Summary
Membrane proteins function in a polarized ionic environment with sodium-rich extracellular and potassium-rich intracellular solutions. Glutamate receptors which mediate excitatory synaptic transmission in the brain show unusual sensitivity to external ions, resulting in an apparent requirement for sodium in order for glutamate to activate kainate receptors. Here we solve the structure of the Na+ binding sites and determine the mechanism by which allosteric anions and cations regulate ligand binding dimer stability, and hence the rate of desensitization and receptor availability for gating by glutamate. We establish a stoichiometry for binding of 2 Na+ to 1 Cl− and show that allosteric anions and cations bind at physically discrete sites with strong electric fields, that the binding sites are not saturated in CSF, and that the requirement of kainate receptors for Na+ occurs simply because other cations bind with lower affinity, and have lower efficacy compared to Na+.
Keywords: Glutamate Receptor, Crystal Structure, Sodium, Cation binding site, Desensitization, Molecular Dynamics
Introduction
The ion channels and signal transduction molecules which play key roles in synaptic function are typically encoded by large gene families with similar structures but diverse functional properties. The physiological significance of such diversity is becoming increasingly well understood, but the underlying molecular mechanisms are generally unknown. The glutamate receptor ion channels (iGluRs) that mediate excitatory transmission are typical examples. They are encoded by a family of 18 genes, named for their selective agonists AMPA, kainate and NMDA, and share a common modular design distinct from other ligand-gated ion channels (Madden, 2002; Mayer, 2006; Mayer and Armstrong, 2004). Individual subtypes exhibit striking differences in their assembly, subcellular targeting, kinetics and modulation, corresponding to varied roles in brain physiology and disease. Kainate receptors, formed from the GluR5-7 and KA1-2 subunits, appear to play specialized roles at particular central synapses. They modulate neurotransmitter release (Chittajallu et al., 1996) and participate in postsynaptic currents that have a wide range of decay rates, from unusually slow (Castillo et al., 1997; Kidd and Isaac, 1999; Vignes and Collingridge, 1997) to conventional rapid excitatory synaptic responses (DeVries and Schwartz, 1999).
Functional studies have established that glutamate receptors are modulated by endogenous ions and small molecules. Such interactions not only control synaptic strength and permit activity dependent changes in receptor function, but could potentially be exploited for the development of novel therapeutic agents. Sites within the pore that regulate binding of permeant ions and blockers are well-characterized, but growing evidence suggests that extracellular allosteric ion binding sites are also critical. We recently established that in kainate receptors a chloride ion is bound in the interface of the active dimer conformation and that the binding of Cl− regulates both desensitization and functional availability of the receptor (Plested and Mayer, 2007). Kainate receptors are also strongly modulated by external monovalent cations (Bowie, 2002; Paternain et al., 2003). Surprisingly, despite their similar structure, AMPA receptors are largely insensitive to changes in external ions. Several recent studies have attempted to address the mechanism of this difference (Paternain et al., 2003; Wong et al., 2007); however, the structure of the cation binding site is unknown, the mechanisms which confer Na+ selectivity have yet to be established, and the mode of action of Na+ remains a mystery.
Here, we have combined functional studies of receptor activity with crystallographic analysis and molecular dynamics simulations to demonstrate that the ligand binding dimer of kainate receptors contains a pair of cation binding sites related by two-fold molecular symmetry. Cations act to stabilize the active dimer assembly. Functional studies establish that a large fraction of kainate receptors are not competent for activation by glutamate at physiological salt concentrations because the ion binding sites are not fully occupied. We reconcile previous reports on the apparent common modulation of kainate receptors by external anions and cations by revealing allosteric coupling between the sodium and chloride binding sites and show that the apparent requirement of kainate receptors for Na+ occurs simply because other cations bind with lower affinity and have lower efficacy.
RESULTS
Cations modulate kainate receptors via the ligand binding core
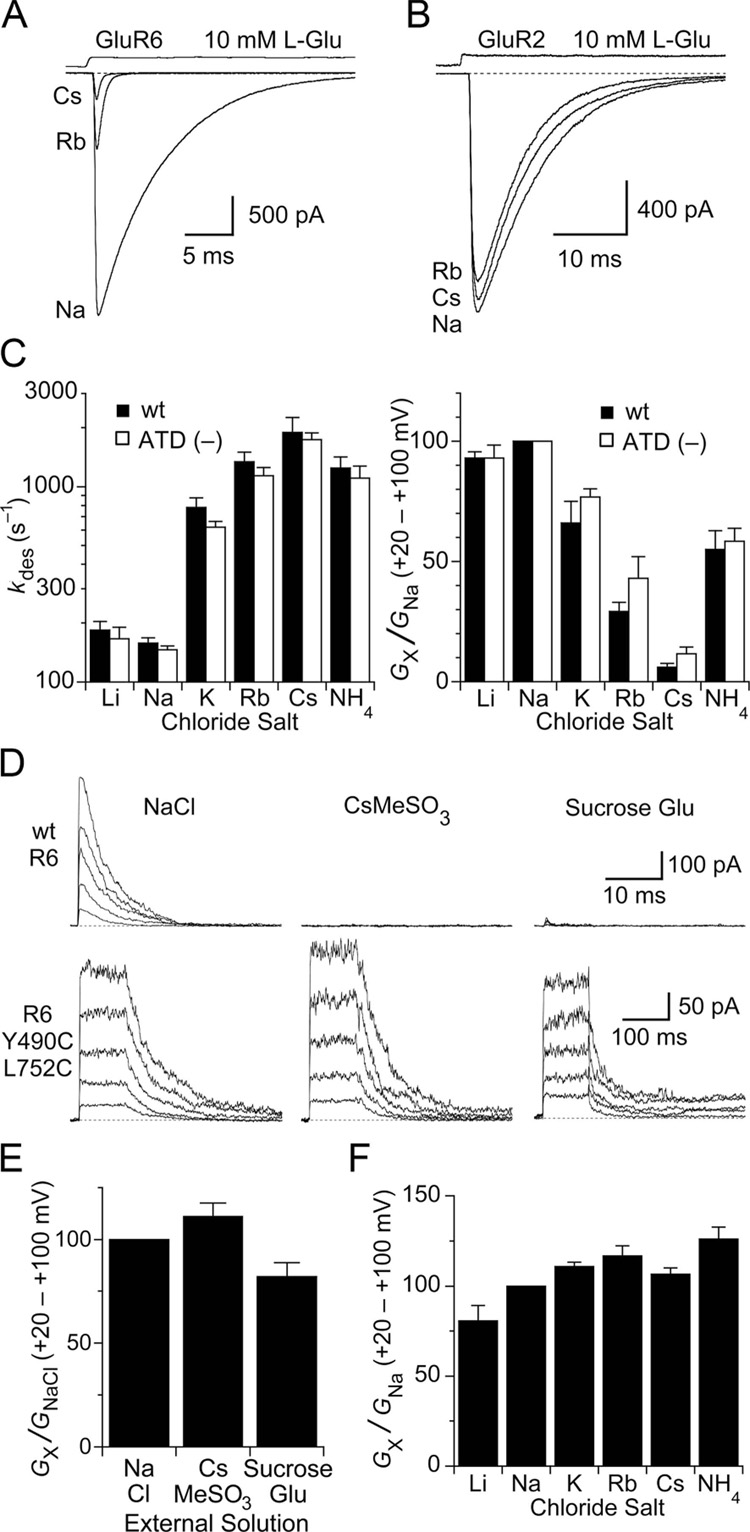
For functional experiments, we used the GluR6 subtype kainate receptor. When extracellular Na+ was replaced by equimolar concentrations of different monovalent cations, the extent of activation, measured as the slope conductance for outward current responses over the range +20 to +100 mV, decreased by up to 94% and the rate of desensitization measured at − 30 mV increased by up to 12-fold (Figure 1A and C). These effects were highly correlated (R = 0.98), suggesting that the mechanisms which control stability of the active state also determine receptor availability for activation by glutamate. The order of efficacy at 150 mM external cation was Na+ > Li+ > K+ > NH4+ > Rb+ > Cs+ (Table 1). Similar to the GluR1 and GluR3 subtypes of AMPA receptor, previously shown to exhibit minimal sensitivity to cations (Bowie, 2002; Paternain et al., 2003), GluR2 subtype AMPA receptors are also insensitive to replacement of Na+ by Rb+ or Cs+ (Figure 1B).
Figure 1. Cations act via sites in the ligand binding domain.
(A) GluR6 responses to 10 mM glutamate are reduced in amplitude and desensitize faster when extracellular Na+ is replaced with Rb+ or Cs+. The effects were fully and rapidly reversible on returning to Na+.
(B) GluR2flip subtype AMPA receptors show little change in glutamate activated responses after exchange of extracellular Na+ by Rb+ or Cs+.
(C) Bar plots summarizing the effects of external monovalent cations on the rate of onset of desensitization (left) and the slope conductance from +20 to +100 mV (right) for responses recorded from wildtype GluR6 and the (ATD–) deletion construct. In all panels, error bars indicate SEM.
(D) Responses to 10 mM glutamate recorded at holding potentials of + 20 to + 100 mV for wild type GluR6 (top row) and the non desensitizing GluR6 Y490C/L752C mutant (bottom row) with NaCl, CsMeSO3 or sucrose in the external solution.
(E) Bar plot of slope conductance measured from + 20 to + 100 mV for the GluR6 Y490C/L752C mutant in NaCl, CsMeSO3 and sucrose solutions.
(F) Bar plot of slope conductance measured from + 20 to + 100 mV for the GluR6 Y490C/L752C mutant in solutions containing Li+, Na+, K+, Rb+, Cs+ or NH4+ as the extracellular monovalent cation.
To identify the domains responsible for the allosteric effects of Na+ on kainite receptors, we studied a truncated GluR6 construct lacking the amino terminal domain (ATD), which is the site of action for some NMDA receptor allosteric modulators (Paoletti et al., 2000; Perin-Dureau et al., 2002). Strikingly, the GluR6 ATD (–) construct had almost identical sensitivity to cation exchange (Figure 1C and Table 1), with a strong correlation between the desensitization rate and the extent of activation (R = 0.99). This result, in combination with the voltage independence and lack of inhibitory effects of intracellular ions (Bowie, 2002), strongly suggests that the functional binding sites for Na+ are located within the ligand binding cores.
External ions are not co-activators of kainate receptors
We previously showed that kainate receptors trapped in their active dimer conformation by intermolecular disulfide cross links between helices D and J (Weston et al., 2006) are insensitive to the allosteric effects of anions (Plested and Mayer, 2007). It has been recently suggested that NaCl acts as a co-activator of kainate receptors because wild type GluR6 responses are abolished when NaCl is replaced by CsMeSO3 (Wong et al., 2006; Figure 1D). However, when desensitization is blocked by cysteine substitution at Y490 and L752, anion and cation sensitivity is lost (Figure 1D lower panel), indicating that kainate receptors can be activated by glutamate even in the absence of NaCl. To avoid effects complicated by ion permeation, we performed our measurements at depolarized membrane potentials where the glutamate activated ion current is carried by ions in the pipette solution. In conditions in which responses of wild-type GluR6 are strongly inhibited, including the replacement of Na+ by Cs+, the replacement of Cl− by MeSO3−, and even in the virtual absence of monovalent inorganic ions, disulfide cross-linked kainate receptors continue to show normal activation by glutamate (Figure 1 D and E). This is inconsistent with co-activation of kainate receptors by glutamate and NaCl, but instead suggests an ion dependent allosteric mechanism that controls desensitization and functional availability of kainate receptors. Further, in contrast to wild-type receptors, glutamate activated currents for cross-linked GluR6 were essentially unchanged when external Na+ was substituted by K+, Rb+ or NH4+ (Figure 1F), except for weak inhibition in Li+ (Table 1), which is the most efficacious cation after Na+ for wild type GluR6. Deactivation kinetics in different cations were similar for the GluR6 L490C/L752C cross linked mutant (Table 1).
Sodium binds with low affinity
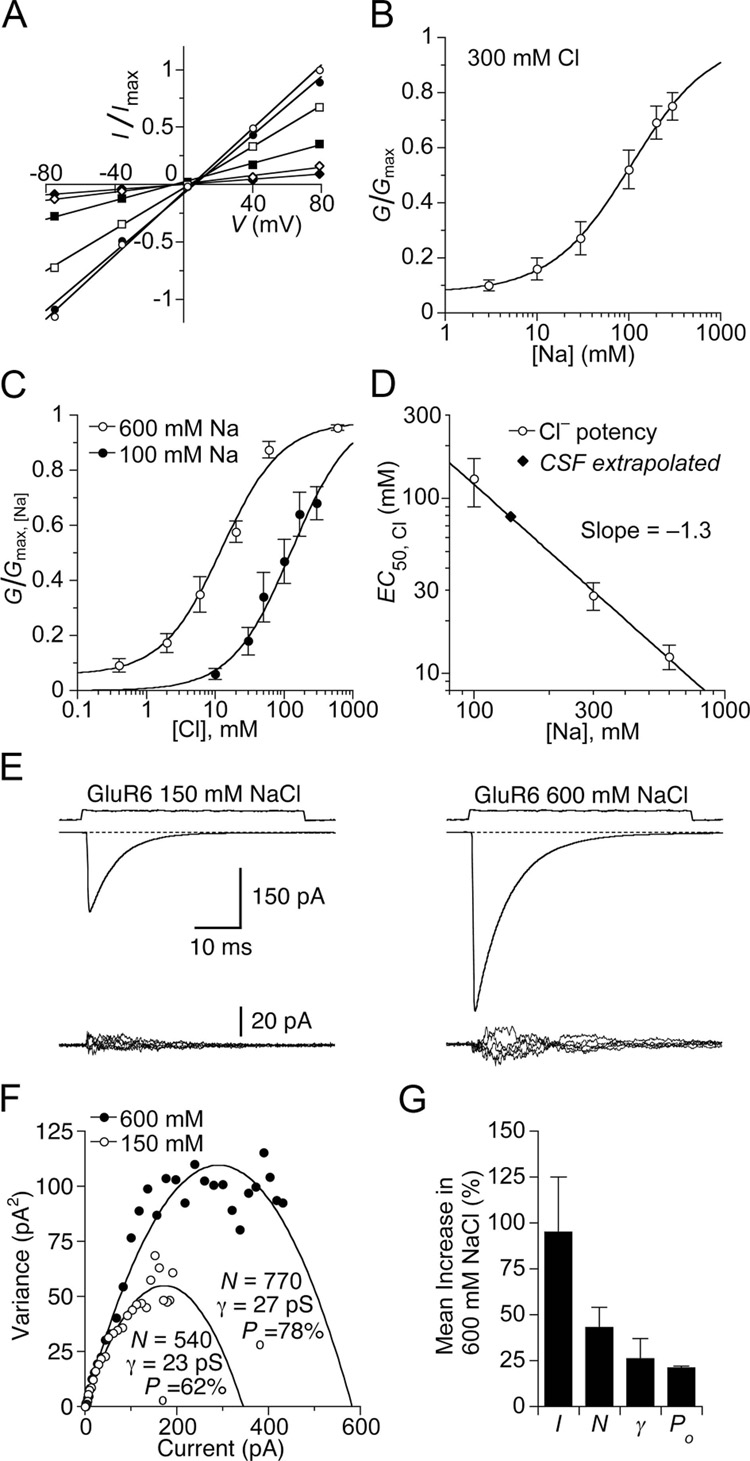
To estimate the apparent affinity of Na+ for GluR6, we measured the slope of the peak current–voltage relationship over the range −80 to +80 mV in solutions containing a range of external Na+ concentrations, from 3 to 300 mM (Figure 2A), with the Cl− concentration maintained at 300 mM using Cs+ as a substitute for Na+. The Na+ concentration required for half-maximal activation was 110 mM (Figure 2B). To test for allosteric coupling between the Na+ and Cl− binding sites, we measured the EC50 for Cl− at lower and higher external Na+ concentrations. In 100 mM Na+ the apparent affinity for Cl− was reduced to 130 ± 40 mM, and in 600 mM Na+ it increased to 11 ± 3 mM (Figure 2C). A plot of the apparent affinity for Cl− versus the external Na+ concentration (Figure 2D), was used to estimate the extent of coupling between binding of Na+ and Cl−. When the EC50 for Cl− was plot on a log-log scale versus Na+ concentration the slope of the fit was −1.3 ± 0.2 (Figure 2D), indicating that there are at least twice as many binding sites for Na+ as for Cl−; that the Na+ and Cl− binding sites are allosterically coupled; and that the stoichiometry of binding is not fixed. Extrapolation of the fit to a physiological Na+ concentration reveals that the apparent affinity for Cl− is likely to be about 90 mM, much closer to the physiological level than estimated previously from measurements performed with 300 mM Na+ (Plested and Mayer, 2007).
Figure 2. The Na+ and Cl− sites are allosterically coupled and not saturated in CSF.
(A) Current-voltage plot for GluR6 responses to 10 mM glutamate recorded in a range of Na+ concentrations (open circles, 300 mM; filled circles, 200 mM; open squares, 100 mM; filled squares, 30 mM; open diamonds, 10 mM; filled diamonds, 3 mM) using Cs+ as a substitute. Glutamate was titrated with CsOH. Slope conductance was fit by linear regression.
(B) Na+ concentration response curve for GluR6 fit with a single binding isotherm (EC50 = 110 ± 50 mM), with a constant to account for the residual current at low ion concentration. Data points show the mean ± SEM for six patches.
(C) Slope conductance for glutamate responses measured as in (A) but with either 100 or 600 mM Na+, with Cl− concentrations varied from 0.3 mM to 600 mM (using MeSO3− as a substitute) fit with single binding isotherms. The apparent affinity for Cl− varies with Na+ concentration: 100 mM Na+ EC50 = 130 ± 40 mM; 600 mM Na+ EC50 = 13 ± 3 mM. Data points represent the mean slope conductance ± SEM for six patches for each curve.
(D) Plot of Cl− EC50 versus Na+ concentration fit by nonlinear regression with a power relation (slope −1.3 ± 0.2). By interpolation, the EC50 for Cl− in physiological saline is about 90 mM. Data points indicate EC50 values and SDs estimated from fits shown in (C); data point at 300 mM is from Plested and Mayer, 2007.
(E) Nonstationary analysis of variance of GluR6 responses to 10 mM glutamate. Means of 60–100 traces (upper row) and 5 consecutive difference current traces (lower row) recorded in 150 and 600 mM NaCl from the same patch. The current is substantially and reversibly increased in 600 mM NaCl, when the membrane potential was adjusted to −20 mV to give the same driving force as for responses recorded in 150 mM NaCl.
(F) Current-variance plot for responses in (E). In 600 mM NaCl, the number of available receptors and open probability increases compared to 150 mM NaCl.
(G) Bar plots for the mean responses of paired observations from six patches; error bars indicate SEM. On average, the current I was increased by 100 ± 30%. The number of receptors N competent for activation by glutamate increased by 43 ± 11%; the conductance γby 26 ± 11 %; and the peak open probability by 21 ± 1%.
Kainate receptors are not fully active at physiological ionic strength
The concentration response analysis for GluR6 modulation by Na+ and Cl− ions suggests that a significant fraction of the ion binding sites are not bound by Na+ or Cl− at physiological ionic strength. Therefore, only about two-thirds of kainate receptors should be competent for activation by glutamate and increasing the external Na+ and Cl− concentration should increase the number of functional receptors.
For individual patches, we collected runs of GluR6 responses in both 150 and 600 mM NaCl. In 600 mM NaCl, we observed a reversible two-fold increase in the current activated by 10 mM glutamate (2.0 ± 0.3, n = 6) compared to responses with 150 mM NaCl (Figure 2E). A non stationary analysis of variance revealed that only a small part of this increase was attributable to increased single channel conductance due to the 4-fold greater ion concentration, most likely because the ion binding sites for permeation have higher affinities than the allosteric ion binding sites. Analysis of the variance versus mean current relationship revealed, in addition, increases in the number of active receptors, and the maximum open probability at the peak of the response to glutamate (Figure 2F). Because the single channel conductance is the best determined parameter obtained from noise analysis (Alvarez et al., 2002), but increases by only 17% compared to the 2-fold increase in current amplitude, we are confident that the greatest part of the increase is an allosteric effect due to a greater number of active receptors combined with increased open probability at the peak of the response (Figure 2G). While it can be difficult to distinguish between effects on receptor number and open probability (Traynelis and Wahl, 1997), we note that, despite the strong statistical negative correlation between estimates of these two variables when fitting the parabolic current variance relation, both exhibited concurrent increases when the salt concentration was increased from 150 to 600 mM, with functional receptor number increasing by 43 ± 11% and peak open probability increasing by 21 ± 1%.
The increase in osmotic pressure to ~1100 mOsm is unlikely to have had non specific effects because the rate of desensitization measured in 150 mM NaCl at an osmotic pressure of 1100 mOsm, kdes 151 ± 7 s−1 (n = 6) was very similar to that measured in standard extracellular solution, 160 ± 10 s−1 (n = 16, P = 0.6), while the rate constant measured in 600 mM NaCl, 124 ± 11 s−1 (n = 6) decreases 1.4-fold (P = 0.02), as expected for an allosteric effect on stability of the active state. Together, these results suggest that at physiological levels of NaCl, the incomplete occupancy of allosteric ion binding sites in the ligand binding core renders at least 1/3 of kainate receptors refractory to activation, and those that are activated have a reduced maximum open probability. Because kainate receptors are not fully bound by Na+, their activity will be sensitive to Na+ fluctuations in the physiological range.
Identification of cation binding sites by surface potential analysis
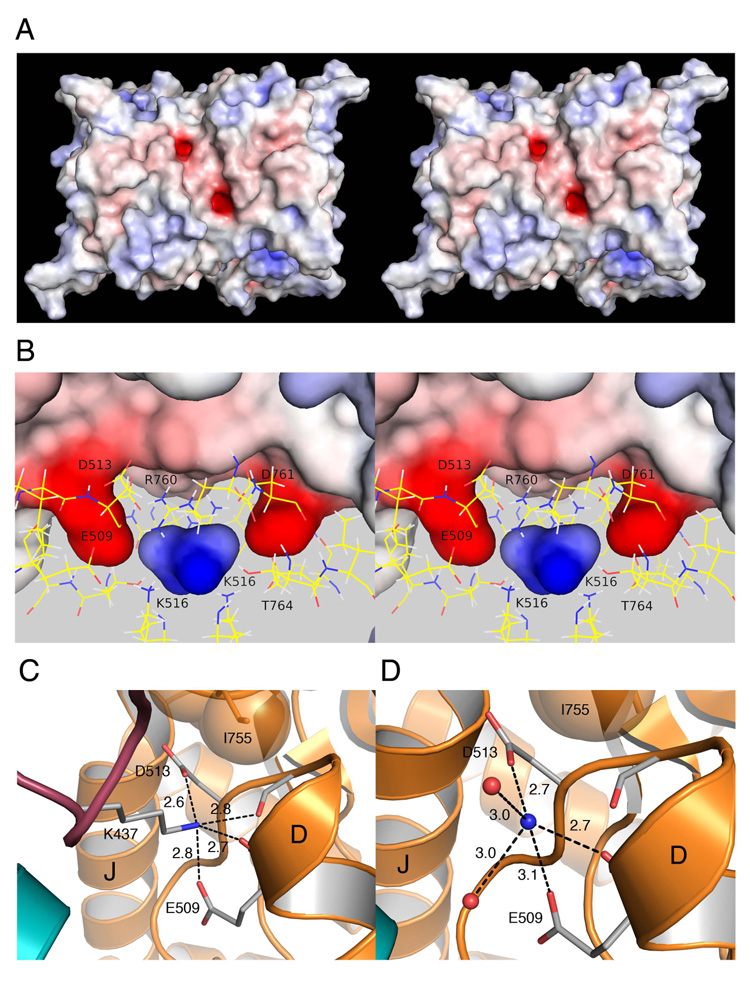
Our crystallographic experiments used the GluR5 subtype kainate receptor for which a range of agonists, partial agonists and antagonists is available, allowing access to a wider range of conformational and crystallographic space than for GluR6. In prior work, GluR5 was crystallized with the competitive antagonist UBP310, but we were unable to identify bound cations in these structures (Plested and Mayer, 2007). We sought different kainite receptor crystal forms and succeeded in crystallizing GluR5 dimer complexes (Table 2) with the partial agonist kainic acid in a solution containing 200 mM Cl− and 575 mM Li+, the most active cation in functional studies after Na+ (Figure 1). Apart from an 18° degree difference in domain closure for each subunit, due to the different conformational states induced by binding of agonists and antagonists (Armstrong and Gouaux, 2000; Mayer et al., 2006), the kainate and UBP310 complex dimers are nearly identical, and when superimposed using domain 1 Cα coordinates the root-mean-square-deviation (rmsd) was only 1.03 Å. A search of the dimer assembly for candidate cation binding sites using GRID (Goodford, 1985) identified one site in each subunit related by two-fold molecular symmetry resulting from dimer assembly. These candidate cation binding sites are generated by two strongly electronegative pockets on the top surface of the dimer, separated by 12 Å (Figure 3A). The pockets penetrate the protein surface by ~4 Å (Figure 3B) and are formed by the side chains of Glu509, Asp513 and Asp761. In combination with the main chain carbonyl oxygen atoms of Glu509 and Ile512, these generate a negative surface potential of −13 kT/e at the base of the pocket, calculated using APBS (Baker et al., 2001). The side chain methyl groups of Thr764 and Ile765 and the guanidinium group of Arg760 form the remainder of the surface of the pocket which is capped by the side chain of Ile755. Between these pockets is an electropositive cavity with a surface potential of +8 kT/e generated by the side chains of Lys516, Arg760 and Thr764 (Figure 3B) which form the previously identified anion binding site (Plested and Mayer, 2007). Notably, the center of the anion binding cavity is only 6 Å from each of the putative cation binding sites and residues which form these sites are interconnected by a complex network of salt bridges, hydrogen bonds, and solvent mediated contacts.
Figure 3. The Na+ and Cl− binding sites are physically discrete.
(A) Stereo view of a surface potential map for the GluR5 kainate complex dimer crystal structure calculated with APBS and contoured from −10 kT/e (red) to +10 kT/e (blue). The view is face on to the plane of the membrane with the N terminus facing upwards. The two electronegative pockets were the most prominent features on the protein surface.
(B) Stereo view of the protein interior with the molecular surface colored by surface potential; side chains forming the cation and anion binding sites drawn in stick configuration; the view is rotated by ≈ 90° from than in A. The electropositive cavity located between the two ‘fingers’ projecting into the protein interior forms the binding site for Cl−.
(C) Crystal structure of the GluR5 complex with the antagonist UBP310 at 1.74 Å resolution (PDB 2F34) showing plugging of the cation binding pocket by the side chain of Lys437 from an adjacent subunit.
(D) Crystal structure of the GluR5 kainate complex with NH4+ in the cation binding site at 1.68 Å resolution; dashed lines indicate protein contacts within hydrogen bond or salt bridge distance from the NH4+ ion. Labels identify helices D and J; in this and subsequent figures the pair of subunits in a dimer assembly are shaded gold and cyan respectively.
Analysis of molecular packing in the GluR5 UBP310 complex, crystallized in the C2221 space group, revealed that each of the putative cation binding sites is occluded by the side chain of Lys437 which projects from loop 1 of two adjacent protein molecules (Figure 3C). The Lys NZ atoms plug the putative cation binding pockets, essentially acting as tethered ammonium ions, and make salt bridges with the side chains of Glu509 and Asp513. In contrast, in the kainate complex, crystallized in the P41212 space group, the putative cation binding sites face solvent channels in the crystal lattice and are free to bind ions. This was established by growing crystals in Li+ and then soaking them for 48 hours in a solution in which Li+ was replaced by NH4+ to mimic conditions which occur in the GluR5 complex with the antagonist UBP310. Using data measured to Bragg spacings of 1.68 Å (Table 2), we observed a strong peak of electron density less than 1Å from the position of the Lys NZ atom in the UBP310 crystal form, which we interpret as a bound NH4+ ion (Fig. 3D). Accordingly, bond-valance analysis for oxygen atoms within 3.5 Å gave values of 1.0 and 0.84 for the NH4+ ions bound to subunits A and B with mean bond distances of 2.94 and 2.99 Å respectively, close to the ideal value of 2.88 Å for tetrahedral coordination (Muller et al., 2003). Although the UBP310 and NH4+ complex structures are similar, there are several interesting differences. The NH4+ ion does not penetrate the pocket as deeply as the tethered Lys side chain, and as a result fails to make a hydrogen bond contact with the carbonyl oxygen atom of Ile512. The NH4+ ion is solvated by a pair of H2O molecules, which are displaced by the methylene groups of the Lys437 side chain which fill up the pocket. Although electrostatic principles and bond-valance analysis suggest that the kainate complex has indeed bound an NH4+ ion, because NH4+ ions and H2O both have 10 electrons it is not possible to unambiguously distinguish between them using X-ray diffraction at 1.68 Å resolution. Furthermore, because in functional studies the rate of onset of desensitization in NH4+ is nearly 8-fold faster than in Na+ (Figure 1C) it is likely that the dimer assembly is less stable for the NH4+ complex.
Crystal structures of the Li+ and Na+ complexes
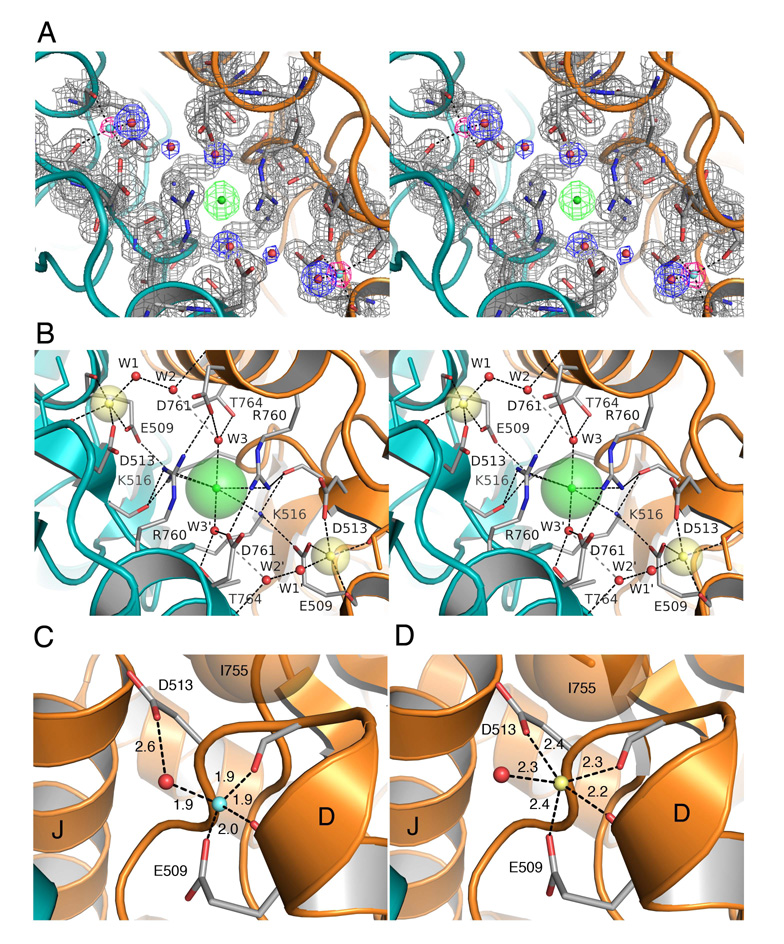
Because the Li+ ion has only two electrons it is much more difficult to detect by X-ray diffraction than other cations. Here, we found that GluR5 kainate dimer assemblies, which crystallized readily in the presence of Li+ and Cl−, diffracted to higher resolution than the other ion complexes studied (Table 2). Despite the weak X-ray scattering power of Li+, during the final stages of refinement using data measured to 1.49 Å resolution, small spheres of electron density emerged in Fo–Fc maps contoured at > 3 σ in the region of the cation binding sites, along with unambiguous electron density in 2mFo-DFc maps for side chains, H2O molecules, and a Cl− ion in the anion binding site (Figure 4A). When the electron density in the cation binding site for each subunit was modeled as a H2O molecule, there were energetically unfavorable close contacts with the protein, and the B values refined to implausibly small values. In contrast, when modeled as Li+ ions (Figure 4C) the mean ion-ligand distance of 1.93 Å (σ = 0.04 Å) was in agreement with the mean value of 1.96 Å (σ = 0.12 Å) obtained from analysis of six structures in the PDB refined at resolutions of 0.98 – 1.62 Å and much shorter than subsequently observed for Na+ and other GluR5 ion complexes (Figure 4D and Figure 6).
Figure 4. High resolution crystal structures of the Li+ and Na+ complexes.
(A) Stereo view of the GluR5 kainate complex with Li+ at 1.49 Å resolution. Electron density for the pair of Li+ ions is shown for an Fo-Fc omit map (pink) contoured at 3.2 σ, with Li+ omitted from the Fc calculation. Electron density for the protein (gray), water molecules (blue), and the Cl− ion (green) is shown for a 2mFo-DFc map contoured at 1.5 σ.
(B) Stereo view of the Na+ complex looking down the molecular two-fold axis of the dimer assembly onto the plane of the membrane; transparent spheres for the bound ions are drawn using Shannon radii; the Na+ and Cl− ions are linked in a network formed by the side chains of Glu509 and Lys516. Numerous salt bridges and hydrogen bonds link the subunits together.
(C) Crystal structure of the GluR5 kainate complex with Li+ in the cation binding site showing tetrahedral coordination of the ion by the side chains of Glu509, Asp 513, the main chain carbonyl oxygen of Glu509, and a H2O molecule, with bond distances in Å.
(D) Crystal structure of the GluR5 kainate complex at 1.72 Å resolution with Na+ in the cation binding site showing 5-fold coordination of the Na+ ion, and block of the 6th coordination site by the side chain of Ile755.
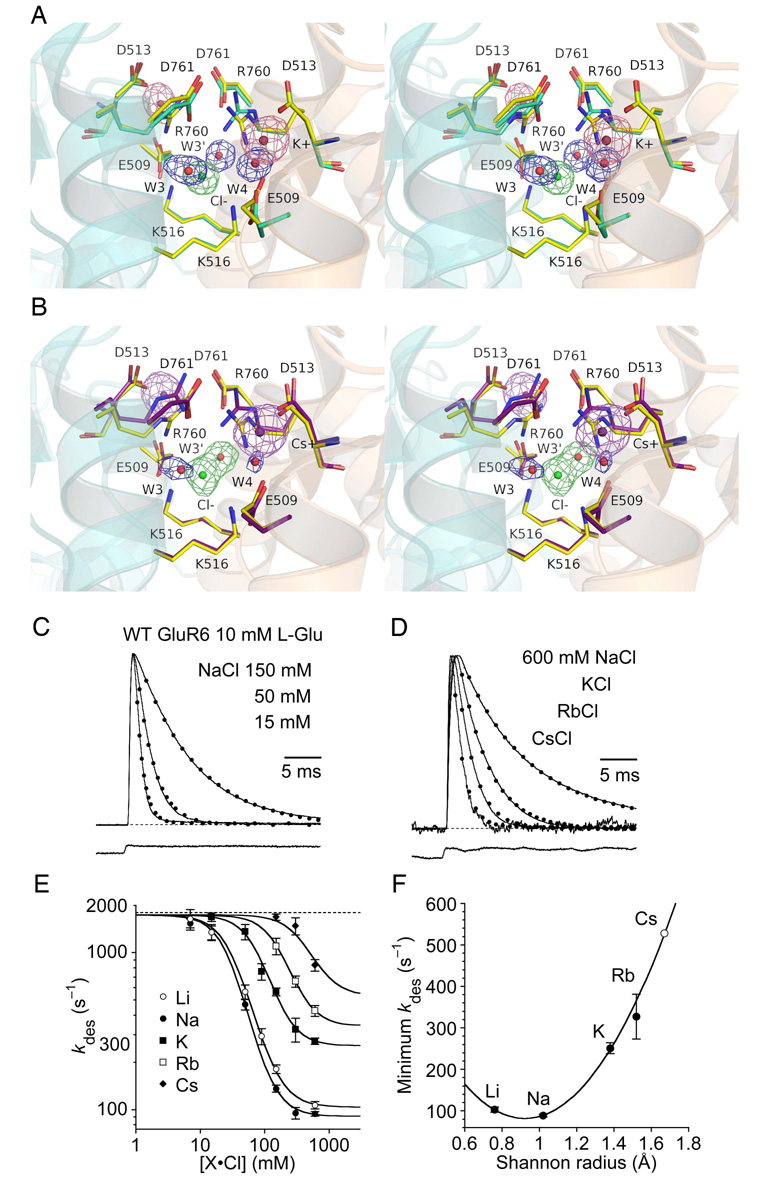
Figure 6. The cation binding site shows weak selectivity for Na+.
(A) Stereo view of the K+ complex crystal structure at 1.72 Å resolution with an Fo-Fc omit map contoured at 3.5 σ for the K+ (pink) and Cl− (green) ions and H2Omolecules (blue) omitted from the Fc calculation. Density for the Cl− ion and its adjacent H2O molecules W3 and W3’ are well resolved as individual peaks. Side chains which form the ion binding sites are drawn in stick representations. The rest of the protein is drawn as a transparent ribbon diagram, with helix J foremost for the left subunit, and helix D foremost for the right subunit. The structure is superimposed on that for the Na+ complex using domain 1 Cα coordinates with an rmsd of 0.20 Å; carbon atoms for the K+ complex side chains are colored pale green and those for the Na+ complex yellow. In the right subunit of the K+ complex the side chain for Arg760 has flipped into an ‘up’ conformation creating a hole filled by W4.
(B) Stereo view of the Cs+ complex crystal structure at 1.97 Å resolution oriented as in A, with an Fo-Fc omit map contoured at 3.5 σ for Cs+ (purple) and Cl− (green) ions and H2O molecules (blue) which were omitted from the Fc calculation. Note that density for the Cl− ion and W3’ is continuous, while that for W3 is weak compared to adjacent atoms. The structure is superimposed on that for the Na+ complex using domain 1 Cα coordinates with an rmsd of 0.19 Å; carbon atoms for the Cs+ complex side chains are colored purple and those for the Na+ complex yellow. In the both subunits of the Cs+ complex the side chain for Arg760 have flipped into an ‘up’ conformation.
(C) Outward currents activated by 10 mM glutamate for wild type GluR6 recorded in the same patch at + 30 mV in 150, 50 and 15 mM external NaCl (sucrose used to maintain osmotic pressure); currents were normalized to the response in 150 mM NaCl and fit with monoexponential decays (dotted lines) with kdes = 120, 410 and 950 s−1 respectively. The bottom trace is the junction current recorded at the end of the experiment.
(D) Outward currents activated by 10 mM glutamate for wild type GluR6 recorded in the same patch with either 600 mM NaCl, KCl, RbCl, or CsCl. The currents were normalized to the response in 600 mM NaCl and fit with monoexponential decays (dotted lines) with kdes = 100, 240, 400 and 675 s−1 respectively. The bottom trace is the junction current recorded at the end of the experiment.
(E) Rate of desensitization with salt concentrations from 6 to 600 mM globally fit by non-linear least squares with a binding isotherm; the curves were constrained to be parallel and to have a common maxima of 1743 s−1; the slope was very close to 2; the freely fitted minima were 103 ± 6, 89 ± 3, 250 ± 10 and 330 ± 50 s−1 for Li+, Na+, K+ and Rb+ respectively. Data points represent the mean ± SEM of the desensitization rate measured in at least five patches.
(F) The minimum rate of desensitization, plotted against Shannon radius, fit with a parabolic function by non-linear regression; the value for Cs+ is extrapolated from the fit. The parabola has a minimum close to the sodium radius, corresponding to kdes, min = 82 ± 3 s−1. Data points represent the minimum fitted rate ± the approximate SD from the fit in (E); we used this relation to fix the minimum desensitization rate in CsCl to be 530 s−1.
We also crystallized GluR5 kainate dimer assemblies in solutions with Na+ as the sole monovalent cation and solved the structure at 1.72 Å resolution (Figure 4B and Table 2). Although, as for NH4+, it is difficult to distinguish Na+ from water on the basis of electron density alone, bond distances for coordination of Na+ by carboxylate and carbonyl oxygen atoms have a characteristic distribution with a mean at 2.42 Å (Harding, 2002), much shorter than the value of 2.8 Å typical for H2O molecules. The mean Na+ ion-ligand distance of 2.32 Å (σ = 0.07 Å) is in excellent agreement with that measured in high resolution crystal structures. Bond valence calculations gave values of 1.09 and 1.15 for the pair of Na+ ions, providing further evidence that they were not H2O molecules (Muller et al., 2003).
Both Li+ and Na+ ions bind to the carboxylate group of Glu509 and to the main chain carbonyl oxygen atoms of Glu509 and Ile512 and are hydrated by a water molecule. The Li+ ion shows 4-fold tetrahedral coordination with a water molecule interspersed between the ion and the carboxylate group of Asp513 (Figure 4C); in contrast the Na+ ion is 5-fold coordinated and interacts directly with Asp513 (Figure 4D). The structure reveals that the hydrophobic side chain of Ile755 caps the cation binding sites and prevents binding of a 6th H2O molecule to the Na+ ion by steric hindrance without interfering with solvation of the Li+ ion. As a result of the greater field strength and smaller size of Li+ compared to Na+ (Shannon radii 0.76 Å and 1.02 Å espectively) the pair of Li+ ions move 0.84 Å and 0.94 Å deeper into the cation binding pockets compared to the Na+ complex, but there is almost no movement of the Cl− ion.
The structures of the Na+ and Li+ GluR5 complexes are nearly identical and superimpose with rmsds of 0.21 Å for Cα atoms. Not surprisingly, due to their close proximity, the anion and cation binding sites in both structures are linked by a complex network of intramolecular and intermolecular bonds (Figure 4B). Particularly striking is the interaction of the amino group of Lys 516 with both the bound Cl− anion and the carboxylate group of Glu509 which binds Na+ or Li+ in the dimer partner subunit. The hydration shells of the Cl− ion and the bound cations are linked by an intermediary H2O molecule W2, which also participates in an intermolecular solvent mediated hydrogen bond network linking the bound cation with helices D and J (Figure 4A and B). Intermolecular salt bridges between Arg760 – Asp761 also link helix J in each subunit with its dimer partner, as observed previously (Plested and Mayer, 2007). However, in the Na+ and Li+ bound GluR5 kainate complexes the χ4 angle of the Arg side chain is rotated by 58° and as a result guanidinium group moves downwards by 1.6 Å, forming a hydrogen bond contact with the main chain carbonyl oxygen of Phe514 in the same subunit (Figure 4A and B). The side chain conformation of Glu509 also differs between the GluR5 kainate complexes with Na+ and Li+ compared to that in the GluR5 UBP310 cyomplex, with rotations of 102°, 168°, and 60° for χ1, χ2 and χ3 which steer the carboxylate group towards the cation. In contrast, the side chains of Asp 513, Lys516, Asp761 and Thr 764 show negligible changes in torsion angle. Associated with the above movements there is a 3° degree tilt of subunits towards each other in the Na+ or Li+ bound complexes compared to the UBP310 structure, resulting in a 1.3 Å movement, measured from the Cα coordinates of Gly557 at the apex of the dimer surface.
Cation binding site mutations alter selectivity and speed desensitization
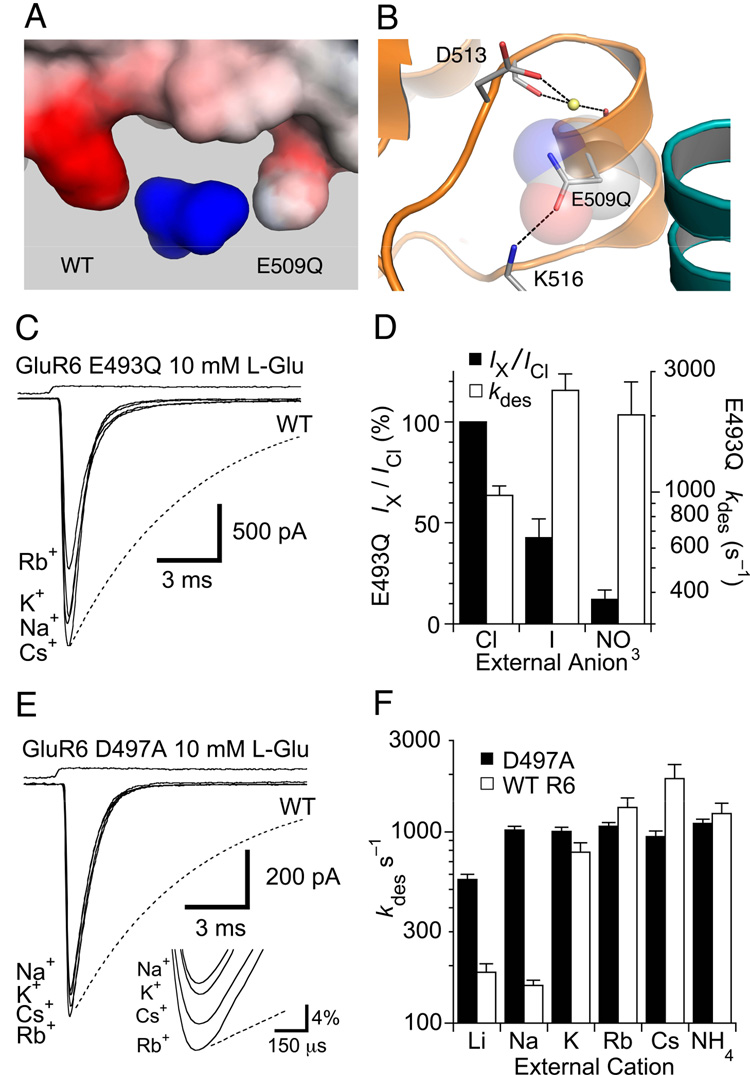
Allosteric coupling between the anion and cation binding sites (Figure 2), combined with their shared structural components (Figure 4), has confounded prior attempts to understand the effects of site directed mutagenesis in the dimer interface of kainite receptors. In the Na+ complex, the bound cation is coordinated by two side chain carboxylate groups and two main chain carbonyl oxygen atoms (Figure 4D). Glu509, which forms the base of the cation binding site, is mostly buried, with a solvent accessible surface area of < 5 Å2, while Asp513, which forms the top of the binding site, is exposed with a solvent accessible surface area of 55 Å2. Surface potential calculations for the isosteric GluR5 E509Q mutant (Figure 5A) reveal a 6 kT/e decrease in surface potential when the mutant side chain is oriented so that its carbonyl oxygen atom makes a hydrogen bond with the side chain of Lys500 that binds the Cl− ion (Figure 5B). In three 20 ns equilibrium MD simulations for E509Q the side chain χ3 angle fluctuated by 27° around a mean value of −121° and never flipped orientation. As a result, the Na+ ion effectively loses a ligand due to replacement of a carboxyl group oxygen by an amide group nitrogen atom.
Figure 5. Cation binding site mutations speed desensitization.
(A) View of the molecular surface of the anion and cation binding sites colored by surface potential for a GluR5 dimer with one wild type and one E509Q mutant subunit, viewed from the protein interior, and contoured from −10 kT/e (red) to +10 kT/e (blue).
(B) The surface potential calculation was performed with the E509Q mutant side chain oriented with its carbonyl group within hydrogen bonding distance of the Lys516 side chain amino group and the amide group facing the Na+ ion.
(C) The equivalent mutation for GluR6 (E493Q) speeds desensitization compared to responses for wild-type GluR6 measured in NaCl (dotted line). Responses measured from the same patch with K+, Rb+ or Cs+ as the external monovalent cation, showed similar rapid desensitization, and the peak amplitude was much less sensitive to cation species than for wild type.
(D) Bar plots summarizing the change in peak amplitude and rate of onset of desensitization for GluR6 E493Q responses measured with Na+ and either chloride, iodide or nitrate as the extracellular anion. The bars show the mean for 5–8 patches, error bars represent SEM.
(E) The GluR6 D497A mutant speeds desensitization compared to responses for wild-type GluR6 measured in NaCl (dotted line). Responses to 10 mM glutamate with Na+, K+, Rb+ or Cs+ measured in the same patch were almost identical in amplitude (see inset), with desensitization rates about six times faster than for wild-type responses in NaCl (dotted line).
(F) Bar plots summarizing the change in peak amplitude for wild type GluR6 and the GluR6 D497A mutant with Li+, Na+, K+, Rb+, Cs+ or NH4+ as the external monovalent cation; error bars represent SEM.
Consistent with the hypothesis that substitution of the carboxylate group by an amide should destabilize cation binding, the equivalent GluR6 mutant E493Q showed a reduced ability to discriminate between cations combined with accelerated desensitization compared to wild type GluR6 (Figure 5C and Table 1). The rate of onset of desensitization with 150 mM Na+ or Li+, 970 ± 90 s−1 (n=16) and 1300 ± 100 s−1 (n= 12) respectively, was 6.5 and 6.1-fold faster than for wild type GluR6, but for other cations kdes was about 1000 s−1 in each case (Table 1). When normalized to values recorded in Na+, E493Q also showed reduced changes in peak conductance (+20 to +100 mV) with an order NH4+ > Na+ ≈ K+ ≈ Cs+ > Li+ > Rb+ that is strikingly different than wild type (Figure 1). Despite its reduced cation sensitivity and faster desensitization rate, E493Q remained sensitive to anions, and the rate of onset of desensitization increased 2.6 and 2.1-fold, to 2500 ± 400 s−1 (n=8) and 2000 ± 700 s−1 (n=5) when Cl− was replaced by I− and NO3−, respectively (Figure 5D), similar to wild type GluR6, for which the rate of onset of desensitization increased 2.3 and 2.7-fold when Cl− was replaced by I− and NO3−.
Mutation to alanine of Asp497, the other acidic amino acid in the GluR6 cation binding site, also increased the desensitization rate (Figure 5E) and reduced sensitivity to cations (Figure 5F and Table 1). Surprisingly, even though the side chain carboxyl group was removed, increasing the diameter of the entrance to the cation binding pocket, the effect on the rate of onset of desensitization was no larger than observed for the isosteric E493Q mutant (Table 1); indeed, for responses in Li+ the rate of desensitization increased only 3.1-fold for GluR6 D497A but 7.2-fold for GluR6 E493Q. The cation selectivity of the peak glutamate activated conductance was also reduced in D497A (Figure 5E) compared to E493Q (Figure 5C and Table 1).
Crystal structures of K+, Rb+ and Cs+ complexes
Prior work suggested that kainate receptors have a unique requirement for Na+, for which only Li+ can act a substitute, while the presence of K+, Rb+ or Cs+ strongly inhibits receptor activity (Bowie, 2002; Paternain et al., 2003). Surprisingly, we observed that for crystals grown in 575 mM Li+ and then soaked for 36–48 hours in solutions with equimolar concentrations of either K+, Rb+ or Cs+, that these ions can bind at the same location as Na+ and Li+ (Figure 6). Because of their large x-ray scattering factor compared to Li+ and water, there is no ambiguity in the electron density maps for these structures, which were refined at resolutions of 1.72 Å for K+, 1.82 Å for Rb+ and 1.97 Å for Cs+ (Table 2). In addition, because Cs has a large anomalous signal,(f ` ` = 3.8 e) at the wavelength of 1Å used for data collection, the identity of Cs+ ions was confirmed by calculation of anomalous difference density maps. The structures were nearly identical to that for Na+ and superimposed with rmsds of 0.20, 0.22 and 0.19 Å for K+, Rb+ and Cs+ respectively, when calculated using domain 1 Cα coordinates, but showed subtle differences which likely have functional significance.
The most striking difference was that the cation binding sites were not saturated in the Rb+ and Cs+ structures. When the bound cations were refined at full occupancy, there were large negative peaks in Fo-Fc maps for both Rb+ and Cs+; manual refinement of ion occupancy using Fo-Fc maps as a guide gave values of 0.65 and 0.70 for the pair of Rb+ ions and 0.50 and 0.70 for Cs+; in contrast, when the bound cations were refined at full occupancy for the Li+, Na+, K+ and NH4+ complexes, Fo-Fc maps in the region of the cation were featureless. Second, the side chain for Arg760 changed conformation in the different complexes. In the Na+ and Li+ complexes, each of the Arg760 side chains in a dimer assumed a ‘down’ conformation, and as a result the guanidinium group moves towards the cation binding site, forming a hydrogen bond with the main chain carbonyl oxygen of Phe514 in the same subunit. In contrast, in the K+, Rb+ and NH4+ structures (Figure 6A), one of the Arg side chains flips to an ‘up’ conformation, moving away from the bound cation; in the Cs+ structure the Arg side chains in both subunits are in an up position (Figure 6B). This movement of Arg760 in the K+, Rb+, Cs+ and NH4+ complex structures opens up a binding site for a H2O molecule which is located within 3.5 Å of the bound cation and which makes hydrogen bond contacts with the carboxylate group of Glu509, the amino group of Lys516 and the main chain amide of Phe514. Third, the main chain peptide bond linking Asp513 with Phe514 adopts different conformations with mean Psi values of either 108 ± 7° (n=5), observed in both subunits in the Li+ complex and one subunit each for the Na+, K+ and NH4+ complexes, or 143 ± 8° (n=9), observed in both subunits in the Rb+ and Cs+ complexes and one subunit each for the Na+, K+, NH4+ complexes. For the K+ and NH4+ bound subunits, the electron density in subunit B clearly revealed both conformations at similar occupancy. As a result of this peptide bond flip, a hydrogen bond made with the hydroxyl group of Thr753 is broken when the carbonyl oxygen group of Asp513 rotates by 35° to the conformation observed in the Rb+ and Cs+ complexes. The dynamic behavior of this peptide bond was also observed in MD simulations. Fourth, in the Cs+ complex, electron density for the bound anion and its associated pair of H2O molecules was diffuse (Figure 6B), while for all of the other ion complexes there were distinct peaks of density for the Cl− ion and two H2O molecules (Figure 4A and Figure 7A). Further, the number of H2O molecules within 3.5 Å of the cations increased compared to the Na+ structure, and the larger cations bound less deeply in the pocket than Na+ due to their increased radius.
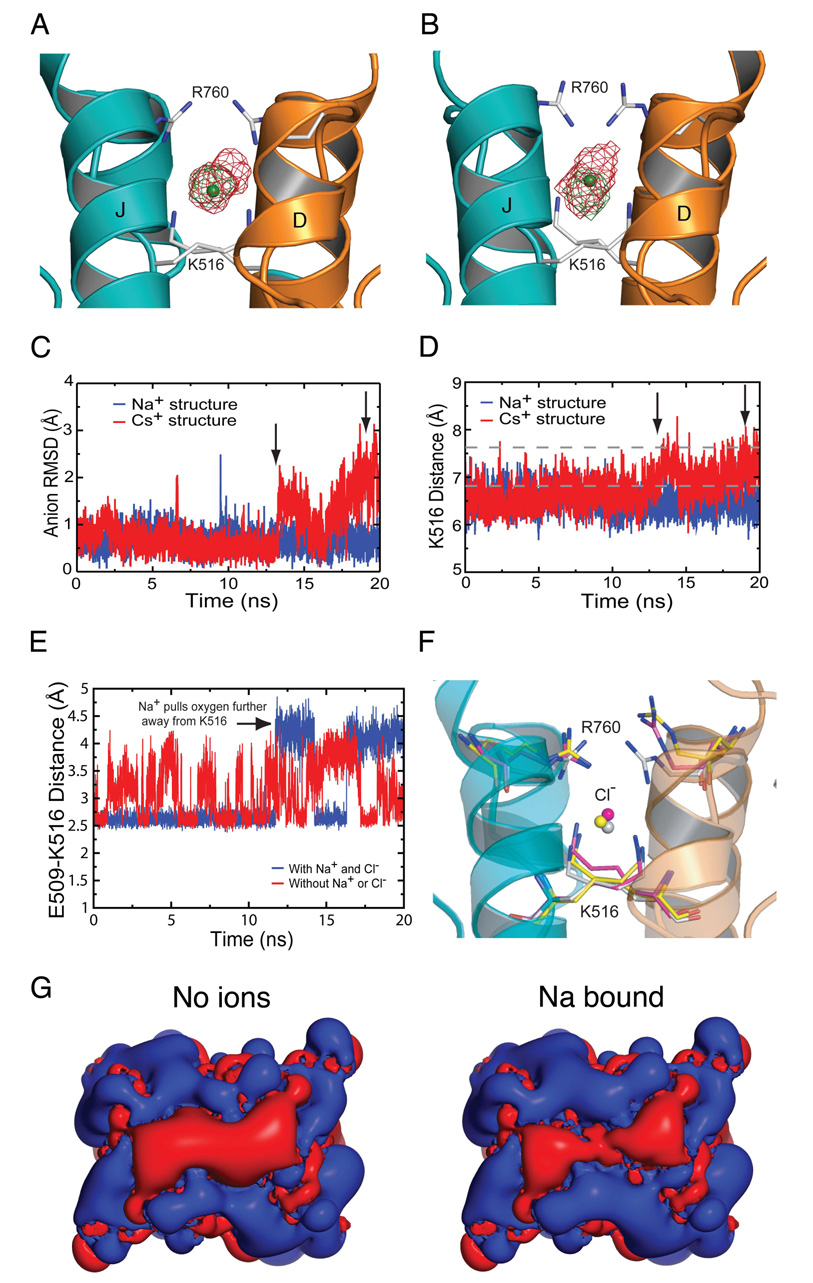
Figure 7. MD simulations reveal an interdependence of anion and cation mobility.
(A) Isocontour map of the Cl− ion position from a simulation of the Na-bound structure with Na+ and Cl− in the binding site (green) and from a simulation with no Na+ present (red).
(B) Isocontour map of the Cl− ion position from a simulation of the Cs-bound structure. The green map is from the data before the Cs+ ion leaves (0–13 ns). The red map is for the whole simulation (0–20 ns).
(C) RMSD of the Cl− ion in the Na-bound simulation (blue) and in the Cs-bound simulation (red). Arrows mark the time points at which the Cs+ ions leave the binding sites, the first at 13 ns and the second at 19 ns.
(D) Distance between the NZ atoms of K516 as a function of time for the Na-bound simulation (blue) and the Cs-bound simulation (red). The increase in distance coincides with the Cs+ ions leaving the protein. Dashed lines indicate the mean RMSD of the Cl− ion for two simulations run with Na+ omitted from the cation binding site.
(E) Distance between E509 OE1 and K516 NZ atoms for the simulation with Na+ ions present (blue) and without Na+ ions (red) in the binding pocket.
(F) Multiple conformations for R760 observed in MD simulations. The R760 structure from subunit A that is most representative of the largest cluster is shown in pink as are the corresponding K516 and Cl− atoms. The equivalent analysis for R760 from subunit B is shown in yellow. The crystal structure conformations for R750 and K516 in the Na+ complex are shown as white sticks.
(G) Isopotential surface maps calculated with APBS and contoured at −0.5 kT/e (red) and +0.5 kT/e (blue); view is the same as Figure 3. The panels on the left and right show the field with the cation site unoccupied and with Na+ bound respectively.
Kainate receptors are functional at high concentrations of K+, Rb+ and Cs+
Because the crystallographic data revealed that at the 575 mM monovalent cation concentration used for crystallization, the binding site showed weak discrimination between cations, we reexamined the ion sensitivity of GluR6 in functional assays over a wide range of concentrations for LiCl, NaCl, KCl, RbCl and CsCl. Responses measured with 15, 50 and 150 mM NaCl are shown in Figure 6C, while Figure 6D shows responses measured with 600 mM NaCl, KCl, RbCl or CsCl. At low salt concentrations the global fit of a binding isotherm to the rate of onset of desensitization converged at a common value of 1743 s−1 (Figure 6E), similar to the value of 1800 ± 200 s−1 measured in an independent experiment in which all monovalent ions were replaced by sucrose (Figure 1D). However, for each ionic species the EC50 differed (Figure 6E), and the minimal rate of desensitization extrapolated to saturating concentrations decreased in the order Cs+ > Rb+ > K+ > Li+ > Na+ (Figure 6F). The EC50 values and rate constants were Cs+ 398 mM, 530 s−1; Rb+ 167 mM, 341 s−1; K+ 77 mM, 255 s−1; Li+ 32 mM, 104 s−1; and Na+ 28 mM, 91 s−1. Thus, the cation binding site is not saturated by Cs+ at the 575 mM concentration used in the ion soaking experiments. A plot of the rate constant of desensitization estimated for saturating ion concentrations versus cation Shannon radius shows that Na+ is the most effective cation (Figure 6F), suggesting that the cation binding site has evolved to function optimally with the ions present in CSF.
Ions and allosteric binding site residues are dynamically coupled
The series of crystal structures solved in this study likely represent low energy conformations of a kainate receptor complex with bound cations and a Cl− ion. To gain insight into the mechanisms underlying allosteric coupling, we performed multiple 20 ns molecular dynamics (MD) simulations for wild type GluR5 and some mutants with different ion combinations (Supplementary information). For simulations using the Na+ bound crystal structure, we observed that the RMSD for the Cl− ion increases from 0.66 ± 0.23 and 0.85 ± 0.45 Å (runs 1 and 2 respectively) to 1.48 ± 0.86 and 1.83 ± 0.55 Å in the absence of Na+, suggesting that occupancy of the cation binding site by Na+ stabilizes anion binding (Figure 7A). In contrast, during simulations performed with Cs+, the Cl− ion shows increased fluctuation, and concurrent with dissociation of Cs+ ions from the binding pocket, the Cl− becomes delocalized (Figure 7B and C). This enhanced mobility of the bound anion is consistent with the diffuse electron density for Cl− observed in the Cs+ crystal structure. Associated with dissociation of Cs+ the distance between the NZ atoms of Lys516 increased, approaching values observed in the simulation with Na+ omitted from the cation binding site (Figure 7D).
We also examined the influence of ions on the structure of their binding sites. After fitting the D and J helices of each frame of the MD trajectory to the initial frame, the combined deviation of atoms in the Na+ binding site, residues Glu509, Asp513 and the backbone carbonyl group of Ile512 was computed. For trajectories started with both Na+ and Cl− bound, the RMSD quickly stabilizes to a value of 1.1 ± 0.2 Å, calculated over the last 10 ns from both subunits from both simulation runs, but for simulations run in the absence of Na+ with only Cl− bound, the RMSD shows greater fluctuation, mean value 1.4 ± 0.5 Å, indicating a higher degree of motion in the binding site in the cation-free state (Supplementary information). Simulations with just Na+ in the cation binding site also gave rise to a higher RMSD, mean 1.3 ± 0.3 Å, while in the absence of any ions the RMSD increased to 1.6 ± 0.4 Å, suggesting that conformational flexibility of the cation binding site, and thus we suggest affinity for Na+, is coupled to the presence of the Cl− ion.
Ion binding site side chains form intermolecular dimer contacts
Glu509 plays a key role in both cation binding and intermolecular contacts across the dimer interface. In the absence of bound cations, the side chain mobility increases and χ3 adopts two distinct conformations (Figure 7E). In subunit A, Glu509 flips between these 10 times for run 1 and 3 times for run 2. This flipping occurs because either of the carboxylate oxygen atoms can form a salt bridge with the NH4+ group of Lys516 in the partner subunit. For subunit B, flipping was less frequent with 1 flip in run 1 and no flips in run 2. The salt bridge between Glu509 and Lys516 is also present in the Na+ complex, but the other carboxylate oxygen of Glu509 now interacts directly with the Na+ ion. As a result, flipping is much less frequent when Na+ is bound, and occurred 2 and 1 times for subunit A, and 0 and 2 times for subunit B in the pair of trajectories. The conformation of Glu509 was dominated by the χ3 dihedral angle, and when flipping did occur the dihedral angles differed in the presence, −74 ± 17° and 117 ± 22°, and absence, −127 ± 22° and 73 ± 37°, of Na+. This difference occurs because the Na+ ion ‘pulls’ Glu509 into the cation binding site, and as a result the oxygen furthest from the NH4+ group of Lys516 moves by approximately 0.5 Å in the presence of Na+. Consistent with the results of physiological experiments and surface potential calculations (Figure 5), the E509Q mutant appears to be unfavorable for stable cation binding since in 6 independent simulations, 3 with Na+ and 3 with K+, cations leave the binding sites within 1 ns. By comparison, Na+ remained bound for the full length of the run in all of the WT simulations. The conformation of Asp513, the second acidic amino acid in the cation binding site, was stable in the Na+ complex, but in the absence of a cation, the carboxyl group flips frequently forming intermittent hydrogen bonds with H2O molecules.
Residues in the lid of the anion binding site are highly mobile
The MD trajectories revealed that Lys516 remains fairly rigid even in the absence of an anion, while the Arg760 side chain is highly mobile independent of the presence of ions. The stability of Lys516 results primarily from a hydrogen bond formed by the NH4+ group with the backbone carbonyl group of Phe514 of the adjacent subunit and from a salt bridge formed between Lys516 and Glu509. These contacts are highly conserved and also occur in AMPA receptor dimers (Armstrong and Gouaux, 2000). In MD simulations for ion-free GluR5 dimers, the NH4+ groups of Lys516 move ~1 Å away from each other, with a mean separation of 7.5 ± 0.3 Å compared to 6.5 ± 0.3 Å for ion bound simulations (Supplementary information). However, no concomitant movement of the backbone was observed, suggesting that Cl− acts to stabilize the interface rather than to pull the monomers together.
Strikingly, in GluR5 crystal structures for different cation complexes and in the UBP310 antagonist complex structure, the Arg760 side chains adopt a range of conformations. In the antagonist complex (Plested and Mayer, 2007) and in the kainite complex with Cs+, the side chains are in the ‘up’ conformation and make bidentate intermolecular salt bridges with Asp761 of the dimer partner subunit. Curiously, this is not observed in the Na+ and Li+ bound structures in which both side chains are in the ‘down’ conformation, while in the K+, Rb+ and NH4+ complex structures only one Arg760 side chain flips into the ‘up’ conformation. This has ramifications for how Cl− might access the anion binding site. To explore the underlying mechanism, we examined simulations with both Na+ and Cl− bound and performed a cluster analysis for each Arg760 side chain (Figure 7F). In all cases, the bidentate bridge with Asp761 was highly dynamic, breaking and reforming repeatedly. Although there was little pattern to the bidentate salt-bridge conformational stability, the absence of allosteric ions produced a consistent increase in the RMSD of Arg760, from 1.6 ± 0.4 Å for simulations with Na+ and Cl− present to 2.6 ± 0.7 Å for simulations with no bound ions.
These results illustrate the extreme mobility of Arg760 compared to the well ordered electron density for this residue observed in crystal structures. Because Arg760 is highly mobile this could provide a route for Cl− entry into an otherwise buried binding site. Our results also indicate that the Arg760–Asp761 bidentate salt-bridge is a transient structure, which in the UBP310 antagonist complex may be favored by crystal packing and favorably placed solute molecules, including a PEG molecule which lies across the dimer interface and fills up the water filled cavity seen in the Na+ complex.
DISCUSSION
Here, we define the mechanism by which Na+ ions modulate kainate receptor activity and solve the structure of the cation binding site. We show that the GluR5 dimer assembly binds Na+ and Cl− with a 2:1 stoichiometry; that the anion and cation binding sites are highly charged, physically discrete, and very dynamic; that the sites are allosterically and structurally coupled; and that occupancy of the ion binding sites is required to maintain kainate receptors in the active conformation. We show that the cation binding site discriminates weakly between cations, but that coupled with differences in efficacy this is sufficient to confer selectivity for Na+ over K+ at physiological ionic strength. Additional calculations, including free energy perturbations, will be required to understand the basis of this selectivity, but these structures show several interesting features relevant to this. Although the cation binding site is exposed to bulk solvent, ions bind in a tunnel where multiple protein ligands substitute for waters of hydration and shield the ion. The tunnel also focuses the electric field in the cation binding site, and thus selectivity for Na+ and Li+ over larger cations would be expected due to the high field strength (Noskov and Roux 2008).
Structural and allosteric coupling of the anion and cation binding sites
Because the isolated ligand binding domains used for structural analysis are uncoupled from the ion channel, we can only speculate on how occupancy of the ion binding sites alters during conformational changes linked to the onset of and recovery from desensitization. In the absence of allosteric cations, kainate receptor dimers have a strong negative electric field which acts as an energy barrier preventing access of anions to their binding site in the dimer interface; binding of Na+ reduces the field, allowing Cl− ions to approach the dimer surface (Figure 7G). The high mobility of the Arg760 side chains observed in MD simulations reveals that access to the anion binding site is much less restricted than suggested by the crystal structures, and it is possible that Cl− ions diffuse in from the top surface of the dimer interface. Another possible route of entry is a solvent filled tunnel that transiently connects the anion and cation binding sites, which in the crystal structures appear as a cavity surrounded by two fingers protruding into the protein interior (Figure 3). During desensitization there are likely to be large movements of the subunits in a dimer interface (Armstrong et al., 2006), which modeling studies show disrupt the anion binding site. Surface potential calculations for a model kainate receptor dimer in the desensitized state based on the corresponding GluR2 structure (Armstrong et al., 2006) indicate that in the desensitized state the cation binding sites are also disrupted, in large part due to solvent exposure of the dimer interface, reducing the field potential that supports cation binding.
Although the anion and cation binding sites are physically discrete, their close proximity and shared structural components have the consequence that mutations to one site perturb the other. In prior work, this led Wong et al. (2007) to conclude that the anion and cation binding sites were not separate and that the ions interacted via an attractive dipole mechanism, which our data reveal is not correct. Some mutations such as GluR6 E493Q do have much stronger effects on cation modulation than anion modulation (Figure 5), while others such as GluR6 R744K have stronger effects on anion modulation (Plested and Mayer, 2007). The basis for this ‘selectivity’ is apparent from inspection of the crystal structures, but cross talk between the sites, an inevitable result of their complex electrostatic profiles and shared structural elements, coupled with the fact that all of the mutations destabilize the dimer complex to different extents, has the consequence that no single mutation exclusively affects modulation by a single ion species.
Cation binding sites in other iGluRs
Crystal structures have now been solved for representative members from each of the major iGluR gene families, except KA1 and KA2. Calculation of surface potential maps for crystal structures of GluR2 homodimers, NR1 homodimers, NR1/NR2 heterodimers, and a delta 2 subunit apo dimer reveals that the highly charged pockets which bind Na+ are present only in kainate receptors. Analysis of GluR5 and GluR6 structures which crystallized as monomers or as oligomeric assemblies which have no biological counterpart, reveals that the highly charged cation binding sites described here are generated only in physiological dimer assemblies in the active conformation. Of interest, the iGluR delta 2 subunit crystallizes as a dimer which binds Ca2+ ions (Naur et al., 2007), and the delta 2 lurcher mutant shows positive allosteric modulation by Ca2+ (Wollmuth et al., 2000). We propose that, as for kainate receptors, dimer stability of delta 2 receptors is regulated by cations, specifically Ca2+, and that this is the mechanism by which Ca2+ facilitates delta 2 subunit activity. Although the cation binding sites in delta receptors share sequence homology with those in kainate receptors, the crystal structures are very different. Surprisingly, the Ca2+ binding site lacks the strong negative potential found in kainate receptors, and the Ca2+ ion is buried in the protein interior. Although the delta2 subunit has Arg and Asp residues at positions equivalent to GluR5 Arg760 and Asp761, both side chains have rotated outwards by ≈ 90° along the axis of molecular two-fold symmetry. As a result, the Ca2+ ion is coordinated by the carboxyl groups of Asp761 from one subunit and the equivalents to Glu509 and Asp513 from the dimer partner, while for kainate receptors all of the protein contacts with Na+ are made by a single subunit.
In the GluR5 Cs+ complex, an additional 11 Cs+ ions were bound to the surface of the protein, in addition to the pair of Cs+ ions located in the cation binding sites. These ‘extra’ Cs+ ions were initially identified by their large electron density peaks in 2Fo-Fc maps and subsequently from inspection of anomalous density difference maps. These extra cation binding sites are weakly electropositive and bind Cs+ ions via interactions with surface exposed carboxylate and carbonyl group oxygen atoms. Like-wise, in the Rb+ and K+ complexes, four additional Rb+ ions and one additional K+ ion were located at the some of the same sites which bound Cs+, while in the other ion complexes these sites frequently contained H2O molecules, which were likely misidentified cations with partial occupancy that could not be discriminated from H2O on the basis of electron density alone. In the Na+ complex crystal structure, bond valence analysis failed to identify any of these H2O molecules as Na+ ions, most likely due to our inability to model complete hydration shells for surrounding water molecules with high B factors (Nayal and Di Cera, 1996). These “extra” ions were also included in the MD simulations and in all cases were found to dissociate from the protein into the solution. Such ‘non-specific’ binding of cations and anions to protein surfaces is a well characterized phenomenon in high resolution crystal structures; is necessary to maintain electroneutrality of the protein surface; and has been used as a method to phase novel structures (Nagem et al., 2001).
Functional significance of allosteric regulation by cations
Our experiments were performed on mammalian kainate receptors, but analysis of gene sequences reveals that the key acidic amino acid side chains and intermolecular salt bridges are present in putative kainate receptors from chick, zebrafish, Xenopus and possibly drosophila (Supplementary Figure 2), suggesting early evolution and conservation of the cation binding site. Although the ligand binding properties of non mammalian iGluRs are poorly defined, and it is unclear whether true AMPA and kainite preferring subtypes exist in invertebrates, our analysis clearly reveals that two families of non-NMDA subtype iGluRs can be distinguished by the presence of either a hydrophobic amino acid at the position equivalent to Ile755 in GluR5 or a conserved lysine, corresponding to kainate and AMPA subtypes of mammalian iGluRs respectively.
Because synaptic membrane proteins function in a chemically polarized ionic environment, with Na+ rich extracellular and K+ rich intracellular compartments and higher extracellular Cl− concentration, the presence of allosteric ion binding sites in kainate receptors is likely to be functionally important. As noted by Paternain et al. (2003), there are large drops in Na+ ion activity during electrical activity which might regulate kainate receptor activity. The K+ rich, low Cl− cytoplasmic ionic environment will favor kainate receptor desensitization, which is required for efficient trafficking to cell surface membranes (Priel et al., 2006; Vivithanaporn et al., 2007). Although our studies were by necessity performed using recombinant receptors, the allosteric action of cations has been observed in studies on native kainate receptors expressed in hippocampal neurons (Paternain et al., 2003). In vivo, such regulation works in conjunction with changes in activity produced by protein kinases and phosphatases and likely will be modulated by accessory proteins which coassemble with many ion channel classes. As a result, much work remains to be done before we will have a full understanding of the allosteric regulation of kainate receptors and other subtypes of iGluRs.
EXPERIMENTAL PROCEDURES
Molecular biology and physiology
Wild-type and mutant glutamate receptors were expressed in HEK-293 cells for outside-out patch recording with rapid solution exchange, as described previously (Plested and Mayer, 2007). Mutations were introduced by overlap PCR and confirmed by sequencing. Brief pulses of glutamate (10 mM) were applied to outside-out patches using a piezo-driven perfusion system. Solution exchange was complete in ~200 µs and patches with exchange times slower than 400 µs (as assessed by junction currents) were discarded. Data were filtered at 2–12 kHz and sampled at 10–50 kHz. Except where noted, the external solution contained (in mM): 0.1 MgCl2, 0.1 CaCl2 5 HEPES and 150 mM XCl (X= Li, Na, K, Rb, Cs, NH4, Tris or NMDG). The internal solution contained (in mM): 115 NaCl, 10 NaF, 0.5 CaCl2, 1 MgCl2, 5 Na4BAPTA, 5 HEPES and 10 Na2ATP, pH 7.3. Further details are described in supplementary information.
Crystallography
The GluR5 S1S2 ligand binding core was overexpressed and purified as previously described (Mayer, 2005) and crystallized as described in supplementary information. The Li complex structure was solved by molecular replacement with PHASER (McCoy et al., 2007) using the GluR6 kainate complex (PDB 1TT1) as a search probe. There were two molecules in the asymmetric unit which coassemble as a dimer. Although the GluR6 search probe and GluR5 refined structures differ in domain closure by 7.2°, automated rebuilding of the molecular replacement solution using ARP/wARP (Morris et al., 2003) traced 249/258 residues in chain A and 229/258 residues in Chain B. The model was then rebuilt with COOT (Emsley and Cowtan, 2004) followed by cycles of restrained refinement with REFMAC5 (Winn et al., 2001), with 3–4 TLS groups per subunit identified by TLSMD (Painter and Merritt, 2006); there was no electron density for the first three residues and the C-terminal glutamate. Structures for the other complexes were solved by rigid body refinement using the Li complex structure stripped of alternative conformations, ligands, ions, and solvent as a starting model, with side chain occupancies for Glu509, Asp513, Arg760 and Asp761 initially set to zero, followed by cycles of manual rebuilding and crystallographic refinement. Additional crystallographic calculations and the generation of maps were performed using the CCP4 suite of programs (Collaborative Computational Project Number 4, 1994). Electrostatic calculations were performed using APBS (Baker et al., 2001), with 150 mM monovalent ion concentrations, assuming dielectrics of 80 for the solution and 2 for protein. Figures were prepared using PYMOL (Delano, 2002).
Molecular dynamics simulations
Molecular dynamics simulations of the Li+, Na+, K+, Rb+ and Cs+ crystal structures were performed using GROMACS 3.3.1 (van der Spoel et al., 2005) with a 2 fs integration time step and a 20 ns duration with two repeats for each structure. Periodic boundary conditions were used and long range electrostatics were treated using the particle mesh Ewald (PME) method (Essmann et al., 1995) with a real-space cutoff of 10 Å. Simulations were performed in an NPT ensemble at 300 K and 1 atm pressure using the Berendsen algorithm (Berendsen et al., 1984). Further details are given in supplementary material.
Supplementary Material
Acknowledgements
We thank Carla Glasser for preparing cDNAs and proteins; the NINDS DNA sequencing facility for support; Peter Freddolino for writing a script to map surface potentials calculated by APBS to atomic coordinates; Drs. S. Heinemann and P. Seeburg for the gift of wild type cDNAs; and Dr. Eric Gouaux for advice and discussion. Synchrotron diffraction data was collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. We thank the UK National Grid Service and the Oxford Supercomputer Centre for computing time. This work was supported by the intramural research program of NICHD, NIH, DHHS (MLM), the Oxford University Clarendon Fund, the Overseas Research Students Awards Scheme (RV), and the Wellcome Trust (PCB).
A.J.R.P designed and performed electrophysiological experiments; M.L.M. performed crystallization experiments and solved the structures; R.V. and P.C.B performed MD simulations; all authors contributed to the interpretation of data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Atomic coordinates and structure factors have been deposited with the Protein Data Bank; accession codes are 3C31 (Li), 3C32 (Na), 3C33 (K), 3C34 (Rb), 3C35 (Cs) and 3C36 (NH4).
References
- Alvarez O, Gonzalez C, Latorre R. Counting channels: a tutorial guide on ion channel fluctuation analysis. Adv Physiol Educ. 2002;26:327–341. doi: 10.1152/advan.00006.2002. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics. 1984;81:3684–3690. [Google Scholar]
- Bowie D. External anions and cations distinguish between AMPA and kainite receptor gating mechanisms. J Physiol. 2002;539:725–733. doi: 10.1113/jphysiol.2001.013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo PE, Malenka RC, Nicoll RA. Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature. 1997;388:182–186. doi: 10.1038/40645. [DOI] [PubMed] [Google Scholar]
- Chittajallu R, Vignes M, Dev KK, Barnes JM, Collingridge GL, Henley JM. Regulation of glutamate release by presynaptic kainate receptors in the hippocampus. Nature. 1996;379:78–81. doi: 10.1038/379078a0. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallog sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- DeVries SH, Schwartz EA. Kainate receptors mediate synaptic transmission between cones and 'Off' bipolar cells in a mammalian retina. Nature. 1999;397:157–160. doi: 10.1038/16462. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. The Journal of Chemical Physics. 1995;103:8577–8593. [Google Scholar]
- Goodford PJ. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- Harding MM. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr D Biol Crystallogr. 2002;58:872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- Kidd FL, Isaac JT. Developmental and activity-dependent regulation of kainate receptors at thalamocortical synapses. Nature. 1999;400:569–573. doi: 10.1038/23040. [DOI] [PubMed] [Google Scholar]
- Madden DR. The structure and function of glutamate receptor ion channels. Nat Rev Neurosci. 2002;3:91–101. doi: 10.1038/nrn725. [DOI] [PubMed] [Google Scholar]
- Mayer ML. Crystal Structures of the GluR5 and GluR6 Ligand Binding Cores: Molecular Mechanisms Underlying Kainate Receptor Selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Armstrong N. Structure and function of glutamate receptor ion channels. Annu Rev Physiol. 2004;66:161–181. doi: 10.1146/annurev.physiol.66.050802.084104. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Ghosal A, Dolman NP, Jane DE. Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- Muller P, Kopke S, Sheldrick GM. Is the bond-valence method able to identify metal atoms in protein structures? Acta Crystallogr D Biol Crystallogr. 2003;59:32–37. doi: 10.1107/s0907444902018000. [DOI] [PubMed] [Google Scholar]
- Nagem RA, Dauter Z, Polikarpov I. Protein crystal structure solution by fast incorporation of negatively and positively charged anomalous scatterers. Acta Crystallogr D Biol Crystallogr. 2001;57:996–1002. doi: 10.1107/s0907444901007260. [DOI] [PubMed] [Google Scholar]
- Naur P, Hansen KB, Kristensen AS, Dravid SM, Pickering DS, Olsen L, Vestergaard B, Egebjerg J, Gajhede M, Traynelis SF, Kastrup JS. Ionotropic glutamate-like receptor delta2 binds D-serine and glycine. Proc Natl Acad Sci U S A. 2007;104:14116–14121. doi: 10.1073/pnas.0703718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayal M, Di Cera E. Valence screening of water in protein crystals reveals potential Na+ binding sites. J Mol Biol. 1996;256:228–234. doi: 10.1006/jmbi.1996.0081. [DOI] [PubMed] [Google Scholar]
- Noskov SY, Roux B. Control of Ion Selectivity in LeuT: Two Na(+) Binding Sites with Two Different Mechanisms. J Mol Biol. 2008;377:804–818. doi: 10.1016/j.jmb.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Perin-Dureau F, Fayyazuddin A, Le Goff A, Callebaut I, Neyton J. Molecular organization of a zinc binding n-terminal modulatory domain in a NMDA receptor subunit. Neuron. 2000;28:911–925. doi: 10.1016/s0896-6273(00)00163-x. [DOI] [PubMed] [Google Scholar]
- Paternain AV, Cohen A, Stern-Bach Y, Lerma J. A role for extracellular Na+ in the channel gating of native and recombinant kainate receptors. J Neurosci. 2003;23:8641–8648. doi: 10.1523/JNEUROSCI.23-25-08641.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin-Dureau F, Rachline J, Neyton J, Paoletti P. Mapping the binding site of the neuroprotectant ifenprodil on NMDA receptors. J Neurosci. 2002;22:5955–5965. doi: 10.1523/JNEUROSCI.22-14-05955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML. Structure and mechanism of kainate receptor modulation by anions. Neuron. 2007;53:829–841. doi: 10.1016/j.neuron.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Priel A, Selak S, Lerma J, Stern-Bach Y. Block of kainate receptor desensitization uncovers a key trafficking checkpoint. Neuron. 2006;52:1037–1046. doi: 10.1016/j.neuron.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wahl P. Control of rat GluR6 glutamate receptor open probability by protein kinase A and calcineurin. J Physiol (Lond) 1997;503:513–531. doi: 10.1111/j.1469-7793.1997.513bg.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. Journal of Computational Chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Vignes M, Collingridge GL. The synaptic activation of kainate receptors. Nature. 1997;388:179–182. doi: 10.1038/40639. [DOI] [PubMed] [Google Scholar]
- Vivithanaporn P, Lash LL, Marszalec W, Swanson GT. Critical roles for the M3-S2 transduction linker domain in kainate receptor assembly and postassembly trafficking. J Neurosci. 2007;27:10423–10433. doi: 10.1523/JNEUROSCI.2674-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MC, Gertler C, Mayer ML, Rosenmund C. Interdomain interactions in AMPA and kainate receptors regulate affinity for glutamate. J Neurosci. 2006;26:7650–7658. doi: 10.1523/JNEUROSCI.1519-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Wollmuth LP, Kuner T, Jatzke C, Seeburg PH, Heintz N, Zuo J. The Lurcher mutation identifies delta 2 as an AMPA/kainate receptor-like channel that is potentiated by Ca(2+) J Neurosci. 2000;20:5973–5980. doi: 10.1523/JNEUROSCI.20-16-05973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AY, Fay AM, Bowie D. External ions are coactivators of kainite receptors. J Neurosci. 2006;26:5750–5755. doi: 10.1523/JNEUROSCI.0301-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AY, MacLean DM, Bowie D. Na+/Cl− dipole couples agonist binding to kainate receptor activation. J Neurosci. 2007;27:6800–6809. doi: 10.1523/JNEUROSCI.0284-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.