Summary
The concerted interconnection between processes driving DNA synthesis, division septum formation and cell wall synthesis and remodeling in rapidly growing bacteria requires precise coordination by signaling mechanisms that are, for the most part, unknown. The YycG (sensor histidine kinase), YycF (response regulator/transcription factor) two-component system of Bacillus subtilis controls the synthesis of enzymes and their inhibitors that function in cell wall remodeling and cell separation. Here it is shown that the YycG sensor histidine kinase is a component of the division septum in growing cells. RT-PCR quantitation of YycF-PO4 regulated gene transcription, in wild type and FtsZ-depleted, septum-less cells, indicated that YycG kinase activity on YycF is dependent on YycG localization to a division septum. The data support a model in which the YycG sensor kinase perceives information at the division septum and regulates the reciprocal synthesis of autolysins and autolysin inhibitors to coordinate growth and division with cell wall restructuring.
Keywords: Bacillus subtilis, YycFG, FtsZ, signal transduction, cell division
Introduction
Two-component and phosphorelay signal transduction systems play a major role in communicating environmental information to the cell and in coordinating cellular processes. Environmental signals are processed through these systems to influence transport and metabolism of catabolites, drugs and other compounds and to control motility systems to direct the chemotactic behavior of the cell, among other uses (reviewed in (Baker et al., 2006; Mascher et al., 2006)). These regulatory systems are key components in cell cycle signaling in Caulobacter crescentus and in sporulation in Bacillus subtilis indicating that they have been adopted to play more crucial roles in cell division and development (reviewed in (Holtzendorff et al., 2006; Piggot and Hilbert, 2004)). However, deletion studies revealed only a few such signaling systems regulate essential cellular processes, thereby rendering the signal transduction system necessary for cellular survival.
One essential signal transduction system, the YycF (response regulator-transcription factor)- YycG (sensor histidine kinase) two-component system has been found to be conserved in most of the Firmicutes but is apparently absent from the genomes of Gram-negative bacteria (Fabret et al., 1999; Ng and Winkler, 2004). Following the initial observation that this system was the only one of 34 two-component signal transduction systems required for cell survival by Bacillus subtilis, the essentiality of orthologs has been confirmed for the important Gram-positive pathogens Staphylococcus aureus, Streptococcus pneumoniae and Enterococcus faecalis (Fabret et al., 1999; Hancock and Perego, 2004; Martin et al., 1999; Ng et al., 2003).
The cellular role of the YycFG system has been assessed in strains where the cellular concentration of the proteins has been depleted by placing these genes under a controllable promoter. The phenotypes arising upon depletion include defects in cell division and cell wall architecture. B. subtilis strains depleted for YycFG form filamentous cells or chains of cells with empty sections (likely a result of cell lysis) whereas over-expression of yycF leads to the formation of mini-cells suggesting some component of cell division was regulated by this system (Fabret and Hoch, 1998). This notion was strengthened by the finding of genes, ftsAZ, important for cell division as part of the YycF regulon (Fukuchi et al., 2000). Subsequent microarray and DNA-binding studies revealed genes coding for autolysins and autolysin activity modulating proteins to be under YycF expression control, thereby helping to explain cell wall defects in the depletion strain (Bisicchia et al., 2007; Howell et al., 2003). Similar types of gene products were identified to be under YycF control in S. aureus and S. pneumoniae as well as fatty acid biosynthesis genes in the latter organism (Dubrac et al., 2007; Mohedano et al., 2005; Ng et al., 2003; Ng et al., 2005). In summary, many of the products of the regulated genes play a role in cell wall metabolism even though the individual target genes differ from species to species.
While the number of genes defined by the regulon is near resolution, the signal input sensed by this system has yet to be defined, and therefore its actual role has been mysterious to this point. Whether YycG responds to a small molecule ligand, as do several other sensor kinases, remains unknown. However, the YycG kinase was shown to interact with two proteins YycH and YycI whose genes are commonly expressed from the same operon as yycFG (Szurmant et al., 2007b). In both yycH and yycI deletion strains YycG activity appears constitutively up-regulated (Szurmant et al., 2005; Szurmant et al., 2007b). Crystal structures of the periplasmic domains of both YycH and YycI proteins revealed a common tertiary fold despite low sequence conservation (Santelli et al., 2007; Szurmant et al., 2006) but a function could not be deduced from the structures. Recent experiments indicate that YycG, YycH and YycI form a membrane bound ternary sensor complex that may serve to integrate signals designed to coordinate cell architecture and cell division (Szurmant et al., 2008). What those signals are and from where they emanate are questions that address the integration of signal transduction in the overall homeostasis mechanisms that synchronize metabolism, biosynthesis and structure into cell division.
The studies reported here establish that the YycG sensor histidine kinase is localized to the division septum in growing cells. Inhibition of septum formation by FtsZ depletion affected the nucleoid location of the YycF response regulator suggesting that the YycG induced phosphorylation of YycF requires interaction with the division septum. RT PCR mRNA quantitation studies in FtsZ depleted cells showed that the formation of the division septum was necessary for YycG phosphorylation of YycF. The data support a model whereby the YycFG system receives its signal at the division septum and constitutes a positive feedback loop that serves to coordinate cell division with cell wall homeostasis.
Results
The YycG sensor histidine kinase co-localizes with FtsZ at the cell division site
The critical function of the YycFG two-component system in the homeostatic control of cell wall and surface constituents raised questions about its regulation; in particular, if the distribution of YycG in the cell membrane might play a role in this control. Initial studies to determine the cellular location of YycG were carried out with green fluorescent protein (GFP) fusions to YycG but the expression of yycG was too low to visualize the GFP. To avoid probable artifacts from over expression of yycG to raise the cellular level of the GFP fusion, we chose to detect YycG with immunofluorescence in normal exponentially growing cells of B. subtilis strain JH642.
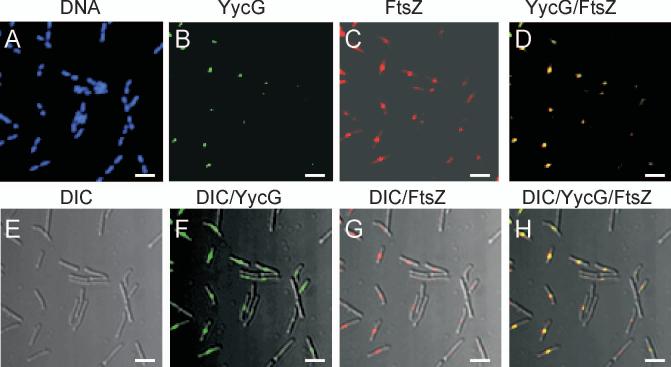
The cellular location of YycG was determined by a specific antibody followed by visualization with a fluorescent-labeled secondary antibody in confocal microscopy. From the images obtained (Fig. 1A-B) it was clear that YycG was located in regions corresponding to potential division sites between DAPI-stained nucleoids. Differential Interference Contrast (DIC) microscopy also revealed the YycG location at mid cell (Fig. 1E-F). In order to confirm the possible division site location of YycG, studies were begun to correlate the localization of YycG with FtsZ (Fig. 1C,G), which is well known to be localized with and crucial for the formation of the division septum (Bi and Lutkenhaus, 1991; Wang and Lutkenhaus, 1993). Overlaying the YycG and FtsZ images revealed that the two proteins co-localized (Fig. 1D,H). To quantify co-localization, 227 cells with visible FtsZ and YycG levels were analyzed for YycG and FtsZ localization to the septum. FtsZ appeared localized in all cells whereas YycG was localized in 224 cells and co-localization was observed in 98.7% of the cell population. Thus the YycG sensor kinase appears to be preferentially localized to the division septum and in the same general region occupied by FtsZ.
Figure 1.
YycG and FtsZ co-localize to the septum in the wildtype B. subtilis strain JH642. YycG (green) and FtsZ (red) proteins were (A-D) visualized immunologically by confocal microscopy and overlain with (E-H) differential interference contrast images, DIC, in exponentially growing cells of JH642 as outlined in Materials and Methods. DNA was visualized by DAPI staining (blue). Bars indicate 5 μm.
YycG localization is dependent upon FtsZ
In order to determine whether the observed localization of YycG was dependent on FtsZ, strain KP444, in which the cellular level of FtsZ could be controlled by the IPTG inducible spac promoter (Beall and Lutkenhaus, 1991), was used (Supplemental Fig. S1). This strain requires IPTG for division septum formation. Experiments designed to lower the cellular concentration of FtsZ were carried out by removal of IPTG from exponentially growing cells and observation of the positions of FtsZ and YycG one and three hours following IPTG removal (Fig. 2). At the earlier time the cells became elongated filaments with the residual FtsZ concentrated at a few possible division sites. However YycG was found spread out in the filament (perhaps in some aggregate or structure) and was not generally associated with a division site and was not concentrated at sites of residual FtsZ (Fig. 2A). At the later time point the remaining FtsZ appeared diffuse in the filaments along with YycG. The cellular level of YycG was unchanged (Fig. 2B). Thus, YycG localization was dependent on FtsZ to form a normal division septum and the two proteins did not co-localize.
Figure 2.
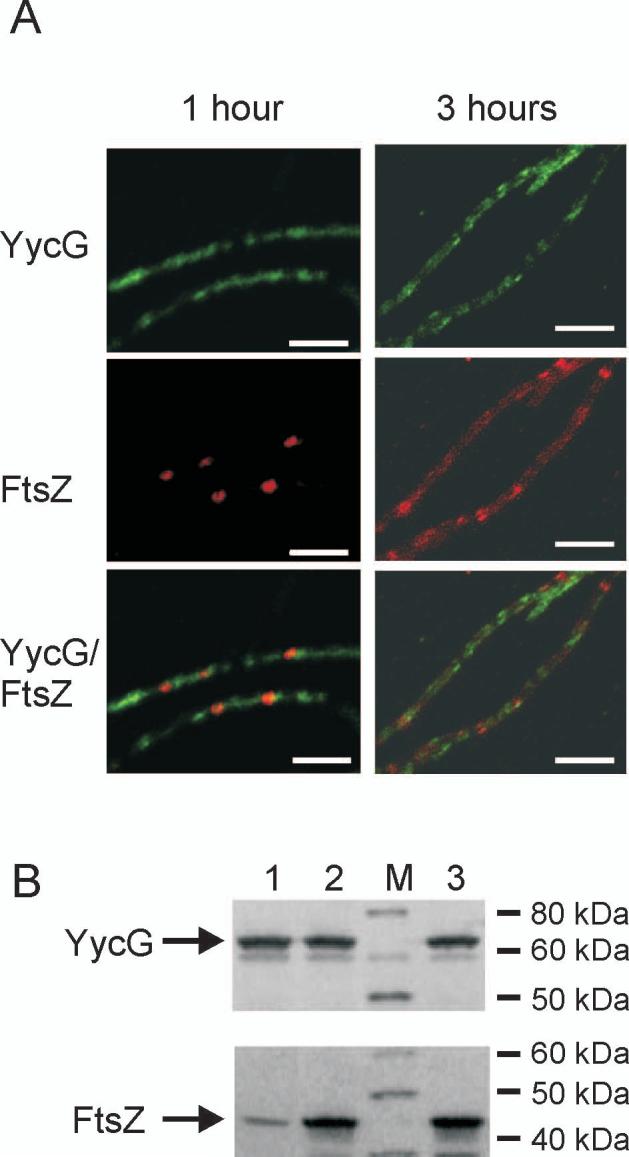
YycG does not localize to the septum in the FtsZ-depleted strain. (A) YycG (green) and FtsZ (red) were visualized immunologically in KP444 cells following 1 and 3 hours growth at 37°C in the absence of IPTG to repress FtsZ expression. Bars indicate 5 μm. (B) Cellular protein levels of YycG and FtsZ were visualized by western blotting using whole cell protein extract derived from strain KP444 grown for one hour either in the absence (lane 1) or presence of 1 mM IPTG (lane 2) or from wild type strain JH642 (lane 3) as reference; Lane M, molecular weight standard.
YycG depletion does not interfere with FtsZ localization
A reciprocal experiment to the one described above was done using strain JH25033 in which the yycG gene was placed under control of the IPTG inducible spac promoter (Supplemental Fig. S1). Depletion of YycG had little effect on growth and after three hours filament formation was not observed consistent with what was previously observed by others utilizing a similar strain (Fukuchi et al., 2000). The majority of the detectable FtsZ protein at this time was still localized to the division septum (Fig. 3A). Quantitative Western immunological analyses revealed a normal level of FtsZ after three hours and greatly diminished cellular concentration of YycG (Fig. 3B). It appears that FtsZ localization is independent of YycG protein levels. However due to the leakiness of the spac promoter sufficient YycG levels remain to sustain growth and although unlikely it cannot be ruled out that in the complete absence of YycG FtsZ localization might be effected.
Figure 3.
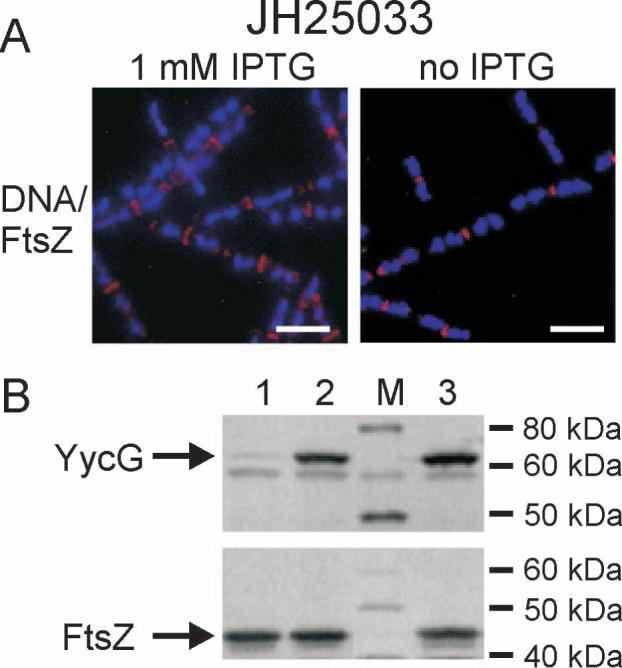
FtsZ localizes to the septum independently of YycG. (A) FtsZ (red) localization was visualized immunologically in strain JH25033 grown three hours either in the presence of 1 mM IPTG (left panel) or in the absence of IPTG (right panel) to induce YycG depletion. DNA (blue) was visualized by DAPI staining. (B) Cellular protein levels of YycG and FtsZ were visualized by western blotting using whole cell protein extract derived from strain JH25033 grown three hours either in the absence (lane 1) or presence of 1 mM IPTG (lane 2) or from wild type strain JH642 (lane3) as reference; Lane M, molecular weight standard.
Co-immunoprecipitation of YycG and FtsZ
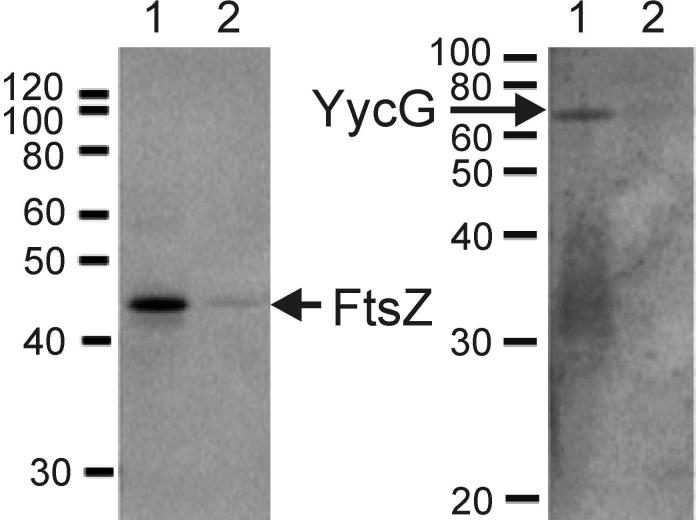
While the co-localization of immunologically detected YycG and FtsZ proteins suggests they are in the same region of the cell at the same time, the results from this technique cannot prove that these proteins interact or that they are part of a common protein complex. In order to further investigate their possible association, immunoprecipitation experiments were undertaken. Growing cells of wild type strain JH642 and the conditional ftsZ expression strain KP444 grown in the absence and presence of IPTG were exposed to formaldehyde to cross-link associated proteins. Extracts were prepared with detergent in a French Press to solubilize both membrane and cytosolic proteins. After mixing the extracts with antibody to FtsZ, the immune-complexes were purified, separated on SDS-PAGE and either YycG or FtsZ detected in a Western analysis. The results of these experiments are shown in Figure 4.
Figure 4.
YycG co-precipitates with FtsZ in an immunoprecipitation assay. Exponentially growing KP444 cells in the presence of IPTG (OD525=0.3) were collected and suspended in media either supplemented with 1 mM IPTG or not. After 1 h, cultures were subjected to cross-linking and immunoprecipitation from cell extracts with purified anti-FtsZ antibodies as described in Material and Methods. FtsZ (left panel) and YycG (right panel) proteins were visualized immunologically by Western blot analysis using anti-YycG or anti-FtsZ antibody, respectively. Lanes 1 an 2 of each gel contain cross-linked sample immunoprecipitated with anti-FtsZ antibody from KP444 grown in the presence of 1 mM IPTG (lane 1) or in its absence (lane 2).
Immune-complexes initially precipitated with anti-FtsZ antibody were assayed for the presence of FtsZ and for co-precipitation of YycG. For KP444 grown in the presence of IPTG both, a clear FtsZ and a clear YycG band could be detected (Fig. 4) and similar results were obtained for the wild type strain JH642 (data not shown). When KP444 was grown in the absence of IPTG FtsZ was barely visible in the immune-complexes and YycG was not detectable. Additional controls with the antibody raised against the sporulation sensor kinase KinA and the DegS sensor kinase were not reactive on the cross-linked immunoprecipitation lanes (data not shown). The presence of YycG in an immune complex precipitated by anti-FtsZ antibody (Fig. 4) was consistent with the immunofluorescence results and suggested that these proteins either interact at the cell division septum or that both of them are part of a larger multiprotein complex present at that site.
YycG localization to the septum is independent of YycH and YycI
YycH and YycI are known to form a ternary complex with YycG (Szurmant et al., 2007b; Szurmant et al., 2008). To investigate a potential involvement of these auxiliary proteins in YycG localization, YycG positioning was probed in a yycHI double deletion strain JH25031 (Fig. S2). No difference in YycG localization was observed when compared to a wild type strain (data not shown) indicating YycG may localize to the division septum even in the absence of YycH and YycI.
Localization of the YycF response regulator to the nucleoid
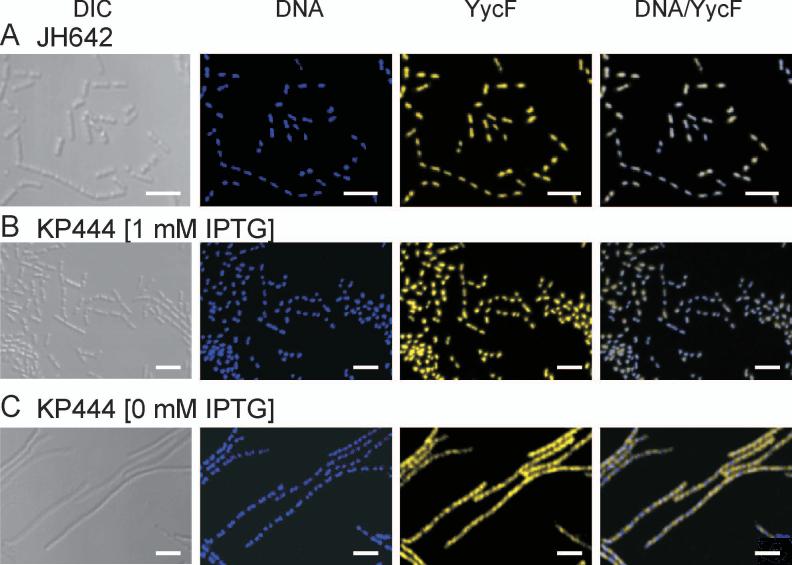
The YycF response regulator is the transcription factor regulated by the YycG sensor kinase. It was of interest to determine the cellular location of YycF, if possible, under the conditions described above. To this end the immunofluorescent technique was used. In the wild type JH642 cells, YycF specific antibody detected YycF associated with the nucleoid. No YycF was found at the septum or other cytoplasmic locations (Fig. 5A). The effect of FtsZ depletion on YycF was determined with the conditional ftsZ expression strain KP444 used previously. In the presence of IPTG the localization of YycF was identical to that observed in JH642 (Fig. 5B). In FtsZ depletion conditions the cells formed filaments as before and YycF was found located throughout the filament and clearly in positions not occupied by the nucleoid (Fig. 5C). Thus the assembly of the Z-Ring by FtsZ and the subsequent septum are required for the observed localization of YycF to the nucleoid. Since binding of YycF to its chromosomal targets is likely to require phosphorylation and FtsZ depletion does not affect either YycF or YycG cellular levels, the results suggest that YycG is inactive as YycF kinase under these conditions.
Figure 5.
YycF localizes to the nucleoid in an FtsZ dependent manner. (A) YycF (yellow) was visualized immunologically in exponentially growing cells of wild type strain JH642. DNA (blue) was visualized by DAPI staining. The two figures were overlaid in DNA/YycF. Similarly DNA and YycF were visualized in the conditional FtsZ mutant strain KP444 grown for three hours at 37°C either in (B) the presence of 1 mM IPTG or (C) in the absence of IPTG to induce FtsZ depletion. DIC is differential interference microscopy.
A similar assay was performed utilizing the conditional yycG expression strain JH25033. Unexpectedly, depletion of YycG upon IPTG removal had no obvious effect on YycF localization (data not shown). As mentioned above as well as by others (Fukuchi et al., 2000) low levels of YycG supplied by the leaky spac promoter are sufficient to sustain growth and it appears these levels are also sufficient to maintain YycF nucleoid localization. YycF localization was also tested in the yycHI deletion strain and did not change in respect to the wild type strain (Supplemental Figure S2).
YycG activation requires a division septum
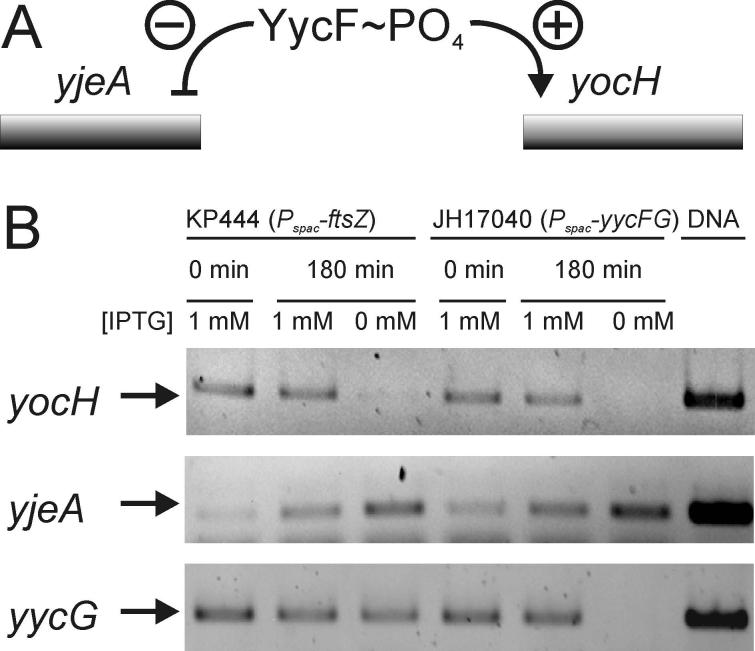
The YycF localization data suggest that YycG is most active as a kinase of YycF when positioned at the division septum. The instability of the aspartyl-phosphate of YycF precludes direct measurement of phosphorylation levels of the response regulator but its transcriptional activity may be determined. To indirectly assess the phosphorylation of YycF, two genes were chosen that require YycF∼PO4 for regulation, yocH and yjeA (Bisicchia et al., 2007). YycF∼PO4 is a positive activator of yocH transcription and a repressor of yjeA transcription. Thus, the messenger RNA levels for these genes should change in opposite directions as a function of the phosphorylation of YycF. As a control, to determine that this indeed happens, RT-PCR measurements of the transcript level of both genes was assayed in a strain, JH17040, in which both YycF and YycG levels were controlled by IPTG (Fig. 6, lanes 4,5 and 6). Shutting off the transcription of these genes by IPTG depletion should result in lowered yocH transcription and increased yjeA transcription. The anticipated result was obtained; both yocH and yycG transcripts were not detectable and the amount of yjeA transcript increased (Fig. 6). The same experiment was carried out in strain KP444 in which ftsZ transcription was regulated by IPTG. Depletion of IPTG and FtsZ in conditions known to prevent division septum formation resulted in identical results for yocH and yjeA transcripts as observed in the control strain (Fig. 6, lanes 1, 2 and 3). The yycG transcript was still produced in these conditions showing that FtsZ depletion did not affect transcription in general. Since the levels of YycG and YycF did not fluctuate under FtsZ depletion and the relevant transcripts mimicked the lowering of YycF∼PO4 levels, it appears that YycG is less active as a kinase of YycF in the absence of a division septum.
Figure 6.
FtsZ depletion mimics YycF∼PO4 depletion in the regulation of the YycFG regulon. (A) A schematic showing regulation of two genes that are under negative (yjeA) or positive (yocH) transcription control by YycF∼PO4 (B) mRNA levels of yocH, yjeA and yycG were determined in an RT-PCR approach. RNA was extracted from strain KP444 grown in the presence of 1 mM IPTG or in its absence to induce FtsZ-depletion, as described in Materials and Methods. Similarly, RNA was extracted from strain JH17040 grown with or without IPTG to observe changes in mRNA levels of the respective genes in response to yycFG depletion. RNA was transcribed into cDNA and amplified by PCR. As a control the respective genes were also amplified directly from genomic DNA (right lane).
Discussion
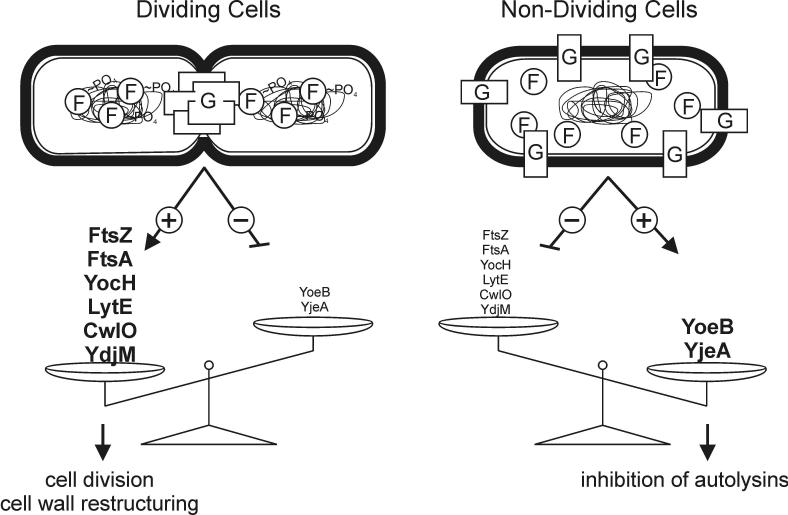
A large number of cellular processes need to be precisely coordinated for a bacterium to repetitively grow and divide as rapidly as it does. A formidable challenge to this process is the need to coordinate internal events, DNA synthesis, septum formation, etc., with extracellular processes, the degradation and resynthesis of the cell wall at the division septum to insure cell separation while maintaining the structural barrier to lysis. The results reported here and elsewhere implicate the YycFG two-component system as a component in this multifactorial signaling process (Bisicchia et al., 2007; Fukuchi et al., 2000). Two-component systems generally control gene expression and it is enlightening to examine the genes that YycF regulates (Bisicchia et al., 2007). YycF∼PO4 is a positive activator of the expression of genes for autolytic enzymes that cleave at several specific sites in the cell wall peptidoglycan matrix. Presumably these enzymes prepare the cell wall for restructuring and cell separation coincident with the synthesis of the division septum. YycF∼PO4 is also a repressor of genes for proteins that inhibit autolytic enzymes, yoeB, and that deacetylate peptidoglycan, yjeA, to inhibit the activity of autolysins. Rapidly growing cells depend on autolytic enzymes for cell division whereas these enzymes require inhibition in slow growth or non-growing conditions (Bisicchia et al., 2007). The ratio of YycF∼PO4/YycF, determines the balance of synthesis of autolysins and their inhibitors. This ratio must be high in rapidly growing cells, to favor autolysin synthesis, and low in non-growing cells to favor synthesis of autolysin inhibitors. Thus, YycF may be thought of as a homeostatic regulator tuned to the division state of cells.
Since the phosphorylation state of YycF depends on the activity of the YycG sensor kinase, YycG kinase becomes an arbitrator of the state of the cell by responding to cellular signals. These signals are most likely recognized by the YycG periplasmic domain, its internal PAS and HAMP domains, and the YycH and YycI proteins with which it associates in a trans-membrane complex (Szurmant et al., 2008). None of these signals are known and both agonists and antagonists probably exist. The experiments reported here show that YycG is located in the cell at the division septum in a position where it is positioned to interpret the state of septum growth and cell division. Moreover, the RT-PCR experiments indicate that location at the division septum is a requirement for YycG phosphorylation of YycF. There are a number of mechanistic and signaling interpretations for this result that could be imagined. The simple, uncomplicated explanation is that YycG is most active as a kinase of YycF when associated with the division septum (Fig. 7).
Figure 7.
Model for a role of YycFG in coordinating cell division with cell wall homeostasis. The present data is consistent with a model in which YycG is activated upon localization to the septum in dividing cells. This activation increases the YycF∼PO4 levels and subsequently leads to enhanced expression of genes involved in cell wall remodeling (yocH, lytE, cwlO and ydjM) and cell division (ftsA and ftsZ) and repression of genes involved in inhibiting cell wall remodeling (yoeB and yjeA). In the absence of septa in non-dividing cells YycG fails to localize, adopts a state of low activity and hence the cellular YycF pool remains unphosphorylated. Under these conditions cell wall remodeling is not required and the expression balance is tipped towards the genes involved in autolysin inhibitory processes. The precise role of the involved genes of the YycF regulon, where known, has been addressed in (Bisicchia et al., 2007).
The question of what drives the localization of YycG to the division septum remains unanswered. Data mentioned here indicate that YycG is still septum localized in mutants lacking both YycH and YycI indicating some domain of YycG must be involved. Equally elusive is the question of what YycG contacts when located at the septum.
Immunoprecipitation experiments showed that FtsZ antibody could coprecipitate YycG from extracts of cells in which the proteins were cross-linked, in vivo, with formaldehyde. Close association between proteins (∼2Å) is needed for cross-linking with this agent, yet substantially more experimentation would be needed to distinguish between a direct interaction of YycG and FtsZ and indirect association with other proteins in common between the two. In any case, YycG joins an ever-growing number of proteins that are being found to localize to the septum in dividing cells since the initial discovery of the Z-ring and subsequent establishment of localization of essential cell divisional proteins to this cellular compartment (reviewed in (Dajkovic and Lutkenhaus, 2006; Goehring and Beckwith, 2005; Margolin, 2005)). In both E. coli and B. subtilis, the divisome assembles in a sequential manner and localization of later proteins to the complex depends on earlier ones (reviewed in (Errington et al., 2003)). Association with the divisome could indicate the origin of signals for YycG activity if it were embedded within the divisome complex.
If the YycG kinase were an integral part of the divisome, required for its assembly, one would anticipate the protein’s essentiality to be at least partially independent of its kinase activity. However Ogasawara and colleagues reported successful disruption of the YycG kinase in a B. subtilis strain over-expressing an assumed constitutively active YycF with a point mutation (D56H) (Fukuchi et al., 2000). The strain was not further analyzed for phenotypes but the evidence suggested that the YycG protein was not essential for divisome assembly or its integrity.
The YycH and YycI auxiliary proteins are known to regulate kinase activity and assemble into a complex with the kinase, independent of FtsZ levels, suggesting that formation of this complex precedes participation in the divisome (unpublished data). Furthermore, neither YycH nor YycI are required for YycG localization to the septa as mentioned above. One can imagine two different mechanisms for YycG activation: one that involves localization of the entire YycGHI sensor complex to the septum and subsequent signal recognition; another in which YycG becomes activated by being physically separated from its regulatory proteins YycH and YycI during cell septation. The former model requires that YycH and YycI co-localize with YycG at the septum and the latter that they do not. This is currently under investigation.
A mechanism by which spatial localization plays a role in the activity of a signal transduction system has been well established for Caulobacter crescentus. Here a complex circuit of proteins is known to regulate the phosphorylation levels and proteolysis of the essential response regulator (RR) CtrA (Biondi et al., 2006; McGrath et al., 2006). Differential localization of the DivJ kinase and PleC phosphatase to opposite cell poles plays a role in asymmetric cell division (Wheeler and Shapiro, 1999). Both proteins act on the intermediary RR DivK, which in turn inhibits phosphorylation of CtrA indirectly via the kinase CckA (Biondi et al., 2006). Indeed there are strong similarities between the B. subtilis YycFG system and the system controlling asymmetric development in C. crescentus. Aside from the fact that they are both essential systems they also regulate expression of operons encoding essential cell divisional proteins, i.e. ftsAZ in B. subtilis and ftsQA in C. crescentus (Fukuchi et al., 2000; Wortinger et al., 2000). Not surprisingly phenotypes for depletion strains include elongated cells in both organisms. A model involving interaction of FtsQA with the DivK RR has recently been proposed (Biondi et al., 2006). Therefore a number of protein interactions and auxiliary proteins render both pathways complicated control circuits of essential processes.
Recently, the MtrAB two-component system that is conserved in the high G+C Gram-positive actinomycetes was shown to be among the group of essential two-component systems (reviewed in (Hoskisson and Hutchings, 2006)). This system appears to serve a role in these organisms analogous to that of YycFG system in the low G+C Gram positive bacteria. Phenotypes observed upon depletion or deletion of this system include elongated cells for Corynebacterium glutamicum (where the system is not essential) and Mycobacterium avium (where only the RR is essential) and an increased sensitivity to antibiotics targeting cell wall synthesis (Cangelosi et al., 2006; Moker et al., 2004). In Mycobacterium tuberculosis (where MtrA and MtrB are both essential) the essential DNA replication gene dnaA appears to be under control of the MtrAB system, and links between mtrAB expression and FtsZ polymerization have also been reported (Fol et al., 2006; Slayden et al., 2006). Furthermore, the conserved lipoprotein LpqB, associated with MtrAB, was proposed to regulate the activity of the MtrB kinase. It is conceivable that this could serve a function similar to that of YycH and YycI in B. subtilis (Hoskisson and Hutchings, 2006).
It now appears that two-component signal transduction systems play a key role in the coordination of cell architecture and cell division in a wide variety of bacteria. Certainly this represents only a glimpse into the complexity of these controls and higher order regulatory processes will unquestionably be revealed.
Experimental Procedures
Culture media and growth conditions
All Escherichia coli strains were grown in Luria-Bertani (LB) broth and all B. subtilis strains were grown in Schaeffer’s Sporulation Medium (SM), in the presence of appropriate antibiotics whenever necessary. Antibiotic concentrations were 100μg/ml ampicillin or 30μg/ml kanamycin for E. coli and 2μg/ml of kanamycin, 5μg/ml of chloramphenicol or 1μg/ml of phleomycin for B. subtilis.
Construction of strains and plasmids
E. coli strains used were DH5α and JM109 for cloning and propagation purposes. All B. subtilis strains were derivatives of JH642 except KP444. Plasmids and strains are listed in table 1.
Table 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevant genotype or description | Source or reference |
|---|---|---|
| Plasmids | ||
| pHY300PLK | E. coli-B. subtilis shuttle vector, Ampr, Tetr | Takara (Japan) |
| pBluescript-KS(+) | cloning vector, Ampr | Stratagene |
| pET28a | his-tag expression vector, Kanr | Navagen |
| pMAL-c2E | MBP-tag expression vector, Ampr | New England Biolabs |
| pJM115 | amyE integration vector for lacZ fusions, Ampr, Kanr | M. Perego, unpublished |
| pJM117 | pMUTIN4 derivative, Ampr, Cmr | M. Perego, unpublished |
| pJM119 | amyE integration vector, Pspac, lacI, Ampr, Kanr | M. Perego, unpublished |
| pJMS1 | pJM117-‘yycF-cat-Pspac | This work |
| pJMS3 | pJM117-‘yycF-cat-Pspac-yycH’ | This work |
| pJMS6 | pJM119-Pspac-yycG, lacI Ampr, Kanr | This work |
| pJT01 | pBluescript-KS(+)-‘yycF-cat-Pspac-yycH’ | This work |
| pJT02 | pHY300PLK-PyycF | This work |
| pJT03 | pBluescript-KS(+)-‘yycF-cat-PyycF-yycH’ | This work |
| pJT04 | YycF-6xhis expression vector | This work |
| pJT05 | 6xhis-YycG expression vector | This work |
| pJT06 | MBP-FtsZ expression vector | This work |
| Strains | ||
| JH642 | pheA1 trpC2 | laboratory stock |
| JH17040 | Pspac-yycFG | (Fabret and Hoch, 1998) |
| KP444 | ftsZ::phleo Pspac-ftsZ | (Beall and Lutkenhaus, 1991) |
| JH25031 | ΔyycHI101 amyE::(PyocH-lacZ aph3A) | (Szurmantet al., 2007b) |
| JH25032 | amyE::(Pspac-yycG-lacZ aph3A lacI) | This work |
| JH25033 | amyE::(Pspac-yycG-lacZ aph3A lacI) yycG::cat PyycF-yycHIJK | This work |
To create an overexpression vector for a C-terminal 6xhis-tagged YycF protein, the yycF gene was PCR amplified with oligonucleotides introducing 5′NcoI and 3′XhoI sites. The resulting fragment was cloned in the respective sites of vector pET28a (Novagen) creating plasmid pJT04. Similarly, the gene fragment coding for the cytoplasmic domain of YycG (codons 213-stop codon) was PCR amplified introducing 5′ NdeI and 3′ BamHI sites and cloned in the same sites of vector pET28a. The resulting plasmid pJT05 expresses an N-terminal 6xhis-tagged cytoplasmic YycG fragment.
To create a maltose binding protein tagged FtsZ overexpression vector, the ftsZ gene was PCR amplified introducing 5′ KpnI and 3′ XbaI sites and the resulting product was cloned in the same sites of vector pMAL-c2E (New England Biolabs), resulting in plasmid pJT6.
Construction of a strain harboring an IPTG inducible yycG gene
The yycG coding sequence and ribosome binding site (B. subtilis 168 coordinates 4150876 to 4152743) was PCR amplified introducing 5′ HindIII and 3′ BamHI sites. The resulting fragment was cloned in the respective sites of the amyE integration vector pJM119 resulting in plasmid pJMS6. B. subtilis strain JH642 was transformed with ScaI linearized vector pJMS6 resulting in double-cross over integration into the amyE locus and producing strain JH25032. This strain harbors an additional copy of yycG under control of the isopropyl-β-d-thiogalactopyranoside (IPTG) inducible spac promoter.
To delete the wild type copy of yycG from strain JH25032, plasmid pJT03 was constructed in a multi-step process. First, the 3′ portion of the yycF gene (coordinates 4152711 to 4153154) was amplified by PCR and blunt-end ligated to ScaI-digested vector pJM117 resulting in pJMS1. Second, the 5′ portion of the yycH gene (coordinates 4150499 to 4150920) was PCR amplified introducing HindIII sites and ligated into HindIII digested pJMS1, producing plasmid pJMS3. Vector pJMS3 was partially digested with EcoRI and PvuII and the 3.2 kbp fragment was subcloned into EcoRI-NaeI-digested pBluescript KS(+) resulting in pJT01.
Next, the promoter region of yycF (coordinates 4153014 to 4153936) was PCR amplified introducing 5′ and 3′ BamHI sites. This 0.9 kbp fragment was digested with BamHI and MboI and the resulting 0.5 kbp fragment was cloned into BglII digested vector pHY300PLK, producing vector pJT02. Lastly, the XbaI-SmaI fragment from pJT02 was subcloned into XbaI-AfeI-digested pJT01 thus replacing the Pspac promoter with the yycF promoter, resulting in pJT03. The BglI linearized pJT03 was used for double-crossover integration in strain JH25032 at the yycG locus, resulting in strain JH25033. This strain features a single IPTG inducible copy of yycG (supplemental Figure S1B).
Production and purification of YycF, YycG and FtsZ
The YycF-6xHis protein was over-expressed in LB medium by IPTG induction of the E. coli BL21(DE3) strain containing pJT04. Cells were broken with a French press, the cell lysate was centrifuged, and the supernatant was applied to a nickel-NTA agarose column (Qiagen). After YycF-6xHis was eluted with a step gradient of imidazole, the protein was dialyzed against YycF buffer (50mM Tris-HCl pH 7.6, 20mM KCl, 5mM β-mercaptoethanol). The solution was applied to Q-sepharose column (Amersham Bioscience), and the protein was eluted with YycF buffer in a gradient of KCl (20 to 500 mM). Finally, the protein was dialyzed against borate buffer (20mM borate, 20mM NaCl, 5 mM β-mercaptoethanol, pH 9.0) to prepare the protein for affinity column purification.
The cytoplasmic region of YycG fused to a histidine-tag (6xHis-YycGC) was induced by IPTG in LB medium from E. coli BL21(DE3) harboring pJT05. The protein was purified with a nickel-NTA agarose column as described for the purification of YycF-6xHis. The eluted 6xHis-YycGC was dialyzed against borate buffer (20mM borate, 50mM NaCl, pH 9.0) to prepare the protein for affinity column purification.
The entire FtsZ protein fused to MalE (MBP-FtsZ) and the maltose binding protein (MBP alone) were over-expressed by IPTG induction from E. coli JM109 containing pJT06 and pMAL-c2E, respectively. Cells harvested by centrifugation were resuspended in TS buffer (20mM Tris-HCl pH 7.4, 0.2M NaCl) with phenylmethylsulfonyl fluoride (PMSF) and were broken with lysozyme and sonication. The cell lysate was centrifuged and the supernatant was applied to an amylose resin column (New England BioLabs). The MBP-FtsZ and MBP proteins were eluted with TS buffer containing 10mM maltose, and then both eluates were dialyzed against borate buffer (20mM borate, 50mM NaCl, pH 9.0).
Affinity purification of anti-YycF, anti-YycG and anti-FtsZ antibodies
For the preparation of the affinity columns for purification of anti-YycF, anti-YycG and anti-FtsZ antibodies, purified YycF-6xHis (4.7mg), 6xHis-YycGC (3.8mg), MBP-FtsZ (0.8mg) and MBP (3.6mg) were cross-linked with 1 ml of Reacti-Gel (6x) Support (PIERCE) according to the manufacturer’s instructions.
Antibodies raised against YycF or YycG were affinity purified from rabbit antiserum as previously described utilizing the appropriate affinity columns (Szurmant et al., 2007a). Similarly, FtsZ antibody was affinity purified from sheep antiserum with the following modification. The eluate from the MBP-FtsZ column, concentrated and dialyzed against PBS was applied to the affinity column cross-linked with MBP in order to purify the IgG reacting with only FtsZ protein. The unbound IgG (anti-FtsZ antibodies) was collected as flow through.
Immunofluorescence microscopy
Wild-type (JH642) and conditional yycG and ftsZ mutant strains were incubated at 37°C in SM with or without 1 mM IPTG, and samples were taken at desired time points. YycG, YycF and FtsZ localization in fixed cells was detected immunologically as previously described (Szurmant et al., 2007a). Primary purified antibodies were used at 1:20 (YycG), 1:50 (YycF) or 1:100 (FtsZ) dilutions. Secondary antibodies were Alexa Fluor goat anti-rabbit IgG or Alexa Fluor 546 donkey anti-sheep IgG (Molecular Probes) used at dilutions of 1:500 and 1:1000, respectively. The samples were observed and photographed with a Bio-Rad Radianece 2100 Rainbow laser scanning confocal microscope (LSCM) (running under Bio-Rad LaserSharp 2000 software) equipped with blue diode/argon/green HeNe/red diode laser and a Nikon TE2000-U microscope (X60 PlanApo oil-immersion objective; numerical aperture 1.4). A series of optical sections collected at 100 nm intervals in the z-axis was used to create the images. All pictures were processed with Adobe Photoshop software.
Immunoprecipitation experiments
B. subtilis cells for immunoprecipitation were prepared as described (Molle et al., 2003). Briefly, cells were grown in 100 ml SM at 37°C until A600 nm = 0.3. Cells were fixed with formaldehyde solution (1.33% final concentration) in 32 mM sodium phosphate buffer at pH 7.4 for 20-30 min. Following washing with phosphate buffered saline (PBS) cells were suspended in IP buffer (PBS containing 10mg/ml of lysozyme, 0.1M EDTA, 1% Triton X-100 and protease inhibitor cocktail [SIGMA]), the suspension was kept at 37°C for 1 h before a passage through a French press. Cell debris was removed by centrifugation and the cell extract was completely cleared by gentle shaking for 30 min followed by an additional centrifugation step.
1 ml of cross-linked cellular extract was subjected to immunoprecipitation by addition of purified antibody raised against FtsZ (1:150), or YycG (1:500) After 1 h incubation at room temperature (FtsZ) or overnight incubation at 4°C (YycG, YycH and YycI) the mixture was cleared by centrifugation, the supernatant was mixed with 40-60 μl of recombinant Protein G agarose (Invitrogen) and gently shaken for 1 h (FtsZ) or 5 h (YycG, YycH and YycI) at room temperature. The resin was washed with PBS containing 1 % Triton X-100 and the immunocomplexes were eluted either with 0.2 M glycine pH 2.2 or by boiling with SDS sample buffer. Glycine eluted samples were neutralized by addition of 1M Tris-HCl (pH 8.0). Samples were used in immunoblot analysis in amounts corresponding to cell density A600 nm = 0.1
Immunoblot analysis
YycG and FtsZ were visualized immunologically as described (Szurmant et al., 2007a). Purified antibodies were used at 1:10,000 dilutions for rabbit anti-YycG antibody and at 1:30,000 dilutions for sheep anti-FtsZ antibody. Secondary horseradish peroxidase-labeled antibodies were used at 1:10,000 dilutions.
RT-PCR
Cells were grown to an OD525nm of 0.3 in the presence of 1 mM IPTG, collected by centrifugation, washed and incubated in fresh media with or without 1 mM IPTG and incubated for an additional 3 h. 15-20 OD525nm units of cells were collected, total RNA isolated, treated with DNAase I and quantified as described except that the cells were lysed with 6 mg/ml lysozyme (Farrell Jr., 2005). RNA (1 μg) was used as template to produce cDNA utilizing SuperScript III Reverse Transcriptase (Invitrogen), following manufactures recommendation in a total volume of 20 μl. 5 μl of the resulting cDNA was used in a standard PCR reaction, and resulting DNA was visualized by agarose gel electrophoresis and ethidium bromide staining. To assure absence of DNA contamination the same procedure was also performed for each pair of oligonucleotides and mRNA preparation in the absence of reverse transcriptase and in all experiments, no PCR product was detected (Supplemental Fig. S3).
Supplementary Material
Acknowledgements
This research was supported in part by grants GM019416 and GM055594 from the National Institute of General Medical Sciences and grant AI055860 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. Oligonucleotide synthesis and DNA sequencing were supported in part by the Stein Beneficial Trust.
We thank William B. Kiosses for help with the confocal microscope, Dr. Dong-Er Zhang for providing access to the fluorescent microscope, Dr. Elizabeth J. Harry for providing anti-FtsZ antibodies, Dr. Cristina Bongiorni for suggestions on purification of YycF and YycG, and Sara Ocon for the construction of the pJMS plasmids.
This is manuscript number 18658 from The Scripps Research Institute.
References
- Baker MD, Wolanin PM, Stock JB. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991;5:447–455. doi: 10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, Devine KM. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol. 2007;65:180–200. doi: 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- Cangelosi GA, Do JS, Freeman R, Bennett JG, Semret M, Behr MA. The two-component regulatory system mtrAB is required for morphotypic multidrug resistance in Mycobacterium avium. Antimicrob Agents Chemother. 2006;50:461–468. doi: 10.1128/AAC.50.2.461-468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajkovic A, Lutkenhaus J. Z ring as executor of bacterial cell division. J Mol Microbiol Biotechnol. 2006;11:140–151. doi: 10.1159/000094050. [DOI] [PubMed] [Google Scholar]
- Dubrac S, Boneca IG, Poupel O, Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189:8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. table of contents.
- Fabret C, Hoch JA. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabret C, Feher VA, Hoch JA. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell RE., Jr. RNA Methodologies, A Laboratory Guide for Isolation and Characterization. Elsevier Academic Press; Burlington: 2005. [Google Scholar]
- Fol M, Chauhan A, Nair NK, Maloney E, Moomey M, Jagannath C, Madiraju MV, Rajagopalan M. Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol Microbiol. 2006;60:643–657. doi: 10.1111/j.1365-2958.2006.05137.x. [DOI] [PubMed] [Google Scholar]
- Fukuchi K, Kasahara Y, Asai K, Kobayashi K, Moriya S, Ogasawara N. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology. 2000;146(Pt 7):1573–1583. doi: 10.1099/00221287-146-7-1573. [DOI] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Hancock LE, Perego M. Systematic inactivation and phenotypic characterization of two-component signal transduction systems of Enterococcus faecalis V583. J Bacteriol. 2004;186:7951–7958. doi: 10.1128/JB.186.23.7951-7958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzendorff J, Reinhardt J, Viollier PH. Cell cycle control by oscillating regulatory proteins in Caulobacter crescentus. Bioessays. 2006;28:355–361. doi: 10.1002/bies.20384. [DOI] [PubMed] [Google Scholar]
- Hoskisson PA, Hutchings MI. MtrAB-LpqB: a conserved three-component system in actinobacteria? Trends Microbiol. 2006;14:444–449. doi: 10.1016/j.tim.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Howell A, Dubrac S, Andersen KK, Noone D, Fert J, Msadek T, Devine K. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol Microbiol. 2003;49:1639–1655. doi: 10.1046/j.1365-2958.2003.03661.x. [DOI] [PubMed] [Google Scholar]
- Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PK, Li T, Sun D, Biek DP, Schmid MB. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J Bacteriol. 1999;181:3666–3673. doi: 10.1128/jb.181.12.3666-3673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Mohedano ML, Overweg K, de la Fuente A, Reuter M, Altabe S, Mulholland F, de Mendoza D, Lopez P, Wells JM. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J Bacteriol. 2005;187:2357–2367. doi: 10.1128/JB.187.7.2357-2367.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moker N, Brocker M, Schaffer S, Kramer R, Morbach S, Bott M. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol Microbiol. 2004;54:420–438. doi: 10.1111/j.1365-2958.2004.04249.x. [DOI] [PubMed] [Google Scholar]
- Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol. 2003;185:1911–1922. doi: 10.1128/JB.185.6.1911-1922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Robertson GT, Kazmierczak KM, Zhao J, Gilmour R, Winkler ME. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol Microbiol. 2003;50:1647–1663. doi: 10.1046/j.1365-2958.2003.03806.x. [DOI] [PubMed] [Google Scholar]
- Ng WL, Winkler ME. Singular structures and operon organizations of essential two-component systems in species of Streptococcus. Microbiology. 2004;150:3096–3098. doi: 10.1099/mic.0.27550-0. [DOI] [PubMed] [Google Scholar]
- Ng WL, Tsui HC, Winkler ME. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J Bacteriol. 2005;187:7444–7459. doi: 10.1128/JB.187.21.7444-7459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Santelli E, Liddington RC, Mohan MA, Hoch JA, Szurmant H. The crystal structure of Bacillus subtilis YycI reveals a common fold for two members of an unusual class of sensor histidine kinase regulatory proteins. J Bacteriol. 2007;189:3290–3295. doi: 10.1128/JB.01937-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayden RA, Knudson DL, Belisle JT. Identification of cell cycle regulators in Mycobacterium tuberculosis by inhibition of septum formation and global transcriptional analysis. Microbiology. 2006;152:1789–1797. doi: 10.1099/mic.0.28762-0. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Nelson K, Kim EJ, Perego M, Hoch JA. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J Bacteriol. 2005;187:5419–5426. doi: 10.1128/JB.187.15.5419-5426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Zhao H, Mohan MA, Hoch JA, Varughese KI. The crystal structure of YycH involved in the regulation of the essential YycFG two-component system in Bacillus subtilis reveals a novel tertiary structure. Protein Sci. 2006;15:929–934. doi: 10.1110/ps.052064406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Fukushima T, Hoch JA. The Essential YycFG Two-Component System of Bacillus subtilis. Methods Enzymol. 2007a;422:396–417. doi: 10.1016/S0076-6879(06)22020-2. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Mohan MA, Imus PM, Hoch JA. YycH and YycI interact to regulate the essential YycFG two-component system in Bacillus subtilis. J Bacteriol. 2007b;189:3280–3289. doi: 10.1128/JB.01936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Bu L, Brooks CL, III, Hoch JA. An essential sensor histidine kinase controlled by transmembrane helix interactions with its auxilliary proteins. Proc Natl Acad Sci U S A. 2008;105:5891–5896. doi: 10.1073/pnas.0800247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lutkenhaus J. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol. 1993;9:435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- Wortinger M, Sackett MJ, Brun YV. CtrA mediates a DNA replication checkpoint that prevents cell division in Caulobacter crescentus. Embo J. 2000;19:4503–4512. doi: 10.1093/emboj/19.17.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.