Abstract
The Hspa1b gene is one of the first genes expressed after fertilization, with expression observed in the male pronucleus as early as the one-cell stage of embryogenesis. This expression can occur in the absence of stress and is initiated during the minor zygotic genome activation. There is a significant reduction in the number of embryos developing to the blastocyte stage when HSPA1B levels are depleted, which supports the importance of this protein for embryonic viability. However, the mechanism responsible for allowing expression of Hspa1b during the minor zygotic genome activation (ZGA) is unknown. In this report, we investigated the role of HSF1 and HSF2 in bookmarking Hspa1b during late spermatogenesis. Western blot results show that both HSF1 and HSF2 are present in epididymal spermatozoa, and immunofluorescence analysis revealed that some of the HSF1 and HSF2 proteins in these cells overlap the 4′,6′-diamidino-2-phenylindole-stained DNA region. Results from chromatin immunoprecipitation assays showed that HSF1, HSF2, and SP1 are bound to the Hspa1b promoter in epididymal spermatozoa. Furthermore, we observed an increase in HSF2 binding to the Hspa1b promoter in late spermatids versus early spermatids, suggesting a likely period during spermatogenesis when transcription factor binding could occur. These results support a model in which the binding of HSF1, HSF2, and SP1 to the promoter of Hspa1b would allow the rapid formation of a transcription-competent state during the minor ZGA, thereby allowing Hspa1b expression.
Keywords: bookmarking, embryogenesis, epididymis, gene regulation, HSF1, HSF2, Hsp70, Hspa1b, sperm, spermatogenesis, testis
Novel binding of HSF1 and HSF2 to the Hspa1b promoter in epididymal spermatozoa may promote expression in the one-cell embryo
INTRODUCTION
In the mammalian embryo, a transition takes place such that maternal control of development is shifted to the zygote in a process termed the zygotic genome activation (ZGA) (reviewed in [1]). This transition can be divided into a minor ZGA, where a small subset of genes including Hspa1b are expressed as early as the one-cell stage, and the major ZGA, which occurs during the two-cell stage and is characterized by a significant burst in both transcription and translation [2–7], with more stringent transcriptional regulation [8–11]. During the minor ZGA, transcription in the one-cell embryo appears to be relatively promiscuous and opportunistic [12, 13], with the majority of transcription occurring in the male pronucleus [14, 15].
The Hspa1b gene is one of the first genes expressed following fertilization, with expression taking place in the absence of stress as early as the one-cell stage of embryogenesis [16, 17]. The importance of Hspa1b during embryogenesis is demonstrated by immunodepletion experiments using HSPA1B antibodies [18]. Those studies demonstrated that reduced levels of HSPA1B lead to a significant reduction in embryos developing to the blastocyte stage. However, despite the importance of HSPA1B for embryonic viability, the mechanism responsible for allowing expression of the Hspa1b gene during the minor ZGA is not known.
In somatic cells, the promoters of a number of genes, including those of the Hspa1b and Myc genes, remain uncompacted and accessible during mitosis [19–23]. The lack of compaction of promoter regions in mitotic cells is referred to as “bookmarking” and is believed to function to permit genes that existed in a transcription-competent state prior to entry into mitosis to be maintained in a form that can be rapidly reassembled into the active state in G1. Recently we have found that in somatic cells the Hspa1b gene is bookmarked during mitosis by the binding of heat shock factor 2 (HSF2) to the heat shock element (HSE) of the Hspa1b promoter [24]. Bookmarking Hspa1b during mitosis allows the rapid expression of this cytoprotective gene in early G1 if the cell encounters stress. Relevant to our study, it has been reported that mice lacking HSF2 display increased embryonic lethality, indicating the importance of this factor for embryogenesis [25].
Heat shock factor 1 (HSF1) is a protein that also binds to the HSE of the Hspa1b promoter during cellular stress and induces expression of Hspa1b (reviewed in [26]). It has been reported that HSF2 interacts with HSF1 [27–29], suggesting the possibility that these two DNA-binding proteins could both be involved in mediating gene bookmarking and facilitating expression of Hspa1b. In addition, expression of Hspa1b during the earliest stages of embryogenesis is HSF1-dependent, although stress is not required [17, 30]. HSF1 is important for embryogenesis since mouse embryos in mothers lacking HSF1 are unable to develop beyond the zygotic stage and exhibit increased embryonic lethality [31–33].
Based on these reports, we hypothesized that HSF1 and HSF2 could be involved in expression of Hspa1b in the male pronucleus of the one-cell embryo. Here we show that HSF1, HSF2, and SP1 are bound to the Hspa1b promoter in mature spermatozoa, which is unusual since transcription has ceased [34–36], chromatin has been reorganized and highly compacted [37], and numerous basal transcription factors, transcriptional regulators, and architectural factors are displaced from chromatin by the point of step 10 spermatids [36]. Considering our previous finding that HSF2 can bookmark the Hspa1b gene in somatic cells, the results presented here suggest a mechanism by which Hspa1b could be expressed in the male pronucleus of the one-cell embryo.
MATERIALS AND METHODS
Animals
All CD-1 mice used in this study were adult males obtained from Harlan (Indianapolis, IN). Animals were maintained in the Division of Laboratory Animal Resources, and studies were performed according to approved Institutional Animal Care and Use Committee guidelines at the University of Kentucky (Lexington, KY) and Louisiana State University (Shreveport, LA).
Generation of Antibodies Against HSF1 and HSF2
Affinity-purified goat polyclonal antibodies to HSF1 and HSF2 were prepared by Bethyl Laboratories (Montgomery, TX). These antibodies were raised against the synthetic peptides TISLLTGSEPPKAKDPTVS and YLCELAPAPLDSDMPLLDS, which correspond to the C-terminal sequences of the mouse HSF1 and HSF2 polypeptides (which are identical to the C-terminal sequences of human HSF1 and HSF2).
Preparation of Sperm Nuclei and Spermatids
Mature sperm was obtained from the caudal epididymides of adult male CD-1 mice by repeatedly puncturing with a 20G needle. Sperm were then gently flushed from the caudal epididymides using PBS. Sperm were pelleted, quickly frozen in liquid nitrogen, and then resuspended in cold PBS to lyse any red blood cells present. Centrifugation of the Percoll (Sigma Chemical Co.) gradient was performed according to established protocols [38] with the following modifications. Sperm, washed three times in cold PBS then resuspended in 1 ml of PBS, were loaded on a discontinuous Percoll gradient created by layering 1.5 ml of 35%, 45%, and 75% Percoll in a 15-ml conical tube. Centrifugation was performed at 700 × g for 30 min. Purity was confirmed via bright field microscopy (Supplemental Fig. 1 available at www.biolreprod.org ). Sperm nuclei were prepared using cetyltrimethylammonium bromide (CTAB) according to published protocols [39] with minor modifications [40]. Immediately prior to the CTAB incubation, the sperm suspension was sonicated for 3–5 sec at 20% power to dissolve clumps and assist in tail removal. Following CTAB incubation, the sperm suspension was passed through siliconized glass wool to remove any remaining tail debris. Enriched populations of mouse early and late spermatids were obtained by centrifugal elutriation performed in the laboratory of Dr. Sidney Grimes (Louisiana State University) according to established protocols [41, 42]. Enriched cell types were cross-linked with 2% paraformaldehyde, quenched with 1× glycine, washed three times with cold 1× PBS, then frozen on dry ice and shipped to us for chromatin immunoprecipitation (ChIP) analysis.
Western Blots
Protein extracts were prepared from sperm nuclei using buffer C (20 mM Hepes [pH 7.9], 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, and 0.5 mM dithiothreitol) followed by brief sonication on ice. Protein extracts were separated on an 8% SDS polyacrylamide gel, and Western blots were performed using goat polyclonal antibodies against HSF1 or HSF2 (described above) according to our established protocols [43–45]. The secondary antibody, α-IgG-horseradish peroxidase, was obtained from an enhanced chemiluminescence kit (Amersham Life Science, Arlington Heights, IL). Samples for Western blots were prepared from at least two different groups of animals, with at least two animals used for each preparation.
Immunofluorescence
Immunofluorescence was performed with goat HSF1 and HSF2 antibodies as well as rabbit HSF1 and HSF2 antibodies according to established protocols in our laboratory [46]. Briefly, nuclei from mouse epididymal spermatozoa were isolated and purified as previously discussed. Purity was confirmed via bright field microscopy. Purified sperm nuclei resuspended in PBS were allowed to adhere to poly-l-lysine (50 μg/ml)-coated coverslips for 15 min, fixed with 4% paraformaldehyde, and permeabilized using 0.5% Triton-X 100/0.5% saponin. Nuclei were probed using goat polyclonal antibodies against HSF1 or HSF2 and rabbit polyclonal antibodies against HSF1 or HSF2. Blots were then incubated with AlexaFluor 488 (Invitrogen)-linked secondary antibodies to visualize HSF1 or HSF2. DNA was visualized using 4′,6′-diamidino-2-phenylindole (DAPI), and images were taken with a Nikon fluorescent microscope with a 100× oil immersion objective and a Nikon spotcam digital imaging camera. Isolation, purification, and immunofluorescence detection were performed twice with each antibody.
Chromatin Immunoprecipitations
All ChIPs were performed at least three times according to established protocols in our laboratory [24] with the following modifications. Precleared chromatin was incubated with 3 μg of goat polyclonal HSF1, goat polyclonal HSF2, or rabbit SP1 antibodies (Santa Cruz Biotechnology, a generous gift from Dr. Dan Noonan) or species-matched control IgG (Sigma) and rotated at 4°C for 16 h. DNA was purified using a QIAquick PCR Purification Kit (Qiagen Inc.) and eluted in 50 μl of 10 mM Tris (pH 8.5). Immunoprecipitated DNA and input samples obtained prior to immunoprecipitation were analyzed by quantitative real time PCR with a Stratagene Mx 4000 system using Brilliant SYBR Green QPCR master mix (Stratagene) and the following primers (shown 5′ to 3′) from Integrated DNA Technologies (Coralville, IA):
Hspa1b: (+) CCGCAACAGTGTCAATAGC, (−) CCTTGAGTAATCGGAGTTGTGG
Hbb-bl: (+) TTGCTCCTCACATTTGCTTCTG, (−) ACTTCATCGGAGTTCACCTTTC
Hist1h4b: (+) ACGAAGCCCGCCATC, (−) TTGGCGTGCTCCGTGTAGGT
Hist1hlt: (+) GCAGTGAGCAGATATGCAAGA, (−) CCAACAGTGATGGGGTAGTG
Samples were checked for specific amplification using dissociation curves analysis software. PCR products were also assayed on polyacrylamide gels with ethidium bromide staining to ensure they were of the expected size. The Ct values were normalized to input DNA (DNA before immunoprecipitation step) and IgG controls using the formula 2∧[(Ct IgG − Ct Input) − (Ct Ab − Ct Input)] (where Ab = HSF1, HSF2, SP1, or IgG). Data is represented as fold differences relative to IgG, which was set to 1. The data shown represent quantitative PCR results from at least three independent sperm purifications and ChIP assays. Error bars represent SEM. Statistical significance was determined using a two-tailed, unpaired t-test.
RESULTS
HSF1 and HSF2 Are Present in Mouse Sperm Nuclei
Based on our hypothesis that HSF2, and possibly HSF1, could function in bookmarking the Hspa1b gene in mature sperm, we assayed for the presence of these factors in mature spermatozoa. To test this we isolated caudal epididymal spermatozoa from adult CD-1 mice and purified the sperm by centrifugation using a Percoll gradient. To minimize any extranuclear protein contamination, the purified sperm were treated with the detergent CTAB in conjunction with a 3- to 5-sec sonication on ice to further disrupt the sperm membrane and facilitate tail removal. Cells were visualized by light microscopy to confirm the complete removal of tails and to confirm the purity of the samples (Supplemental Fig. 1). Protein extracts were prepared from sperm nuclei and assayed by Western blots using antibodies against HSF1 and HSF2 (Fig. 1). The results presented in Figure 1 are representative of the banding patterns observed from two independent mouse sperm protein isolations and Western blots and indicates that both HSF1 and HSF2 are present in mature spermatozoa. We observed two bands present in the HSF1 blot (top panel) migrating at approximately 80 kDa that likely represent the α- and β-splice variants of this protein and/or different phosphorylation states [47, 48]. The single band observed for HSF2 (bottom panel) migrated at approximately 70 kDa and is consistent with previous reports [48].
FIG. 1.
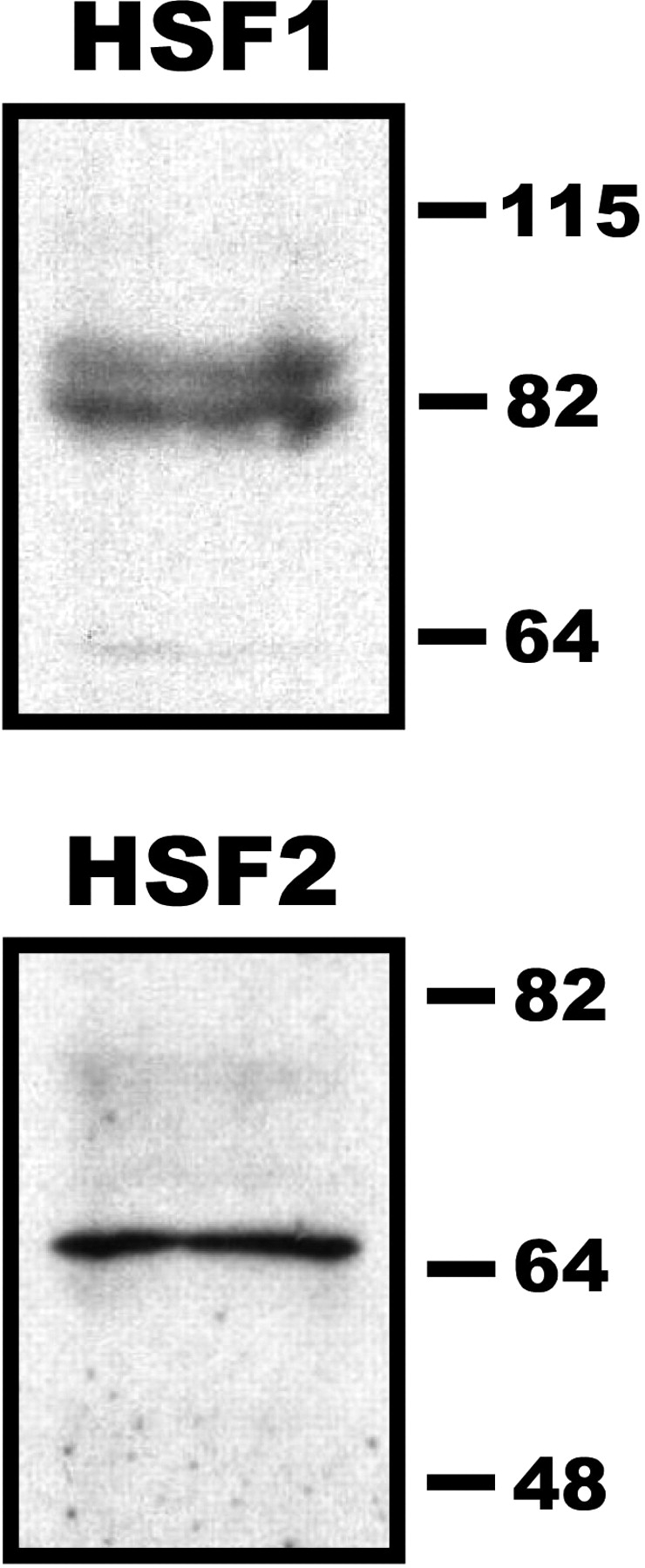
HSF1 and HSF2 are present in mouse sperm nuclei. Protein extracts prepared from mouse sperm nuclei were separated on an 8% SDS polyacrylamide gel and assayed by Western blot using goat polyclonal antibodies against HSF1 (top panel) or HSF2 (bottom panel). Prestained protein markers were used to indicate molecular mass (kDa). Images are representative of results obtained from extracts prepared from at least two different sets of animals (at least two animals per group).
Localization of HSF1 and HSF2 in Sperm Nuclei
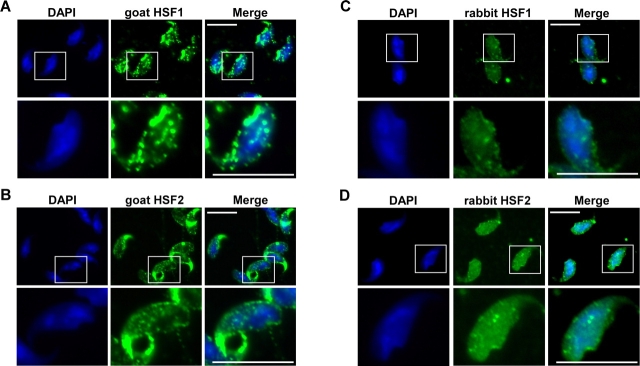
Based on our Western blot results we wanted to determine specifically where HSF1 and HSF2 were localized in mature spermatozoa and whether they exhibit any colocalization with DNA. Sperm nuclei were prepared as described for Western blots to minimize any extranuclear contamination. Nuclei were allowed to adhere to poly-l-lysine-coated coverslips for 15 min at room temperature. Immunofluorescence analysis was then performed using goat HSF1 or HSF2 polyclonal antibodies followed by incubation with AlexaFluor 488-linked secondary antibodies (Fig. 2, A and B, middle panels). DAPI staining was used to visualize DNA (left panels). In the merged images (right panels), HSF1 and HSF2 were observed within the DNA-containing region of mature spermatozoa. In addition to staining in the nuclear periphery, we consistently observed punctate staining of HSF1 and HSF2 overlapping the DNA staining. To further confirm the colocalization of HSF1 and HSF2 with DAPI-stained DNA, we repeated the sperm purification and immunofluorescence as described above but used different, previously characterized HSF1 and HSF2 rabbit polyclonal antibodies (Fig. 2, C and D, respectively). The results shown in Figure 2 are representative images from multiple sperm purifications and visualizations. The detection of HSF1 and HSF2 using goat polyclonal antibodies appeared to display slightly brighter punctate labeling compared to the rabbit HSF1 and HSF2 polyclonal antibodies, but the general labeling pattern of HSF1 and HSF2 is very similar.
FIG. 2.
Immunofluorescent localization of HSF1 and HSF2 in mouse sperm. Purified mouse sperm nuclei were allowed to adhere to poly-l-lysine-coated coverslips, fixed with paraformaldehyde, and permeabilized. Nuclei were probed using goat polyclonal antibodies against HSF1 (A, middle lane) or HSF2 (B, middle lane) and rabbit polyclonal antibodies against HSF1 (C, middle lane) or HSF2 (D, middle lane). Primary antibodies were detected with AlexaFluor 488-linked secondary antibodies to visualize HSF1 or HSF2. DNA was visualized using DAPI staining (left lanes), and images were taken with a Nikon fluorescent microscope with a 100× oil immersion objective and a Nikon spotcam digital imaging camera. White boxes highlight specific cells that are enlarged (below indicated image) to show further detail. Images shown are representative of the staining patterns observed from two independent experiments. Bars = 5 μm.
ChIP Analysis of HSF1 and HSF2 in Mature Sperm
Western blots and immunofluorescence confirmed that HSF2, a factor known to bookmark the Hspa1b gene, and HSF1, a factor required for the stress-induced expression of Hspa1b, are both present in mature spermatozoa, and each factor colocalizes with DNA. Based on these results, we hypothesized that one or both of these factors could be bound to the Hspa1b promoter in mature spermatozoa as part of a mechanism for allowing the preferential expression of Hspa1b in the male pronucleus of the one-cell embryo. To test this, we performed ChIP assays on caudal spermatozoa using polyclonal antibodies against HSF1, HSF2, and goat IgG as a negative control antibody (Fig. 3, A and B). We assayed the binding of HSF1 and HSF2 to the promoter of the Hspa1b gene as well as the promoters of the β-globin Hbb-bl and histone Hist1h4b genes (two negative control genes lacking recognizable HSE promoter elements). DNA fragments precipitated by the indicated antibodies were assayed by quantitative real-time PCR. The results indicate that binding of HSF1 and HSF2 to the Hspa1b promoter is statistically higher (P < 0.01) than to the β-globin and Hist1h4b gene promoters, indicating that these two HSFs are present on the Hspa1b promoter in the DNA of mature spermatozoa. The goat HSF1 and HSF2 antibodies specifically precipitate Hspa1b-containing DNA (black bars) compared with the goat IgG negative control, which displays a lack of specificity for Hspa1b-containing DNA (gray bars).
FIG. 3.
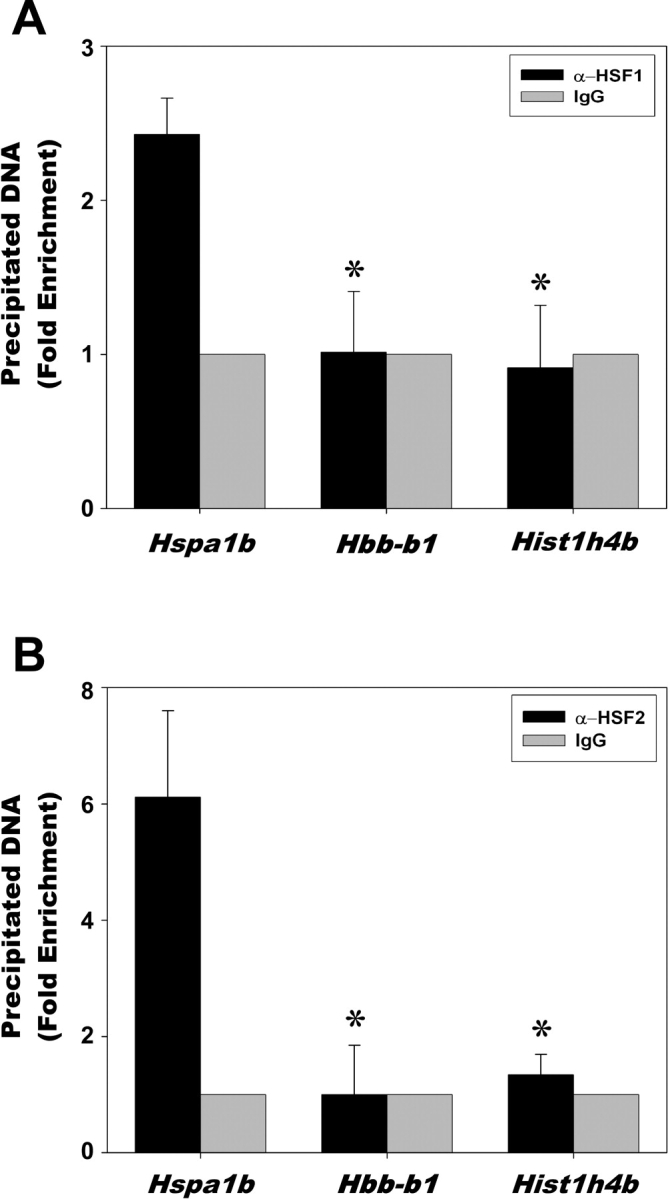
ChIP analysis of HSF1 and HSF2 binding in mature spermatozoa. Purified mouse caudal epididymal spermatozoa were crosslinked, lysed, sonicated, and subjected to immunoprecipitation using polyclonal antibodies against HSF1 (A) or HSF2 (B). Goat IgG was used as a negative control antibody and set to =1 (no binding). The precipitated DNA fragments were subjected to quantitative PCR using primers that amplified the proximal promoters of the Hspa1b, β-globin Hbb-b1, and histone Hist1h4b genes. Asterisk (*) indicates a statistically significant difference (P < 0.01) between the Hspa1b and the controls Hbb-b1 and Hist1h4b.
ChIP Analysis of SP1 in Mature Sperm
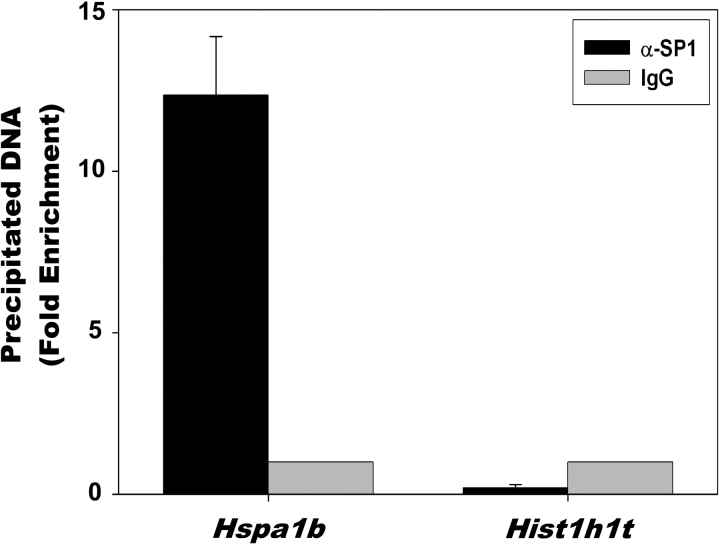
Based on previous results suggesting that the transcription factor SP1 is important for expression of Hspa1b in early-stage embryogenesis, we performed ChIP assays on caudal spermatozoa using rabbit polyclonal antibodies against SP1, and rabbit IgG as a negative control antibody (Fig. 4). We assayed the binding of SP1 to the promoter of the Hspa1b gene as well as the promoter of the testis-specific histone Hist1hlt gene using quantitative PCR. The Hist1hlt gene is expressed exclusively in mid- to late pachytene spermatocytes, with expression regulated in part by the binding of SP1 to the proximal promoter of Hist1hlt. We found that SP1 bound to the Hspa1b promoter approximately 12-fold more than to the H1fnt promoter. The specificity of the SP1 antibodies for precipitating Hspa1b-containing DNA (black bars) is demonstrated by the lack of Hspa1b-containing DNA precipitated by rabbit IgG negative control (gray bars). The binding of SP1 to the Hist1hlt promoter was much lower than that observed for IgG; however, this was not due to any detectable problem associated with the amplification, since input DNA was amplified equally well using primers that recognize the Hspa1b promoter and Hist1hlt promoter (Supplemental Fig. 2).
FIG. 4.
ChIP analysis of SP1 binding in mature spermatozoa. Purified spermatozoa from mouse caudal epididymides were cross-linked, lysed, sonicated, and subjected to immunoprecipitation using polyclonal antibodies against SP1. Rabbit IgG was used as a negative control antibody and set to =1 (no binding). The precipitated DNA fragments were subjected to quantitative PCR using primers that amplified the proximal promoters of the Hspa1b or Hist1h1t genes. There was a statistically significant difference (P < 0.001) between the results for Hspa1b and Hist1h1t.
ChIP Analysis of HSF2 in Spermatids
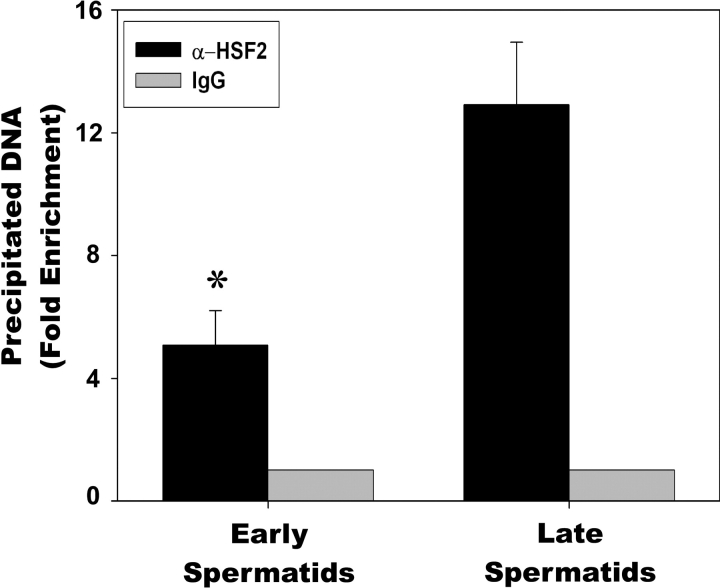
Once we determined that HSF1, HSF2, and SP1 could reside on the Hspa1b promoter in mature sperm, we reasoned that these factors were most likely not binding in sperm due to the high level of chromatin compaction. Therefore, we began investigating earlier spermatogenic cells, including early and late spermatids. Since HSF2 is the most likely factor contributing to the “open” chromatin conformation of Hspa1b in mature sperm, we performed ChIP assays in early and late spermatids using goat HSF2 antibodies (Fig. 5). From four independent ChIP assays, we consistently found that HSF2 bound to the Hspa1b promoter nearly 2.5-fold higher in late spermatids than early spermatids. The specificity of the HSF2 antibodies for precipitating Hspa1b-containing DNA (black bars) is demonstrated by the lack of Hspa1b-containing DNA precipitated by goat IgG-negative control (gray bars).
FIG. 5.
ChIP analysis of HSF2 binding in spermatids. Early spermatids and late spermatids from mouse testes were isolated by centrifugal elutriation, cross-linked, sonicated, and subjected to immunoprecipitation using goat polyclonal antibodies against HSF2. Goat IgG was used as a negative control antibody and set to =1 (no binding). Precipitated DNA fragments were subjected to quantitative PCR using primers that amplified the proximal promoter of Hspa1b. Asterisk (*) indicates a statistically significant difference (P < 0.01) between early spermatids and late spermatids.
DISCUSSION
There has been a significant amount of research attempting to elucidate the biological role(s) of HSF1 and HSF2. Some of this research has used mice with mutations in these genes to identify their function. There is good agreement in the literature regarding the importance of HSF1 and HSF2 for male and female fertility [25, 32, 49–51]. However, the role of these factors during embryogenesis is not as clear. One report has implicated Hsf1 mutations in increased prenatal lethality in mice [33]. Another report also found that mice lacking HSF1 displayed a number of phenotypes including prenatal lethality, the severity and lethality of which was found to be influenced by the genetic background of the Hsf1 mutant mice [50]. Mutations of Hsf have also been studied in Drosophila with embryonic lethality at the 1st or 2nd larval instar [52]. Although there are likely to be non-Hsp gene targets for HSF1 and HSF2 in early embryos, the embryonic defects associated with a lack of these factors could be attributed to a lack of regulation of the Hspa1b. The importance of HSF2 during embryogenesis has also been studied in mice. One report found that Hsf2 mutant mice were viable with normal life spans and behavioral functions, suggesting that Hsf2 was not essential for embryonic development [53]. However, other reports found that Hsf2 mutant mice displayed an increase in embryonic lethality [25, 32].
The differences observed for Hsf1 and Hsf2 mutant mice could be attributed to the genetic background of the mice, a phenomenon that has been observed in other studies [54, 55]. Another possible explanation is that these factors are able to compensate for each other, as HSF1 and HSF2 are highly homologous (72% identical in DNA-binding domain [56]) and both have been shown to bind to HSE promoter elements. Consistent with this hypothesis, previous work showed that mice lacking both HSF1 and HSF2 have more severe defects than mice lacking HSF1 or HSF2 individually [49]. Also, a recent report has demonstrated an interplay between HSF1 and HSF2 and a potential role for HSF2 in the HSF1-mediated induction of major heat shock genes [29].
Due to our interest in fully understanding the roles of HSF1 and HSF2 during spermatogenesis and early embryogenesis, we began investigating the possibility that HSF1 and HSF2 were functioning to bookmark the cytoprotective Hspa1b gene to provide a mechanism by which Hspa1b could be expressed in early embryos in the event of cellular stress. In this report we show that both HSF1 and HSF2 are present in mature spermatozoa and are bound to the Hspa1b promoter. We have also found that SP1 is bound to the Hspa1b promoter. These observations are novel since transcription has ceased and the chromatin has been reorganized and compacted to a level that is approximately 6-fold more compact than chromatin found in mitotic cells [37]. The reason for the transcriptional silence in mature sperm could be the removal of many transcription factors from the highly compacted DNA. In support of this hypothesis, a previous study showed that as transcription ceases, a number of basal transcription factors, transcriptional regulators, and architectural factors are displaced from chromatin beginning in step 7 spermatids, with no detectible binding observed by the point of step 10 spermatids [36]. The presence of HSF1 and HSF2 on the Hspa1b promoter provides an interesting mechanism by which the Hspa1b gene could be rapidly and preferentially expressed in the male pronucleus during early embryogenesis. We have previously shown that HSF2 can bookmark the Hspa1b gene in mitotic cells [24]. Furthermore, HSF1 is important for embryogenesis [33, 50] and is required for the expression of Hspa1b during early embryogenesis [17]. With the opportunistic and essentially unregulated transcription in the one-cell embryo [12, 13], the presence of HSF1 and HSF2 on the Hspa1b promoter would facilitate the rapid assembly of any necessary transcriptional machinery on the Hspa1b promoter versus other promoters lacking bound factors at this stage of embryogenesis. Another group using immunolocalization techniques reported that HSF2 was consistently pronuclear in one-cell embryos in the absence of stress [17], which is consistent with our findings. They also found that HSF1 was cytoplasmic in the absence of stress, but it is likely that the HSF1 bound to the Hspa1b promoter in the male pronucleus would be below the level of detection by immunolocalization.
Recently our laboratory has shown that HSF2 can bind to the promoters of Hspb2, Hsp90aa1, and Fos in mitotic cells [57]. Other genes, many of which are members of the heat shock superfamily, are also bound by HSF1 and HSF2 [58]. This suggests that HSF1 and HSF2 may have a larger role in the expression of other genes important for early-stage embryonic development.
For many years mature spermatozoa were considered inert cells with the single function of delivering paternal DNA to the ovum. However, as more studies of spermatozoa emerge, it appears that a number of important processes are occurring in spermatozoa. Bookmarking the Hspa1b gene in spermatozoa makes particular sense given the crucial need for molecular chaperones to handle the large number of proteins that are translated during embryogenesis and the various stresses that can occur, including osmotic and pH changes. The ability to rapidly express the HSPA1B protein would clearly promote embryonic viability.
Supplementary Material
Acknowledgments
We would like to thank Dr. Sidney Grimes for kindly providing centrifugally elutriated mouse germ cells and Dr. Dan Noonan for providing SP1 antibodies. We would also like to thank other members of our laboratory for insightful discussions during these studies.
Footnotes
1Supported by a Lalor Foundation postdoctoral grant and a National Institutes of Health (NIH) HD grant (F32HD050043) to D.C.W and NIH grants GM61053 and GM64606 to K.D.S.
REFERENCES
- Minami N, Suzuki T, Tsukamoto S.Zygotic gene activation and maternal factors in mammals. J Reprod Dev 2007; 53: 707–715. [DOI] [PubMed] [Google Scholar]
- Bellier S, Chastant S, Adenot P, Vincent M, Renard JP, Bensaude O.Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J 1997; 16: 6250–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN.The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J 1982; 1: 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett SK, Webb M, Maro B, Johnson MH.Meiosis II, mitosis I and the linking interphase: a study of the cytoskeleton in the fertilised mouse egg. Cytobios 1985; 43: 295–305. [PubMed] [Google Scholar]
- Latham KE, Garrels JI, Chang C, Solter D.Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one- and two-cell stages. Development 1991; 112: 921–932. [DOI] [PubMed] [Google Scholar]
- Nothias JY, Majumder S, Kaneko KJ, DePamphilis ML.Regulation of gene expression at the beginning of mammalian development. J Biol Chem 1995; 270: 22077–22080. [DOI] [PubMed] [Google Scholar]
- Taylor KD, Piko L.Patterns of mRNA prevalence and expression of B1 and B2 transcripts in early mouse embryos. Development 1987; 101: 877–892. [DOI] [PubMed] [Google Scholar]
- Majumder S, Miranda M, DePamphilis ML.Analysis of gene expression in mouse preimplantation embryos demonstrates that the primary role of enhancers is to relieve repression of promoters. EMBO J 1993; 12: 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas E, Linney E, Hassell J, DePamphilis ML.The need for enhancers in gene expression first appears during mouse development with formation of the zygotic nucleus. Genes Dev 1989; 3: 1493–1506. [DOI] [PubMed] [Google Scholar]
- Rothstein JL, Johnson D, DeLoia JA, Skowronski J, Solter D, Knowles B.Gene expression during preimplantation mouse development. Genes Dev 1992; 6: 1190–1201. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML.Regulation of gene expression in preimplantation mouse embryos: effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Dev Biol 1991; 147: 403–414. [DOI] [PubMed] [Google Scholar]
- Choo KB, Chen HH, Liu TY, Chang CP.Different modes of regulation of transcription and pre-mRNA processing of the structurally juxtaposed homologs, Rnf33 and Rnf35, in eggs and in pre-implantation embryos. Nucleic Acids Res 2002; 30: 4836–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Svoboda P, Schultz RM, Stein P.Regulation of zygotic gene activation in the preimplantation mouse embryo: global activation and repression of gene expression. Biol Reprod 2001; 64: 1713–1721. [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM.Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181: 296–307. [DOI] [PubMed] [Google Scholar]
- Beaujean N, Bouniol-Baly C, Monod C, Kissa K, Jullien D, Aulner N, Amirand C, Debey P, Kas E.Induction of early transcription in one-cell mouse embryos by microinjection of the nonhistone chromosomal protein HMG-I. Dev Biol 2000; 221: 337–354. [DOI] [PubMed] [Google Scholar]
- Christians E, Campion E, Thompson EM, Renard JP.Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription. Development 1995; 121: 113–122. [DOI] [PubMed] [Google Scholar]
- Fiorenza MT, Bevilacqua A, Canterini S, Torcia S, Pontecorvi M, Mangia F.Early transcriptional activation of the hsp70.1 gene by osmotic stress in one-cell embryos of the mouse. Biol Reprod 2004; 70: 1606–1613. [DOI] [PubMed] [Google Scholar]
- Matwee C, Kamaruddin M, Betts DH, Basrur PK, King WA.The effects of antibodies to heat shock protein 70 in fertilization and embryo development. Mol Hum Reprod 2001; 7: 829–837. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK.Gene bookmarking: keeping the pages open. Trends Biochem Sci 2005; 30: 605–610. [DOI] [PubMed] [Google Scholar]
- Christova R, Oelgeschlager T.Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol 2002; 4: 79–82. [DOI] [PubMed] [Google Scholar]
- John S, Workman JL.Bookmarking genes for activation in condensed mitotic chromosomes. Bioessays 1998; 20: 275–279. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C.Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 1995; 83: 29–38. [DOI] [PubMed] [Google Scholar]
- Michelotti EF, Sanford S, Levens D.Marking of active genes on mitotic chromosomes. Nature 1997; 388: 895–899. [DOI] [PubMed] [Google Scholar]
- Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge OK, Sarge KD.Mechanism of hsp70i gene bookmarking. Science 2005; 307: 421–423. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhang J, Moskophidis D, Mivechi NF.Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis 2003; 36: 48–61. [DOI] [PubMed] [Google Scholar]
- Akerfelt M, Trouillet D, Mezger V, Sistonen L.Heat shock factors at a crossroad between stress and development. Ann N Y Acad Sci 2007; 1113: 15–27. [DOI] [PubMed] [Google Scholar]
- Alastalo TP, Hellesuo M, Sandqvist A, Hietakangas V, Kallio M, Sistonen L.Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J Cell Sci 2003; 116: 3557–3570. [DOI] [PubMed] [Google Scholar]
- He H, Soncin F, Grammatikakis N, Li Y, Siganou A, Gong J, Brown SA, Kingston RE, Calderwood SK.Elevated expression of heat shock factor (HSF) 2A stimulates HSF1-induced transcription during stress. J Biol Chem 2003; 278: 35465–35475. [DOI] [PubMed] [Google Scholar]
- Ostling P, Bjork JK, Roos-Mattjus P, Mezger V, Sistonen L.Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J Biol Chem 2007; 282: 7077–7086. [DOI] [PubMed] [Google Scholar]
- Christians E, Michel E, Adenot P, Mezger V, Rallu M, Morange M, Renard JP.Evidence for the involvement of mouse heat shock factor 1 in the atypical expression of the HSP70.1 heat shock gene during mouse zygotic genome activation. Mol Cell Biol 1997; 17: 778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E, Davis AA, Thomas SD, Benjamin IJ.Maternal effect of Hsf1 on reproductive success. Nature 2000; 407: 693–694. [DOI] [PubMed] [Google Scholar]
- Kallio M, Chang Y, Manuel M, Alastalo TP, Rallu M, Gitton Y, Pirkkala L, Loones MT, Paslaru L, Larney S, Hiard S, Morange M, et al. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J 2002; 21: 2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ.Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem 1998; 273: 7523–7528. [DOI] [PubMed] [Google Scholar]
- Monesi V.Ribonucleic acid synthesis during mitosis and meiosis in the mouse testis. J Cell Biol 1964; 22: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monesi V, Geremia R, D'Agostino A, Boitani C.Biochemistry of male germ cell differentiation in mammals: RNA synthesis in meiotic and postmeiotic cells. Curr Top Dev Biol 1978; 12: 11–36. [DOI] [PubMed] [Google Scholar]
- Zheng J, Xia X, Ding H, Yan A, Hu S, Gong X, Zong S, Zhang Y, Sheng HZ.Erasure of the paternal transcription program during spermiogenesis: the first step in the reprogramming of sperm chromatin for zygotic development. Dev Dyn 2008; (in press). [DOI] [PubMed]
- Ward WS, Coffey DS.DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod 1991; 44: 569–574. [DOI] [PubMed] [Google Scholar]
- Hishinuma M, Sekine J.Separation of canine epididymal spermatozoa by Percoll gradient centrifugation. Theriogenology 2004; 61: 365–372. [DOI] [PubMed] [Google Scholar]
- Balhorn R, Gledhill BL, Wyrobek AJ.Mouse sperm chromatin proteins: quantitative isolation and partial characterization. Biochemistry 1977; 16: 4074–4080. [DOI] [PubMed] [Google Scholar]
- Maione B, Pittoggi C, Achene L, Lorenzini R, Spadafora C.Activation of endogenous nucleases in mature sperm cells upon interaction with exogenous DNA. DNA Cell Biol 1997; 16: 1087–1097. [DOI] [PubMed] [Google Scholar]
- Grabske RJ, Lake S, Gledhill BL, Meistrich ML.Centrifugal elutriation: separation of spermatogenic cells on the basis of sedimentation velocity. J Cell Physiol 1975; 86: 177–189. [DOI] [PubMed] [Google Scholar]
- vanWert JM, Wolfe SA, Grimes SR.Testis-specific expression of the rat histone Hlt gene in transgenic mice. Biochemistry 1995; 34: 8733–8743. [DOI] [PubMed] [Google Scholar]
- Hilgarth RS, Hong Y, Park-Sarge OK, Sarge KD.Insights into the regulation of heat shock transcription factor 1 SUMO-1 modification. Biochem Biophys Res Commun 2003; 303: 196–200. [DOI] [PubMed] [Google Scholar]
- Lubert EJ, Sarge KD.Interaction between protein phosphatase 2A and members of the importin beta superfamily. Biochem Biophys Res Commun 2003; 303: 908–913. [DOI] [PubMed] [Google Scholar]
- Xing H, Mayhew CN, Cullen KE, Park-Sarge OK, Sarge KD.HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J Biol Chem 2004; 279: 10551–10555. [DOI] [PubMed] [Google Scholar]
- Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD.Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem 2001; 276: 18513–18518. [DOI] [PubMed] [Google Scholar]
- Goodson ML, Sarge KD.Regulated expression of heat shock factor 1 isoforms with distinct leucine zipper arrays via tissue-dependent alternative splicing. Biochem Biophys Res Commun 1995; 211: 943–949. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI.Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol 1993; 13: 1392–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ying Z, Jin X, Tu N, Zhang Y, Phillips M, Moskophidis D, Mivechi NF.Essential requirement for both hsf1 and hsf2 transcriptional activity in spermatogenesis and male fertility. Genesis 2004; 38: 66–80. [DOI] [PubMed] [Google Scholar]
- Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ.HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J 1999; 18: 5943–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Huang L, Zhang J, Moskophidis D, Mivechi NF.Targeted disruption of hsf1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible Hsp molecular chaperones. J Cell Biochem 2002; 86: 376–393. [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Mortin MA, Wu C.Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J 1997; 16: 2452–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Christians E, Forster M, Xiao X, Connell P, Plumier JC, Zuo X, Richardson J, Morgan S, Benjamin IJ.Heat shock transcription factor 2 is not essential for embryonic development, fertility, or adult cognitive and psychomotor function in mice. Mol Cell Biol 2002; 22: 8005–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagutelli X.Effect of the genetic background on the phenotype of mouse mutations. J Am Soc Nephrol 2000; 11(suppl 16):S101–S105. [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, Barnard JA, Yuspa SH, et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 1995; 269: 230–234. [DOI] [PubMed] [Google Scholar]
- Schuetz TJ, Gallo GJ, Sheldon L, Tempst P, Kingston RE.Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc Natl Acad Sci U S A 1991; 88: 6911–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson DC, Skaggs HS, Sarge KD.HSF2 binds to the Hsp90, Hsp27, and c-Fos promoters constitutively and modulates their expression. Cell Stress Chaperones 2007; 12: 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein ND, Chen WC, Kingston RE, Myers RM.Transcriptional regulation and binding of heat shock factor 1 and heat shock factor 2 to 32 human heat shock genes during thermal stress and differentiation. Cell Stress Chaperones 2004; 9: 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.