Abstract
Synthetic genetic array analyses identify powerful genetic interactions between a thermosensitive allele (sec14-1ts) of the structural gene for the major yeast phosphatidylinositol transfer protein (SEC14) and a structural gene deletion allele (tlg2Δ) for the Tlg2 target membrane-soluble N-ethylmaleimide-sensitive factor attachment protein receptor. The data further demonstrate Sec14 is required for proper trans-Golgi network (TGN)/endosomal dynamics in yeast. Paradoxically, combinatorial depletion of Sec14 and Tlg2 activities elicits trafficking defects from the endoplasmic reticulum, and these defects are accompanied by compromise of the unfolded protein response (UPR). UPR failure occurs downstream of Hac1 mRNA splicing, and it is further accompanied by defects in TOR signaling. The data link TGN/endosomal dynamics with ceramide homeostasis, UPR activity, and TOR signaling in yeast, and they identify the Sit4 protein phosphatase as a primary conduit through which ceramides link to the UPR. We suggest combinatorial Sec14/Tlg2 dysfunction evokes inappropriate turnover of complex sphingolipids in endosomes. One result of this turnover is potentiation of ceramide-activated phosphatase-mediated down-regulation of the UPR. These results provide new insight into Sec14 function, and they emphasize the TGN/endosomal system as a central hub for homeostatic regulation in eukaryotes.
INTRODUCTION
Eukaryotic cells control signal transduction by compartmentalizing membrane surfaces so that signaling reactions are registered with high spatial and temporal precision. Phosphoinositides (PIPs) represent major components of membrane-associated signaling systems across the Eukaryota (Fruman et al., 1998; Strahl and Thorner, 2007). PIPs are well suited for diverse roles in signaling because of their chemical heterogeneity—a heterogeneity that permits “on-demand” formation of unique membrane domains upon which specific biological reactions are localized. In addition to the information defined by PIP identity, there exist additional layers of functional specification for PIP signaling at the level of on-demand PIP synthesis. Recent work identifies phosphatidylinositol/phosphatidylcholine (PtdIns/Ptd Cho)-transfer proteins (PITPs) as important coincidence detectors that stimulate PtdIns kinase activity in response to specific metabolic cues (Ile et al., 2006; Schaaf et al., 2008). Sec14, the major yeast PITP, is the founding member of the uniquely eukaryotic Sec14-protein superfamily whose large diversity is apparent in even the simplest eukaryotic cells (Phillips et al., 2006). Such an evolutionary expansion of the Sec14-superfamily argues for a high degree of functional specification for Sec14-like proteins. This conjecture has large implications for how PIP signaling can be further diversified, and it is supported by experimental data in both yeast and higher plants (Routt et al., 2005; Vincent et al., 2005; Schaaf et al., 2008).
Structural studies reveal, for the first time, the mechanisms by which Sec14 and other Sec14-like PITPs regulate lipid-modifying enzymes. Crystallographic and functional studies indicate these proteins couple a sensor function with an interfacial channeling of PtdIns to PtdIns-kinases (Schaaf et al., 2008). This level of control is essential in cells because of the poor activity of PtdIns-kinases on liposomal PtdIns substrates, and it requires heterotypic exchange of PtdIns and PtdCho by Sec14 in order to stimulate PIP synthesis. The structural design of the Sec14 molecule is intriguing in that its PtdIns and PtdCho binding substrates occupy surprisingly distinct, but nonetheless overlapping, sites within the PITP. The data are best explained by mechanisms where Sec14-like PITPs function as biochemical nanoreactors with coincidence–detection (i.e., sensor) power. Thus, Sec14-like PITPs have the capacity to stimulate on-demand synthesis of specific PIPs in response to specific physiological cues (Ile et al., 2006; Schaaf et al., 2008).
Although crystallographic and biophysical studies are illuminating how Sec14 and Sec14-like proteins operate at the level of the single molecule (Sha et al., 1998; Phillips et al., 1999; Smirnova et al., 2006, 2007; Ryan et al., 2007; Schaaf et al., 2008), a precise understanding of the physiological roles for Sec14 remains incomplete. Genetic studies amply demonstrate that Sec14 coordinates phospholipid (PL) metabolism with membrane trafficking (Bankaitis et al., 1990; Cleves et al., 1991a,b; McGee et al., 1994; Skinner et al., 1995; Fang et al., 1996; Xie et al., 1998; Rivas et al., 1999; Li et al., 2002). Based on both biochemical and morphological criteria, the membrane trafficking block associated with Sec14 deficiencies is assigned to the level of vesicle formation on yeast late-Golgi (i.e., trans-Golgi network [TGN]) membranes (Novick et al., 1980; Bankaitis et al., 1989; Cleves et al., 1991b). Yet, various data report Sec14 involvement may be required for only a subset of post-Golgi trafficking pathways (Cleves et al., 1991b; Fang et al., 1996; Schaaf et al., 2008). Given recent demonstrations that secretory cargo are packaged into distinct vesicle populations (Harsay and Bretscher,1995), and, given the suggestion that some pathways for exocytosis emanate from endosomal compartments (Harsay and Schekman, 2002), it is apparent that the Sec14 execution point(s) is not precisely defined.
Herein, we report the use of synthetic genetic array (SGA) analyses to further address how Sec14 functions in the yeast secretory pathway. The data highlight Tlg2, a target membrane-soluble N-ethylmaleimide-sensitive factor attachment protein receptor (t-SNARE) that regulates TGN/endosome dynamics, as a particularly robust genetic “interactor” with sec14-1ts mutations. Consistent with the genetic interaction data, analyses of Snc1 vesicle (v)-SNARE and plasma membrane-vacuole trafficking reveal significant defects in endosomal trafficking pathways in Sec14-deficient yeast strains. Paradoxically, we find yeast double mutants deficient in both Sec14 and Tlg2 are strongly defective in protein transport from the endoplasmic reticulum (ER). This ER-export defect is accompanied by failure of the unfolded protein response (UPR) and compromised target of rapamycin (TOR) signaling. Lipidomic and biochemical experiments identify deranged ceramide homeostasis, and activity of the Sit4 ceramide-activated phosphatase, as a significant basis for stress response failure. We propose that Sec14 insufficiencies, when combined with loss of Tlg2 activity, derange TGN/endosome dynamics—and sphingolipid (SL) trafficking—to such an extent that abnormal turnover of complex SLs occurs in the TGN/endosomal system. Inappropriate SL catabolism expands ceramide pools with the consequence that ceramide-activated phosphatase pathways are abnormally activated. These findings provide new insight into Sec14 function in yeast, and they further emphasize the involvement of TGN/endosomes as central hubs for control of intracellular homeostatic processes in eukaryotic cells.
MATERIALS AND METHODS
Yeast Strains and Media
Yeast complex medium and synthetic complete media were prepared as described previously (Sherman et al., 1983). Methods for transforming yeast with plasmids and linear DNA fragments were based on published protocols (Ito et al., 1983; Rothstein, 1983; Sherman et al., 1983). Fine chemicals were obtained from Sigma-Aldrich (St. Louis, MO), except where specified. Restriction endonucleases were supplied by New England Biolabs (Ipswich, MA). 35S-Translabel was purchased from MP Biomedicals (Irvine, CA). The genotypes of relevant yeast strains are listed in Table 1. Plasmids used are listed in Supplemental Table S1.
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| CTY1-1A | Mat a, sec14-1, ura3-52, his3-200, lys2-801 |
| CTY2-1 Cα | Mat α, sec14-1, ade2-101 |
| CTY83 | Mat a, sec6-4, ura3Δ |
| CTY159 | Mat a, sec14-1, kes1-1, ura3-52, his3-200, lys2-801 |
| CTY160 | Mat a, sec14-1, cki1-1, ura3-52, his3-200, lys2-801 |
| CTY182 | Mat a, ura3-52, his3-200, lys2-801 |
| CTY1568 | Mat α, stt4-4ts, ura3Δ, lys2-801, trp1-1 |
| CTY1626 | Mat α, pik1-101, ade2-101 his3Δ-1, ura3Δ |
| CTY1793 | Mat α, his3Δ-1, leu2Δ, ura3Δ, met15Δ, can1Δ::pMFA1-HIS3 |
| CTY1916 | Mat a, snc2Δ::KanMX4 his3Δ-1, leu2Δ, met15Δ, ura3Δ |
| CTY1918 | Mat a, tlg2Δ::KanMX4, his3Δ-1, leu2Δ, met15Δ, ura3Δ |
| CTY1919 | Mat a, gsg1Δ::KanMX4 his3Δ-1, leu2Δ, met15Δ, ura3Δ |
| CTY1920 | Mat a, sec14-1, tlg2Δ::KanMX4, ade2-101 his3Δ-1, leu2Δ, ura3Δ |
| CTY1958 | CTY159 tlg2Δ::KanMX4 |
| CTY1960 | Mat α, stt4-4ts, tlg2Δ::KanMX4, ura3Δ, lys2-801, trp1-1 |
| CTY1961 | Mat α, pik1-101, tlg2Δ::KanMX4, ade2-101 his3Δ-1, ura3Δ |
| CTY1962 | Mat α, sec14-1, snc2Δ::KanMX4, ade2-101, his3Δ-1, leu2Δ, ura3Δ |
| CTY1963 | CTY160 tlg2Δ::KanMX4 |
| CTY1964 | CTY1793 sec14-1::URA3 |
| CTY1967 | Mat α, sec14-1, gsg1Δ::KanMX4, ade2-101, his3Δ-1, leu2Δ, ura3Δ, met15Δ |
| CTY1968 | CTY1920 lag1Δ::HIS3 |
| CTY1969 | CTY1920 isc1Δ::HIS3 |
| CTY1970 | CTY1920 ppn1Δ::HIS3 |
| CTY1971 | CTY1920 sit4Δ::HIS3 |
| CTY1972 | Mat α, sec14-1, tlg2Δ::KanMX4 |
Genetic Manipulations
To generate the sec14-1ts::URA3 allele, a 739-base pairs ′sec14-1ts fragment was amplified from pCTY468 with the use of oligonucleotides SEC14-A (5′-AAAGAATTCCTGCGAAAAAT GGAGGAAGGATTATGGTACCG-3′) and SEC14-B (5′-CCCGCATGCCCCAGATCTCGT TACTTAGAACTCCTCTTTTCTCTCTCG-3′) that are clamped with EcoRI and BglII sites, respectively (underlined in primer sequences). The ‘sec14-1ts polymerase chain reaction (PCR) product, bounded by nucleotides +389 to +1128, includes the temperature-sensitive mutation and sec14-1ts sequence that lies downstream of the sec14-1ts termination codon. The resulting ‘sec14-1ts fragment was digested at its unique EcoRI and BglII sites, and the digestion product was subcloned into pRE247 to yield plasmid pRE853.
The yeast URA3 gene was amplified with oligonucleotides URA3-A (5′-CCCAGATCTGAAGAGTATTGAGAAGGGCAACGG-3′) and URA3-B (5′-CCCGAGCTCGGTTCTGGCGAGGTATTGGATAGTTCC-3′) from pCTY468 and clamped with BglII and SacI sites (underlined in primer sequences). The URA3 PCR product was ligated into plasmid pRE853, in flanking configuration to ‘sec14-1ts to create plasmid pRE860. Oligonucleotides SEC14-C (5′-CCCAGATCTCCCGAGCTCCGAGAGAGAAAAGAGGAGTTCTAAGTAACG-3′) and SEC14-D (5′-CCCGCATGCGGCTTGTGATACAAAACGGCTCAACCG-3′) were used to amplify a 690-base pair ‘sec14-1ts fragment clamped with SacI and SphI sites (underlined). The ‘sec14-1ts PCR product, bounded by nucleotide sequence +998 to +1688, was introduced adjacent to URA3 in pRE860 to yield pRE861. This plasmid contains the sec14-1ts::URA3 allele, which was excised from pRE861 by an EcoRI and SphI digest, transformed into CTY1793, and incorporated via homologous recombination to create CTY1964. Integration events were confirmed by PCR and unselected ts growth phenotypes at 37°C that were rescued by SEC14.
Deletion of the LAG1, ISC1, PPN1, and SIT4 coding regions used disruption cassettes consisting of yeast HIS3 as selectable marker flanked by 35–40 nucleotides homologous to the targeted gene. Disruption cassettes were designed such that, following integration into the yeast genome, the entire open reading frame coding region of the target gene was replaced with HIS3. Correct recombinants were validated by diagnostic PCR. A comprehensive list of primers used in these experiments is available from us by request.
Synthetic Genetic Array
SGA analysis was performed as described previously (Tong et al., 2001, 2004). Briefly, the sec14-1ts::URA3 strain CTY1964 was robotically crossed against an array of 4672 individual knockouts of nonessential genes to generate sec14-1ts double mutant arrays. The resulting sec14-1ts double mutants were then screened for genetic interactions. Tetrad analyses confirmed synthetic sec14-1ts genetic interactions of interest.
Fluorescence Microscopy
N-[3-Triethylammoniumpropyl]-4-[p-diethylaminophenylhexatrienyl] pyridinium dibromide (FM4-64) staining was performed as described by Vida and Emr (1995). Cells were grown to mid-logarithmic phase in synthetic defined medium at 30°C and offered FM4-64 (Invitrogen, Carlsbad, CA) for 10 min. Labeling was terminated by intoxicating cells with a NaN3/NaF cocktail (1 mM each). For green fluorescent protein (GFP)-Snc1 and FYVE-dsRed imaging cells were also cultured in synthetic defined medium at 30°C and shifted to 37°C as appropriate. Cells were viewed with an E600 microscope (Nikon, Tokyo, Japan) equipped with a 512 × 512 back-illuminated frame-transfer charge-coupled device camera (Princeton Instruments, Trenton, NJ) and MetaMorph software (Molecular Devices, Sunnyvale, CA) was used to capture images.
Transmission Electron Microscopy
Yeast were grown to mid-logarithmic phase (OD600 nm = 0.3), and cultures were shifted to 37°C for 2 h. Ten OD600 nm units of cells were isolated and fixed in 3% glutaraldehyde. Cells were spheroplasted in the presence of Zymolyase, stained with 2% osmium tetroxide and 2% uranyl acetate, dehydrated in a 50, 70, 90% ethanol series, and washed in 100% ethanol and 100% acetone, respectively. On embedding in Spurr's resin, cell pellets were incubated at 60°C for 48 h. Sections were prepared as described previously (Adamo et al., 2001) and visualized at 80 kV on a Tecnai 12 electron microscope (FEI, Hillsboro, OR). Images were captured using Gatan micrograph 3.9.3 software (Gatan, Pleasanton, CA).
Metabolic Labeling and Immunoprecipitation
The appropriate strains were grown in minimal media lacking methionine and cysteine to mid-logarithmic phase (OD600 nm = 0.5). Where indicated, cultures were shifted to 30, 33.5, or 37°C for 2 h and radiolabeled with [35S]-amino acids (100 μCi/ml, Translabel; New England Nuclear). Chase was initiated by introduction of unlabeled methionine and cysteine (2 mM each, final concentration) for the specified time, and chase was terminated by addition of trichloroacetic acid (5% wt/vol, final concentration). Immunoprecipitation and resolution of carboxypeptidase Y (CPY) (Cleves et al., 1991; Young et al., 2000) and hemagglutinin (HA)-Hac1I (Chapman and Walter, 1997) by SDS-polyacrylamide gel electrophoresis (PAGE) and autoradiography were performed as described previously.
Determination of LacZ Activity
Yeast strains were transformed with plasmid pJT30 (Wilkinson et al., 2000), and transformants were selected for uracil prototrophy. Overnight cultures were diluted to an OD600 nm of 0.1 and incubated at 30°C for 4 h in media selective for uracil prototrophy. Where specified, cells were shifted to 37°C for 2 h. Cells were subsequently isolated, and resuspended in 5 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 10 mM MgSO4, 50 mM 2-mercaptoethanol, pH 7.0). Aliquots (0.8 ml) were collected, cells were permeabilized in 50 μl of 0.1% (wt/vol) SDS and 100 μl of CHCl3, and samples were equilibrated to 30°C. Assays were initiated by addition of 160 μl of o-nitrophenyl-galactopyranoside (4 mg/ml stock solution in Z buffer) and incubated at 30°C for 20 min. Reactions were terminated by addition of 400 μl of 1 M Na2CO3, pH 9.0, the OD420 nm was measured, and LacZ activity (U) was calculated by multiplying OD420 nm/OD600 nm by 1000.
Reverse Transcription (RT)-PCR
Total RNA was isolated from Saccharomyces cerevisiae by glass bead lysis, and cDNA was synthesized with SuperScript II reverse transcriptase by using oligo(dT)12-18 amplifying primers (Invitrogen). Endogenous HAC1U/HAC1I, KAR2, and ACT1 mRNAs were detected by PCR by using Hac1 cDNA_F/Hac1 cDNA_R, Kar2 cDNA_F/Kar2 cDNA_R, and Act1 cDNA_F/Act1 cDNA_R primer sets, respectively (oligonucleotide primer sequences available on request).
Microarray Experiments
Total RNA was isolated by glass bead lysis, and RNA integrity was determined using the RNA 6000 Nano LabChip kit and 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Cyanine (Cy)3- and Cy5-labeled cDNAs were synthesized from total RNA by direct labeling (Agilent Technologies). Array experiments were also performed with dye switch. Microarray hybridizations were performed using a Yeast 4x 44k oligonucleotide array slide (015072; Agilent Technologies). Microarrays were hybridized overnight, washed, dried, and scanned on a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA). A Lowess normalization procedure was performed to adjust the Cy3 and Cy5 channels, the image files were analyzed with Gene Spring GX 7.3.1 software (Agilent Technologies), and the log10 ratio of Cy5/Cy3 was reported for each gene.
Ceramidomic Analysis
Intracellular ceramide levels were measured using normal phase high-performance liquid chromatography coupled to atmospheric pressure chemical mass spectrometry (Bielawski et al., 2006). Neutral lipids were isolated from yeast extracts generated by pooling three independent cultures of each yeast strain that had been grown at 30°C or shifted to 37°C for 2 h. Electrospray ionization-tandem mass spectrometry analysis of ceramides and internal standards was performed on a TSQ 7000 triple quadruple mass spectrometer (Thermo Fisher Scientific, Waltham, MA) operating in a multiple reaction monitoring positive ionization mode. Calibration curves for purposes of quantification were constructed by plotting peak area ratios of synthetic standards. Ceramides were normalized to total organic phosphate levels.
SL Analyses
Cells were cultured overnight at 30°C in SD minimal media supplemented with 20 μCi/ml [2-3H]myo-inositol (20 Ci/mmol; American Radiolabeled Chemicals, St. Louis, MO) (6 ml of culture volume). Cultures were then shifted to 37°C for 2 or 3 h, as appropriate, and the experiment was terminated with trichloroacetic acid (final concentration, 5%) and chilling on ice. After twice washing cell pellets with water, SLs were isolated by two consecutive extractions with 1 ml of extraction buffer (diethylether:95% ethanol:water:pyridine, 5:15:15:1, vol/vol) for 60 min at 60°C (Hanson and Lester, 1980). Lipids were deacylated by in methanolic 0.1 M NaOH at 37°C for 1 h, and reactions were terminated with 200 μl of 1 M acetic acid. The extracts were dried overnight under vacuum. SLs were reconstituted in 300 μl of lipid wash solvent (1-butanol:petroleum ether:ethyl formate, 20:4:1, vol/vol) and back extracted three times with double distilled H2O. SLs were dried to a film under N2 gas, reconstituted in 50 μl of CHCl3, and 20-μl samples were resolved by one-dimensional thin layer chromatography (TLC) by using 20-cm Whatman HP-K plates developed with CHCl3:CH3OH:4.2 N ammonia (40:10:1). Plates were sprayed with ENH3ANCER (PerkinElmer Life and Analytical Sciences, Boston, MA) and exposed to film at −80°C.
Mass spectrometric analyses of inositol SLs was performed according to Guan and Wenk (2006), with the following modifications. Dimyristoyl-PtdCho, dimyristoyl-phosphatidylethanolamine, dioctyl-PtdIns, dimyristoyl-phosphatidylserine, and C19:0-ceramide were added as internal standards. Quantification of individual molecular species was carried out using multiple reaction monitoring with a 4000 Q-Trap mass spectrometer (Applied Biosystems, Foster City, CA). Typically, 25 μl of samples were injected for analysis. In these experiments, the first quadrupole, Q1, was set to pass the precursor ion of interest to the collision cell, Q2, where it underwent collision induced dissociation. The third quadruple, Q3, was set to pass the structure-specific product ion characteristic of the precursor lipid of interest. Each individual ion dissociation pathway was optimized with regard to collision energy to minimize variations in relative ion abundance due to differences in rates of dissociation (Guan and Wenk, 2006). Lipid mass was calculated relative to relevant internal standards. Comparison of the means of wild-type and individual genotypes from three independent experiments was performed. The quantities of lipids are expressed as ion intensities relative to wild-type levels, converted to a log10 scale, and represented as a heat map. The difference in levels of individual lipid species between wild-type and specified mutants was determined statistically using the Kruskal–Wallis test. All lipid standards were obtained from Avanti Polar Lipids (Alabaster, AL), with the exceptions of C19:0 ceramide, which was obtained from Matreya (State College, PA), and dioctyl-PtdIns, which was from Echelon Biosciences (Salt Lake City, UT).
Online Supplemental Material
Supplemental Figure S1 describes time courses for establishment of morphological manifestations of secretory pathway dysfunction in sec14-1ts and sec6-4ts mutant, respectively. Supplemental Figure S2 describes time courses of CPY maturation in sec14-1ts tlg2Δ double mutants. Supplemental Figure S3 documents sensitivity of sec14-1ts tlg2Δ yeast to inositol deprivation and dithiothreitol (DTT) challenge. Supplemental Figure S4 provides evidence that Hac1I stability is not compromised in sec14-1ts tlg2Δ yeast. Supplemental Figure S5 documents the effects of genetic ablation of Isc1 and Ppn1 function on ceramide mass. Supplemental Table S1 lists the plasmids used in this study. Supplemental Table S2 lists the genes that, when deleted, show synthetic genetic interactions with sec14-1ts. Supplemental Table S3 lists genes whose expression is most affected (positively and negatively) in sec14-1ts tlg2Δ double mutants.
RESULTS
Identification of Synthetic Interactions with sec14-1ts by SGA
A high-throughput SGA (Tong et al., 2001) was performed in which sec14-1ts was queried against deletion alleles for each of the 4672 nonessential yeast genes. Two different types of experiments were conducted. First, we sought deletions that restore viability to sec14-1ts mutants at 37°C (i.e., “bypass Sec14” mutants). Those searches yielded only already recognized bypass Sec14 mutations, indicating previous screens are likely saturated (data not shown). Second, we sought deletions that exacerbate sec14-1ts growth phenotypes at 30°C. Results of three independent SGA analyses were compared, and interactions consistently scored in all three analyses were pooled into a final list. By this criterion, individual deletions in 206 nonessential genes yielded potential synthetic interactions with sec14-1ts at 30°C.
The SGA screen reported genes known previously to exhibit strong synthetic interactions with sec14-1ts. These genes include structural genes for the Arf1 GTPase (ARF1), the Gcs1 Arf-GAP (GCS1), the Sec14-like protein Sfh3 (SFH3), phospholipase D (SPO14), and the Trs85 (GSG1) subunit of the large transport protein particle (TRAPPII) involved in a Ypt31/32-dependent trafficking from the TGN/endosomal system (Supplemental Table S2). Given that Sec14 potentiates the activities of two essential yeast PtdIns 4-OH kinases (Stt4 and Pik1) and that these kinases interface with activity of the Mss4 PtdIns-4-P 5-OH kinase in generating PtdIns-4,5-P2, an overlap between the genes identified in the sec14-1ts SGA screen with those identified in pik1ts (Sciorra et al., 2005), stt4ts (Tabuchi et al., 2006), and mss4ts (Audhya et al., 2004) SGA screens was expected. The representation of genes involved in the protein kinase C/mitogen-activated protein kinase cell integrity pathway (e.g., ROM2 and RHO1) is consistent with Sec14 involvement in Stt4- and Mss4-dependent PtdIns-4,5-P2 synthesis that controls this circuit. Similarly, genes of the ADP ribosylation factor (ARF), Arl, and Ypt31/32 Rab GTPase cycles (ARF1, GCS1, ARL3, GYP1, and GSG1) support the functional collaboration between Sec14 and Pik1, among other possibilities.
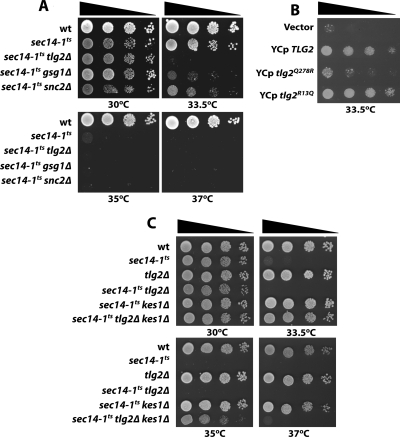
Of particular interest, the SGA analyses identified two genes encoding core components of the membrane trafficking machinery not previously encountered in sec14-1ts interaction screens. These included the TLG2 t-SNARE and SNC2 v-SNARE, and these SNAREs are genuine binding partners with execution points in the TGN/endosomal system (Abeliovich et al., 1998). Reconstruction of each double mutant by meiotic cross and tetrad analysis confirmed the SGA results. Relative strengths of synthetic interaction for sec14-1ts with individual tlg2Δ, gsg1Δ, and snc2Δ alleles are shown in Figure 1A. Of these synthetic interactions, the one interaction involving sec14-1ts and tlg2Δ was particularly severe. The double mutant failed to grow at all at 33.5°C, normally a semipermissive temperature for sec14-1ts single mutants.
Figure 1.
Synthetic genetic interactions. (A) The sec14-1ts allele exhibits strong synthetic interactions with tlg2Δ, gsg1Δ, and snc2Δ (genotypes indicated at left). Indicated double mutants were generated by meiotic segregation, and they were derived from tetrads with four viable spores and where all markers segregated in a Mendelian manner. Cells were spotted in a 10-fold dilution series on YPD and incubated at either permissive (30°C), semipermissive (33.5°C), or restrictive (35 and 37°C) temperatures for sec14-1ts strains. Growth results were recorded after 48 h. (B) Loss of Tlg2 SNARE activity exacerbates sec14-1ts associated growth defects. Growth properties of sec14-1ts tlg2Δ cells carrying YCp(URA3), YCp(TLG2), YCp(tlg2Q278R), and YCp (tlg2R13Q) were determined. Cells were spotted in a 10-fold dilution series on uracil-free minimal medium and incubated at 33.5°C for 72 h. (C) tlg2Δ compromises kes1Δ-mediated bypass Sec14. Yeast strains of indicated genotype were spotted in 10-fold dilution series on YPD and incubated at 30, 33.5, 35, or 37°C as indicated. Growth results were recorded after 48 h.
Functional rescue experiments were performed to determine what specific properties of Tlg2 contribute to the observed genetic interaction. The possibility that loss of Tlg2 t-SNARE function was the primary defect from the Tlg2 perspective was tested by expression of a tlg2Q278R allele in sec14-1ts tlg2Δ yeast. The mutant Tlg2Q278R carries an inactivating missense substitution in the zero-layer SNARE interaction motif (Abeliovich et al., 1998; Fasshauer et al., 1998). As a specificity control, the effects of Tlg2R13Q expression were also examined. The R13Q substitution disrupts Tlg2 binding to Vps45, but it does not compromise Tlg2 t-SNARE activity (Abeliovich et al., 1999; Bryant and James, 2003). Tlg2Q278R expression failed to rescue growth of the sec14-1ts tlg2Δ double mutant at 33.5°C, whereas Tlg2R13Q was fully competent to do so (Figure 1B). We conclude it is specific loss of Tlg2 t-SNARE function that exacerbates sec14-1ts growth defects.
Finally, the effect of tlg2Δ on bypass Sec14 was tested by introducing kes1Δ and cki1Δ alleles into sec14-1ts tlg2Δ strains. Although growth of sec14-1ts tlg2Δ kes1Δ mutants was restored at 33.5°C, the strain grew poorly at 35°C and not at all at 37°C (Figure 1C). Bypass Sec14 mechanisms involving inactivation of the cytidine diphosphate-choline pathway for PtdCho biosynthesis were nullified by tlg2Δ given sec14-1ts tlg2Δ cki1 triple mutants failed to grow at 33.5°C (data not shown).
Endosomal Defects in sec14-1ts Mutants
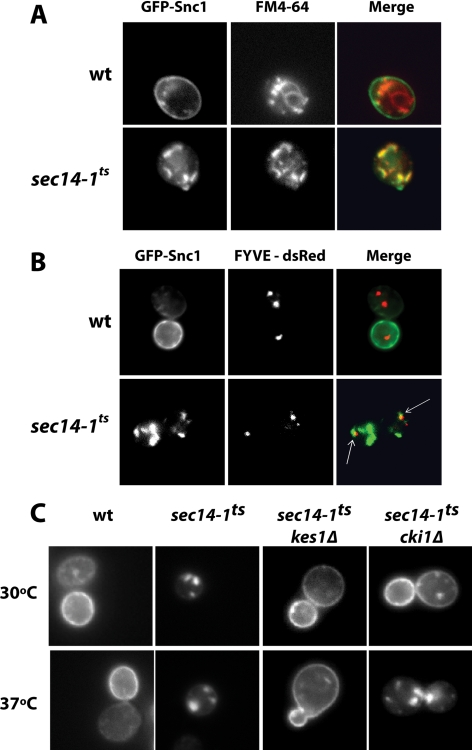
The Tlg2 SNARE interacts with various binding partners, all of which are annotated as proteins of the TGN/endosomal system (Abeliovich et al., 1998, 1999; Holthuis et al., 1998; Gavin et al., 2002). Functionally, Tlg2 facilitates the targeting/fusion of endosome-derived vesicles with late Golgi compartments and homotypic fusion of TGN membranes (Abeliovich et al., 1998; Holthuis et al., 1998; Coe et al., 1999; Brickner et al., 2001; Kama et al., 2007). This raised the possibility that Tlg2 and Sec14 cooperate in regulation of TGN/endosomal dynamics. To investigate the recycling pathway for a plasma membrane protein, we followed the distribution of the exocytic v-SNARE Snc1 tagged with GFP. Snc1 traffics from the TGN to the cell surface and is recycled back to the TGN through the endosomal system for additional rounds of exocytic transport. In >90% of wild-type cells GFP-Snc1 localized nearly exclusively to the plasma membrane (Figure 2A). By contrast, even at the permissive temperature of 30°C, sec14-1ts mutants displayed obvious deficits in GFP-Snc1 localization. Very little GFP-Snc1 accumulation is recorded at the plasma membrane. Rather, the majority of the GFP-Snc1 is sequestered in an intracellular pool characterized by punctate compartments (Figure 2A).
Figure 2.
Endocytic trafficking is defective in sec14-1ts mutants. (A) GFP-Snc1 accumulates in intracellular compartments in sec14-1ts mutants. Wild-type and isogenic sec14-1ts yeast harboring YCp(GFP-SNC1) were grown to mid-logarithmic growth phase in uracil-free minimal medium at 30°C. Cells were incubated with FM4-64 (10 μM) for 10 min after which cells were intoxicated with NaN3/NaF (10 mM final, each). Cells were washed twice with NaN3/NaF (10 mM final, each), and FM4-64 and GFP-Snc1 profiles were imaged using a Nikon E600 microscope. GFP-Snc1, FM4-64 and merge profiles are shown, as indicated. (B) GFP-Snc1 and FYVE-dsRed localization profiles are shown. Wild-type and isogenic sec14-1ts yeast harboring YCp(GFP-SNC1) were grown to mid-logarithmic growth phase in uracil-free minimal medium at 30°C. GFP-Snc1 and FYVE-dsRed profiles were imaged using a Nikon E600 microscope. GFP-Snc1, FYVE-dsRed, and merge profiles are shown, as indicated. Arrows highlight examples of where GFP-Snc1 and FYVE-dsRed compartments are juxtaposed. (C) GFP-Snc1 localization in bypass Sec14 mutant yeast. Yeast strains with the indicated genotypes harboring YCp(GFP-SNC1) were grown to mid-logarithmic growth phase in uracil-free minimal medium at 30°C and then either maintained at 30°C or shifted to 37°C for 60 min. The GFP-Snc1 profile was imaged using a Nikon E600 microscope.
Endocytic tracer experiments using the lipophilic dye FM4-64 identify these GFP-Snc1 compartments as endosomes. Under conditions where cells are pulsed with FM4-64 for 10 min at 30°C, wild-type yeast efficiently internalize the FM4-64 into punctate compartments, and a significant fraction of the dye reaches the limiting vacuolar membrane. Very little colocalization is recorded for GFP-Snc1 and FM4-64 under these conditions (Figure 2A). By contrast, >75% of the GFP-Snc1 compartments in isogenic sec14-1ts mutants are labeled with FM4-64 at 30°C, and no obvious FM4-64 labeling of the vacuolar membrane is observed (Figure 2A). Similar results were obtained after 1 h shift of sec14-1ts cells to 37°C (data not shown).
We also probed the relationship between the intracellular site of GFP-Snc1 accumulation and compartments decorated with FYVE-dsRed; a sensor for 3-OH phosphorylated PtdIns derivatives that serve as independent markers for endosomal compartments (Corvera et al., 1999; Kutateladze et al., 1999). As expected, very little colocalization was recorded for GFP-Snc1 and FYVE-dsRed in isogenic wild-type cells. In isogenic sec14-1ts strains at 30°C, some 30% of the GFP-Snc1 puncta either colocalized with, or lay immediately adjacent to, compartments labeled with FYVE-dsRed (Figure 2B). These collective data indicate Sec14-deficient mutants are strongly defective in trafficking of GFP-Snc1 from endosomal compartments—even at permissive temperatures.
With regard to how bypass Sec14 mutations affect GFP-Snc1 trafficking defects, an obvious plasma membrane GFP-Snc1 pool was registered in >90% of the sec14-1ts kes1 and sec14-1ts cki1 cells imaged at 30°C (Figure 2C). In the sec14-1ts kes1 mutants, an essentially wild-type GFP-Snc1 localization profile was restored. In sec14-1ts cki1 mutants, however, a significant pool of GFP-Snc1 remained sequestered in punctate intracellular compartments. When bypass Sec14 mutants were shifted to 37°C (a permissive growth temperature for both strains—even though a normally lethal Sec14 deficiency is imposed), GFP-Snc1 exhibited a plasma membrane localization profile only in the sec14-1ts kes1 mutant. The GFP-Snc1 reporter was predominantly trapped in cytosolic puncta in the sec14-1ts cki1 strain (Figure 2C). More than 90% of the sec14-1ts cki1 cells imaged showed no detectable plasma membrane GFP-Snc1 at 37°C—even though the cells grow actively under these conditions. Thus, GFP-Snc1 trafficking defects in Sec14-deficient yeast are efficiently remediated only by kes1 bypass Sec14 alleles, and these data suggest that kes1- and cki1-mediated bypass Sec14 pathways occur by different mechanisms.
Acute Inactivation of Sec14 Results in Accumulation of Pleiomorphic Membrane Structures.
The sec14-1ts mutation exerts differential effects on certain exocytic and vacuolar trafficking pathways. sec14-1ts yeast are strongly defective for invertase exocytosis when challenged with restrictive temperatures (37°C), but are not as defective for CPY trafficking to the vacuole (Cleves et al., 1991b; Fang et al., 1996; Schaaf et al., 2008). In both cases, however, the accumulated pool of cargo presents terminal α-1,3-mannose linkages on its N-linked glycosyl chains, thereby reporting complete maturation of glycosyl chains on cargo glycoproteins whose trafficking itinerary is disrupted in Sec14-deficient yeast (Bankaitis et al., 1989; Cleves et al., 1991b). When coupled with the potent sec14-1ts/tlg2Δ synthetic interactions, and demonstrations that efficient endosomal function requires Sec14, these findings raise questions of whether Sec14 exerts direct effects on TGN egress of specific classes of cargo, or whether it exerts indirect effects on exocytosis as a result of defects in TGN/endosomal dynamics (e.g., recycling of v-SNARES from endosomes to the TGN). Because depletion of Snc1- and Snc2 v-SNAREs leads to accumulation of docking/fusion-defective secretory vesicles in yeast (Protopopov et al., 1993), this morphological phenotype provides a basis for distinguishing between these possibilities.
Using thin-section electron microscopy as assay in a time course after shift to 37°C, we find the early manifestations of Sec14 deficiency reflect accumulation of small stacks or toroid arrangements of pleiomorphic membranes (Supplemental Figure S1A). Accumulation of these structures becomes more extreme upon extended incubation of sec14-1ts mutants at 37°C (Supplemental Figure S1A). At no point do we detect obvious accumulation of secretory vesicles, as would be expected if defects in TGN/endosomal dynamics in sec14-1ts mutants simply resulted in formation of v-SNARE (i.e., Snc1 and Snc2)-depleted secretory vesicles. For purpose of comparison, the morphological phenotype associated with accumulation of fusion-incompetent secretory vesicles is provided by electron microscopy of sec6-4ts mutants at 37°C (Supplemental Figure S1B). Interestingly, sec14-1ts mutants do not exhibit an obvious proliferation of pleiomorphic intracellular compartments (or vesicles) at 30°C—the endosomal defects notwithstanding.
CPY Trafficking in sec14-1ts tlg2Δ Double Mutants
To further investigate the basis of the synthetic interaction between sec14-1ts and tlg2Δ, we examined CPY trafficking through the TGN/endosomal pathway in yeast. As indicated above, CPY transit from the TGN to the vacuole is only modestly affected by inactivation of Sec14. Delivery of CPY to the vacuole is kinetically delayed, but essentially all of the CPY ultimately transits to its vacuolar destination and is appropriately processed to its mature active form (Cleves et al., 1991b; Fang et al., 1996). One potential mechanism for the synthetic interaction between sec14-1ts and tlg2Δ is both gene products participate in independent pathways that impinge on membrane trafficking, and the combined defects exert a more complete trafficking block. Alternatively, both Sec14 and Tlg2 defects may partially inactivate the same trafficking pathway, in which case the combined defects levy a stronger effect. Both scenarios predict exacerbated CPY transport defects from the TGN of sec14-1ts tlg2Δ double mutants.
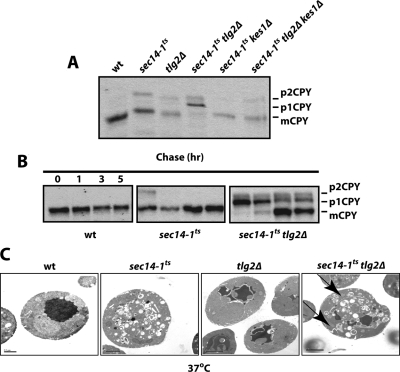
The CPY trafficking itinerary is readily followed in pulse-chase radiolabeling experiments. The core-glycosylated ER form (p1 CPY) chases to p2 CPY that presents fully matured glycosyl chains and is diagnostic of CPY pools transiting the TGN/endosomal systems. On delivery to the vacuole, p2 CPY is proteolytically cleaved to generate the mature vacuolar form (mCPY). As shown in Figure 3A and Supplemental Figure S2A, precursor CPY forms chase rapidly to mCPY in wild-type strains and in tlg2Δ and sec14-1ts single mutants strains at 30°C. Although mild kinetic defects were scored for p2 CPY conversion to mCPY in tlg2Δ and sec14-1ts yeast at 37°C, CPY trafficking was far more defective in sec14-1ts tlg2Δ double mutants at 30, 33.5, and 37°C (Figure 3A and Supplemental Figures S2, A and B). Paradoxically, the p1 CPY precursor form was the primary species that accumulated at 37°C in sec14-1ts tlg2Δ double mutants, and p1 CPY accumulation was conspicuous even at 33.5°C (Figure 3A and Supplemental Figures S2, A and B). In experiments where fate of the p1 CPY accumulated at 37°C was monitored after temperature downshift to 25°C, only poor conversion to p2- or mCPY was scored. Indeed, a significant fraction of p1 CPY accumulated in sec14-1ts tlg2Δ cells at 37°C persisted throughout a 5-h chase at the 25°C permissive temperature (Figure 3B). The defects in ER trafficking were general. ER forms of pro-α-factor accumulated in sec14-1ts tlg2Δ double mutants at 37°C (Supplemental Figure S2C).
Figure 3.
Early secretory block in sec14-1ts tlg2Δ yeast. (A) CPY trafficking is blocked at the ER in sec14-1ts tlg2Δ double mutants, and exit from the ER is restored in the isogenic sec14-1ts tlg2Δ kes1Δ strain. Yeast cultures were grown in minimal media at 25°C, shifted to 37°C for 2 h, and radiolabeled with [35S]-amino acids for 30 min. Chase (10 min) was initiated by introducing excess unlabeled methionine and cysteine, and it was terminated with ice-cold trichloroacetic acid (final concentration, 5%). The core glycosylated p1 CPY, the fully modified p2 CPY, and the mCPY are indicated at right. Genotypes are at top. (B) Early secretory block in sec14-1ts tlg2Δ mutants is poorly reversible. Yeast strains with the indicated genotypes were grown in minimal media at 25°C, shifted to 37°C for 2 h, and radiolabeled with [35S]-amino acids for 30 min. Chase was initiated by addition of excess unlabeled methionine and cysteine, and cultures were concomitantly shifted to 25°C. Chase at 25°C was for 1, 3, and 5 h, as indicated. (C) Thin-section electron microscopy. Yeast strains with the indicated genotypes were cultured to early logarithmic growth phase in YPD medium at 30°C. The cultures were then shifted to the 37°C for an additional 2 h. Cells were fixed, embedded in Spurr's resin, stained with uranyl acetate, and imaged using a transmission electron microscope (20 kV). Representative images are shown (bar, 1 μm). Distended ER is highlighted by arrows.
sec14-1ts tlg2Δ Double Mutants Are Defective in Protein Egress from the ER
The poor reversibility of the p1 CPY maturation block in sec14-1ts tlg2Δ strains could, in principle, arise from collapse of mechanisms for recycling of essential TGN components from the endosomal system to the TGN. Such failures may result in wholesale defects in retention of glycosyltransferases in the Golgi of sec14-1ts tlg2Δ double mutants at 37°C and would manifest themselves as a maturation block without a bona fide trafficking block. Alternatively, a genuine defect in protein trafficking from early stages of the secretory pathway (i.e., the ER) could potentially be imposed in sec14-1ts tlg2Δ double mutants. Support for this possibility was forthcoming from thin-section electron microscopy analyses. We found tlg2Δ single mutants incubated at 30 and 37°C were morphologically normal, whereas sec14-1ts single mutants incubated at restrictive temperatures characteristically accumulated the toroid structures that represent defective cargo-engorged late secretory organelles (Figure 3C). Examination of thin section electron micrographs of sec14-1ts tlg2Δ double mutants revealed both the accumulation of toroid structures typical of sec14-1ts mutants shifted to 37°C and a noticeable distension of ER membranes. Distended ER was observed in essentially all sec14-1ts tlg2Δ cells examined, and this phenotype resembled that recorded for yeast inactivated for the Sec12 nucleotide exchange factor for the Sar1 GTPase, i.e., a factor directly required for transport of newly synthesized glycoproteins from the ER (d'Enfert et al., 1991; Barlowe and Schekman, 1993).
ER Defects and UPR Integrity
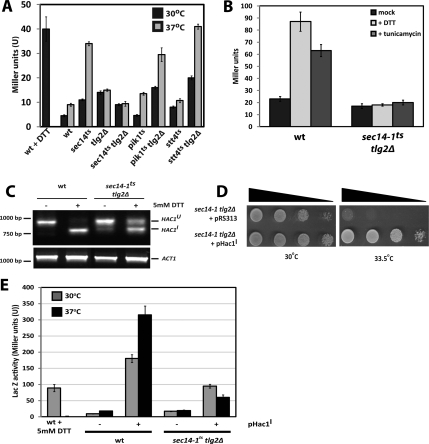
sec14-1ts tlg2Δ double mutants grow very poorly in the absence of inositol and display marked sensitivities to DTT in minimal medium (Supplemental Figure S3). Because these phenotypes are hallmarks of defects in ER stress responses (Nikawa and Yamashita, 1992; Cox et al., 1993; Kohno et al., 1993), we examined whether the ER trafficking block in the sec14-1ts tlg2Δ double mutant was a manifestation of ER stress. To that end, the integrity of the UPR was assessed in sec14-1ts tlg2Δ cells and related to UPR activity in the corresponding single mutants and wild-type controls. UPR activity was monitored in two ways. The first assay uses a yeast centromeric vector (pJT30)-based lacZ reporter placed under transcriptional control of a yeast UPR enhancer (Wilkinson et al., 2000). This system reports the ultimate transcriptional readout of UPR signaling and is therefore sensitive to defects at any stage of the UPR pathway. Second, an RT-PCR strategy that distinguishes unspliced HAC1 mRNA (996-base pair product) from spliced message (744-base pair product) was used. Ire1 is an ER membrane protein that splices HAC1 mRNA so that a functional Hac1 transcription factor can be translated (Cox and Walter, 1996, Sidrauski et al., 1996; Sidrauski and Walter, 1997; Chapman and Walter, 1997). This second assay surveys the integrity of events that lie upstream of the HAC1 mRNA processing execution point of the UPR.
The LacZ assay reported that challenge of wild-type yeast with DTT stimulated UPR activity some fivefold relative to basal, as expected. In agreement with previous reports (Chang et al., 2002), the UPR was induced 2- and 6.5-fold over basal in sec14-1ts single mutants cultured at 30 and 37°C, respectively (Figure 4A). Shift of the sec14-1ts tlg2Δ strain to 37°C, however, failed to evoke a robust UPR. Interestingly, UPR failure was not detected in pik1ts tlg2Δ or stt4ts tlg2Δ double mutants that are defective in the Pik1 and Stt4 PtdIns 4-OH kinases whose activities are stimulated by Sec14 in vivo (Hama et al., 1999; Rivas et al., 1999; Schaaf et al., 2008) (Figure 4A). The UPR defects in sec14-1ts tlg2Δ double mutants were not a specific result of using elevated temperature as stressor. These were also apparent under conditions of chemical insult with either DTT or tuncamycin. Such challenges potently induced UPR activity in UPR-competent yeast (8-fold and 6-fold relative to untreated cells, respectively; Figure 4B); yet, UPR-regulated LacZ activity was not induced in sec14-1ts tlg2Δ cells at 37°C (Figure 4B).
Figure 4.
UPR activity in sec14-1ts tlg2Δ double mutants. (A) Down-regulation of the UPR in sec14-1ts tlg2Δ relative to parental sec14-1tscells. β-Galactosidase expression was followed as reporter of UPR activity in yeast strains of the indicated genotype that harbor the pJT30 YCp(UPRE::LACZ) plasmid where LACZ transcription is under control of the KAR2 enhancer (Wilkinson et al., 2000). Cells were grown at the permissive temperature (30°C) or shifted to the restrictive temperature (37°C), for sec14-1ts, 2 h before analysis. As control for UPR activity, wild-type yeast were treated with DTT (final concentration, 5 mM) for 60 min before assay. (B) The UPR cannot be activated in sec14-1ts tlg2Δ yeast by challenge with ER stress agents. Yeast strains of indicated genotype were cultured, and UPR activity was assayed, as described in A, with the modification that cultures were either mock treated, challenged with DTT (5 mM), or challenged with 10 μg/ml tunicamycin 1 h before analysis. (C) UPR silencing is downstream of Ire1p function. cDNA was synthesized from total RNA isolated from either wild-type or sec14-1ts tlg2Δ cells grown in the presence or absence of 5 mM DTT. In all cases, the yeast strains were grown at 30°C overnight and shifted to 37°C for 2 h before DTT challenge. Endogenous HAC1 and ACT1 mRNA levels were monitored by PCR as reporters of UPR activity and as normalizing factor, respectively. (D) HAC1I expression rescues sec14-1ts tlg2Δ-associated growth defects. The sec14-1ts tlg2Δ double mutant was transformed with either vector control (pRS313) or YCp HAC1I (pRC43), as indicated, transformants were spotted in 10-fold dilution series on YPD, and incubated at either permissive (30°C) or restrictive (33.5°C) temperatures. Growth was scored after 72 h. (E) Hac1I transcriptional activation potency is reduced in sec14-1ts tlg2Δ cells. UPR-dependent β-galactosidase expression (from the pJT30 reporter plasmid) for wild-type and sec14-1ts tlg2Δ yeast in the absence or presence of HAC1I expression (sustained by the YCpHAC1I plasmid pRC43). Cells were grown at the permissive temperature (30°C) and challenged with the indicated temperatures for 2 h before analysis. As standard for UPR activity, wild-type yeast were challenged with DTT (final concentration, 5 mM) for 1 h before assay.
Inability to induce the UPR in sec14-1ts tlg2Δ double mutants was not associated with failure to splice HAC1 mRNA. A predominant 996-base pair PCR product (corresponding to HAC1U mRNA) was amplified from cDNA synthesized from RNA isolated from unstressed wild-type and sec14-1ts tlg2Δ controls (Figure 4C), signifying a quiescent UPR. By contrast, a spliced 744-base pair product diagnostic of an active UPR was amplified from cDNA derived from both DTT-challenged wild-type yeast and sec14-1ts tlg2Δ double mutants challenged with DTT at 37°C. We do note HAC1 splicing seemed suboptimal in sec14-1ts tlg2Δ double mutants as evidenced by the persistence of some unspliced HAC1 message relative to wild-type controls (Figure 4C).
The demonstration that HAC1 splicing is executed in sec14-1ts tlg2Δ yeast at 37°C signifies the UPR is initiated but that it fails primarily downstream of HAC1 mRNA processing. This finding is in accord with the results of experiments where sec14-1ts tlg2Δ yeast were transformed with pRC43 (pHAC1I)—an expression plasmid encoding a modified HAC1 mRNA that does not require mRNA splicing to program synthesis of an active Hac1 translation product (Chapman and Walter, 1997). HAC1I expression effected only a modest rescue of the UPR (Figure 4D)—even in the face of normal accumulation of the Hac1 protein (Supplemental Figure S4)—and this partial effect was a result of reduced potency for Hac1 in transcriptional activation (Figure 4E).
Transcriptional Profiling of the Larger UPR
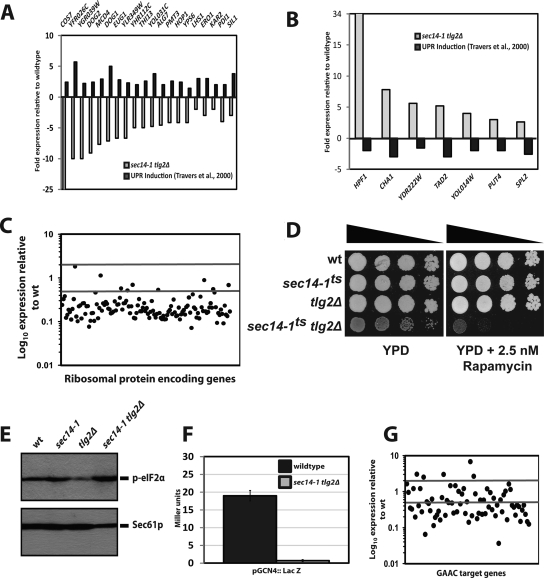
The UPR induces elevated expression of 381 target genes in S. cerevisiae (Travers et al., 2000). Transcriptional profiling was performed to investigate whether UPR targets are uniformly silenced in sec14-1ts tlg2Δ yeast, or whether the effects are more limited. Whole genome microarray analyses (σ ≥ 3) demonstrated that some 30% of the genes whose expression is induced by the UPR were induced upon DTT challenge at 37°C in sec14-1ts tlg2Δ double mutants. However, 24% of the 381 UPR genes were inappropriately down-regulated, and expression of the remaining 46% was neither induced nor repressed more than threefold. Thus, a substantial aspect of the UPR fails in sec14-1ts tlg2Δ yeast. In agreement with the UPRE::LACZ data, transcriptional profiling reported diminished KAR2 expression in sec14-1ts tlg2Δ cells (Figure 5A). Inspection of the gene cluster whose expression is inappropriately down-regulated under stress conditions in sec14-1ts tlg2Δ cells prominently features genes whose products function in the ER (e.g., EUG1, LHS1, SIL1, KAR2, and ERO1; Figure 5A). Attenuation of ER stress response gene expression is consistent with the perturbation of ER function observed in sec14-1ts tlg2Δ yeast.
Figure 5.
Transcriptional profiling of sec14-1ts tlg2Δ yeast. Gene expression profiles were compared for sec14-1ts tlg2Δ (Cy5) double mutants relative to wild-type yeast (Cy3) by using Gene Spring version 7.3.1 software (Agilent Technologies). (A and B) Aberrant regulation of the UPR regulon in sec14-1ts tlg2Δ mutants. The expression profile of a selection of genes whose expression is either up-regulated (A) or down-regulated (B) by the UPR (as reported by Travers et al., 2000) is shown. Light gray bars present the expression of genes in sec14-1ts tlg2Δ mutants relative to wild-type, whereas dark gray bars represent gene expression in wild-type cells treated with DTT (5 mM) for 60 min relative to mock-treated wild-type cells (Travers et al., 2000). (C) TOR signaling is defective in sec14-1ts tlg2Δ mutants. The expression profile is shown for genes encoding ribosomal structural proteins (RP). Expression maps show the log10 expression profile of genes, after Lowess normalization, of sec14-1ts tlg2Δ (Cy5) double mutants relative to wild-type yeast (Cy3). Analysis was performed using Gene Spring version 7.3.1 software (Agilent Technologies). Gray lines represent ± twofold expression difference. (D) sec14-1ts tlg2Δ mutants are hypersensitive to rapamycin. Yeast strains of the indicated genotype were spotted in a 10-fold dilution series on either YPD alone or YPD supplemented with 2.5 nM rapamycin. Cells were grown at 30°C and growth was scored after 72 h. (E) Partial elevation of phosphor-eIF2α levels in sec14-1ts tlg2Δ cells. Yeast total protein lysates were prepared from yeast strains of the indicated genotype, resolved by SDS-PAGE and blotted to nitrocellulose. Blots were probed with either anti-phospho eIF2α or anti-Sec61p antibody. (G) Down-regulation of the general amino acid control (GAAC) response in sec14-1ts tlg2Δ mutants. The expression profile is shown for genes encoding amino acid biosynthetic enzymes. Expression maps show the log10 expression profile of genes, after Lowess normalization, of sec14-1ts tlg2Δ (Cy5) double mutants relative to wild-type yeast (Cy3). Analysis was performed using Gene Spring version 7.3.1 software (Agilent Technologies). Gray lines represent ± 2-fold expression difference. (F) Gcn4 is not derepressed in sec14-1ts tlg2Δ mutants. β-Galactosidase expression was followed as reporter of GCN4 derepression in yeast strains of the indicated genotype that harbor the p180 YCp(GCN4::LACZ) plasmid where LACZ transcription and translation is under GCN4 regulated control (Hinnebusch, 1993, 1997). Cells were grown at the permissive temperature (30°C) and then shifted to the restrictive temperature (37°C), for sec14-1ts, 2 h before analysis.
Another feature of the yeast UPR is down-regulation, by twofold or greater, of another set of ≈150 genes (Travers et al., 2000). The transcriptional profiling data report this aspect of the UPR also fails in sec14-1ts tlg2Δ double mutants. For example, expression of the HPF1, CHA1, TAD2, PUT4, and SPL2 genes was elevated at least twofold in sec14-1ts tlg2Δ yeast incubated at 37°C, regardless of whether or not DTT challenge was imposed (Figure 5B). In sum, the transcriptional profiling data report a broad failure of the UPR in sec14-1ts tlg2Δ double mutants.
Activity of the Hsf and TOR Pathways in sec14-1 tlg2Δ Mutants
We expanded the transcriptional profiling to consider derangements in addition to those scored for the UPR. Microarray analyses (σ ≥ 3 threshold) reported expression of ca. 479 genes was elevated, and transcription of 450 genes was down-regulated, in sec14-1ts tlg2Δ double mutants relative to wild-type controls. When a σ ≥ 4 threshold is imposed in the analyses, the numbers fall to 242 and 260 genes, respectively. We focus on the data set generated by use of the more stringent filter. Supplemental Table S3A identifies the top 20 annotated genes whose expression is induced ≥10.2-fold in sec14-1ts tlg2Δ double mutants. Within this group are four (BTN2, RPN4, HSP42, and HSP82) whose expression is dependent on the Hsf1 transcription factor. Inspection of the 260 gene list for which expression is induced fourfold or more in sec14-1ts tlg2Δ double mutants reveals a prominent representation of other stress-response genes whose transcription is normally activated by Hsf1 (e.g., YDJ1, SSA1, SSA4, HSP104, HSP26, UBI4, PIR1, PIR3, YAP1801, and BUD7). Thus, Hsf1-dependent transcriptional regulation is strongly potentiated in sec14-1ts tlg2Δ double mutants, perhaps as compensatory response to an ineffective UPR (Liu and Chang, 2008).
Supplemental Table S3B lists 20 genes whose expression is reduced less than ninefold in sec14-1ts tlg2Δ strains. Representation of genes encoding proteins of the large and small ribosomal subunits is conspicuous in this list (RPS22A, RPS7A, RPS4A, RPS18B, RPS11A, RPS10A RPS11B, RPL20A RPL16B, RPL22A, RPL20B, and RPL14A). The cluster of genes whose expression is diminished fourfold or less in sec14-1ts tlg2Δ yeast counts ≈100 genes encoding ribosomal subunits and genes of the “Ribi” regulon (Chen and Powers, 2006; e.g., ADE4, ADE5, ADE6, ADE7, ADE17, LYS9, LYS12, URA1, URA3, URA4, and URA7). Expression of most of these genes is subject to potent enhancement via activity of protein kinase A (PKA) and TOR pathways that transmit cell proliferative signals in yeast (Chen and Powers, 2006). The signatures of defective PKA/TOR signaling in sec14-1ts tlg2Δ double mutants are consistent with a highly active Hsf1-dependent transcriptional program given Hsf1 antagonizes PKA/TOR signaling (Bandhakavi et al., 2008).
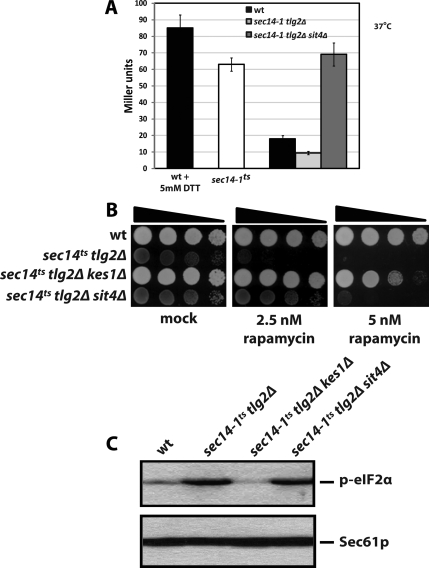
Although PKA activity is not obviously affected in sec14-1ts tlg2Δ double mutants (data not shown), TOR pathway signaling is powerfully compromised in those same strains. Two lines of additional evidence speak to this conclusion. First, growth of sec14-1ts tlg2Δ mutants is exquisitely sensitive to rapamycin (Figure 5D). Even very low concentrations of the drug (2.5 nM) exert strong growth inhibitory effects. Second, sec14-1ts tlg2Δ yeast exhibited significantly elevated levels of the phospho-form of eukaryotic initiation factor 2α (eIF2α) (Figure 5E). This is an expected consequence of compromised TOR activity because TOR inhibits the Gcn2 eIF2α kinase, which, in turn, phosphorylates eIF2α (Hinnebusch, 1993, 1997).
Elevated phospho-eIF2α did not extend to derepressed Gcn4 synthesis in sec14-1ts tlg2Δ double, however, as would normally be expected (Hinnebusch, 1993, 1997). Using a reporter construct in which both LacZ transcription and translation is subject to GCN4 5′ UTR control (pGCN4:: LacZ), we recorded an ≈30-fold reduction of LacZ expression in sec14-1ts tlg2Δ double mutants relative to congenic wild-type strains at 37°C (Figure 5F). Consistent with defective Gcn4 production, general amino acid control gene expression was diminished in sec14-1ts tlg2Δ double mutants at 37°C (Figure 5G). The unusual Gcn4 translation defects (in the face of elevated phospho-eIF2 levels) have an interesting and relevant precedent. These are also observed in yeast with PtdIns-4-phosphate deficits, indicating this phosphoinositide promotes translation initiation in yeast (Cameroni et al., 2006). Given Sec14 potentiates both Stt4- and Pik1-mediated PtdIns-4-phosphate synthesis (Hama et al., 1999; Phillips et al., 1999; Rivas et al., 1999; Schaaf et al., 2008), we interpret the Gcn4 translation defects observed in sec14-1ts tlg2Δ double mutants at 37°C to primarily stem from reduced PtdIns-4-phosphate levels. The precise mechanism notwithstanding, such Gcn4 translational defects raised interesting possibilities for UPR failure in sec14-1ts tlg2Δ yeast, given Gcn4 is an essential component of the UPR (Patil et al., 2004). We find that efficient ectopic Gcn4 expression in sec14-1ts tlg2Δ yeast does not resuscitate the quiescent UPR, however (data not shown).
Derangement of Ceramide Homeostasis in sec14-1ts tlg2Δ Yeast
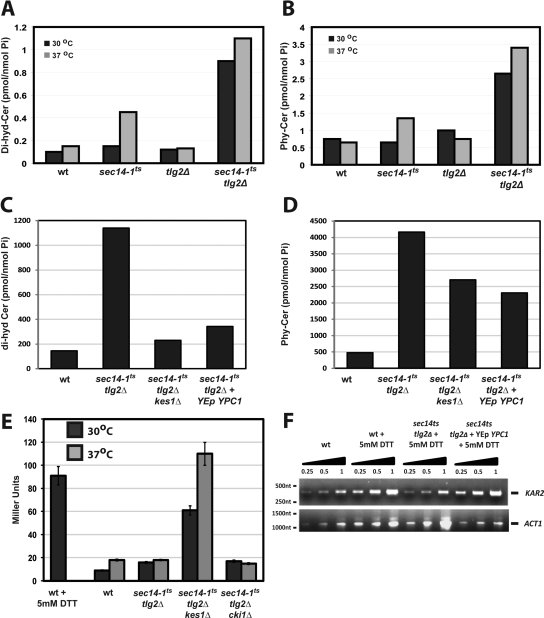
Genes encoding enzymes relevant to ceramide and SL metabolism were well represented in the sec14-1ts SGA list (Supplemental Table S2). We therefore examined the possibility that the UPR defect in sec14-1ts tlg2Δ double mutants derived from some other aspect of lipid metabolism. Quantitative lipidomic approaches were used to compare ceramide compositions of sec14-1ts tlg2Δ cells relative to wild-type, sec14-1ts, and tlg2Δ controls incubated at either 30 or 37°C. In pooled samples of wild-type yeast cultured at 30°C, total dihydro- and phytoceramide mass was measured at 0.15 pmol/nmol Pi and 0.4 pmol/nmol Pi, respectively (Figure 6, A and B). These values were not dramatically altered when the wild-type strain was shifted to 37°C. Similar values were obtained for isogenic tlg2Δ mutants at 30 and 37°C. In the sec14-1ts mutant, both ceramide species were present at wild-type levels at 30°C. However, shift to 37°C for 2 h resulted in approximately fourfold and threefold increases in dihydro- and phytoceramide mass, respectively (Figure 6, A and B). The elevations in ceramide load, however, were much more pronounced in sec14-1ts tlg2Δ double mutant yeast. Total dihydro- and phytoceramide levels were elevated some nine- and sevenfold, respectively, at 30°C. Elevation in the mass for these species was even more dramatic after shift to 37°C for 2 h. Dihydro- and phytoceramide mass each increased some 10-fold relative to basal wild-type levels at 30°C (Figure 6, A and B).
Figure 6.
Derangements in ceramide homeostasis in sec14-1ts tlg2Δ yeast correlate with UPR silencing. Dihydroceramide (A) and phytoceramide (B) levels are elevated in sec14-1ts tlg2Δ yeast. Quantitative lipidemic approaches were used to measure endogenous ceramides in yeast of indicated genotype. Lipids were extracted from three pooled cultures (30 OD600 nm) of each yeast strain grown overnight at 30°C and either maintained at 30°C, or shifted to 37°C for 2 h, before lipid extraction. Bulk dihydroceramides and phytoceramides are represented as femtomoles of ceramide per nanomole of Pi. (C and D) Derangements in dihydro- and phytoceramides are corrected in sec14-1ts tlg2Δ yeast by genetic ablation of KES1 or by increased dosage of the Ypc1 ceramidase. Endogenous ceramide profiles are shown for sec14-1ts tlg2Δ kes1Δ and sec14-1ts tlg2Δ YEp(YPC1) yeast relative to wild-type and sec14-1ts tlg2Δ strains. Culture conditions and parameters for lipid extraction are as described in A and B. (E) Functional ablation of Kes1 restores UPR competence to sec14-1ts tlg2Δ yeast. β-Galactosidase assays were performed on wild-type, sec14-1ts tlg2Δ, sec14-1ts tlg2Δ kes1Δ, and sec14-1ts tlg2Δ cki1Δ yeast cells harboring the YCp(UPRE::LACZ) UPR-reporter plasmid. Cells were grown at the permissive temperature (30°C) overnight, and then they were shifted to the restrictive temperature (37°C), for 2 h before assay. As positive control for UPR activity, wild-type yeast were challenged with 5 mM DTT for 1 h before assay. (F) Increased Ypc1 ceramidase activity restores UPR activity to sec14-1ts tlg2Δ yeast. cDNA was synthesized from total RNA isolated from mock treated wild-type yeast, and from wild-type, sec14-1ts tlg2Δ, and sec14-1ts tlg2Δ YEp(YPC1) strains challenged with 5 mM DTT. In all cases, the indicated strains were grown at 30°C overnight and shifted to 37°C for 2 h before DTT challenge. KAR2 and ACT1 mRNA levels were monitored by PCR as reporters of UPR activity and as normalizing factor, respectively.
Detailed examination of the ceramidomics data revealed that increases in total dihydroceramide content of sec14-1ts tlg2Δ cells resulted from a general elevation across the spectrum of dihydroceramide species. Mass increases in very long chain molecular species (C22-Cer–C26-Cer), and in C18/C18:1-Cer, were especially pronounced, however, and they were of greatest quantitative significance (Table 2). More specific alterations in phytoceramide molecular species were measured for sec14-1ts tlg2Δ yeast. The large elevation in total phytoceramide mass in sec14-1ts tlg2Δ cells was accounted for by elevations in mass of the very long chain (C24-Cer, C26-Cer, and C28-Cer) and C18-Cer molecular species (Table 3). Levels of shorter chain and unsaturated phytoceramides were either unchanged, or they were reduced, in sec14-1ts tlg2Δ double mutants relative to wild type (Table 3).
Table 2.
Dihydroceramide mass measurements
| Dihydroceramide (fmol/nmol Pl) |
||||
|---|---|---|---|---|
| wt | 37°C |
sec14-1ts Δtlg2 | ||
| sec14-1ts | Δtlg2 | |||
| C12-Cer | 2 | 2 | 1 | 3 |
| C14-Cer | 1 | 2 | 2 | 3 |
| C16-Cer | 5 | 8 | 4 | 35 |
| C18-Cer | 5 | 22 | 6 | 104 |
| C18:1-Cer | 5 | 13 | 4 | 40 |
| C20-Cer | 11 | 85 | 12 | 133 |
| C20:1-Cer | 16 | 36 | 7 | 28 |
| C22-Cer | 67 | 217 | 71 | 294 |
| C22:1-Cer | 17 | 37 | 11 | 52 |
| C24-Cer | 2 | 15 | 6 | 152 |
| C24:1-Cer | 2 | 1 | 1 | 7 |
| C26-Cer | 11 | 22 | 5 | 277 |
| C26:1-Cer | 1 | 2 | 1 | 11 |
| Total | 145 | 462 | 131 | 1139 |
Table 3.
Phytoceramide mass measurements
| Phytoceramide (fmol/nmol Pl) |
||||
|---|---|---|---|---|
| wt | 37°C |
sec14-1ts Δtlg2 | ||
| sec14-1ts | Δtlg2 | |||
| C14-Cer | 18 | 20 | 9 | 30 |
| C16-Cer | 161 | 70 | 144 | 57 |
| C18-Cer | 36 | 24 | 32 | 51 |
| C18:1-Cer | 42 | 29 | 29 | 19 |
| C20-Cer | 4 | 10 | 6 | 71 |
| C20:1-Cer | 10 | 21 | 17 | 101 |
| C22-Cer | 19 | 33 | 15 | 20 |
| C22:1-Cer | 34 | 19 | 26 | 9 |
| C24-Cer | 76 | 175 | 81 | 326 |
| C24:1-Cer | 41 | 28 | 39 | 10 |
| C26-Cer | 22 | 541 | 94 | 3060 |
| C26:1-Cer | 2 | 12 | 2 | 45 |
| C28-Cer | 11 | 336 | 8 | 363 |
| Total | 476 | 1318 | 502 | 4162 |
Intracellular Ceramide Load and the UPR
Defects in ceramide homeostasis in sec14-1ts tlg2Δ yeast correlate with UPR failure, as indicated by the consequences of functional ablation of the Kes1 sterol-binding protein on UPR competence and ceramide load in sec14-1ts tlg2Δ cells. Analyses of Kes1 involvement in ceramide regulation were prompted by demonstrations that the sec14-1ts tlg2Δ kes1Δ triple mutant regained the ability to traffic p1 CPY to the late stages of the secretory pathway (see above). Endogenous dihydroceramide levels were substantially corrected in sec14-1ts tlg2Δ kes1Δ cells relative to isogenic sec14-1ts tlg2Δ strains. Dihydro- and phytoceramide mass was elevated 1.6-fold and 7-fold, respectively in the sec14-1ts tlg2Δ kes1Δ triple mutant (compared with 10-fold for both species in sec14-1ts tlg2Δ controls; Figure 6, C and D). Reductions in bulk ceramide reflected general decreases in all dihydroceramides—from reductions in the very long chain molecular species (Figure 6C) and from reductions in C24-Cer and C18:1-Cer phytoceramides (Figure 6D). These effects were accompanied by UPR reactivation in sec14-1ts tlg2Δ kes1Δ triple mutants (Figure 6E). Indeed, these triple mutants exhibited a constitutively high level of UPR activity. By contrast, sec14-1ts tlg2Δ cki1 triple mutants exhibited a quiescent UPR (Figure 6E).
The striking increases in ceramide mass in sec14-1ts tlg2Δ yeast, when coupled with restoration of nearly normal dihydroceramide profiles and UPR competence by Kes1 inactivation, suggested ceramides contribute to UPR silencing. To examine this possibility more directly, Ypc1 ceramidase expression was increased in sec14-1ts tlg2Δ double mutants in attempts to enzymatically relieve the large ceramide load. Ypc1 hydrolyzes both dihydro- and phytoceramide species (Mao et al., 2000a,b). Quantitative ceramidomics demonstrated that both dihydro- and phytoceramide levels were reduced in sec14-1ts tlg2Δ yeast strains by increasing Ypc1 dosage. Again, the most striking effect was observed for the dihydroceramides (4- and 2.5-fold, respectively; Figure 6, C and D), with the quantitatively most pronounced decrease in the C24-Cer and C18:1-Cer molecular species. Reductions in phytoceramide mass came primarily at the expense of the C24-Cer and C18:1-Cer molecular species.
Overexpression of Ypc1 efficiently rescued the UPR in sec14-1ts tlg2Δ double mutants, as evidenced by restoration, with increased YPC1 gene dosage, of elevated KAR2 mRNA expression in the face of DTT challenge at 37°C (Figure 6F). These results, when taken with the kes1Δ ceramidomics, indicate a causal relationship between ceramide mass and UPR failure in sec14-1ts tlg2Δ yeast. The specific diminution in dihydroceramide mass evoked by increased Ypc1 expression links UPR rescue with reduced load of the very dihydroceramide species that show the largest mass increase in the UPR-incompetent parental sec14-1ts tlg2Δ strain.
Inositol SL Catabolism and UPR Silencing
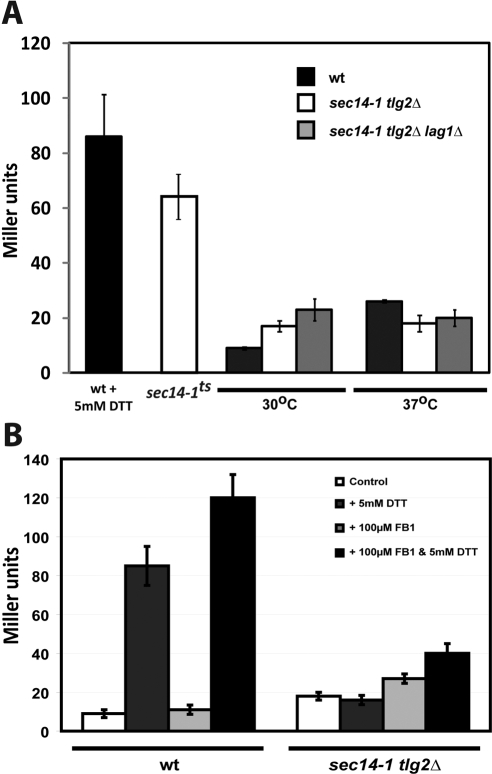
Several mechanisms could account for elevation of ceramide mass in sec14-1ts tlg2Δ double mutants: 1) increased de novo ceramide synthesis, 2) diminished ceramide utilization, or 3) excess ceramide production via accelerated degradation of complex SLs. Both genetic and pharmacological data argue against a significant contribution via elevated de novo ceramide synthesis. Neither genetic ablation of the structural gene for the major yeast ceramide synthase (LAG1; Guillas et al., 2001; Schorling et al., 2001), nor general inhibition of ceramide synthase activities with fumonisin B1, resuscitated the UPR in sec14-1ts tlg2Δ yeast (Figure 7, A and B).
Figure 7.
Ceramide synthesis and regulation of the UPR in sec14-1ts tlg2Δ yeast. (A) β-Galactosidase expression was followed as reporter of UPR activity in wild-type, sec14-1ts tlg2Δ, and sec14-1ts tlg2Δ lag1Δ yeast that harbor the pJT30 YCp(UPRE::LACZ) plasmid where LACZ transcription is under control of the KAR2 enhancer. Cells were grown at the permissive temperature (30°C) or shifted to the restrictive temperature (37°C) for sec14-1ts, 2 h before analysis. As control for UPR activity, wild-type yeast were treated with DTT (final concentration, 5 mM) for 60 min before assay. (B) β-Galactosidase UPR reporter assays were performed on wild-type and sec14-1ts tlg2Δ yeast strains carrying YCp(UPRE::LACZ). The yeast strains were grown at 30°C to mid-logarithmic growth phase, harvested and subcultured into fresh synthetic complete growth medium either supplemented with dimethyl sulfoxide or with fumonisin B1 (100 μM) and cultured for an additional 6 h. Cultures were subsequently shifted to 37°C 2 h before assay. Where indicated, yeast were challenged with 5 mM DTT (final concentration) for 1 h before analysis.
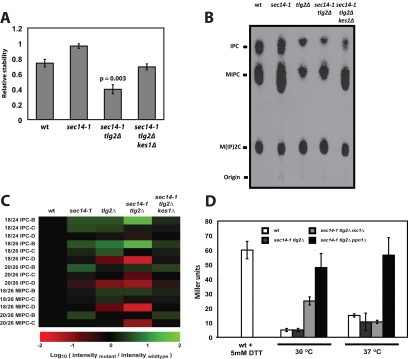
Chase experiments indicate inositol-SL degradation was accelerated ≈1.8-fold upon a 3-h shift to 37°C in sec14-1ts tlg2Δ double mutants relative to wild-type and sec14-1ts yeast. Functional ablation of Kes1 reset the rates of inositol-SL turnover in sec14-1ts tlg2Δ double mutants at 37°C to wild-type values (Figure 8A). Additional radiolabeling experiments showed that the fully modified mannosyl-di-inositolphosphorylceramide [M(IP)2C] was present in normal quantities in sec14-1ts tlg2Δ double mutants (arguing against decreased ceramide use models) and that inositolphosphorylceramide (IPC) and mannosyl inositolphosphorylceramide (MIPC) species were abnormally reduced in those cells (Figure 8B). These data identify IPC and MIPC as the most labile species. Again, incorporation of kes1Δ into the sec14-1 tlg2Δ genetic background restored a substantially wild-type SL profile (Figure 8B). Finally, consistent with the conclusions drawn from radiolabeling experiments, IPC and MIPC lipidomic profiling by mass spectrometry confirmed that both IPC and MIPC levels are compromised in tlg2Δ, and particularly in sec14-1 tlg2Δ mutants.
Figure 8.
Ceramide homeostasis and enhanced catabolism of inositol SLs. (A) Inositol-SLs are labile in sec14-1ts tlg2Δ double mutants. Wild-type, sec14-1ts, sec14-1ts tlg2Δ, and sec14-1ts tlg2Δ kes1Δ yeast were radiolabeled to steady state with 3[H]inositol (20 μCi/ml) at 30°C. Subsequently, cells were washed and resuspended in glucose-SD media and incubated for 3 h at 37°C in the presence of unlabeled inositol (12 μM). Lipids were extracted from cells and subject to alkaline methanolysis. Inositol-SLs were quantified by liquid scintillation counting and relative inositol-SL stability for each strain is expressed as the ratio of [3H] after chase:[3H] before chase. Values represent averages (and standard deviations) from three independent experiments. (B) Inositol SL profiles are altered in tlg2Δ mutants. Wild-type, sec14-1ts, tlg2Δ, sec14-1ts tlg2Δ, and sec14-1ts tlg2Δ kes1Δ yeast were labeled to steady state with [3H]inositol (20 μCi/ml). After a 2-h shift to the restrictive temperature for the sec14-1ts allele (37°C), lipids were extracted from 5 OD600 nm of cells and subject to alkaline methanolysis. TLC profiles for the inositol-SLs for yeast strains of indicated genotype are shown and each lipid assignment (indicated at left) was determined by Rf. (C) Quantitative inositol SLomics. Complex SLs were extracted from 25 OD600 of yeast and analyzed by mass spectrometry. Lipid mass was calculated by comparison with internal standards. Each SL species is represented as the log10 of the mutant:wild-type ion intensity ratio. Species shown in red report decreased mass in mutant relative to wild-type, whereas green indicates increase. (D) Incorporation of a ppn1Δ allele into the sec14-1ts tlg2Δ genetic background restores UPR activity. UPR activity was monitored via the UPRE::LACZ reporter in wild-type, sec14-1ts tlg2Δ, sec14-1ts tlg2Δ isc1Δ, and sec14-1ts tlg2Δ ppn1Δ yeast strains as indicated. Yeast were cultured at the permissive temperature (30°C) to mid-logarithmic growth phase and then shifted to the restrictive temperature (37°C) for 2 h before analysis. As control for UPR activity, wild-type yeast were challenged with DTT (final concentration, 5 mM) for 60 min before assay.
Several species of IPC and MIPC are found in S. cerevisiae, and these vary according to the extent of hydroxylation of the C26 fatty acid: IPC-B/MIPC-B are unhydroxylated, IPC-C/MIPC-C are monohydroxylated, and IPC-D/MIPC-D are dihydroxylated (Lester and Dickson, 1993; Hechtberger et al., 1994; Dunn et al., 1998). IPC-C and MIPC-D represent the major species. IPC-B and MIPC-B represent minor forms. Marked reductions in IPC-C, IPC-D, MIPC-C, and MIPC-D mass were recorded in tlg2Δ mutants relative to both wild-type and sec14-1ts yeast (Figure 8C). These reductions were most pronounced in sec14-1ts tlg2Δ double mutants. Conversely, both IPC-B and MIPC-B mass was increased in tlg2Δ and sec14-1ts tlg2Δ mutants relative to wild-type and sec14-1ts strains (Figure 8C). These elevations, however, were insufficient to counter the IPC-C/MIPC-C and IPC-D/MIPC-D deficits. Again, by this assay, the IPC and MIPC deficits recorded sec14-1ts tlg2Δ mutants were corrected by kes1Δ (Figure 8C).
Phospholipase Isc1 degrades long chain IPC and MIPC species to ceramides (Sawai et al., 2000; Kitagaki et al., 2007). Introduction of isc1Δ into sec14-1ts tlg2Δ yeast evoked rather modest reductions in ceramide load in those cells (Supplemental Figure 5A). A ppn1Δ allele was also introduced into sec14-1ts tlg2Δ yeast. This effort was motivated by similarities between the vacuolar PPN1 endopolyphosphatase (Kumble and Kornberg, 1996; Shi and Kornberg, 2005), with sphingomyelinases (http://db.yeastgenome.org/cgi-bin/protein/domainPage.pl?dbid=S000002860). Although we have yet to demonstrate Ppn1 phospholipase activity, ppn1Δ incorporation into sec14-1ts tlg2Δ strains reduced ceramide load to the same extent as did isc1Δ (Supplemental Figure 5B). With regard to the UPR, no restorative effect was recorded at 37°C in sec14-1ts tlg2Δ isc1Δ cells relative to the isogenic sec14-1ts tlg2Δ parent (Figure 8D). However, a fivefold elevation in UPR activity was measured in sec14-1ts tlg2Δ ppn1Δ mutants relative to the sec14-1ts tlg2Δ parent strain, i.e., levels comparable with those recorded for sec14-1ts strains at 37°C (Figure 8D). These data suggest Ppn1 somehow contributes to expansion of a ceramide pool that compromises the UPR.
Ceramide Exerts Its Effects on Stress Responses through the Sit4 Phosphatase
Ceramide is a potent signaling mediator in eukaryotic cells, and it exerts its effects in part through ceramide activated protein kinases and phosphatases (Nickels and Broach, 1996; Dickson et al., 2006). In yeast, the major catalytic subunit of ceramide-activated protein phosphatase is encoded by SIT4. We tested whether elevations in endogenous ceramide represses the UPR via enhanced Sit4 activity in sec14-1ts tlg2Δ double mutants. Indeed, sit4 Δ rescued UPR activity in sec14-1ts tlg2Δ yeast. A sevenfold elevation in UPR activity was measured in sec14-1ts tlg2Δ sit4Δ cells relative to the sec14-1ts tlg2Δ parental strain at 37°C, i.e., sit4Δ elevated UPR activity in the sec14-1ts tlg2Δ yeast to the scale recorded for sec14-1ts cells at 37°C.
With regard to TOR pathway activity, sec14-1ts tlg2Δ kes1Δ, and sec14-1ts tlg2Δ sit4Δ yeast were significantly more resistant to rapamycin than was the sec14-1ts tlg2Δ strain (Figure 9B). However, the elevated levels of phospho-eIF2α characteristic of the parental double mutant were reduced by functional ablation of only Kes1—loss of Sit4 function had no such effect (Figure 9C).
Figure 9.
Ceramide effects on the UPR and the Sit4 phosphatase. (A) Incorporation of sit4Δ into the sec14-1ts tlg2Δ genetic background restores the UPR. β-Galactosidase expression was followed as reporter of UPR activity in wild-type, sec14-1ts tlg2Δ and sec14-1ts tlg2Δ sit4Δ yeast that harbor YCp(UPRE::LACZ). Cells were grown at the permissive temperature (30°C) to mid-logarithmic growth phase and then shifted to 37°C for 2 h before analysis. As control for UPR activity, wild-type yeast were treated with DTT (final concentration, 5 mM) for 1 h before assay. (B) Rapamycin sensitivity in sec14-1ts tlg2Δ sit4Δ and sec14-1ts tlg2Δ kes1Δ mutants. Yeast (genotypes indicated) were spotted in 10-fold dilution series on either YPD or YPD supplemented with 2.5 or 5 nM rapamycin. Cells were incubated at 30°C and growth was scored after 72 h. (C) eIF2α phosphorylation is reduced in sec14-1ts tlg2Δ kes1Δ cells. Lysates were prepared from yeast strains of the indicated genotype, fractionated by SDS-PAGE, and blotted to nitrocellulose. Blots were probed with anti-phospho-eIF2α or anti-Sec61p antibody.
DISCUSSION
Herein, we describe synthetic interactions for the sec14-1ts allele with individual deletion of structural genes for the Tlg2 t-SNARE, the Snc2 v-SNARE, and the Trs85 subunit of the TRAPPII complex. Tlg2 dysfunction is particularly detrimental to growth of sec14-1ts mutants, and this synthetic interaction reflects loss of Tlg2 SNARE activity. The collective data indicate a complex role for Sec14 in the regulation of TGN/endosomal membrane trafficking, and they reveal new insight into how TGN/endosomal system activity is interdigitated with pathways for control of cellular homeostasis. We demonstrate that dual Sec14/Tlg2 insufficiencies levy unexpected defects in membrane trafficking from the ER that are associated with failure of the UPR. Compromised TOR signaling is also registered. Moreover, we find that combined Sec14/Tlg2 defects evoke dramatic increases in ceramide mass with consequent compromise of the UPR, in part via Sit4 protein phosphatase activity. These data extend studies linking nutrient sensing with membrane traffic through the TGN/endosomal system (Chen and Kaiser, 2003; Wedaman et al., 2003; Cameroni et al., 2006; Yan et al., 2006; Puria et al., 2008; Rohde et al., 2008) and identify ceramide as a quality control reporter for late secretory pathway function.
Sec14 and the TGN/Endosomal System
Assignment of the Sec14 execution point to the level of vesicle budding from the TGN is based on three lines of evidence: 1) the acquisition of terminal glycosyl modifications on cargo proteins whose trafficking is disturbed (Stevens et al., 1982; Cleves et al., 1991b), 2) the colocalization of a peripheral membrane Sec14 pool with the Kex2 protease (Cleves et al., 1991b; Redding et al., 1991; Brickner et al., 2001), and 3) the accumulation of exocytic cargo in pleiomorphic organelles under Sec14-insufficient conditions (Novick et al., 1980; Whitters et al., 1994). The synthetic phenotypes associated with coupling of tlg2Δ and sec14-1ts alleles, when coupled with the disturbed GFP-Snc1 and FM4-64 trafficking from endosomal compartments in sec14-1ts mutants, argue Sec14 exhibits what is operationally an endosomal execution point.
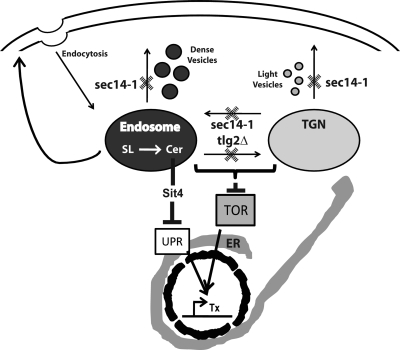
TGN/endosomal dynamics are intrinsically complex because proteins annotated as components of the exocytic machinery also score as regulators of the TGN/endosomal system, e.g., the Ypt31/32 Rabs and their downstream effectors (Cai et al., 2005; Chen et al., 2005). An endosomal execution point for Sec14 suggests several (and not mutually exclusive) possibilities for how Sec14 coordinates lipid metabolism with exocytic membrane trafficking. It also has implications for mechanisms for bypass Sec14 (to be treated elsewhere). First, Sec14 may participate in retrieval of key components of the exocytic protein machinery (i.e., v-SNARES) from endosomes to the TGN (Figure 10). Synthetic interactions between sec14-1ts and tlg2Δ, and sec14-1ts and mutations in the ARF pathway (Yanagisawa et al., 2002; Li et al., 2002), are consistent with this view, given the evidence of ARF involvement in retrograde membrane trafficking systems for cargo recycling/retrieval (Letourneur et al., 1994; Gilchrist et al., 2006; Robinson et al., 2006). Counter to the predictions of this model, we find the GFP-Snc1 trafficking defects of sec14-1ts mutants are manifest at permissive temperatures, that bypass Sec14 does not strictly correlate with remediation of endosomal defects, and that the terminal morphologies of sec14-1ts yeast are inconsistent with v-SNARE “depletion” models.
Figure 10.
Sec14 function in the TGN/endosomal dynamics. Plausible execution points for Sec14 in trafficking from the TGN to the plasma membrane, from endosomes to the plasma membrane, and within the TGN/endosomal system are indicated by hatched X. Details are described in the text. Catastrophic defects in TGN/endosomal function result in inositol SL (SL) turnover in TGN/endosomal compartments and enhanced generation of ceramide (Cer) therein. Expanded TGN/endosomal Cer pools act via Sit4 to depress the transcriptional (Tx) components of the UPR. Endosome/TGN trafficking defects additionally result in defective TOR signaling.
Second, Sec14 may primarily regulate formation of anterograde vesicles (Figure 10). These could represent endosome-derived vesicles for trafficking to the plasma membrane, or TGN-derived vesicles for trafficking to an endosomal compartment. With regard to the former scenario, reports that certain cargo (e.g., invertase) is packaged into endosome-derived vesicles that ferry cargo to the plasma membrane (Harsay et al., 2002) are consistent with the bulk of our imaging data. Invertase accumulation in nonvesicular compartments of sec14-1ts cells also supports models proposing a block in anterograde secretory vesicle biogenesis. With regard to TGN to endosome trafficking, the ARF pathway and PtdIns-4-P cooperate in potentiating trafficking from the TGN to endosomes (Demmel et al., 2008). Because Sec14 interfaces with both ARF activity and PtdIns-4-P production, this scenario remains tenable.
Third, we cannot exclude the possibility that Sec14 plays a direct role in homotypic TGN and/or endosome fusion and that such defects result in defects in vesicle biogenesis from the TGN/endosomal system. Presently, a resolution of the various possibilities is complicated by 1) yeast cells require only minute amounts of Sec14 for viability (Salama et al., 1990; Phillips et al., 1999; Schaaf et al., 2008) and 2) the sec14-1ts gene product is not completely devoid of function—even at 37°C (Cleves et al., 1989, 1991b; Ryan et al., 2007). Thus, in vivo experiments are subject to difficulties in interpreting threshold requirements for Sec14-dependent functions at any given step in the secretory pathway. Difficulties in unambiguous distinction of endosomal compartments from TGN, and different endosomal compartments from each other, pose additional complications. Resolution of these questions requires in vitro reconstitution of Sec14 function in TGN/endosomal dynamics. For the specific purposes of this report, we leave consideration of the Sec14 execution point(s) at regulation of TGN/endosomal dynamics. We focus on novel homeostatic defects associated with compromise of the TGN/endosomal system.
Failure of the UPR and TOR Signaling Pathways
The sec14-1ts/tlg2Δ genetic interaction reveals that grave compromise of TGN/endosomal dynamics in sec14-1ts tlg2Δ yeast attenuates the UPR (Figure 10). Such UPR silencing evokes a powerful, and only poorly reversible, block in protein trafficking from the ER in sec14-1ts tlg2Δ double mutants. Early steps of the UPR are properly registered in sec14-1ts tlg2Δ double mutants as evidenced by Ire1-mediated splicing of HAC1 mRNA. The Hac1 polypeptide is also produced, demonstrating translation of spliced HAC1 mRNA occurs. Thus, Hac1 failure is assigned to the transcriptional activation step. Interestingly, a functional UPR elevates expression of numerous genes whose products exhibit TGN/endosomal execution points (Travers et al., 2000), indicating the UPR is intimately tuned to TGN/endosomal function.
Transcriptional profiles of sec14-1ts tlg2Δ mutants indicate TOR signaling is also sensitive to TGN/endosomal function, consistent with growing evidence that central components of the TOR pathway either reside on TGN/endosomal membranes or are otherwise compromised by membrane trafficking defects through that membrane system (Bertram et al., 2000; Chen and Kaiser, 2003; Wedeman et al., 2003; Cameroni et al., 2006; Aronova et al., 2007; Puria et al., 2008; Rohde et al., 2008). Together, these findings define the TGN/endosomal system as a central hub for control of intracellular homeostatic signaling.
Deranged Ceramide Homeostasis
Our data implicate ceramide as a signal that silences the UPR in sec14-1ts tlg2Δ double mutants because dihydro- and phytoceramides are elevated in sec14-1ts tlg2Δ yeast. Restoration of the UPR in these double mutant strains is realized by reduction of the ceramide load—either via ectopic ceramidase expression or by Kes1 inactivation. Ceramidomic analyses implicate the dihydroceramides as the most likely candidates for agents that silence the UPR. How is ceramide mediating such effects? The evidence identifies the Sit4 ceramide-activated protein phosphatase as significant conduit for linking deranged ceramide homeostasis to ER stress response failure in sec14-1ts tlg2Δ yeast. That is, Sit4 acts as a ceramide “sensor.” The net result is inappropriate down-regulation of key nuclear signaling pathways.
The data suggest the UPR-active ceramide pool is generated in sec14-1ts tlg2Δ yeast primarily as a result of enhanced catabolism of IPCs and MIPCs (Figure 10). We posit the relevant catabolism occurs in TGN/endosomes, and that ceramide pools so generated hold potent signaling power. The data also forecast ceramide as a quality control reporter for trafficking status of the TGN/endosomal system, a system ideally suited to sense the functional status of post-Golgi exocytic, endocytic and vacuolar membrane trafficking pathways. Last, we suggest that proper trafficking of SLs into and thru the TGN/endosomal system is important in setting the proper gain for the ceramide signaling that emanates from TGN/endosomes. With ceramide as rheostat, subtle defects in TGN/endosomal function can influence ER/nuclear activities upon modest elevations in ceramide mass. However, these same responses can also be compromised when ceramide homeostasis is catastrophically deranged (in loose analogy to a death signal?).
Why is the TOR pathway compromised in sec14-1ts tlg2Δ mutants at 37°C? Other than coupling surveillance of TGN/endosomal activity to TOR signaling, our results do not provide clear insight. Elevated ceramide associated with severe defects in TGN/endosome trafficking may contribute to defects in the proximal stages of TOR signaling. If so, our data indicate this occurs without an obligate Sit4 involvement, and compromise of at least the early stages of TOR signaling may occur independently from mechanisms that link ceramides and the UPR.
Kes1 and the TGN/Endosomal System
There exists a relationship between sterol sensing and SL distribution in yeast (Malathi et al., 2004). In that regard, the role of the Kes1 oxysterol binding protein in SL and ceramide metabolism is intriguing given sterol and phosphoinositide binding are essential properties for Kes1 function in the TGN/endosomal system in vivo (Fang et al., 1996; Li et al., 2002; Im et al., 2005). Genetic ablation of Kes1 evokes bypass Sec14 (Fang et al., 1996), and this mechanism for bypass Sec14 is uniquely efficient in remediating the endosomal defects of sec14-1ts mutants (see Results). When coupled with reports that Kes1 is required for efficient transport of a “raft-reporter” from the TGN (Proszynski et al., 2005), these data suggest Kes1 organizes membrane surfaces in the TGN/endosomal system so that vesicle biogenesis and SL metabolism/trafficking can be appropriately coordinated. Although Kes1 defects potentiate Pik1 PtdIns 4-OH kinase-mediated accumulation of PtdIns-4-P (Li et al., 2002, Fairn et al., 2007; Schaaf et al., 2008), the effects of kes1Δ on PtdIns-4-P homeostasis do not easily explain the restoration of stress responses in sec14-1ts tlg2Δ double mutants. Rather, we propose Kes1 inactivation disorganizes SL/sterol domains, thereby facilitating SL escape from a Sec14- and Tlg2-deficient compartment. Such escape contributes to normalization of the SL/ceramide metabolic derangements associated with defective TGN/endosomal trafficking.
Finally, recent evidence indicates that a mammalian ceramide transfer protein shuttles ceramide from the ER directly to the TGN in a manner that involves the ER-localized FFAT motif receptor VAP (Hanada et al., 2003; Kawano et al., 2006). It was therefore a matter of considerable interest whether yeast VAP (Scs2; Kagiwada et al., 1998) plays any role in functional interactions between the TGN/endosome system and the UPR. Deletion of SCS2 has no obvious effect on UPR silencing in sec14-1ts tlg2Δ double mutants at 37°C, scs2Δ does not itself evoke bypass Sec14, nor does scs2Δ compromise mechanisms for bypass Sec14 (data not shown). Thus, we find no evidence to indicate an obligate Scs2 requirement in linkage of ceramide-dependent TGN/endosomal events with regulation of ER/nuclear stress responses.
Supplementary Material
ACKNOWLEDGMENTS
V.A.B. dedicates this paper to the memory of Beth Jones—a professional in every sense of the word. We also acknowledge the University of North Carolina Lineberger Comprehensive Cancer Center Genome Analysis and Nucleic Acids Core facilities. We acknowledge our UNC colleagues Doug Cyr, Pat Brennwald, Guendolina Rossi, and Mara Duncan for helpful discussions and reagents; Jonathan Weissman (University of California-San Francisco) for helpful suggestions in the early phases of this work; Yusuf Hannun, Lina Obeid, and Besim Ogretmen (Medical University of South Carolina) for helpful discussion; Paul Herman (The Ohio State University) for advice and reagents for assessing PKA pathway function; and Hal Mekeel for training and assistance with electron microscopy. Fluorescence imaging experiments were performed at the University of North Carolina Michael Hooker Microscopy Facility. We are grateful to Jacek Bielawski and Alicja Bielawska (Medical University of South Carolina) for performing the quantitative ceramidomic profiling of our yeast strains of interest. We are also grateful to Peter Walter (University of California-San Francisco) for anti-Ire1 antibody and for the HAC1I expression plasmid, to Randy Schekman (University of California-Berkeley) for Sec61 antibody, to Todd Graham (Vanderbilt University) for FYVE-dsRed plasmids, to Ben Glick (University of Chicago) for Sec7-dsRed plasmids, to Alan Hinnebusch (National Institutes of Health) for helpful advice and for plasmid p180, to Chris Beh (Simon Fraser University) for GFP-Snc1 plasmid, and to Colin Stirling (University of Manchester, United Kingdom) for anti-CPY antibody and for the UPRE-LACZ reporter plasmid (pJT30). This work was supported by National Institutes of Health grant GM-44530 (to V.A.B.). K. T. is a postdoctoral trainee of the National Institutes of Health Molecular Mycology and Pathogenesis training grant (5T32-AI052080), and G. S. is a postdoctoral fellow of the Deutsche Forschungsgemeinshaft (SCHA 1274/1-1). C. B. and R. B. were supported by grants from the Canadian Institutes of Health Research and Genome Canada through the Ontario Genomics Institute (to C. B.). X. G. and M.R.W. were supported by Competitive Research Program award 2007-04 from the Singapore National Research Foundation (to M.R.W.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0426) on August 27, 2008.
REFERENCES
- Abeliovich H., Darsow T., Emr S. D. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 1999;18:6005–6016. doi: 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H., Grote E., Novick P., Ferro-Novick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J. Biol. Chem. 1998;273:11719–11727. doi: 10.1074/jbc.273.19.11719. [DOI] [PubMed] [Google Scholar]
- Adamo J., Moskow J. J., Gladfelter A. S., Viterbo D., Lew D. J., Brennwald P. J. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J. Cell Biol. 2001;155:581–592. doi: 10.1083/jcb.200106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronova S., Wedaman K., Anderson S., Yates J., III, Powers T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin and membrane trafficking in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;18:27779–27794. doi: 10.1091/mbc.E07-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., Loewith R., Parsons A. B., Gao L., Tabuchi M., Zhou H., Boone C., Hall M. N., Emr S. D. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 2004;23:3747–3757. doi: 10.1038/sj.emboj.7600384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandhakavi S., Xie H., O'Callaghan B., Sakurai H., Kim D. H., Griffin T. J. Hsf1 activation inhibits rapamycin resistance and TOR signaling in yeast revealed by combined proteomic and genetic analysis. PLoS ONE. 2008;3:e1598. doi: 10.1371/journal.pone.0001598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis V. A., Malehorn D. E., Emr S. D., Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis V. A., Aitken J. R., Cleves A. E., Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- Barlowe C., Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Bertram P. G., Choi J. H., Carvalho J., Ai W., Zeng C., Chan T.-F., Zheng X.F.S. Tripartite regulation of Gln3p by TOR, Ure2, and phosphatases. J. Biol. Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- Bielawski J., Szulc Z. M., Hannun Y. A., Bielawska A. Simultaneous quantitative analysis of bioactive SLs by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Brickner J. H., Blanchette J. M., Sipos G., Fuller R. S. The Tlg SNARE complex is required for TGN homotypic fusion. J. Cell Biol. 2001;155:969–978. doi: 10.1083/jcb.200104093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant N. J., James D. E. The Sec1p/Munc18 (SM) protein, Vps45p, cycles on and off membranes during vesicle transport. J. Cell Biol. 2003;161:691–696. doi: 10.1083/jcb.200212078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Zhang Y., Pypaert M., Walker L., Ferro-Novick S. Trs120p mediates traffic from the early endosome to a late Golgi compartment. J. Cell Biol. 2005;171:823–833. doi: 10.1083/jcb.200505145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., De Virgilio C., Deloche O. Phosphatidylinositol 4-phosphate is required for translation initiation in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:38139–38149. doi: 10.1074/jbc.M601060200. [DOI] [PubMed] [Google Scholar]
- Chang H. J., Jones E. W., Henry S. A. Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics. 2002;162:29–43. doi: 10.1093/genetics/162.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R. E., Walter P. Translational attenuation mediated by an mRNA intron. Curr. Biol. 1997;7:850–859. doi: 10.1016/s0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- Chen E. J., Kaiser C. A. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 2003;161:333–347. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. C., Powers T. Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr. Genet. 2006;49:281–293. doi: 10.1007/s00294-005-0055-9. [DOI] [PubMed] [Google Scholar]
- Chen S. H., Chen S., Tokarev A. A., Liu F., Jedd G., Segev N. Ypt31/32 GTPases and their novel F-box effector protein Rcyl regulate protein recycling. Mol. Biol. Cell. 2005;16:178–192. doi: 10.1091/mbc.E04-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves A. E., Novick P J., Bankaitis V. A. Mutations in the SACI gene suppress defects in yeast Golgi and yeast actin function. J. Cell Biol. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves A. E., McGee T. P., Bankaitis V. A. Phospholipid transfer proteins: a biological debut. Trends Cell Biol. 1991a;1:30–34. doi: 10.1016/0962-8924(91)90067-j. [DOI] [PubMed] [Google Scholar]
- Cleves A. E., McGee T. P., Whitters E. A., Champion K. M., Aitken J. R., Dowhan W., Goebl M., Bankaitis V. A. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991b;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe J. G., Lim A. C., Xu J., Hong W. A role for Tlg1p in the transport of proteins within the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:2407–2423. doi: 10.1091/mbc.10.7.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvera S., D'arrigo A., Stenmark H. Phosphoinositides in membrane traffic. Curr. Opin. Cell Biol. 1999;11:460–465. doi: 10.1016/S0955-0674(99)80066-0. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Shamu C. E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Demmel L., et al. The clathrin adaptor Gga2p is a phosphatidylinositol 4-phosphate effector at the Golgi exit. Mol. Biol. Cell. 2008;19:1991–2002. doi: 10.1091/mbc.E06-10-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Enfert C., Barlowe C., Nishikawa S., Nakano A., Schekman R. Structural and functional dissection of a membrane glycoprotein required for vesicle budding from the endoplasmic reticulum. Mol. Cell. Biol. 1991;11:5727–5734. doi: 10.1128/mcb.11.11.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Sumanasekera C., Lester R. L. Function and metabolism of SLs in Saccharomyces cerevisiae. Prog. Lipid Res. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Dunn T. M., Haak D., Monaghan E., Beeler T. J. Synthesis of monohydroxylated inositolphosphorylceramide (IPC-C) in Saccharomyces cerevisiae requires Scs7p, a protein with both a cytochrome b5-like domain and a hydroxylase/desaturase domain. Yeast. 1998;14:311–321. doi: 10.1002/(SICI)1097-0061(19980315)14:4<311::AID-YEA220>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Fairn G. D., Curwin A. J., Stefan C. J., McMaster C. R. The oxysterol binding protein Kes1p regulates Golgi apparatus phosphatidylinositol-4-phosphate function. Proc. Natl. Acad. Sci. USA. 2007;104:15352–15357. doi: 10.1073/pnas.0705571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M., Kearns B. G., Gedvilaite A., Kagiwada S., Kearns M., Fung M.K.Y., Bankaitis V. A. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Sutton R. B., Brunger A. T., Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman D. A., Meyers R. E., Cantley L. C. Phosphoinositide kinases. Annu. Rev. Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., Bösche M., Krause R., Grandi P., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gilchrist A., et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Guan X. L., Wenk M. R. Mass spectrometry-based profiling of phospholipids and SLs in extracts from Saccharomyces cerevisiae. Yeast. 2006;23:465–477. doi: 10.1002/yea.1362. [DOI] [PubMed] [Google Scholar]
- Guillas I., Kirchman P. A., Chuard R., Pfefferli M., Jiang J. C., Jazwinski S. M., Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H., Schnieders E. A., Thorner J., Takemoto J. Y., DeWald D. Direct involvement of phosphatidylinositol-4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:34294–34301. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- Hanson B. A., Lester R. L. The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J. Lipid Res. 1980;21:309–315. [PubMed] [Google Scholar]
- Harsay E., Bretscher A. Parallel secretory pathways to the cell surface in yeast. J. Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E., Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J. Cell Biol. 2002;156:271–285. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hechtberger P., Zinser E., Saf R., Hummel K., Paltauf F., Daum G. Characterization, quantification and subcellular localization of inositol-containing SLs of the yeast, Saccharomyces cerevisiae. Eur. J. Biochem. 1994;225:641–649. doi: 10.1111/j.1432-1033.1994.00641.x. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Gene-specific translational control of the yeast Gcn4 gene by phosphorylation of eukaryotic initiation factor 2. Mol. Microbiol. 1993;10:215–223. doi: 10.1111/j.1365-2958.1993.tb01947.x. [DOI] [PubMed] [Google Scholar]
- Hinnebush A. G. Translational regulation of yeast Gcn4. A window on factors that control initiator t-RNA binding to the ribosome. J. Biol. Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- Holthuis J. C., Nichols B. J., Pelham H. R. The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol. Biol. Cell. 1998;9:3383–3397. doi: 10.1091/mbc.9.12.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ile K. E., Schaaf G., Bankaitis V. A. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat. Chem. Biol. 2006;2:576–583. doi: 10.1038/nchembio835. [DOI] [PubMed] [Google Scholar]
- Im Y. J., Raychaudhuri S., Prinz W. A., Hurley J. H. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Muratat K., Kimura A. Transformation of intact yeast cells with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S., Hosaka K., Murata M., Nikawa J., Takatsuki A. The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism. J. Bacteriol. 1998;180:1700–1708. doi: 10.1128/jb.180.7.1700-1708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kama R., Robinson M., Gerst J. E. Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol. Cell. Biol. 2007;27:605–621. doi: 10.1128/MCB.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M., Kumagai K., Masahiro Nishijima, Hanada K. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAP-interacting FFAT motif of CERT. J. Biol. Chem. 2006;281:30279–30288. doi: 10.1074/jbc.M605032200. [DOI] [PubMed] [Google Scholar]
- Kitagaki H., Cowart L. A., Matmati N., Vaena de Avalos S., Novgorodov S. A., Zeidan Y. H., Bielawski J., Obeid L. M., Hannun Y. A. Isc1 regulates SL metabolism in yeast mitochondria. Biochim. Biophys. Acta. 2007;1768:2849–2861. doi: 10.1016/j.bbamem.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K., Normington K., Sambrook J., Gething M. J., Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumble K. D., Kornberg A. Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J. Biol. Chem. 1996;271:27146–27151. doi: 10.1074/jbc.271.43.27146. [DOI] [PubMed] [Google Scholar]
- Kutateladze T. G., Ogburn K. D., Watson W. T., de Beer T., Emr S. D., Burd C. G., Overduin M. Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol. Cell. 1999;3:805–811. doi: 10.1016/s1097-2765(01)80013-7. [DOI] [PubMed] [Google Scholar]
- Lester R. L., Dickson R. C. SLs with inositolphosphate-containing head groups. Adv. Lipid. Res. 1993;26:253–274. [PubMed] [Google Scholar]
- Letourneur F., Gaynor E. C., Hennecke S., Démollière C., Duden R., Emr S. D., Riezman H., Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Li X.M. P., Rivas M., Fang J., Marchena B., Mehotra A., Chaudhary L., Feng G. D., Prestwich, Bankaitis V. A. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chang A. Heat shock response relieves ER stress. EMBO J. 2008;27:1049–1059. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K., et al. Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C-related protein reveals a primordial role in subcellular SL distribution. J. Cell Biol. 2004;164:547–556. doi: 10.1083/jcb.200310046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C., Xu R., Bielawska A., Obeid L. M. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J. Biol. Chem. 2000a;275:6876–6884. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- Mao C., Xu R., Bielawska A., Szulc Z. M., Obeid L. M. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J. Biol. Chem. 2000b;275:31369–31378. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- McGee T. P., Skinner H. B., Whitters E. A., Henry S. A., Bankaitis V. A. A phosphatidylinositol transfer protein controls the phosphatidylcholine content of yeast Golgi membranes. J. Cell Biol. 1994;124:273–287. doi: 10.1083/jcb.124.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels J. T., Broach J. R. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- Nikawa J., Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol. Microbiol. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Patil C. K., Li H., Walter P. Gen4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004;2:E246. doi: 10.1371/journal.pbio.0020246. 1208–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S. E., et al. Yeast Sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol. Cell. 1999;4:187–197. doi: 10.1016/s1097-2765(00)80366-4. [DOI] [PubMed] [Google Scholar]
- Phillips S. E., Vincent P., Rizzieri K., Schaaf G., Gaucher E. A., Bankaitis V. A. The diverse biological functions of phosphatidylinositol transfer proteins in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2006;41:1–28. doi: 10.1080/10409230500519573. [DOI] [PubMed] [Google Scholar]
- Proszynski T. J., et al. A genome-wide visual screen reveals a role for SLs and ergosterol in cell surface delivery in yeast. Proc. Natl. Acad. Sci. 2005;102:17981–17986. doi: 10.1073/pnas.0509107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopov V., Govindan B., Novick P., Gerst J. E. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in. S. cerevisiae. Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- Puria R., Zurita-Martizez S. A., Cardenas M. E. Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2008;105:7194–7199. doi: 10.1073/pnas.0801087105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K., Holcomb C., Fuller R. S. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J. Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas M. P., Kearns B. G., Xie Z., Guo S., Sekar M. C., Hosaka K., Kagiwada S., York J. D., Bankaitis V. A. Relationship between altered phospholipid metabolism, diacylglycerol, bypass Sec14, and the inositol auxotrophy of yeast sac1 mutants. Mol. Biol. Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M., Poon P. P., Schindler C., Murray L. E., Kama R., Gabriely G., Singer R. A., Spang A., Johnston G. C., Gerst J. E. The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol. Biol. Cell. 2006;17:1845–1858. doi: 10.1091/mbc.E05-09-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J. R., Bastidas R., Puria R., Cardenas M. E. Nutritional control via Tor signaling in Saccharomyces cerevisiae. Curr. Opin. Microbiol. 2008;11:153–160. doi: 10.1016/j.mib.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Routt S. M., Ryan M. M., Tyeryar K., Rizzieri K., Roumanie O., Brennwald P. J., Bankaitis V. A. Nonclassical PITPs activate phospholipase D via an Stt4p-dependent pathway and modulate function of late stages of the secretory pathway in vegetative yeast cells. Traffic. 2005;6:1157–1172. doi: 10.1111/j.1600-0854.2005.00350.x. [DOI] [PubMed] [Google Scholar]
- Ryan M. M., Temple B.R.S., Phillips S. E., Bankaitis V. A. Conformational dynamics of the major yeast phosphatidylinositol transfer protein Sec 14, Insights into the mechanisms of PL exchange and diseases of Sec14-like protein deficiencies. Mol. Biol. Cell. 2007;18:1928–1942. doi: 10.1091/mbc.E06-11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama S. R., Cleves A. E., Malehorn D. E., Whitters E. A., Bankaitis V. A. Cloning and characterization of the Kluyveromyces lactis SEC14: a gene whose product stimulates Golgi secretory function in S. cerevisiae. J. Bacteriol. 1990;172:4510–4521. doi: 10.1128/jb.172.8.4510-4521.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H., Okamoto Y., Luberto C., Mao C., Bielawska A., Domae N., Hannun Y. A. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:39793–39798. doi: 10.1074/jbc.M007721200. [DOI] [PubMed] [Google Scholar]
- Schaaf G., Ortlund E., Tyeryar K., Mousley C., Ile K., Woolls M., Garrett T., Raetz C.R.H., Redinbo M., Bankaitis V. A. The functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the Sec14-superfamily. Mol. Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciorra V. A., Audhya A., Parsons A. B., Segev N., Boone C., Emr S. D. Synthetic gene array analysis of the PtdIns 4-kinase Pik1 identifies components in a Golgi-specific Ypt31/rab-GTPase pathway. Mol. Biol. Cell. 2005;15:2038–2047. doi: 10.1091/mbc.E04-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorling S., Vallée B., Barz W. P., Riezman H., Oesterhelt D. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha B., Phillips S. E., Bankaitis V. A., Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol transfer protein Sec14. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hinks J. B. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1983. pp. 1–113. [Google Scholar]
- Shi X., Kornberg A. Endopolyphosphatase in Saccharomyces cerevisiae undergoes post-translational activations to produce short-chain polyphosphates. FEBS Lett. 2005;579:2014–2018. doi: 10.1016/j.febslet.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Sidrauski C., Cox J. S., Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Sidrauski C., Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Skinner H. B., McGee T. P., McMaster C., Fry M. R., Bell R. M., Bankaitis V. A. Phosphatidylinositol transfer protein stimulates yeast Golgi secretory function by inhibiting choline-phosphate cytidylyltransferase activity. Proc. Natl. Acad. Sci. USA. 1995;92:112–116. doi: 10.1073/pnas.92.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova T., Chadwick T. G., MacArthur R., Poluekov O., Song L., Ryan M., Schaaf G., Bankaitis V. A. The chemistry of PL binding by the Saccharomyces cerevisiae phosphatidylinositol transfer protein Sec14 as determined by electron paramagnetic resonance spectroscopy. J. Biol. Chem. 2006;281:34897–34908. doi: 10.1074/jbc.M603054200. [DOI] [PubMed] [Google Scholar]
- Smirnova T., Chadwick T. G., van Tol J., Ozarowski A., Poluektov O., Schaaf G., Ryan M. M., Bankaitis V. A. Local polarity and hydrogen bonding inside the Sec14 PL-binding cavity: high-field multifrequency studies. Biophys. J. 2007;92:3686–3695. doi: 10.1529/biophysj.106.097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T. H., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Strahl T., Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M., Audhya A., Parsons A. B., Boone C., Emr S. D. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in SL regulation. Mol. Cell. Biol. 2006;26:5861–5875. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Wedaman K. P., Reinke A., Anderson S., Yates J., III, McCaffery J. M., Powers T. The Tor kinases are in distinct membrane associated protein complexes in S. cerevisiae. Mol. Biol. Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar and endosomal dynamics in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent P., Chua M., Nogue F., Fairbrother A., Mekheel H., Xu Y., Allen N., Bibikova T. N., Gilroy S., Bankaitis V. A. A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J. Cell Biol. 2005;168:801–812. doi: 10.1083/jcb.200412074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedaman K. P., Reinke A., Anderson S., Yates J. III, McCaffery J. M., Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. J. Biol. Chem. 2003;275:35727–35733. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitters E. A., McGee T. P., Bankaitis V. A. Purification and characterization of a late Golgi compartment from Saccharomyces cerevisiae. J. Biol. Chem. 1994;269:28106–28117. [PubMed] [Google Scholar]
- Wilkinson B. M., Tyson J. R., Reid P. J., Stirling C. J. Distinct domains within yeast Sec61p involved in post-translational translocation and protein dislocation. J. Biol. Chem. 2000;275:521–529. doi: 10.1074/jbc.275.1.521. [DOI] [PubMed] [Google Scholar]
- Xie Z., Fang M., Rivas M. P., Faulkner A., Sternweis P. C., Engebrecht J., Bankaitis V. A. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc. Natl. Acad. Sci. USA. 1998;95:12346–12351. doi: 10.1073/pnas.95.21.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Shen G., X., Jiang Y. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 2006;25:3546–3555. doi: 10.1038/sj.emboj.7601239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa L., Marchena J., Xie Z., Li X., Poon P. P., Singer R., Johnston G., Randazzo P. A., Bankaitis V. A. Activity of specific lipid-regulated ARFGAPs is required for Sec14p-dependent Golgi secretory function in yeast. Mol. Biol. Cell. 2002;13:2193–2206. doi: 10.1091/mbc.01-11-0563.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B. P., Craven R. A., Reid P. J., Willer M., Stirling C. J. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2000;20:262–271. doi: 10.1093/emboj/20.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.