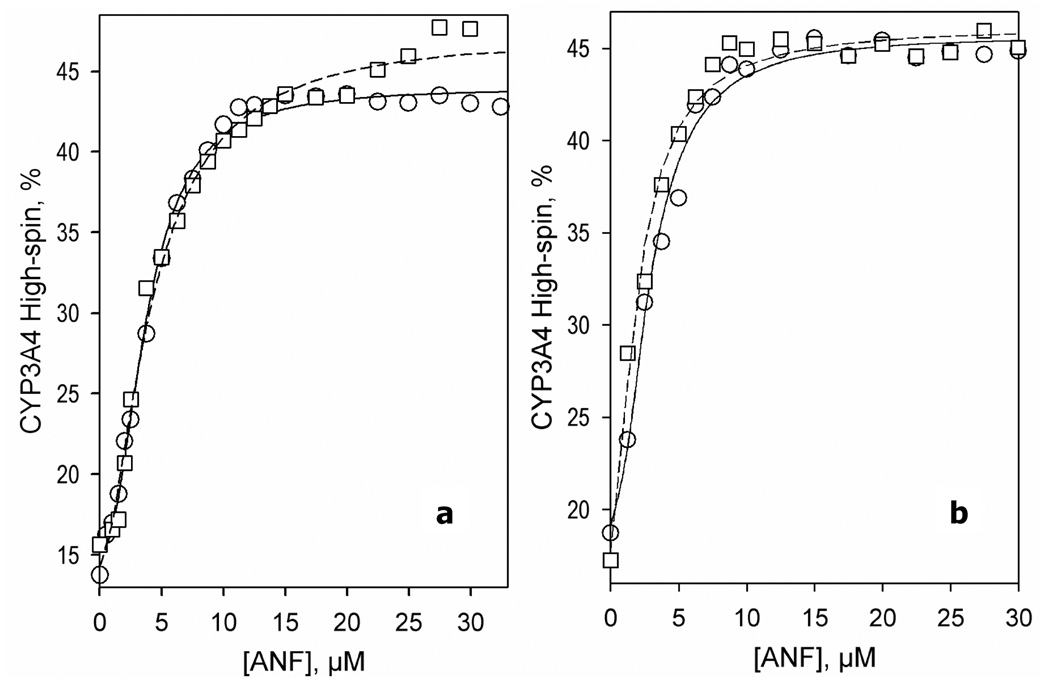
Figure 3.
Interactions of ANF with CYP3A4, CYP3A4(C58,C64), and their BADAN-modified derivatives monitored by the substrate-induced spin shift. A series of absorbance spectra of 1 µM heme protein were recorded at different ANF concentrations, and the percent of high spin content versus the substrate concentration was plotted. The lines show fitting of these data sets to the Hill equation with S50 = 3.7 and n = 2.2 for CYP3A4 (a, circles, solid line), S50= 4.2 and n = 1.7 for CYP3A4-BADAN (a, squares, dashed line), S50= 2.9 and n = 2.0 for CYP3A4(C58,C64) (b, circles, solid line), and S50 = 2.0 and n = 1.6 for CYP3A4(C58,C64)-BADAN (b, squares, dashed line). Experimental conditions as indicated in Figure 1.
Table 1.
Parameters of the interaction of CYP3A4 and its derivatives with substrates
| Protein | Parameter | Substrate | ||
|---|---|---|---|---|
| BCT | 1-PB | ANF | ||
| CYP3A4 | S50 or KD, µM | 0.65 ± 0.33 | 13.2 ± 2.9 | 4.8 ± 2.2 |
| n | n/a | 1.6 ± 0.20 | 1.9 ± 0.3 | |
| ΔFh(%)a | 40 ± 15 | 40 ± 8 | 36 ± 8 | |
| CYP3A4-BADAN | S50 or KD, µM | 0.68 ± 0.48 | 8.0 ± 0.3 | 4.7 ± 0.9 |
| n | n/a | 1.4 ± 0.3 | 1.5 ± 0.5 | |
| ΔFh(%)a | 30 ± 8 | 42 ± 7 | 30 ± 6 | |
| CYP3A4(C58,C64) | S50 or KD, µM | 0.71 ± 0.34 | 8.7 ± 2.7 | 2.7 ± 0.5 |
| n | n/a | 1.6 ± 0.2 | 2 ± 0.1 | |
| ΔFh(%)a | 39 ± 7 | 36 ± 7 | 32 ± 9 | |
| CYP3A4(C58,C64)-BADAN | S50 or KD, µM | 0.62 ± 0.13 | 7.5 ± 1.6 | 2.2 ± 0.3 |
| n | n/a | 1.7 ± 0.6 | 1.5 ± 0.2 | |
| ΔFh(%) | 40 ± 8 | 31 ± 2 | 28 ± 4 | |
The amplitude of the substrate-induced changes in the high-spin content. In the absence of substrate the content of the high-spin state was within the range of 12–18% for all enzyme variants.
The values shown in the table are determined from the fitting of the titration curves to the Hill equation (for 1-PB and ANF) or to the equation for the isotherm of bimolecular association (for BCT). The results represent the averages of two to four individual measurements. The deviations correspond to the confidence intervals calculated for p = 0.05.
All four enzyme variants also exhibited cooperativity in their interactions with 1-PB, while showing no cooperativity with BCT, which is also consistent with our earlier results (30, 44, 45). As shown in Table 1, the parameters of the interactions of CYP3A4, CYP3A4(C58,C64), and their BADAN-labeled derivatives with substrates show no significant effect of cysteine depletion of the enzyme or modification with BADAN.
Substrate-induced changes in the fluorescence of BADAN incorporated into CYP3A4
While addition of TST caused virtually no effect on the fluorescence of CYP3A4(C58,C64)-BADAN, interactions of the enzyme with BCT or 1-PB caused a 5–10% increase in the fluorescence intensity of the probe (data not shown). However, the low amplitude of these changes makes them poorly reproducible and allows no interpretation in terms of the changes in the concentration of the enzyme-substrate complex. Importantly, no substrate-induced displacement of the position of the BADAN fluorescence band was observed with any of these substrates, indicating no changes in the polarity of the probe environment upon substrate binding.
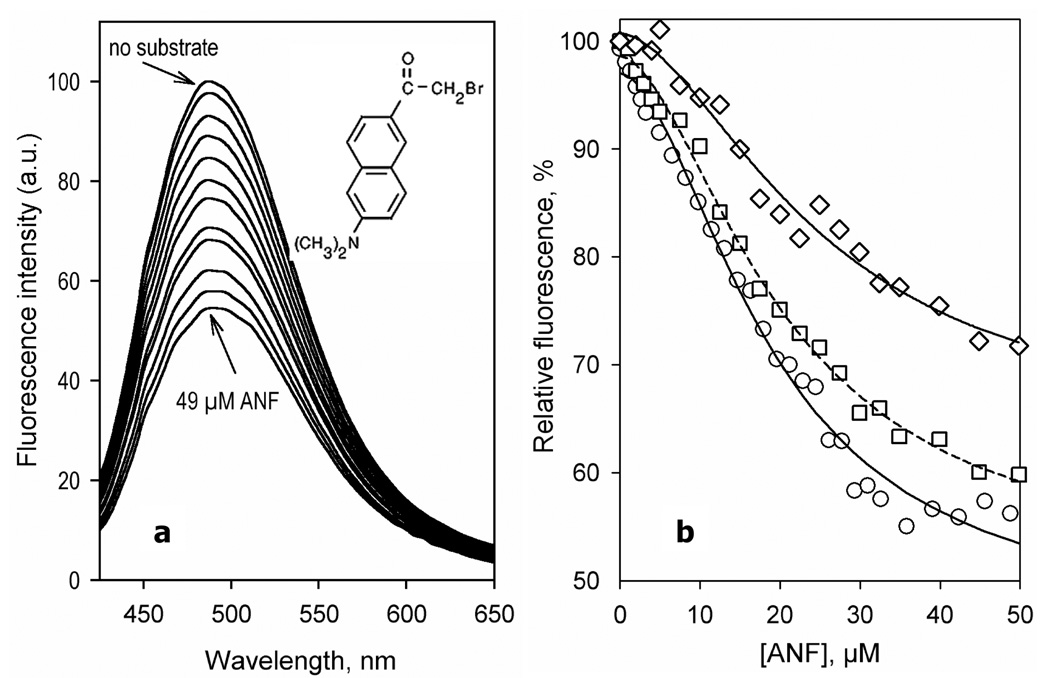
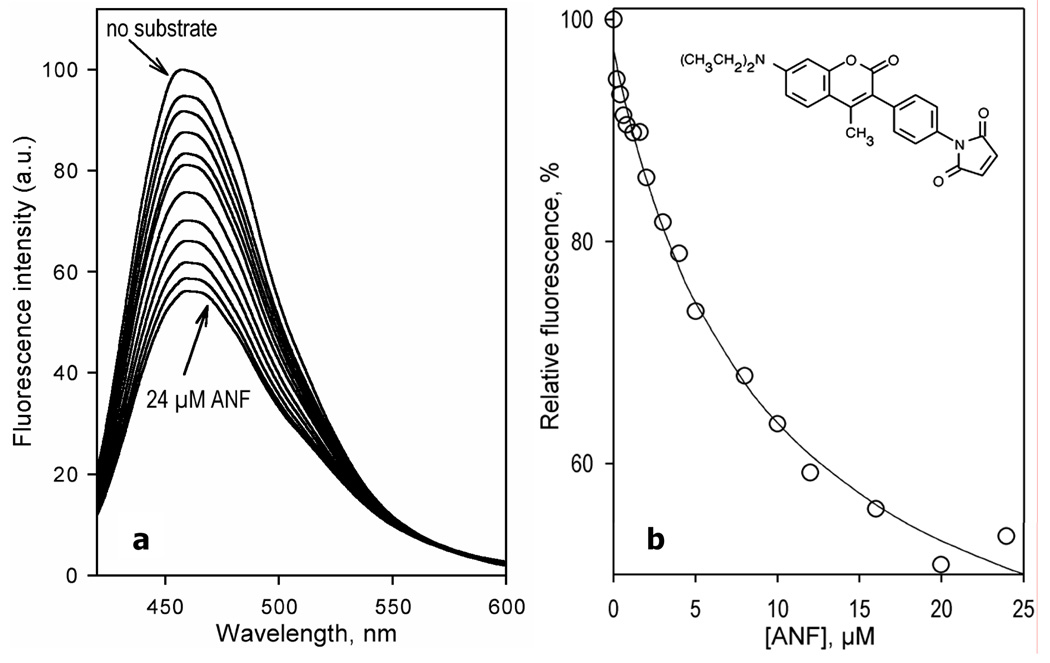
In contrast to the other substrates, addition of ANF to CYP3A4(C58,C64)-BADAN resulted in a substantial decrease in the fluorescence intensity (Fig.4). Similar to the observations with BCT, TST, and 1-PB, the position of the band remains unchanged upon ANF binding. The dependence of the intensity of fluorescence of BADAN on the concentration of ANF can be approximated with the Hill equation (Fig. 4b) with the values of S50 and Hill coefficient of 18.2 ± 0.7 µM and 1.7 ± 0.1, respectively and the maximal amplitude of the fluorescence decrease of 49 ± 6 % (Table 2).
Figure 4.
Interaction of CYP3A4(C58,C64)-BADAN with ANF monitored by the changes in the fluorescence of the probe. (a) A series of emission spectra of 3 µM enzyme were recorded at 0, 1.3, 3.3,, 6.5, 9.8, 13, 16, 20, 23, 26, 31 and 49 µM ANF. (b) Relative fluorescence intensity versus substrate concentration for CYP3A4(C58,C64)-BADAN and for the enzyme pre-incubated with 1-PB or TST. The lines show fitting of the data sets to the Hill equation with S50 = 18.8 µM and n = 1.8 for CYP3A4(C58,C64) (circles, solid line), S50 = 20.5 µM and n = 1.8 for the enzyme pre-incubated with 15 µM 1-PB (squares, dashed line), and S50 = 25 µM and n = 1.7 (diamonds, solid line) for the enzyme pre-incubated with 100 µM TST. Experimental conditions as indicated in Figure 3.
Table 2.
Interactions of ANF with CYP3A4 modified with BADAN, CPM or mBBr monitored by the fluorescence of the probe
| Protein | Parameter | Added compound | ||
|---|---|---|---|---|
| none | 1-PB, 15 µM | TST, 100 µM | ||
| CYP3A4(C58,C64)-BADAN | S50, µM | 18.2 ± 0.7 | 23.7 ± 3.5 | 21.4 ± 7.2 |
| n | 1.7 ± 0.1 | 1.8 ± 0.3 | 1.6 ± 0.2 | |
| Δ I, %a | − 49 ± 8 | − 55 ± 6 | − 32 ± 12 | |
| CYP3A4(C58,C64)-CPM | S50, µM | 13.7 ± 4.0 | n/d | n/d |
| n | 1.1 ± 0.25 | n/d | n/d | |
| Δ I, %a | −60 ± 10 | n/d | n/d | |
| CYP3A4(C58,C64)-mBBr | S50, µM | 12.3 ± 1.4 | n/d | n/d |
| n | 1.1 ± 0.3 | n/d | n/d | |
| Δ I, %a | 25 ± 16 | n/d | n/d | |
Maximal relative change in the intensity of fluorescence of the probe.
All parameters given in the table represent the averages of two to four individual measurements. The deviations represent the confidence intervals calculated for p = 0.05.
Thus, the concentration dependence of the effect of ANF on BADAN fluorescence is clearly different from that on the spin state of CYP3A4. The substrate-induced spin shift was almost complete upon addition of 10 µM ANF and remained unchanged at ANF concentrations higher than 15 µM (Fig. 2, Fig. 3). In contrast, the most prominent effect of ANF on the fluorescence of BADAN is exerted in the range of 10 – 30 µM. Therefore, ANF-induced changes in BADAN fluorescence reveal an additional binding process, which is silent in terms of the modulation of the spin equilibrium of the enzyme.
Importantly, with all four substrates studied here the changes in the fluorescence of the wild type CYP3A4-BADAN were quite similar to those observed in BADAN-labeled CYP3A4(C58,C64). The effect of 1-PB, BCT, and TST on the fluorescence of the probe in CYP3A4-BADAN was negligibly small (data not shown). In contrast, addition of ANF resulted in an important decrease in the intensity of fluorescence, revealing a process which obeys the Hill equation with an S50 of 13.0 ± 1.0 µM and n value of 1.8 ± 0.1.
It should be noted, however, that the cooperativity observed in the fluorometric titration of BADAN-labeled enzyme with ANF may be apparent. Since the fluorescence of BADAN is lessened by FRET to the heme, displacement of the Soret absorbance band of CYP3A4 due to the substrate-induced spin shift, which reduces the overlapping of the emission band of the donor (BADAN) with the absorbance band of acceptor (heme), would cause an increase in the intensity of fluorescence of the donor due to decreased efficiency of FRET. This effect is likely to be responsible for the low-amplitude increase in the intensity of BADAN fluorescence caused by 1-PB and BCT. Therefore, it seems likely that the S-shape of the plot of the fluorescence of CYP3A4(C58,C64)-BADAN or CYP3A4-BADAN vs. ANF concentration is caused by overlapping of the above effect of spin-state-modulating binding events with a decrease in fluorescence caused by ANF-specific low affinity binding.
Effect of TST and 1-PB on the interactions of BADAN-labeled CYP3A4(C58,C64) with ANF
The data presented above indicate the presence of an additional binding site for ANF, which is silent in terms of the substrate-induced spin shift, and raise the question whether other substrates can bind at this presumed “effector” site, despite exerting no effect of their own on BADAN fluorescence. To address this question we performed a fluorometric titration of CYP3A4(C58,C64)-BADAN with ANF in the presence of TST or 1-PB. The concentrations of these compounds (100 µM and 15 µM respectively) were chosen to be above their respective S50 values. The presence of these substrates does not affect the interactions of ANF with its putative low-affinity binding site (Table 2, Fig. 4b), as indicated by a lack of any significant effect of TST or 1-PB on the S50 or n values derived from the changes in fluorescence observed in the titrations of CYP3A4(C58,C64)-BADAN with ANF in the presence of 1-PB (Fig. 4b, squares) or TST (Fig. 4b, diamonds). However, TST but not 1-PB decreased the amplitude of ANF-induced fluorescence change from 42 ± 8 % to 32 ± 12 %.
These results lead to a conclusion that ANF possesses at least three different binding sites in CYP3A4. Two of these sites are involved in the cooperative binding reflected in ANF-induced spin transition and at least one additional site is revealed in the low-affinity interactions detectable by the fluorescent probes. In this context it is important to note that absorbance titration experiments also revealed no competition of TST and 1-PB with ANF, as the addition of these substrates had no significant effect on the S50 or n value characteristic of the ANF-induced spin transition (data not shown). Therefore, none of the three implied ANF-binding sites appear to overlap with those involved in the interactions of the enzyme with 1-PB and TST.
The effect of ANF on the fluorescence of the probe in CYP3A4(C58,C64) labeled with CPM and mBBr
One of possible mechanisms of the decrease of the intensity of BADAN fluorescence without any solvatochromic shift of the emission band may be based on the changes in the efficiency of FRET from BADAN to the heme of the enzyme. In this case similar changes should occur with other probes capable of FRET to the heme. Therefore, we studied the effect of ANF on the fluorescence of two other labels - CPM and mBBr attached to CYP3A4(C58,C64).
The CPM-labeled protein (CYP3A4(C58,C64)-CPM) has an extensive fluorescence band centered at 462 nm, which undergoes a considerable decrease in amplitude upon the interaction of the enzyme with ANF (Fig. 5). Titration curves obtained in these experiments obey the Hill equation with an S50 value of 13.7 ± 4.0 µM and Hill coefficient of 1.1 ± 0.25 (Fig. 5, Table 2). The lack of cooperativity of ANF binding to the low affinity site in CYP3A4(C58,C64)-CPM supports our interpretation that the apparent cooperativity of the interactions with CYP3A4(C58,C64)-BADAN observed in fluorometric titrations with ANF is due to the displacement of the P450 absorbance band upon the ANF-induced spin shift. Since the fluorescence band of CPM is shifted by ~30 nm towards shorter wavelength, the change in the overlapping of the emission band of the probe with the absorbance band of P450 upon low-to-high spin transition is considerably less important in this case.
Figure 5.
Interaction of CYP3A4(C58,C64)-CPM with ANF monitored by the changes in the fluorescence of the probe. (a) A series of emission spectra of 1.5 µM CYP3A4(C58,C64)-CPM recorded at 0, 0.4, 1.2, 2, 3, 4, 5, 8, 10, 12, 16 and 24 µM of ANF. (b) Relative fluorescence intensity versus substrate concentration. The lines show the fitting of the data sets to the Hill equation with S50 = 13 µM and n = 1.1. Experimental conditions as indicated in Figure 3.
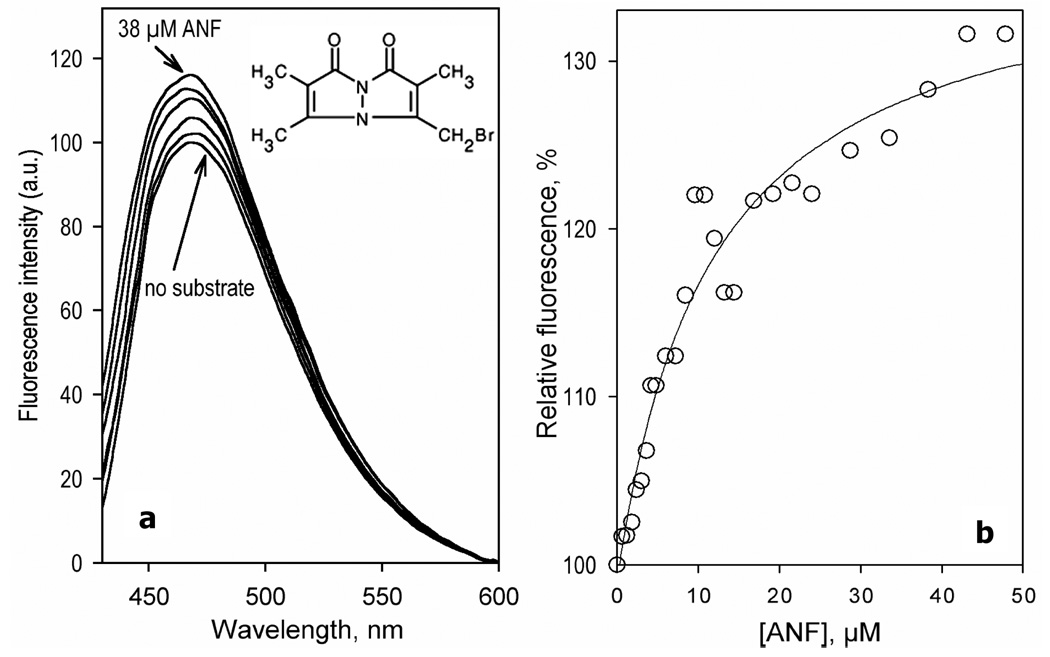
CYP3A4(C58,C64) labeled with mBBr has a fluorescence intensity close to that of CYP3A4(C58,C64)-BADAN, which was quite unexpected, as bimane-based probes are known to have considerably lower fluorescence than dimethylaminonaphthalene-based labels (50). A broad fluorescence band of mBBr in CYP3A4 is positioned around 480 nm and has considerable overlapping with the absorbance band of the heme. Thus, if the effect of ANF on the fluorescence of CYP3A4(C58,C64)-BADAN and CYP3A4(C58,C64)-CPM reflects changes in the efficiency of FRET, a similar result should occur with CYP3A4(C58,C64)-mBBr. However, addition of ANF to CYP3A4(C58,C64)-mBBr resulted in ~25% increase in the fluorescence of the label. The titration curves observed in this case reveal no cooperativity (n = 1.1 ± 0.3) and were characterized by an S50 value of 12.3 ± 1.4 µM (Fig. 6), which is consistent with that determined with two other labels. Therefore, although the experiments with CYP3A4(C58,C64)-mBBr confirmed the presence of an additional ANF binding site, they oppose the suggestion that the effect of ANF is based on the modulation of FRET to the heme.
Figure 6.
Interaction of CYP3A4(C58,C64)-mBBr with ANF monitored by the changes in the fluorescence of the probe. (a) A series of emission spectra of 1.5 µM enzyme recorded at 0, 2.4, 6, 12, 24, and 38 µM of ANF. (b) Relative fluorescence intensity versus substrate concentration. The lines show the fitting of the data sets to the Michaelis-Menten equation with KM = 12 µM. Experimental conditions as indicated in Figure 3.
Effect of ANF on the interactions of CYP3A4 with TST and 1-PB
The low-affinity ANF-binding site in CYP3A4 appears to be specific for ANF, as no competition with 1-PB and TST was observed in our fluorometric titration experiments. To explore the relationship between this site and heterotropic activation of the enzyme, it was important to probe the effect of the binding of ANF to the low-affinity site on CYP3A4 interactions with other substrates.
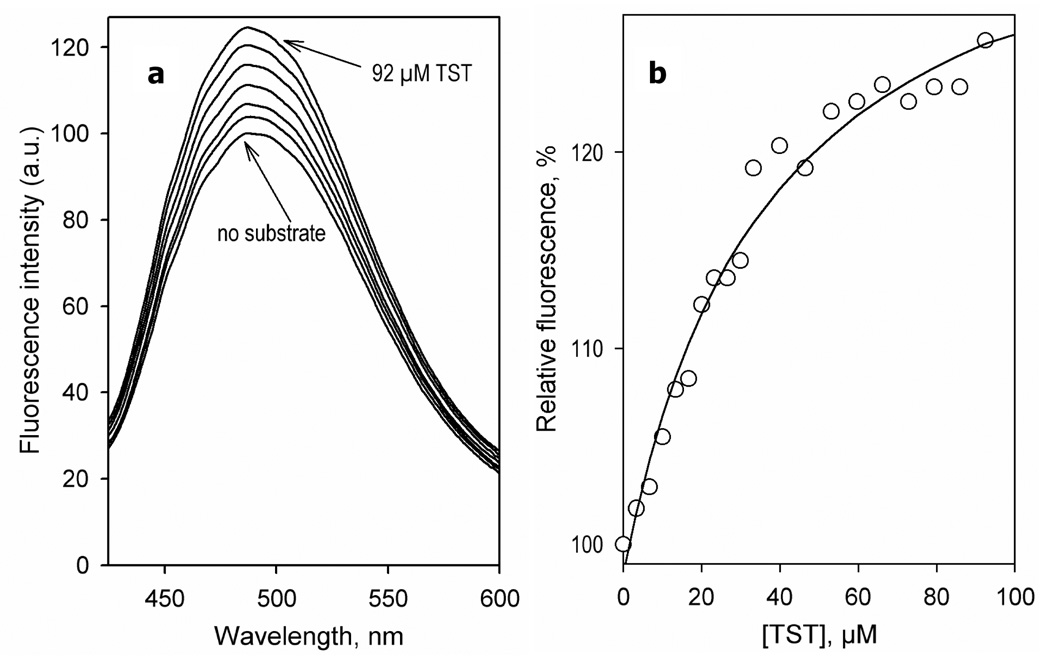
Although TST alone showed virtually no effect on the intensity of BADAN fluorescence, its addition to CYP3A4(C58,C64)-BADAN preincubated with 25 µM ANF resulted in a considerable increase in the intensity of fluorescence of the probe, which was accompanied by a distinct shift of the fluorescence maximum by up to 6–7 nm towards shorter wavelengths (Fig. 7). The TST-dependent changes in the intensity of BADAN may be approximated with the Michaelis-Menten equation with the maximal amplitude of the fluorescence increase of 34 ± 7% and Km of 42 ± 9 µM, which is consistent with the values determined earlier from absorbance titration experiments with the wild type enzyme (47).
Figure 7.
Effect of TST on the fluorescence of CYP3A4(C58,C64)-BADAN in the presence of 25 µM ANF. (a) A series of emission spectra of CYP3A4(C58,C64)-BADAN (3 µM) in the presence of 25 µM ANF recorded at 0, 10, 20, 30, 40, 66, and 92 µM TST. (b) Changes in the relative intensity of fluorescence of CYP3A4(C58,C64)-BADAN upon addition of testosterone in the presence of 25 µM ANF. The line show the fitting of the data set to the Michaelis-Menten equation with KM = 38 µM and the maximal amplitude of fluorescence increase of 37 %. Experimental conditions as indicated in Figure 3.
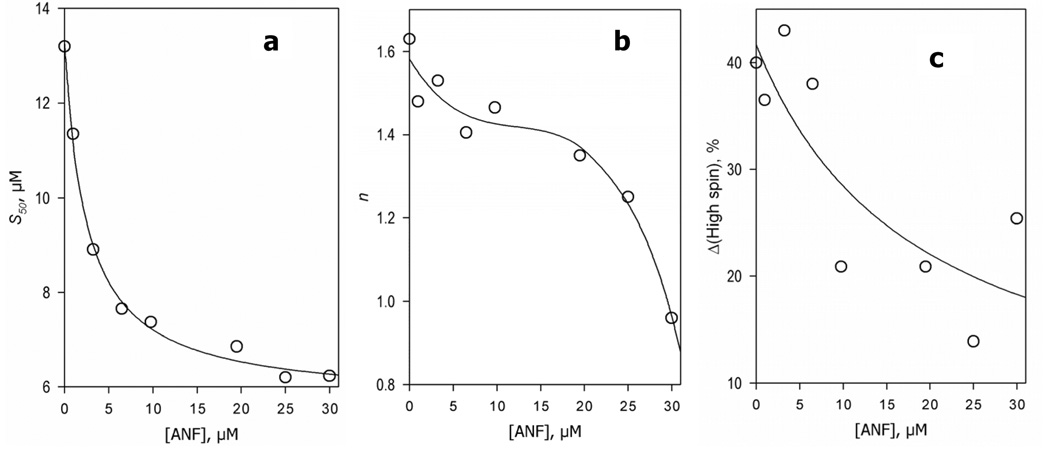
We also studied the effect of increasing concentrations of ANF on the interactions of the wild type CYP3A4 with 1-PB monitored by absorbance spectroscopy (Fig. 8). We chose 1-PB for these experiments, as it exhibits considerably higher affinity to CYP3A4 and higher amplitude of the substrate-induced spin shift than TST. Importantly, the affinity of the enzyme for 1-PB is clearly increased in the presence of ANF, and the dependence of S50 on the concentration of ANF may be approximated by a hyperbolic equation with the apparent KM value of 2.6 µM (Fig. 8a), which is close to the value of S50 characteristic for the binding of ANF to CYP3A4 detected by a substrate-induced spin shift. At the same time, the Hill coefficient for 1-PB binding remains almost unaffected at up to 10 µM ANF but decreases when the concentrations become close to the S50 characteristic of the low-affinity binding site revealed in our fluorescence studies (13 – 18 µM, Table 2). The homotropic cooperativity with 1-PB is completely abolished (i.e., n = 1) at 30 µM ANF.
Figure 8.
Effect of increasing concentrations of ANF on the parameters of CYP3A4 interactions with 1-PB monitored by the substrate-induced spin shift. Each data point shown on the graphs represents an average of the results of 2–6 individual experiments. The typical confidence interval (p = 0.05) calculated for these estimates was about ± 20% of the respective values. Solid lines shown in panels a and c represent the results of the fitting of the data sets to the Michaelis-Menten equation (KM values of 2.5 µM and 18 µM, respectively). The line in panel b represents the fitting of the data set by a third-order polynomial. The curve is given solely to visualize the general trend of the points and has no conceptual meaning.
Assessing the efficiency of FRET from CPM and BADAN to the heme by H2O2-dependent heme depletion
In order to probe more thoroughly how FRET to the heme may affect the bound probe we studied the changes in fluorescence upon heme destruction in CYP3A4(C58,C64)-BADAN, CYP3A4(C58,C64)-CPM and CYP3A4(C58,C64)-mBBr. Mild treatment of P450 enzymes with hydrogen peroxide may be used for quantitative removal of the heme from the protein, while having minimal impact on the polypeptide chain (56, 57). Therefore, an increase in the intensity of fluorescence of the probes in the course of heme depletion by hydrogen peroxide may be used to probe the efficiency of FRET to the heme of the enzyme. We monitored the changes in the absorbance of P450 and in the fluorescence of the label simultaneously, which was possible due to a custom design of a remote sample chamber of the fluorometer and implementation of a rapid scanning CCD spectrometer in combination with a computer-controlled beam switch (see Materials and Methods).
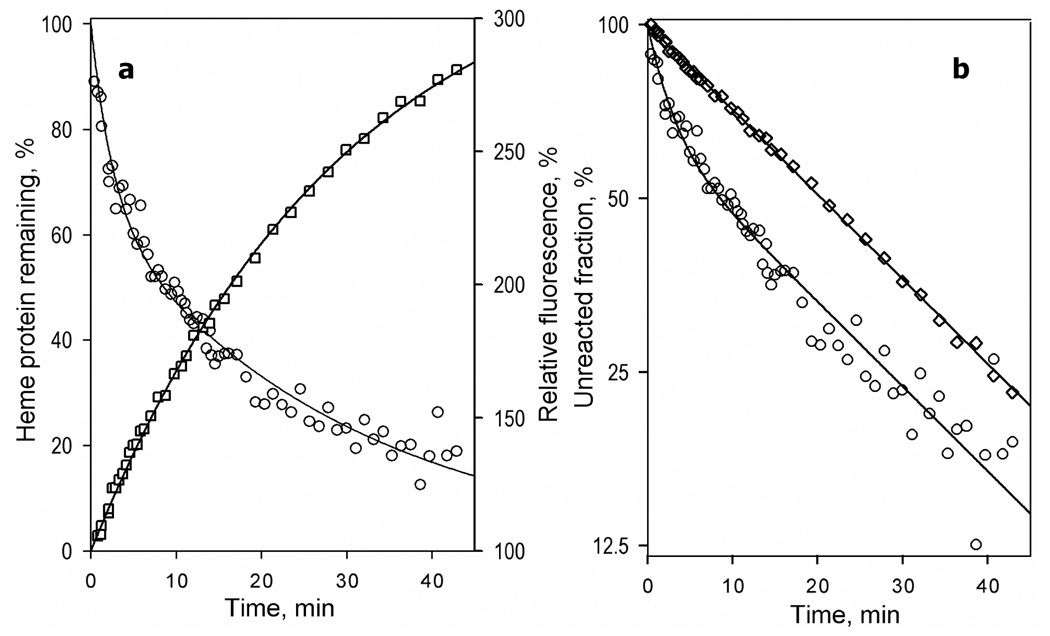
The time course of the changes in the fluorescence of the probe and the content of heme protein after addition of 60 mM H2O2 to a solution of CYP3A4(C58,C64)-CPM is shown in Fig. 9. Consistent with our earlier results (58), the kinetics of heme depletion in CYP3A4 was bi-exponential. The two phases of the process exhibited rate constants differing by an order of magnitude, with the fast phase constituting about 60% of the total reaction amplitude (Fig. 9; Table 3). The kinetic parameters of the heme depletion in the labeled CYP3A4(C58,C64) were similar to those determined earlier for the wild type enzyme (58) and showed no dependence on the nature of the label (BADAN, CPM, or mBBr). Therefore the biphasicity of the heme depletion appears to represent an inherent feature of the enzyme rather than being induced by incorporation of the label.
Figure 9.
Kinetics of H2O2-dependent heme depletion (circles) and the corresponding changes in the intensity of fluorescence of the label (squares) in CYP3A4(C58,C64)-CPM in linear (a) and semi-logarithmic (b) coordinates. Conditions: 1.5 µM CYP3A4(C58,C64)-CPM and 60 mM H2O2 in 0.1 M Na-HERES buffer, 1mM DTT, 1mM EDTA.
Table 3.
Kinetic parameters of H2O2-dependent heme depletion of CYP3A4(C58,C64) labeled with BADAN, CPM, or mBBr
| Protein | Substrate | k1 | k2 | F1a | F1b (fluor.) | Efficiency of FRET, %c |
|---|---|---|---|---|---|---|
| CYP3A4(C58,C64)-BADAN | none | 0.16 ± 0.05 | 0.027 ± 0.024 | 66 ± 16 | 0 | 79 ± 1 |
| ANFd | 0.27 ± 0.10 | 0.032 ± 0.010 | 59 ± 13 | 0 | 72 ± 6 | |
| CYP3A4(C58,C64)-CPM | none | 0.38 ± 0.10 | 0.044 ± 0.011 | 60 ± 30 | 0 | 69 ± 9 |
| ANF | 0.29 ± 0.03 | 0.038 ± 0.10 | 66 ± 23 | 8 ± 2 | 73 ± 4 | |
| CYP3A4(C58,C64)-mBBr | none | 0.25 ± 0.1 | 0.034 ± 0.010 | 47 ± 20 | 8 ± 3 | 65 ± 10 |
| ANF | 0.18 ± 0.10 | 0.025 ± 0.010 | 42 ± 7 | 15 ± 2 | 65 ± 13 |
The fraction of the fast phase in the kinetics of heme depletion.
The fraction of the fast phase in the kinetics of H2O2-dependent fluorescence increase.
Apparent efficiency of FRET to the heme calculated from the amplitude of H2O2-dependent fluorescence increase determined by the fiiting of the kinetic curve to a biexponential equation.
50 µM.
All parameters given in the table represent averages of two to four individual measurements. The deviations represent the confidence intervals calculated for p = 0.05.
Surprisingly, the kinetics of the changes in the fluorescence of the probes was considerably different from the kinetics of the heme loss. In contrast to the biphasic kinetics of heme depletion, the increase in fluorescence of CYP3A4(C58,C64)-CPM (Fig. 9b) and CYP3A4(C58,C64)-BADAN upon heme removal was monoexponential. In the case of the enzyme labeled with mBBr, which revealed an increase rather than decrease in fluorescence intensity of the label upon titration with ANF, the kinetics of H2O2-dependent fluorescence increase was bi-exponential, but the amplitude of the fast phase was considerably lower than that observed in the kinetics of heme depletion (Table 3). As shown in Fig 9b, complete heme depletion in resulted in ~4-fold increase in the intensity of fluorescence of CYP3A4(C58,C64)-CPM (Fig. 9b). Correspondingly, apparent efficiency of FRET to the heme in this case was calculated to be of 69 ± 9 %. Similar values of the FRET efficiency were determined for CYP3A4(C58,C64)-BADAN and CYP3A4(C58,C64)-mBBr (Table 3).
Addition of 50 µM ANF had no significant effect on the kinetics of heme depletion and fluorescence increase (Table 3). Moreover, ANF had no effect on the amplitude of FRET determined from these experiments (Table 3). Therefore, in agreement with the conclusion made above, these experiment show that the modulation of the efficiency of FRET from the probe to the heme of the enzyme is not involved in the mechanism of ANF-induced changes in the fluorescence of the probes. Furthermore, these results are consistent with distribution of the pool of the enzyme between two different fractions. One of these fractions in CYP3A4(C58,C64)-BADAN and CYP3A4(C58,C64)-CPM appears to have the fluorescence of the label completely quenched. The bimane probe in CYP3A4(C58,C64)-mBBr appears to be fluorescent in both subpopulations, although these fractions reveal considerably different quantum yield of the fluorescence of the probe.
Probing the location of the label in CYP3A4(C58,C64) by constructing cysteine-depleted quintuple mutants
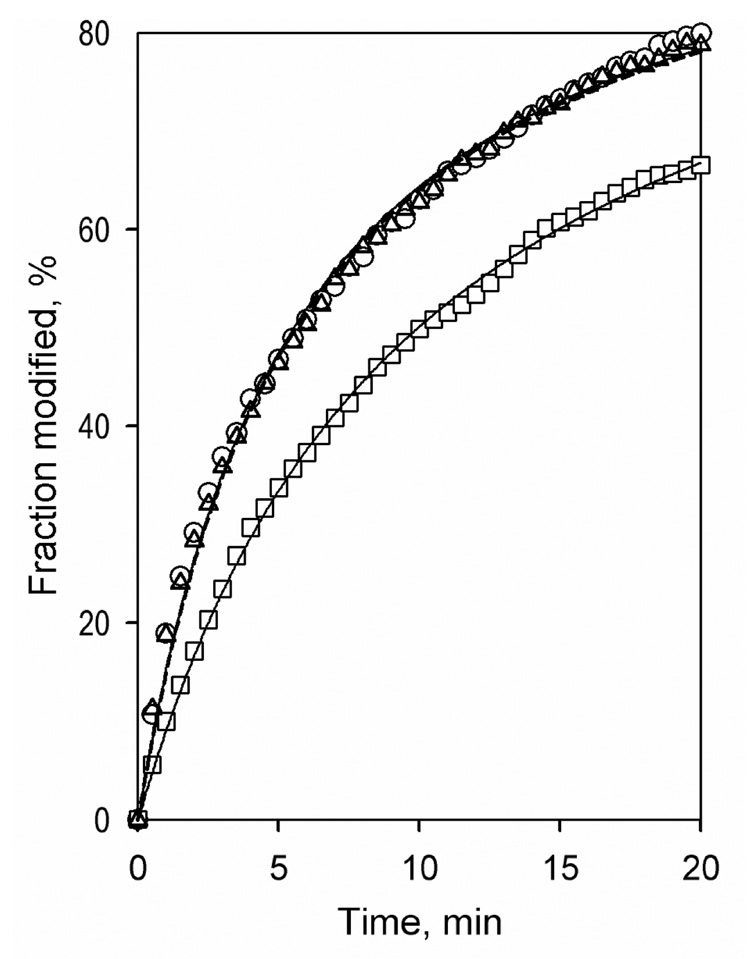
Mechanistic interpretation of the ANF-dependent changes in the fluorescence of the probes introduced into CYP3A4(C58,C64) mutant requires identification of the exact position of the labeling. To identify this position we created two quintuple mutants - CYP3A4(C58) and CYP3A4(C64), each bearing only one potentially accessible cysteine residue. In both cases expression and purification of the mutants resulted in yields comparable to the wild-type enzyme. Both mutants exhibited Soret region absorbance spectra similar to the wild type enzyme and did not reveal any considerable P420 content. While the kinetics of labeling of CYP3A4(C64) by BADAN was similar to that observed with CYP3A4(C58,C64), the modification of CYP3A4(C58) was considerably slower (Fig. 10). Recovery of the protein after modification of CYP3A4(C58) by BADAN was as low as 25%, as compared to 75% recovery observed with both CYP3A4(C64) and CYP3A4(C58,C64) mutants. We attribute this loss of labeled CYP3A4(C58) to a denaturation of the enzyme caused by modification of Cys-58. Moreover, as revealed by the spectra of the CO-complex of the ferrous enzyme, BADAN-modified CYP3A4(C58) contained ~75% of the inactivated P420 form of the enzyme, while CYP3A4(C64)-BADAN contained only ~12% P420, which is similar to CYP3A4(C58,C64)-BADAN. Furthermore, while the position of the emission band of CYP3A4(C64)-BADAN was similar to that observed with the quadruple mutant (488 nm with excitation at 385 nm), the band of CYP3A4(C58) is shifted to 496 nm, which is indicative of a more polar environment of the label in this case.
Figure 10.
Kinetics of modification of CYP3A4(C58,C64) (circles, solid line), CYP3A4(C58) (rectangles, solid line) and CYP3A4(C64) (triangles, dashed line) by BADAN. Conditions: 10 µM heme protein and 12 µM BADAN in 0.1 M Na-Hepes buffer, pH 7.4, 10% glycerol, 0.2% Igepal CO-630 under Argon atmosphere and a constant stirring. The reaction was monitored by increase in fluorescence at 500 nm (excitation at 400 nm). The lines show the approximation of the experimental results with a second order kinetic equation with the rate constants of 18.0, 10.0 and 17.8 mM−1 min−1 for CYP3A4(C58,C64), CYP3A4(C58), and CYP3A4(C64), respectively.
Titration of CYP3A4(C64)-BADAN with ANF and TST showed that, while TST has no effect on the fluorescence of the probe, the addition of ANF resulted in a considerable decrease in the fluorescence intensity, similar to CYP3A4(C58,C64)-BADAN. Dependence of the intensity of fluorescence of BADAN on the concentration of ANF obeys the Hill equation with S50 = 20.6 ± 3.7 µM, Hill coefficient of 1.7 ± 0.4, and maximal amplitude of fluorescence decrease of 40 ± 11% (the values represent the averages of four repetitive measurements). Similar to the quadruple mutant, addition of TST to CYP3A4(C64)-BADAN in the presence of 25 µM ANF resulted in up to a ~25% increase in the fluorescence of the probe, which reveals a testosterone binding process obeying the Michaelis-Menten equation with KM of 30 µM. Furthermore, the results of experiments on H2O2-dependent heme depletion of CYP3A4(C64)-BADAN were also similar to those obtained with the BADAN-modified quadruple mutant. Specifically, the kinetics of the fluorescence increase upon H2O2-dependent heme depletion of CYP3A4(C64)-BADAN was monoexponential (k = 0.04 s−1), in contrast to bi-exponential kinetics of the heme loss. The apparent efficiency of FRET from BADAN to the heme in CYP3A4(C64)-BADAN was around 62%.
Therefore, the remarkable similarity in the behavior of CYP3A4(C64)-BADAN to that of CYP3A4(C58,C64)-BADAN, together with the slow rate of modification of CYP3A4(C58) by BADAN and instability of the resulting adduct provides convincing evidence that Cys-64 is the site of preferential modification of CYP3A4(C58,C64) by BADAN and presumably the other labels.
Discussion
In this study we employ modification of CYP3A4 with thiol-reactive fluorescent probes in combination with cysteine-depleting mutagenesis as a versatile strategy to explore substrate-induced conformational transitions and their possible involvement in the mechanisms of cooperativity. In a recent study of P450eryF we successfully applied a FRET-based version of this approach to elucidate the mechanisms of the substrate-induced spin shift and cooperativity (49). In contrast to P450eryF, which has only one cysteine residue (Cys-154) accessible to modification with thiol-reactive probes, CYP3A4 possesses six potentially reactive cysteines, namely Cys-58, Cys-64, Cys-98, Cys-239, Cys-377 and Cys-468. The CYP3A4(C58,C64) variant of the enzyme bearing C98S, C239S, C377S and C468A substitutions described here was obtained in the course of our work targeted to engineering an enzyme completely depleted of reactive cysteines, which would represent a useful template for further site-directed mutagenesis and chemical modification. In our follow-up studies to CYP3A4(C58,C64) we also elaborated CYP3A4(C58) and CYP3A4(C64) quintuple mutants, which are also described in this report.
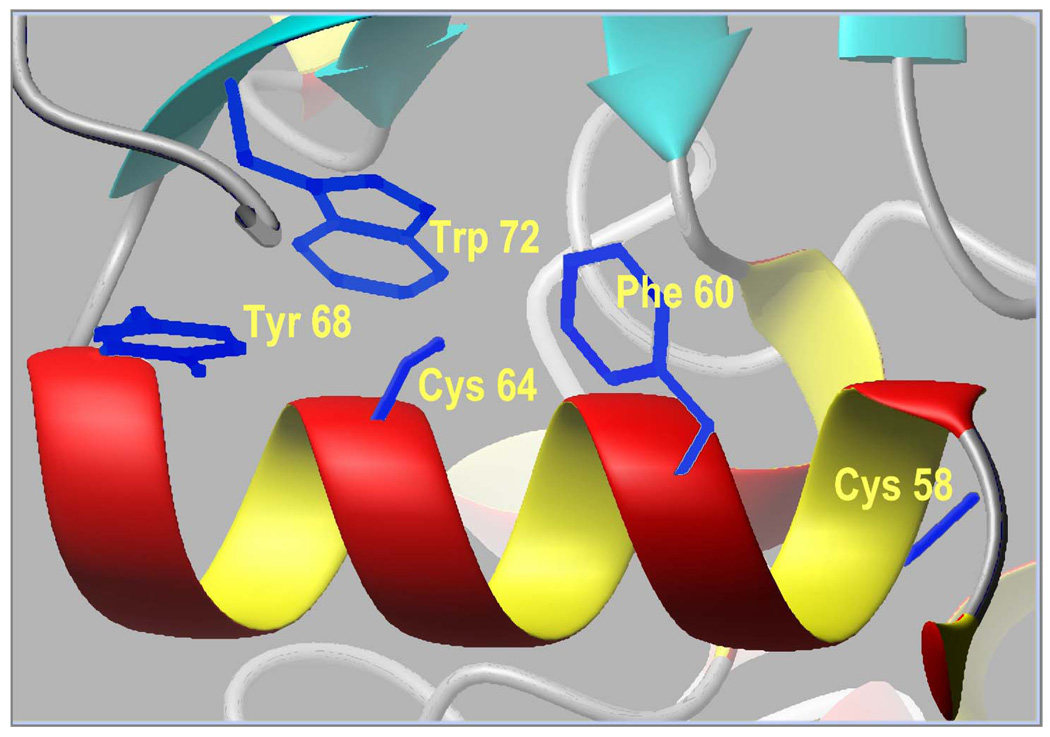
The two potentially reactive cysteines, Cys-58 and Cys-64, in CYP3A4(C58,C64) are both located in α-helix A. Based on analysis of the CYP3A4 crystal structure (54, 55) these cysteine residues might be expected to be less accessible than the four that were deleted, suggesting that the reactivity of CYP3A4(C58,C64) with thiol-reactive compounds would be abolished or considerably lower than the wild type. However, the rate of modification of the quadruple mutant with CPM, BADAN or BODYPY-FL iodacetamide was comparable to that of the wild type. At the same time, incorporation of more than one molecule of a thiol-reactive probe into this mutant results in its denaturation. Therefore, we conclude that one of the two remaining reactive cysteine residues in the mutant is readily accessible to modification. According to CYP3A4 crystal structures we suggest that the modification of CYP3A4(C58,C64) occurs at Cys-64, which is oriented towards the surface of the protein and has neighboring aromatic rings of Phe-60, Tyr-68, and Trp-72, which may facilitate accommodation of a hydrophobic dye (Fig. 11). In contrast, modification of Cys-58, which is oriented towards the protein core and faces the loop preceeding β1–4, appears to be impeded by steric constrains and is likely to result in protein denaturation. In agreement with this inference, we demonstrated that the modification of CYP3A4(C58) with BADAN is accompanied by a loss of ~75% of the protein during purification due to denaturation. Furthermore, the low amounts of CYP3A4(C58)-BADAN that can be recovered show ~75% conversion to the P420 state, which was not observed with the quadruple mutant. In contrast, the properties of CYP3A4(C64)-BADAN were similar to those observed with CYP3A4(C58,C64)-BADAN. Therefore, the modification of CYP3A4(C58,C64) with SH-reactive probes may be identified with confidence as taking place at Cys-64 .
Figure 11.
Fragment of structure of cytochrome P450 3A4 (PDB entry 1TQN) (54) illustrating the neighborhood of Cys-58 and Cys-64.
Interestingly, the interaction of CYP3A4(C58,C64)-BADAN with TST, BCT, or 1-PB has no substantial effect on either the amplitude or position of the fluorescence band of BADAN. This observation suggests the absence of significant conformational changes in the protein, since virtually any transition in the proximity of the label would change the polarity of its environment and inevitably result in displacement of the emission band of BADAN (40–43). In contrast to TST, BCT, and 1-PB, addition of ANF caused a 40% decrease in the fluorescence of BADAN, although the position of the band remains unchanged (Fig. 4). This observation reveals a binding site for ANF, which is characterized by an S50 value around 18 µM. This value is considerably higher than that associated with the ANF-dependent spin transition (2.2 µM), which shows prominent cooperativity. The presence of this low-affinity binding site for ANF is also confirmed in our experiments with two other labels – CPM and mBBr. Thus, CYP3A4 appears to possess two binding sites revealed in the ANF-induced spin shift and a distinct low-affinity binding site, any or all of which may be involved in the heterotropic cooperativty observed with this compound.
The existence of multiple ANF-binding sites has been proposed in many studies (46–48, 59, 60), although direct evidence for a third ANF-binding site in CYP3A4 was lacking.. As the most important increase in the affinity of CYP3A4 for 1-PB is observed at concentrations of ANF below 5 µM (Fig. 8a), this effect may be attributed to ANF binding to the high-affinity site(s). However, a further increase in ANF concentration caused a considerable decrease in the Hill coefficient for 1-PB binding (Fig. 8b), such that the homotropic cooperativity of 1-PB binding is abolished at concentrations of ANF >30 µM. This range of concentrations of ANF is consistent with a role of the third (low-affinity) binding site. Thus, the effects of ANF on 1-PB binding to CYP3A4 suggest that all three ANF-binding sites may be pertinent to heterotropic cooperativity, but that the role of the newly discovered low-affinity site is distinct from that of the sites revealed in the ANF-induced spin shift
The involvement of the low-affinity ANF binding site in the modulation of interactions of CYP3A4 with substrates is also confirmed by the results of our fluorometric titrations of CYP3A4(C58,C64)-BADAN with TST. In contrast to the interactions of CYP3A4(C58,C64)-BADAN with TST alone, which have no effect on BADAN fluorescence, the binding of TST in the presence of ANF causes an increase in fluorescence of up to 34%. At the same time TST does not compete for the low-affinity ANF binding site, as the presence of TST has no effect on S50 and n values for the binding of ANF determined in our fluorometric titrations. These results imply therefore that the binding of TST induces additional rearrangement in the structure of the ANF-bound enzyme, which causes an increase in the intensity of fluorescence of the probe, while the binding of TST alone is incapable of inducing these changes.
Further insight into the mechanism of action of ANF requires understanding of the physical nature of the effect of this substrate on the fluorescence of BADAN and CPM. Theoretically, this effect may be caused by a direct interaction of ANF with the probe. However, according to the known crystal structures of CYP3A4 (36, 54, 55), the highly ordered and tightly packed protein structure around the site of modification (Fig. 11) imposes important steric constraints on ANF binding in this region, especially once the cysteine residue is modified with a fluorescent probe. Another explanation is that the binding of ANF induces a long-range perturbation of the protein structure, resulting in increased efficiency of FRET from BADAN to the heme due to changes in the distance between the label and the heme and/or in the orientation of the label relative to the heme ring. Although the effect of ANF observed in CYP3A4(C58,C64)-BADAN is reproduced in CYP3A4(C58,C64)-CPM (Fig. 5), ANF increases rather than decreases the fluorescence intensity of the protein labeled with mBBr (Fig. 6). This finding and results of experiments on the effect of heme depletion on the intensity of fluorescence of the three labels disproves the modulation of the efficiency of FRET as a basis for the ANF-induced changes in fluorescence intensity. Although the intensity of the fluorescence of all three labels is increased upon destruction of the heme moiety by H2O2, and the efficiency of FRET calculated from these experiments (Table 3) is consistent which the expected values according to the Förster equation, addition of ANF does not exert any effect on the efficiency of FRET to the heme (Table 3).
Nonetheless, the heme depletion experiments provide crucial mechanistic insights. Consistent with our earlier observation (58), biexponential kinetics of heme destruction suggests persistent conformational heterogeneity of CYP3A4, such that the protein appears to be distributed between two populations with different accessibility of the heme moiety to H2O2. Furthermore, the striking difference between the time course of heme depletion and of the fluorescence increase (Fig. 9) suggests that the fraction of the protein undergoing heme depletion in the fast phase has the label completely quenched, so that the fast phase is not reflected in the kinetics of the fluorescence increase. Inspection of the closest neighborhood of Cys-64 provides a possible source for this quenching. As illustrated in Fig. 11, Cys-64 is surrounded by three aromatic residues – Phe-60, Tyr-68, and Trp-72. The effect of quenching of fluorescence by the interactions of fluorophores with tyrosine and, most importantly, tryptophan due to photoinduced electron transfer (PET) from these aromatic amino acid residues is well documented (61–63). Functionality as electron acceptors in PET has been demonstrated for both coumarins (64), dimethylaminonaphthalene (65), and bimane (66). Therefore, we suggest that the fraction of labeled CYP3A4(C58,C64) represented in the fast phase of the heme depletion has the fluorescent probe quenched due to PET from Trp-72 or, less likely, Tyr-68. Importantly, in the case of mBBr this effect is partially abolished (Table 3). That may indicate that the distance between Trp-72 and the bimane label, which is the smallest label used in this study, may be too large for efficient PET, and that the fluorescence of the sample reflects the emission from both fractions of the enzyme (though to different extents). This also explains why the ANF-induced decrease in the fluorescence of the probe with CPM and BADAN was not reproduced with mBBr.
Close proximity between Trp-72 and the fluorescent probe also suggests that CPM, BADAN or mBBr could serve as extremely efficient acceptors for FRET from this tryptophan residue. Consistent with this expectation, the spectrum of excitation of CYP3A4(C58,C64)-BADAN reveals a clear maximum at 278, indicating the excitation of the label through FRET from tryptophan (Fig. 1). Importantly, the position of the maximum of the emission of the probe exhibits clear dependence on the wavelength of excitation. Excitation at the maximum of its longer-wavelength band (384 nm) results in an emission maximum of BADAN at 488 nm. However, in the case of excitation at 280 nm, where both tryptophan and BADAN have comparable extinction coefficients, the emission band becomes broader and is displaced to 496 nm. A maximal red shift of the emission band of BADAN (to 507 nm) is observed upon excitation around 310 – 315 nm, which corresponds to the position of the minimum in the spectrum of direct excitation of the BADAN probe. Therefore, we conclude that the fraction of the enzyme, which has the fluorescence of the label quenched due to PET from tryptophan, has the label in a more polar environment than the highly fluorescent fraction.
Our results clearly demonstrate the presence of an additional ANF-binding site that is distinct from the two sites reflected in the ANF-induced spin shift. Our results are consistent with the involvement of the changes in the efficiency of PET to CPM and BADAN probes incorporated at Cys-64 from the neighboring Trp-72 and/or Tyr-68 residues. In addition, we demonstrated that two stable conformers of the enzyme revealed in the biphasic kinetics of H2O2-dependent heme depletion show a dramatic difference in the intensity of the fluorescence of the probe incorporated at Cys-64. Therefore, the pool of CYP3A4 appears to be distributed into two persistent conformers that differ in conformation in the region of α-helix A and its surroundings, which is reflected in different polarity in the environment of BADAN introduced in this region and also apparently affects the interactions of this label with neighboring aromatic tryptophan and/or tyrosine residues.
The above evidence suggests the involvement of conformational heterogeneity of CYP3A4 in the observed effect of ANF. One possible mechanism of the ANF-mediated decrease in fluorescence of the BADAN and CPM probes is modulation by this effector of the partitioning between two conformers of the enzyme. This interpretation is consistent with the hypothesis of Koley and co-authors that the mechanism of action of ANF involves redistribution of the protein between two conformers with different substrate specificity and different accessibility of the heme pocket (21, 25, 38). Another possibility is that the binding of ANF quenches the fluorescence of the label in the fluorescent conformer by some local changes in the proximity of the label that increase the probability of PET from Trp-72 and/or Tyr-68 to the label without any changes in the partitioning of fluorescent and completely quenched conformers.
It should be noted that none of the three putative ANF-binding events reveal competition for the sites involved in the interactions of the enzyme with 1-PB or TST. This leads to the suggestion the ANF-binding sites are all distinct from those involved in 1-PB and TST binding. It appears improbable that one and the same molecule of CYP3A4 possess more than three distinct binding sites for substrates and effectors. To our understanding, our observations are best explained by the suggestion that at least some of these sites are located in distinct subunits of a CYP3A4 oligomer, and that the mechanism of action of ANF involves modulation of subunit interactions. This hypothesis is consistent with our earlier conclusion that the oligomers of CYP3A4 consist of distinct conformers having different position of spin equilibrium and possessing different substrate binding properties (37). Therefore, the low-affinity ANF-binding site discovered in this study may be located either on the same molecule of the enzyme as the other two ANF-binding sites, or situated on another subunit of the oligomer. Moreover, this binding site may be formed by the interface of interactions of two subunits in the oligomer. The latter possibility is favored by a recent X-ray structure of a CYP3A4 dimer crystallized in the presence of TST, which shows the binding of two molecules of testosterone in the dimerization interface (67), and earlier obvservation of two molecules of palmitic acid in the dimerization interface of P450 2C8 (68). Therefore, we believe that further insight into the mechanisms of heterotropic cooperativity with ANF requires consideration of the oligomer of CYP3A4 as a functional unit of substrate binding.
Acknowledgements
The authors are grateful to Dr. Sandra Graham and Dr. Yonghong Zhao for their help in structural interpretation of the results of this study.
ABBREVIATIONS AND TEXTUAL FOOTNOTES
- CYP3A4
cytochrome P450 3A4
- ANF
α-naphthoflavone
- BADAN
6-bromoacetyl-2-dimethylaminonaphthalene
- CPM
7-diethylamino-3-(4’-maleimidylphenyl)-4-methylcoumarin
- mBBr
monobromobimane
- BCT
bromocriptine
- TST
testosterone
- CHAPS
3-[(3-cholamido-propyl)dimethylammonio]-1-propanesulfonate
- DTT
dithiothreitol
- Hepes
N-[2-hydroxyethylpiperazine-N'-[2-ethanesulfonic acid]
- 1-PB
1-pyrenebutanol
- CYP3A4(C58, C64)
a C98S/C239S/C377S/C468A quadruple mutant of CYP3A4 bearing two accessible cysteines; CYP3A4(C58) and CYP3A4(C64) quintuple mutants obtained by adding to CYP3A4(C58, C64) the C64A and C58T replacements, respectively.
Footnotes
This research was supported by NIH grant GM54995, Center grant ES06676, and research grant H-1458 from the Robert A. Welch Foundation.
References
- 1.Atkins WM. Non-Michaelis-Menten kinetics in cytochrome P450-catalyzed reactions. Annu. Rev. Pharmacol. Toxicol. 2005;45:291–310. doi: 10.1146/annurev.pharmtox.45.120403.100004. [DOI] [PubMed] [Google Scholar]
- 2.Atkins WM. Implications of the allosteric kinetics of cytochrome P450s. Drug Discovery Today. 2004;9:478–484. doi: 10.1016/S1359-6446(04)03072-7. [DOI] [PubMed] [Google Scholar]
- 3.Yoon MY, Campbell AP, Atkins WM. "Allosterism" in the elementary steps of the cytochrome P450 reaction cycle. Drug Metab. Rev. 2004;36:219–230. doi: 10.1081/dmr-120033998. [DOI] [PubMed] [Google Scholar]
- 4.Ekins S, Stresser DM, Williams JA. In vitro and pharmacophore insights into CYP3A enzymes. Trends Pharm. Sci. 2003;24:161–166. doi: 10.1016/s0165-6147(03)00049-x. [DOI] [PubMed] [Google Scholar]
- 5.Khan KK, Liu H, Halpert JR. Homotropic versus heterotopic cooperativity of cytochrome P450eryF: A substrate oxidation and spectral titration study. Drug Metab. Disp. 2003;31:356–359. doi: 10.1124/dmd.31.4.356. [DOI] [PubMed] [Google Scholar]
- 6.Hutzler JM, Tracy TS. Atypical kinetic profiles in drug metabolism reactions. Drug Metab. Disp. 2002;30:355–362. doi: 10.1124/dmd.30.4.355. [DOI] [PubMed] [Google Scholar]
- 7.Ekins S, Ring BJ, Binkley SN, Hall SD, Wrighton SA. Autoactivation and activation of the cytochrome P450s. Int. J. Clin. Pharm. Ther. 1998;36:642–651. [PubMed] [Google Scholar]
- 8.Korzekwa KR, Krishnamachary N, Shou M, Ogai A, Parise RA, Rettie AE, Gonzalez FJ, Tracy TS. Evaluation of atypical cytochrome P450 kinetics with two-substrate models: evidence that multiple substrates can simultaneously bind to cytochrome P450 active sites. Biochemistry. 1998;37:4137–4147. doi: 10.1021/bi9715627. [DOI] [PubMed] [Google Scholar]
- 9.Miller GP, Guengerich FP. Binding and oxidation of alkyl 4-nitrophenyl ethers by rabbit cytochrome P450 1A2: Evidence for two binding sites. Biochemistry. 2001;40:7262–7272. doi: 10.1021/bi010402z. [DOI] [PubMed] [Google Scholar]
- 10.Hutzler JM, Wienkers LC, Wahlstrom JL, Carlson TJ, Tracy TS. Activation of cytochrome P4502C9-mediated metabolism: mechanistic evidence in support of kinetic observations. Arch. Biochem. Biophys. 2003;410:16–24. doi: 10.1016/s0003-9861(02)00665-3. [DOI] [PubMed] [Google Scholar]
- 11.Locuson CW, Gannett PM, Tracy TS. Heteroactivator effects on the coupling and spin state equilibrium of CYP2C9. Arch. Biochem. Biophys. 2006;449:115–129. doi: 10.1016/j.abb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Hummel MA, Gannett PM, Aguilar JS, Tracy TS. Effector-mediated alteration of substrate orientation in cytochrome P450 2C9. Biochemistry. 2004;43:7207–7214. doi: 10.1021/bi036158o. [DOI] [PubMed] [Google Scholar]
- 13.Tracy TS, Hutzler JM, Haining RL, Rettie AE, Hummel MA, Dickmann LJ. Polymorphic variants (CYP2C9*3 and CYP2C9*5) and the F114L active site mutation of CYP2C9: effect on atypical kinetic metabolism profiles. Drug Metab. Disp. 2002;30:385–390. doi: 10.1124/dmd.30.4.385. [DOI] [PubMed] [Google Scholar]
- 14.Ariyoshi N, Miyazaki M, Toide K, Sawamura Y, Kamataki T. A single nucleotide polymorphism of CYP2B6 found in Japanese enhances catalytic activity by autoactivation. Biochem. Biophys. Res. Commun. 2001;281:1256–1260. doi: 10.1006/bbrc.2001.4524. [DOI] [PubMed] [Google Scholar]
- 15.Ingelman-Sundberg M, Johansson I, Hansson A. Catalytic properties of the liver microsomal hydroxylase system in reconstituted phospholipid vesicles. Acta Biol. Med. Germ. 1979;38:379–388. [PubMed] [Google Scholar]
- 16.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Harlow GR, Halpert JR. Analysis of human cytochrome P450 3A4 cooperativity: construction and characterization of a site-directed mutant that displays hyperbolic steroid hydroxylation kinetics. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueng YF, Kuwabara T, Chun YJ, Guengerich FP. Cooperativity in oxidations catalyzed by cytochrome P450 3A4. Biochemistry. 1997;36:370–381. doi: 10.1021/bi962359z. [DOI] [PubMed] [Google Scholar]
- 19.Tang W, Stearns RA. Heterotropic cooperativity of cytochrome P450 3A4 and potential drug-drug interactions. Curr. Drug Metab. 2001;2:185–198. doi: 10.2174/1389200013338658. [DOI] [PubMed] [Google Scholar]
- 20.Shou M, Grogan J, Mancewicz JA, Krausz KW, Gonzalez FJ, Gelboin HV, Korzekwa KR. Activation of cyp3a4 - evidence for the simultaneous binding of 2 substrates. Biochemistry. 1994;33:6450–6455. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- 21.Koley AP, Buters JTM, Robinson RC, Markowitz A, Friedman FK. Differential mechanisms of cytochrome P450 inhibition and activation by alpha-naphthoflavone. J. Biol. Chem. 1997;272:3149–3152. doi: 10.1074/jbc.272.6.3149. [DOI] [PubMed] [Google Scholar]
- 22.Galetin A, Clarke SE, Houston JB. Quinidine and haloperidol as modifiers of CYP3A4 activity: Multisite kinetic model approach. Drug Metab. Disp. 2002;30:1512–1522. doi: 10.1124/dmd.30.12.1512. [DOI] [PubMed] [Google Scholar]
- 23.Domanski TL, Liu J, Harlow GR, Halpert JR. Analysis of four residues within substrate recognition site 4 of human cytochrome P450 3A4: role in steroid hydroxylase activity and alpha- naphthoflavone stimulation. Arch. Biochem. Biophys. 1998;350:223–232. doi: 10.1006/abbi.1997.0525. [DOI] [PubMed] [Google Scholar]
- 24.Schwab GE, Raucy JL, Johnson EF. Modulation of rabbit and human hepatic cytochrome P-450-catalyzed steroid hydroxylations by alpha-naphthoflavone. Mol. Pharmacol. 1988;33:493–499. [PubMed] [Google Scholar]
- 25.Koley AP, Robinson RC, Markowitz A, Friedman FK. Drug-drug interactions: effect of quinidine on nifedipine binding to human cytochrome P450 3A4. Biochem. Pharmacol. 1997;53:455–460. doi: 10.1016/s0006-2952(96)00836-2. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig E, Schmid J, Beschke K, Ebner T. Activation of human cytochrome P-450 3A4-catalyzed meloxicam 5'-methylhydroxylation by quinidine and hydroquinidine in vitro. J. Pharmacol. Exp. Ther. 1999;290:1–8. [PubMed] [Google Scholar]
- 27.Ngui JS, Chen Q, Shou MG, Wang RW, Stearns RA, Baillie TA, Tang W. In vitro stimulation of warfarin metabolism by quinidine: Increases in the formation of 4'- and 10-hydroxywarfarin. Drug Metab. Disp. 2001;29:877–886. [PubMed] [Google Scholar]
- 28.Atkins WM, Wang RW, Lu AYH. Allosteric behavior in cytochrome P450-dependent in vitro drug-drug interactions: A prospective based on conformational dynamics. Chem. Res. Toxicol. 2001;14:338–347. doi: 10.1021/tx0002132. [DOI] [PubMed] [Google Scholar]
- 29.Schrag ML, Wienkers LC. Topological alteration of the CYP3A4 active site by the divalent cation Mg2+ Drug Metab. Disp. 2000;28:1198–1201. [PubMed] [Google Scholar]
- 30.Davydov DR, Halpert JR, Renaud JP, Hui Bon Hoa G. Conformational heterogeneity of cytochrome P450 3A4 revealed by high pressure spectroscopy. Biochem. Biophys. Res. Commun. 2003;312:121–130. doi: 10.1016/j.bbrc.2003.09.247. [DOI] [PubMed] [Google Scholar]
- 31.Davydov DR, Botchkareva AE, Kumar S, He YQ, Halpert JR. An electrostatically driven conformational transition is involved in the mechanisms of substrate binding and cooperativity in cytochrome P450eryF. Biochemistry. 2004;43:6475–6485. doi: 10.1021/bi036260l. [DOI] [PubMed] [Google Scholar]
- 32.Isin EM, Guengerich FP. Kinetics and thermodynamics of ligand binding by cytochrome P450 3A4. J. Biol. Chem. 2006;281:9127–9136. doi: 10.1074/jbc.M511375200. [DOI] [PubMed] [Google Scholar]
- 33.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. An open conformation of mammalian cytochrome P4502B4 at 1.6-angstrom resolution. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J. Biol. Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J. Biol. Chem. 2006;281:5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 36.Ekroos M, Sjцgren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13684–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davydov DR, Fernando H, Baas BJ, Sligar SG, Halpert JR. Kinetics of dithionite-dependent reduction of cytochrome P450 3A4: Heterogeneity of the enzyme caused by its oligomerization. Biochemistry. 2005;44:13902–13913. doi: 10.1021/bi0509346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koley AP, Robinson RC, Friedman FK. Cytochrome P450 conformation and substrate interactions as probed by CO binding kinetics. Biochimie. 1996;78:706–713. doi: 10.1016/s0300-9084(97)82528-x. [DOI] [PubMed] [Google Scholar]
- 39.Koley AP, Dai RK, Robinson RC, Markowitz A, Friedman FK. Differential interaction of erythromycin with cytochromes P450 3A1/2 in the endoplasmic reticulum: A CO flash photolysis study. Biochemistry. 1997;36:3237–3241. doi: 10.1021/bi962110h. [DOI] [PubMed] [Google Scholar]
- 40.Macgregor RB, Weber G. Estimation of the polarity of the protein interior by optical spectroscopy. Nature. 1986;319:70–73. doi: 10.1038/319070a0. [DOI] [PubMed] [Google Scholar]
- 41.Kawski A, Kuklinski B, Bojarski P. Ground and excited state dipole moments of BADAN and ACRYLODAN determined from solvatochromic shifts of absorption and fluorescence spectra. Z. Naturforsch. A. 2001;56:407–411. [Google Scholar]
- 42.Raguz M, Brnjas-Kraljevic J. Resolved fluorescence emission spectra of PRODAN in ethanol/buffer solvents. J. Chem. Information Modeling. 2005;45:1636–1640. doi: 10.1021/ci050166p. [DOI] [PubMed] [Google Scholar]
- 43.Hiratsuka T. ATP-induced opposite changes in the local environments around Cys(697) (SH2) and Cys(707) (SH1) of the myosin motor domain revealed by the prodan fluorescence. J. Biol. Chem. 1999;274:29156–29163. doi: 10.1074/jbc.274.41.29156. [DOI] [PubMed] [Google Scholar]
- 44.Davydov DR, Fernando H, Halpert JR. Variable path length and counter-flow continuous variation methods for the study of the formation of high-affinity complexes by absorbance spectroscopy. An application to the studies of substrate binding in cytochrome P450. Biophys. Chem. 2006;123:95–101. doi: 10.1016/j.bpc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Fernando H, Halpert JR, Davydov DR. Resolution of multiple substrate binding sites in cytochrome P450 3A4: the stoichiometry of the enzyme-substrate complexes probed by FRET and Job's titration. Biochemistry. 2006;45:4199–4209. doi: 10.1021/bi052491b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domanski TL, He YA, Harlow GR, Halpert JR. Dual role of human cytochrome P450 3A4 residue Phe-304 in substrate specificity and cooperativity. J. Pharmacol. Exp. Ther. 2000;293:585–591. [PubMed] [Google Scholar]
- 47.Hosea NA, Miller GP, Guengerich FP. Elucidation of distinct ligand binding sites for cytochrome P450 3A4. Biochemistry. 2000;39:5929–5939. doi: 10.1021/bi992765t. [DOI] [PubMed] [Google Scholar]
- 48.Domanski TL, Halpert JR. Analysis of mammalian cytochrome P450 structure and function by site-directed mutagenesis. Curr. Drug Metab. 2001;2:117–137. doi: 10.2174/1389200013338612. [DOI] [PubMed] [Google Scholar]
- 49.Davydov DR, Botchkareva AE, Davydova NE, Halpert JR. Resolution of two substrate-binding sites in an engineered cytochrome P450eryF bearing a fluorescent probe. Biophys. J. 2005;89:418–432. doi: 10.1529/biophysj.104.058479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Invitrogen - Molecular Probes_Inc. The Handbook: A Guide to Fluorescent Probes and Labeling Technologies. 2006 Web edition: http://probes.invitrogen.com/handbook/sections/0200.html.
- 51.Davydov DR, Deprez E, Hui Bon Hoa G, Knyushko TV, Kuznetsova GP, Koen YM, Archakov AI. High-Pressure-Induced Transitions in Microsomal Cytochrome P450 2B4 in Solution - Evidence for Conformational Inhomogeneity in the Oligomers. Arch. Biochem. Biophys. 1995;320:330–344. doi: 10.1016/0003-9861(95)90017-9. [DOI] [PubMed] [Google Scholar]
- 52.Renaud JP, Davydov DR, Heirwegh KPM, Mansuy D, Hui Bon Hoa G. Thermodynamic studies of substrate binding and spin transitions in human cytochrome P450 3A4 expressed in yeast microsomes. Biochem. J. 1996;319:675–681. doi: 10.1042/bj3190675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen B, Doneanu CE, Gartner CA, Roberts AG, Atkins WM, Nelson SD. Fluorescent photoaffinity labeling of cytochrome P450 3A4 by lapachenole: Identification of modification sites by mass spectrornetry. Biochemistry. 2005;44:1833–1845. doi: 10.1021/bi048228c. [DOI] [PubMed] [Google Scholar]
- 54.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-angstrom resolution. J. Biol. Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 55.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 56.Uvarov VY, Tretiakov VE, Archakov AI. Heme maintains catalytically active structure of cytochrome P-450. FEBS Lett. 1990;260:309–312. doi: 10.1016/0014-5793(90)80131-2. [DOI] [PubMed] [Google Scholar]
- 57.Pikuleva IA, Lapko AG, Chashchin VL. Functional reconstitution of cytochrome P-450scc with hemin activated with Woodward's reagent K. Formation of a hemeprotein cross-link. J. Biol. Chem. 1992;267:1438–1442. [PubMed] [Google Scholar]
- 58.Kumar S, Davydov DR, Halpert JR. Role of cytochrome b5 in modulating peroxide-supported CYP3A4 activity: Evidence for a conformational transition and cytochrome P450 heterogeneity. Drug Metab. Disp. 2005;33:1131–1136. doi: 10.1124/dmd.105.004606. [DOI] [PubMed] [Google Scholar]
- 59.He YA, Roussel F, Halpert JR. Analysis of homotropic and heterotropic cooperativity of diazepam oxidation. Arch. Biochem. Biophys. 2003;409:92–101. doi: 10.1016/s0003-9861(02)00484-8. [DOI] [PubMed] [Google Scholar]
- 60.Harlow GR, Halpert JR. Alanine-scanning mutagenesis of a putative substrate recognition site in human cytochrome P450 3A4. Role of residues 210 and 211 in flavonoid activation and substrate specificity. J. Biol. Chem. 1997;272:5396–5402. doi: 10.1074/jbc.272.9.5396. [DOI] [PubMed] [Google Scholar]
- 61.Doose S, Neuweiler H, Sauer M. A close look at fluorescence quenching of organic dyes by tryptophan. Chemphyschem. 2005;6:2277–2285. doi: 10.1002/cphc.200500191. [DOI] [PubMed] [Google Scholar]
- 62.Marme N, Knemeyer J-P, Sauer M, Wolfrum J. Inter- and Intramolecular Fluorescence Quenching of Organic Dyes by Tryptophan. Bioconjugate Chem. 2003;14:1133–1139. doi: 10.1021/bc0341324. [DOI] [PubMed] [Google Scholar]
- 63.Nakashima K, Tanida S, Miyamoto T, Hashimoto S. Photoinduced electron transfer from indolic compounds to 1-pyrenemethanol in polystyrene latex dispersions. J. Photochem. Photobiol. A. 1998;117:111–117. [Google Scholar]
- 64.Kumbhakar M, Sukhendu N, Haridas P, Sapre A, Mukheriee T. Photoinduced intermolecular electron transfer from aromatic amines to coumarin dyes in sodium dodecyl sulphate micellar solutions. J. Chem. Phys. 2003;119:388. [Google Scholar]
- 65.Wang YH, Zhu MZ, Lio T, Guo QX. The electron Transfer Reaction between p-Nitrobenzoates and N-Dimethylnaphthalene. Chinese Chem. Lett. 2003;14:159–162. [Google Scholar]
- 66.Mansoor SE, Mchaourab HS, Farrens DL. Mapping proximity within proteins using fluorescence spectroscopy. A study of T4 lysozyme showing that tryptophan residues quench bimane fluorescence. Biochemistry. 2002;41:2475–2484. doi: 10.1021/bi011198i. [DOI] [PubMed] [Google Scholar]
- 67.He YA, Gajiwala KS, Wu M, Parge H, Burke B, Lee CA, Wester MR. Crystal structure of human CYP3A4 in complex with testosterone. 16-th International Symposium on Microsomes and Drug Oxidations (MDO 2006), Program and Abstract Book; Budapest, Hungary. 2006. p. 114. [Google Scholar]
- 68.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. Structure of human microsomal cytochrome P4502C8 - Evidence for a peripheral fatty acid binding site. J. Biol. Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]