Abstract
Reepithelialization during cutaneous wound healing involves numerous signals that result in basal keratinocyte activation, spreading, and migration, all linked to a loosening of cell–cell adhesion structures. The transcription factor Slug is required for this process, and EGF treatment of human keratinocytes induced activating phosphorylation of Erk5 that coincides with slug transcription. Accordingly, ectopic activation of Erk5 led to increased Slug mRNA levels and faster wound healing, whereas keratinocyte migration was totally blocked by Erk5 pathway inhibition. Expression of a shRNA specific for Erk5 strongly diminished Erk5 levels in keratinocytes and significantly decreased their motility response to EGF, along with induction of Slug expression. These Erk5-deprived keratinocytes showed an altered, more compact morphology, along with disruption of desmosome organization. Accordingly, they displayed an altered ability to form cell aggregates. These results implicate a novel EGFR/Erk5/Slug pathway in the control of cytoskeleton organization and cell motility in keratinocytes treated with EGF.
INTRODUCTION
Wound healing is a multistep process involving an initial inflammatory response that provides a large number of cytokines and growth factors (Martin et al., 1992), including ligands for the epidermal growth factor receptor (EGFR). These extracellular stimuli, together with mechanical stimuli, activate migration and reepithelialization in basal and suprabasal keratinocytes. Keratinocytes at the leading edge of a wound undergo a phenotypic conversion that includes a dramatic reorganization of the cytoskeleton and associated junctional structures (Krawczyk, 1971; Paladini, 1996). Intermediate filaments retract from the cell surface and attached desmosomes and hemidesmosomes are dissolved. Partial or complete dissolution of the basement membrane zone occurs, associated with an obvious loss of cell polarity. These changes are accompanied by profound alterations in the actin-based cytoskeleton and an increase in migratory activity (Stenn and Depalma, 1988). Overall, reepithelialization can be described as a partial epithelial-mesenchymal transition (EMT) involving cells that are both cohesive and motile (Arnoux et al., 2005). During EMT, epithelial cells progressively adopt mesenchymal characteristics and become motile. This process occurs during developmental stages: gastrulation, heart formation, neural crest migration, somitogenesis, and palate formation. Slug and Snail, members of the Snail family of transcription factors, appear to be involved in these developmental processes and have been linked to the EMT in most EMT models (Nieto, 2002). Slug is also required for epithelial cell motility in wound healing, and Slug-deficient mice do not reepithelialize in an ex vivo assay (Savagner et al., 2005). Slug is expressed by basal keratinocytes (Savagner et al., 2005; Turner et al., 2006) and can regulate integrins and E-cadherin in this cellular context (Turner et al., 2006; Zhang et al., 2003). Similar to its relative Snail, overexpression of Slug can directly repress the E-cadherin promoter in various transformed cell lines. Slug and Snail genes have been linked to carcinoma progression in several cancer types (Come et al., 2006; Elloul et al., 2005; Shih et al., 2005). However, the role of Slug during cancer progression is not clear and probably involves distinct mechanisms, as it is physiologically coexpressed with E-cadherin in several cell types, including keratinocytes (Come et al., 2006).
Slug is induced by several intracellular signaling pathways, including those activated by EGF, FGFs, TGFβ, and oncogenic Ras (Edme et al., 2002; Romano and Runyan, 2000). In the context of wound healing, EGF is particularly relevant because the EGF receptor is up-regulated during this process. Activation of this receptor contributes significantly to the migratory and invasive potential of keratinocytes (Brown et al., 1989; Chernoff and Robertson, 1990; Andree et al., 1994; Hudson and McCawley, 1998). This receptor induces all the major mitogen-activated protein (MAP) kinase cascades, namely the Erk1/2, Erk5 (Kato et al., 1997), p38, and JNK pathways, where Erk1/2 and Erk5 appear particularly important for proliferation of epithelial cells. Erk1/2 activation, but not that of Erk5, was found to mediate migration of epithelial cells during wound healing in Madin-Darby canine kidney (MDCK) cells (Matsubayashi et al., 2004). On the other hand, other studies concluded that the Erk5 pathway is necessary for EGF-induced morphogenesis in MDCK cells (Karihaloo et al., 2001). Thus, the role of Erk5 in epithelial movement is disputed.
In this article, we examine the signaling pathways that drive EGF induction of Slug expression and cell migration during wound healing. An in vitro assay based on immortalized keratinocytes was used to identify which MAP kinase pathway function downstream of the EGFR. We find that the Erk5 cascade controls Slug promoter activity and plays an important role in reepithelialization, in part through regulation of the actin cytoskeleton.
MATERIALS AND METHODS
Plasmids
A pGL3-Basic Luciferase Reporter Vector (Promega, Madison, WI) is used to assess the activity of a 2800-bp mouse Slug promoter previously described (Conacci-Sorrell et al., 2003). A human Slug promoter was a kind gift from P. Wade (National Institute of Environmental Health Sciences, NIH, Research Triangle Park, NC; Fujita et al., 2003). The pCMV-RasV12 vector (Nissen et al., 2001) encodes a constitutively active form of the Ras protein (G12V). The pcDNA3 expression vectors for wild-type Erk5 or the constitutively active forms for Mek1 (MEK1SS3) and Mek5 (S313D/T317D), termed Mek5D, have been described previously (Brunet et al., 1994; Mulloy et al., 2003). Dominant-negative MAP kinases for the Erk5 pathway, namely Erk5AEF (T218A and Y220F) and Mek5A (S311A and T315A), were generously provided by Dr. J.-D. Lee (The Scripps Research Institute, La Jolla, CA; Kato et al., 1997). The pCMV-βGal pBabe vector was used as described (Mulloy et al., 2003). The plasmid pCDNA3.H.Slug was a kind gift from T. Ip (University of Massachusetts Medical School, Worcester, MA; Hemavathy et al., 2000).
Reagents and Antibodies
HaCaT cells were pretreated with EGFR inhibitor AG1478 (Calbiochem, La Jolla, CA) and Erk inhibitor PD184352 (provided by P. Cohen, Dundee, Scotland) for 1 h at concentrations indicated in the text and then induced with EGF (Sigma, St. Louis, MO). The following primary antibodies were used: goat anti-Slug and goat anti-ERK5 agarose conjugate (G18 and sc-1284 AC, respectively; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-Erk1/2 and Erk5 and phosphospecific rabbit anti-Erk1/2 (Cell Signaling Technology, Beverly, MA), mouse monoclonal anti-paxillin (BD Transduction Laboratories, Lexington, KY), mouse monoclonal antidesmoplakin (American Research Products, Belmont, MA), rabbit pan-specific anti-keratin (Zymed, South San Francisco, CA). A rabbit antibody anti-Slug (Abcam, Cambridge, MA) with increased sensitivity was used for Figures 6 and 7.
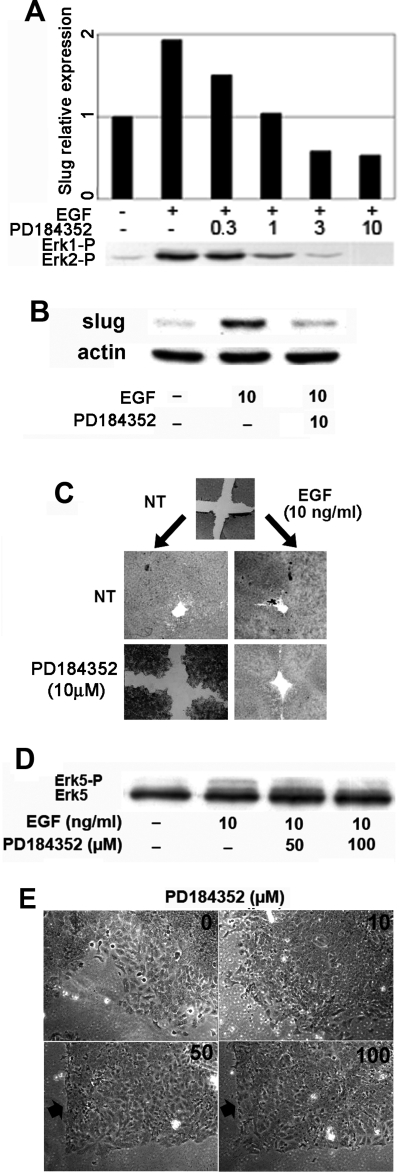
Figure 6.
Pharmacological inhibition of Erks affects Slug expression and reepithelialization by HaCaT cells. (A) Serum-starved HaCaT cells were pretreated for 1 h with the indicated concentration of PD184352 (μM) and then induced for 15 min with 10 ng/ml EGF (+). Slug RNA levels were analyzed and quantified as described in the legend to Figure 2. Protein extracts were loaded on SDS-PAGE and transferred to nitrocellulose, and phosphoERK1/2 was detected by immunoblotting. (B) Serum-starved HaCaT cells were pretreated for 1 h with PD184352 and then induced for 2 h with 10 ng/ml EGF as indicated. Protein levels were analyzed and quantified as described in the legend to Figure 3, using β-actin as the reference. (C) HaCaT cells were grown on 24-well plate to high confluency in 10% FBS. Cell monolayers were equivalently wounded and pretreated with 10 μM PD184352 for 1 h followed by stimulation with 10 ng/ml EGF as indicated for 52 h. (D) Serum-starved HaCaT cells were pretreated for 1 h with 50 or 100 μM PD184352 and then induced for 15 min with 10 ng/ml EGF as indicated. Erk5 was visualized by immunoblotting as described in the legend to Figure 5. (E) HaCaT cells were prepared as in C and pretreated for 1 h with the indicated concentrations of PD184352 before the addition of 10 ng/ml EGF. Wound edge is shown after 52 h culture. When 50 or 100 μM PD184352 was applied to the cells, the wound edge remained essentially stationary (arrow), but cells remained alive.
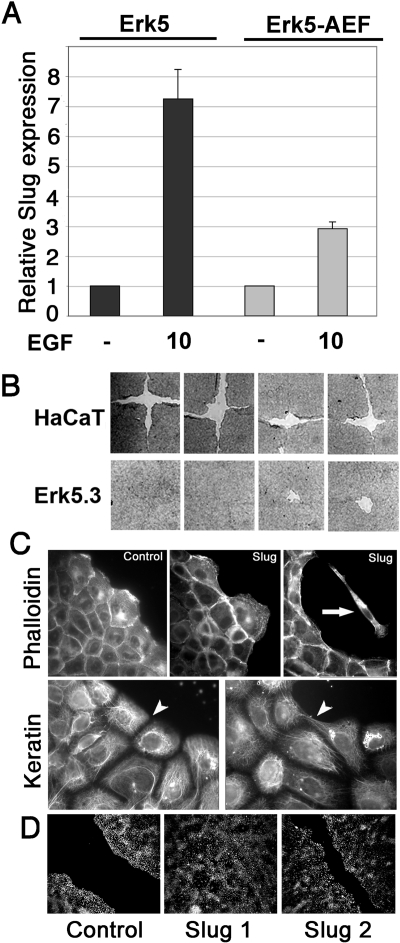
Figure 7.
Erk5 potentiates EGF-driven Slug induction and reepithelialization by HaCaT cells. (A) Pools of transfectants stably expressing Erk5 or Erk5-AEF were serum-starved overnight and then stimulated with 10 ng/ml EGF for 1 h. RNAs were extracted, and Slug RNA levels were determined by quantitative RT-PCR as described in the legend to Figure 3. (B) Wound-healing assay using wt HaCaT cells or the Erk5-overexpressing clone Erk5.3, as described in the legend to Figure 4C. Four separate experiments are shown. (C) HaCaT clones that overexpress Slug or generated with the vector alone (control) were analyzed for actin (phalloidin) and keratin mesh organization. Some cells detached from the aggregates and migrated as individual cells (arrow). Keratin organization was more homogeneous in control than in Slug-overexpressing clones (arrowhead). (D) Wound-healing assay using control or Slug-overexpressing clones Slug1 and Slug 2. Cells were treated with 10 ng/ml EGF and photographed 48 h after wounding.
Cell Culture and Treatment
The Chinese hamster lung fibroblast cell line CCL39 was cultivated as described (Mulloy et al., 2003). The immortalized human keratinocyte cell line HaCaT was a kind gift from Prof. Fusenig (University of Heidelberg, Heidelberg, Germany; Boukamp et al., 1992) and were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), glutamine, and antibiotics. All cells were grown at 37°C in a humidified 5% CO2 incubator. Stable clones were obtained after magnetofection (Oz Biosciences, Marseille, France) on HaCaT cells. Polyclonal cell populations were selected with 1 mg/ml G418 (Invitrogen, Carlsbad, CA). Monoclones were obtained by limit dilution. Treatments were realized 3 d after seeding 106 cells onto a six-well plate. Serum was withdrawn from culture medium overnight before treatment. Cells were incubated for 1 h with AG1478 or PD184352 before EGF stimulation. Except when noticed otherwise, protein were extracted after 15-min treatment to study Erk5, after 2-h treatment for Slug protein expression, and after 1 h for RNA extraction.
Preparation of Skin Explants
Briefly, the Slug-LacZ mouse line was initially generated by inframe insertion of the β-galactosidase gene into the zinc finger coding region of the Slug gene (Jiang et al., 1998) and was graciously provided by Dr. Thomas Gridley (The Jackson Laboratory, Bar Harbor, ME). Animals were treated in accordance with institutional guidelines (Institut National de la Sante et de la Recherche Medicale). Assays were conducted according to Mazzalupo et al. (2002). Areas of dorsal skin were shaved, disinfected, and spread flat onto a culture dish. A 4-mm sterile punch was used to excise explants that were individually placed into 24-well culture plate. Explants were partially immerged in 0.2 ml culture medium at 37°C, 5% CO2. After overnight incubation, explants were totally submerged in culture medium. Medium was changed twice a week. Medium was prepared as follows: DMEM supplemented with 10% FBS, triiodothyronine (20 ng/ml), transferrin (5 μg/ml), cholera toxin (0.1 nM), hydrocortisone (0.4 μg), gentamicin (25 μg/ml), and penicillin (60 μg/ml). EGF (10 ng/ml) was added in some cases, as indicated.
Reporter Gene Assays
CCL39 cells were transfected using Fugene 6 (Roche, Indianapolis, IN) as described (Mulloy et al., 2003). Briefly, 2 × 105 cells/well were plated onto a six-well plate. One day later, a plasmid encoding a luciferase gene driven by a 2800-base pair mouse Slug promoter (250 ng) was cotransfected with mutant constructs. pCMV-βGal (50 ng) was cotransfected to monitor transfection efficiency.
Total amount of DNA transfected was kept constant by adding empty vector (pBabe). After 16 h, culture medium was changed to 0.5% FBS. Two days after transfection, cells were lysed in a lysis buffer (Promega) and the activities of β-galactosidase and luciferase (Promega) were measured as described (Mulloy et al., 2003). To correct for transfection efficiency, the luciferase activity was reported to β-galactosidase expression in every duplicate and expressed as a fold induction relative to control. Experiments were repeated at least three times in duplicate.
Western Blot Analysis
The cells were lysed in a buffer containing 10 mM Tris, pH 7.7, 1% SDS (wt/vol), 5 mM EDTA, 10 mM β-glycerophosphate, 10 mM sodium pyrophosphate, and a protease inhibitor cocktail (Complete, Roche). Cell debris were sedimented (13,000 × g, 5 min) and discarded. The protein content of the soluble fractions was determined with the BCA Protein assay Reagent kit (Pierce, Rockford, IL). Protein extracts (40 μg) were resolved by 7.5% or 12% SDS-PAGE gel as appropriate, and transferred to a nitrocellulose membrane (Amersham, Piscataway, NJ). Membranes were blocked by incubation with 5% nonfat dry milk in TBST for 1 h at room temperature and probed with primary antibodies diluted into the same solution or 5% bovine serum albumin in TBST at 4°C overnight. Immune complexes were detected by chemiluminescence with HRP-conjugated secondary antibodies.
ERK5 Kinase Assays
The assays were performed according to the protocol of Seyfried et al. (2005). HaCaT cells were deprived of serum for 18 h before induction for 30 min with either 10 ng/ml EGF or 20% FBS. The culture plate was placed on ice, and the cells washed once with ice-cold Krebs-Ringer HEPES-buffered saline (50 mM HEPES, pH 7.4, 130 mM NaCl, 5 mM KCl, 5 mM MgCl2, and 1.5 mM CaCl2). Cells were lysed in Triton lysis buffer (TLB; 20 mM Tris, pH 7.4, 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 10% glycerol, 0.1 mM orthovanadate, 0.1 mM 4-(2-aminoethyl)benzenesulfonyl floride (AEBSF), 2.5 μg/ml leupeptin, 2.5 μg/ml aprotinin, and 2.5 μg/ml pepstatin). Extracts were clarified by centrifugation (14,000 × g for 10 min at 4°C). The concentration of soluble proteins was quantified by the Bradford method (Sigma). Cell lysate, 450 μg, was incubated with anti-ERK5-Agarose (sc-1284, lot no. G302) for 3 h at 4°C. Immune complexes were washed twice with TLB and twice with kinase buffer (KB; 25 mM HEPES, pH 7.4, 25 mM MgCl2, 25 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mM orthovanadate, 0.1 mM AEBSF, 2.5 μg/ml leupeptin, 2.5 μg/ml aprotinin, and 2.5 μg/ml pepstatin). The immune complexes were incubated for 15–30 min at RT with shaking in KB containing 1 μg glutathione S-transferase (GST)-MEF2C fixed on glutathione-agarose, 23 μM ATP, and 4 μCi [γ-32P]ATP (3000 Ci/mmol). The agarose complexes were washed twice with TLB, and proteins were analyzed by SDS-PAGE. Radioactive GST-MEF2C was visualized by phosphorimagery of the Coomassie-stained, dried gel on a Typhoon 9200.
Quantitative RT-PCR
Total RNA was isolated from treated cells with RNA extraction kit (Macherey-Nagel, Hoerd, France). cDNA synthesis was carried out in a final volume of 20 μl of first-strand buffer (Invitrogen) containing 1 μg of total RNA, 10 U/μl SuperScriptII reverse transcriptase (Invitrogen), 0.25 μg of random hexamer, and 10 mM concentration of each deoxynucleoside triphosphate. The following human forward and reverse primers were used for specific amplifications: 36B4, GTGATGTGCAGCTGATCAAGACT and GATGACCAGCCCAAAGGAGA; Slug, CCCTGAAGATGCATATTCGGAC and CTTCTCCCCCGTGTGAGTTCTA; and Erk5, ACGAGTACGAGATCATCGAGACC and GGTCACCACATCGAAAGCATTAGG. cDNAs diluted at 1/30 were mixed with a solution containing primers and SYBR-Green mix (Applied Biosystems, Courtaboeuf, France) and were tested with AbiPrism7000. All the amplifications were done in a final volume of 25 μl using standard PCR conditions (40 cycles with an annealing/elongation step at 60°C). Results are derived from the average of at least two independent experiments or RT-QPCR estimations. Gene expression was reported relative to housekeeping gene 36B4.
Immunofluorescence
HaCaT were grown on multichamber glass slide (Lab-TekII, Nunc, Naperville, IL), fixed, and permeabilized in 4% formaldehyde + 0.5% Triton, or cooled methanol (−20°C) for keratins and desmoplakin labeling. After permeabilization and 15 min wash in PBS + 0.05% Tween, primary antibodies were incubated for 1 h in a 10% goat serum PBS solution. Secondary antibodies were incubated in some cases with fluorescein-labeled phalloidin (Sigma) and DAPI in a 10% goat serum PBS solution for 30 min at room temperature. Phase-contrast microscopy and immunofluorescence of cell cultures was performed using a Leica DM IRB inverted microscope (Leica Microsystems, Deerfield, IL) and images were acquired with a CoolSNAP HQ camera (Roper Scientific, Tucson, AZ). Representative cell aggregates are shown on the figures, based on at least three independent experiments.
In Vitro Reepithelialization Assay
Denudation zones were created by pushing the narrow end of a sterile P1000 plastic pipette tip through the quiescent, contact-inhibited, HaCaT keratinocyte monolayer. After wound healing, cells were fixed in methanol and colored with eosin and methylene blue using the RAL 555 coloration kit (Research Associates Laboratory, Dallas TX). Relative reepithelialization was quantified by the difference between uncovered wound area at t = 0 and t = 24 h, as calculated using Image J software (http://rsb.info.nih.gov/ij/; NIH, Bethesda, MD). The value is reported relative to SH-Control HaCaT cells. Experiments were performed with or without mitomycin with similar results. When applied, mitomycin was added to the culture medium 1 h before wounding for 24 h. Mitomycin activity was checked independently and found to totally block HaCaT cell proliferation at the concentration used: 0.8 μg/ml.
Short Hairpin RNA and Infection
Erk5 and nonspecific control short hairpin RNA (shRNA) lentiviral particles were purchased from Sigma. HaCaT cells were seeded the day before infection. The next day, cells were infected with an average 2 lentiviral particles per cell in a 20% FBS medium containing 8 μg/ml polybrene. Selection started 48 h after infection by incubation with 1 mg/ml puromycin (Sigma). Down-regulation of Erk5 expression was confirmed by immunoblotting compared with actin protein level. The following shRNA sequence called E275 is used for specific Erk5 expression down-regulation: CCGGGCCAAGTACCATGATCCTGATCTCGAGATCAGGATCATGGTACTTGGCTTT TT. A nontarget shRNA control sequence (shControl) contains mismatches to any known human or mouse genes and so serves as a negative control: CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTT TT.
Estimation of Cell–Cell Distance
After fixation, cells were incubated with DAPI for 30 min as described previously. Contiguous cells growing in close aggregates were spotted by phase-contrast or phalloidin staining. Distances between nuclei centers, defined as an arbitrary point within the nucleus central area, from which measures would be established, were computed using Openlab software (Improvision, Lexington, MA). Several measurements were made for each nuclear center point to neutralize potential interference from the nuclei center localization. At least 100 measurements were performed for each cell type to calculate an average distance for untreated cells or cells transfected with SH-Control or SH-Erk5. The data were evaluated using Student's t test.
Cell–Cell Adherence Assay
Cell–cell adherence was estimated using the original method developed by M. Takeichi (Takeichi, 1977). Briefly, cells were suspended in DMEM supplemented with 10% FBS, glutamine, antibiotics, and 20 mM HEPES buffer using trypsin (0.01%). Cells were plated on albumin-coated dishes and incubated on a gyratory shaker (100 rpm) for 180 min at 37°C. After fixation with glutaraldehyde (5% in PBS), cell aggregates were counted on an inverted microscope and sorted into four size classes, based on the number of cells involved in aggregates. Percentage of cells involved in each class was calculated for each cell type.
RESULTS
EGF Activates Slug Expression
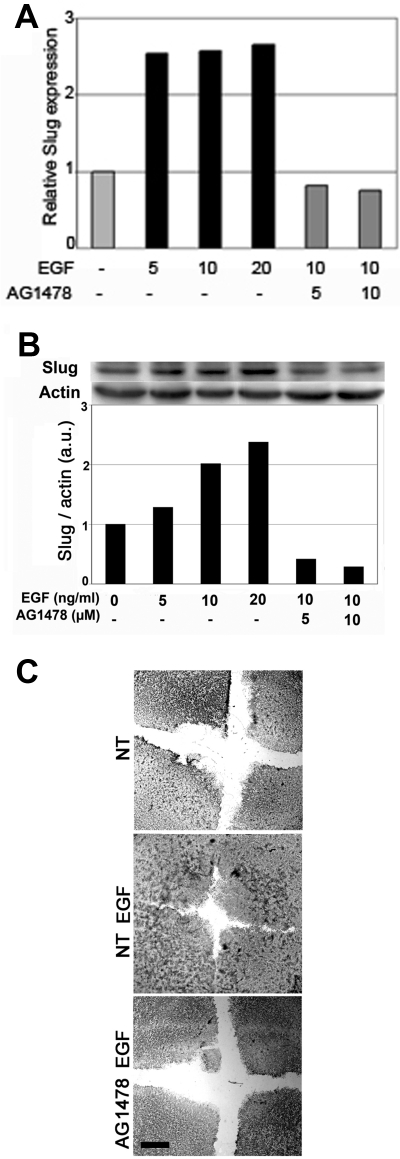
The transcription factor Slug is required for cutaneous reepithelialization and is known to induce desmosome internalization (Savagner et al., 1997); therefore, we investigated the effect of EGF on Slug RNA and protein levels in keratinocytes. Cells were serum-starved overnight as the expression of Slug can be regulated by serum factors (Savagner et al., 1997). EGF induced a two- to threefold increase in Slug mRNA 30 min after treatment (Figure 1A) and a corresponding increase in Slug protein levels (Figure 1B). Slug mRNA induction was already maximal at low doses of EGF and was blocked by AG1478, a specific inhibitor of EGFR (Figure 2A). Interestingly, there was a dose-dependent increase in Slug protein levels 2 h after EGF treatment that was again inhibited by AG1478 (Figure 2B). The relative decrease of slug expression level after AG1478 treatment suggests EGFR were activated by distinct pathways. Accordingly, AG1478 also blocked EGF-induced migration and reepithelialization in the in vitro wound-healing assay. These results suggest that Slug induction is an important step downstream of the EGFR in this cell system (Figure 2C).
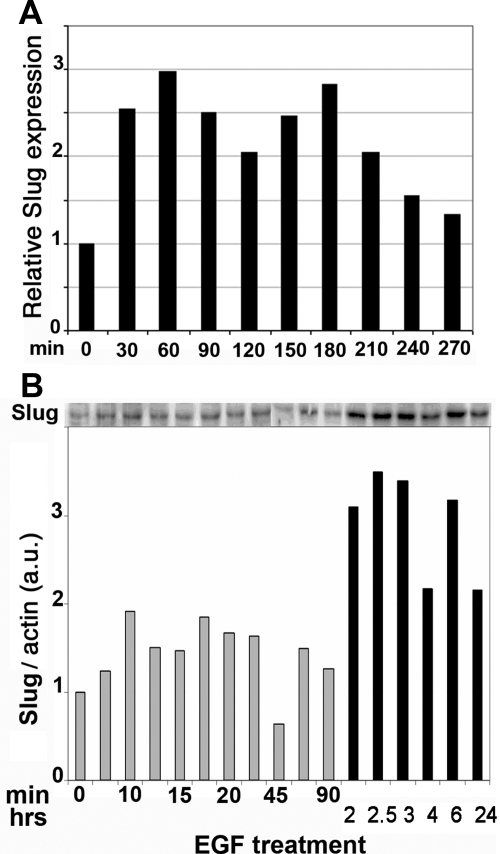
Figure 1.
EGF induces Slug expression. Serum-starved HaCaT cells were stimulated with 10 ng/ml EGF for the indicated time before extracting RNA and protein. (A) RNA levels were measured by quantitative RT-PCR. Slug RNA levels were calculated relative to the gene 36B4, using the value 2ΔCT where ΔCT is the number of cycles between Slug and 36B4. Slug RNA levels are represented as fold induction relative to serum-starved cells. (B) Protein extracts (40 μg) were loaded on SDS-PAGE and transferred to nitrocellulose, and Slug was detected and quantified by immunoblotting, using β-actin as the reference for protein levels. Two gels were needed to span the kinetics. Protein levels were quantified from the films using NIH Image software. Abbreviation used: a.u., arbitrary unit.
Figure 2.
The EGFR mediates EGF-induced Slug expression and reepithelialization by immortalized keratinocytes. Serum-starved cells were pretreated for 1 h with the indicated concentration of the specific EGFR inhibitor AG1478 and then stimulated with the indicated concentrations of EGF for 48 h. RNA (A) and protein (B) levels were analyzed and quantified as described in the legend to Figure 3, using β-actin as the reference for protein levels. (C) Confluent HaCaT cells were wounded, and reepithelialization was evaluated 28 h later in untreated cells (NT), cells incubated with 10 ng/ml EGF alone (NT EGF) or with AG1478 (AG 1478 EGF). Scale bar, 100 μm.
EGF-induced Reepithelialization Requires Slug Expression
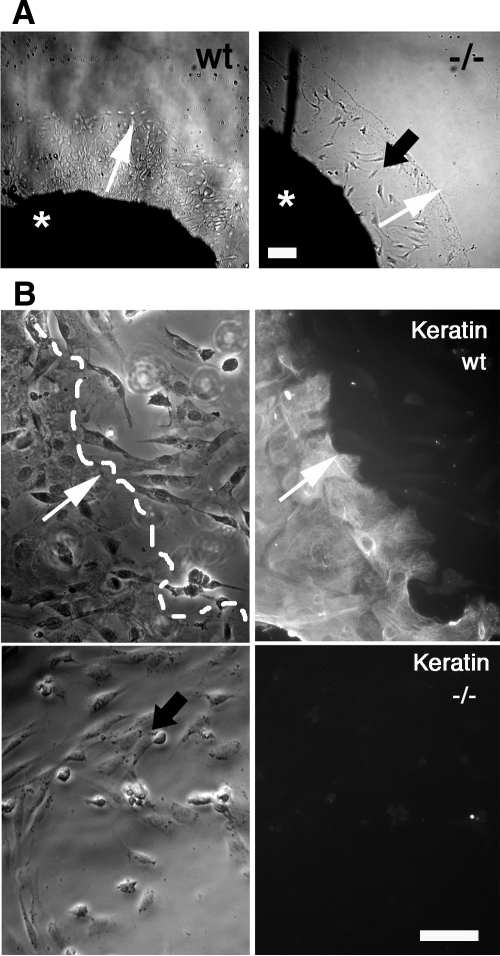
We used an ex vivo assay to more readily monitor the role of Slug in EGF-mediated reepithelialization. Briefly, skin explants were cultured on culture dishes to allow keratinocyte delamination and migration from the explants. As expected from previous observations ex vivo and in vivo (Chernoff and Robertson, 1990; Andree et al., 1994), we found that EGF accelerated cell delamination and migration (data not shown). To evaluate the contribution of Slug to this EGF-driven process, we performed the same experiment using skin explants from Slug-LacZ homozygote (−/−) mice previously described (Savagner et al., 2005) in presence of 10 ng/ml EGF. Although some mesenchymal cells migrated out of these explants, no keratinocytes could be detected by keratin immunostaining, even in the presence of EGF (Table 1 and Figure 3). The keratin expression pattern did not vary significantly between Slug-LacZ homozygote (−/−) and wild-type mouse, as seen on explants and histological sections (data not shown). These results suggest that Slug is necessary for EGF-induced reepithelialization. Overall, our results reveal a direct link between the EGF–EGFR pathway and the function of Slug in reepithelialization.
Table 1.
Mice skin explants from WT or Slug-deficient mice (−/−)
| WT | WT EGF | −/− | −/− EGF | |
|---|---|---|---|---|
| No. of explants | 12 | 12 | 12 | 12 |
| No. of explants displaying ex vivo keratinocyte reepithelialization | 11 | 9 | 0 | 0 |
Explants were maintained ex vivo to allow keratinocyte reepithelialization with or without added EGF, and 12 explants were used for each mouse. Migrating keratinocytes were identified by keratin immunolocalization (see Figure 3). Experiment was repeated four times with similar results.
Figure 3.
Keratinocytes from Slug−/− mice fail to spread and migrate from skin explants, even in the presence of EGF. (A) Skin explants from a wild-type (wt) and a Slug-LacZ homozygote mouse (−/−) were cultured ex vivo with 10 ng/ml EGF. (B) Phase-contrast micrographs and keratin (CK) immunolocalization in cells migrating from wild-type and Slug−/− skin explants. Keratinocytes are identified by keratin reactivity. The results shown are representative of four separate experiments. Black arrow, mesenchymal cells negative for keratin immunolabeling; white arrow; direction of keratinocyte migration; broken white line, keratinocyte migrating front; *, skin explant. Scale bar, 25 μm.
The Erk5 Pathway Is Implicated in the Regulation of Slug Promoter
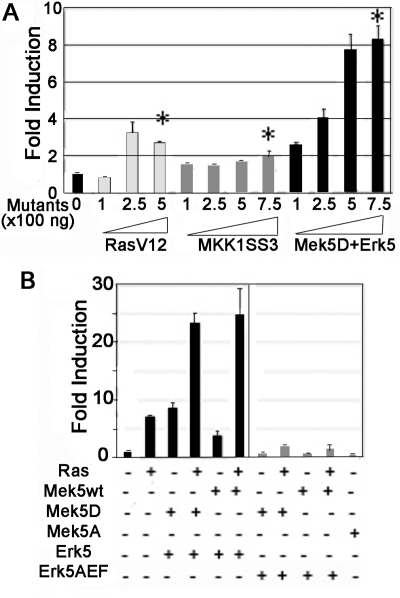
To identify the pathways mediating EGF induction of Slug expression, we used a reporter gene where luciferase expression is controlled by a 2800-base pair DNA fragment spanning the mouse Slug promoter, as previously described (Conacci-Sorrell et al., 2003). We transfected this reporter gene with expression vectors that encode constitutively active forms of molecules that regulate different MAPK cascades. These experiments were performed in the hamster lung epithelial cell line CCL39 for reasons of transfection efficiency and low background. Oncogenic Ras, namely the RasV12 mutant, led to a significant threefold increase in Slug promoter activity (Figure 4A). To selectively activate the Erk1/2 cascade, we used a constitutively active mutant of Mek1, Mek1SS3 (Lavoie et al., 1996). A high amount of this expression vector induced a moderate, statistically significant induction of the Slug reporter gene (Figure 4A). Selective activation of the JNK and p38 cascades by overexpression of constitutively active MKK7 and MKK3, respectively, also led to a moderate but significant induction of the Slug promoter (Supplemental Figure S1). Interestingly, ectopic activation of the Erk5 pathway by coexpression of a constitutively active mutant of Mek5 (MEK5D) together with wtErk5 had a stronger effect, resulting in a dramatic eightfold increase in Slug promoter activity (Figure 4A), which suggested its potential functional relevance. This increase was blocked by coexpressing Mek5D with Erk5AEF, a dominant negative mutant, in CCL39 cells (Figure 4B; Mulloy et al., 2003). Interestingly, the Erk5 cascade participates in Ras-driven activation of the Slug reporter gene, because cotransfection of either Mek5wt or Mek5D together with Erk5 strongly enhanced Slug reporter induction by RasV12 (Figure 4B, left panel). Importantly, the effect of RasV12 on the Slug promoter was strongly diminished by cotransfecting Erk5AEF or the inactive Mek5 mutant Mek5A (Figure 4B, right panel). These results suggest a specific role for the Erk5 pathway in potently activating the Slug promoter both on its own and downstream of activated Ras in the heterologous context of CCL39 cells. They also indicate a weak induction upon ectopic activation of Erk1/2, confirming a previous study (Conacci-Sorrell et al., 2003). We obtained similar results using a reporter gene driven by the human Slug promoter (data not shown).
Figure 4.
A reporter gene driven by the Slug promoter is strongly and specifically activated by the Erk5 pathway. (A) CCL39 cells were cotransfected with 250 ng of the reporter gene prSlug-Luc and expression vectors for RasV12, MK1SS3, Mek5D+Erk5 at 100, 250, 500, and 750 ng (1, 2.5, 5, and 7.5, respectively) as indicated. (B) CCL39 cells were transfected with 250 ng prSlug-Luc and the indicated mix of expression vectors (250 ng each). Erk5-AEF expresses a dominant negative mutant of Erk5. The fold induction was calculated relative to the activity of prSlug-Luc alone. The transfections were performed in duplicate and are representative of three independent experiments. *Significantly different from the control transfection (paired Student's test, p < 0.05).
EGF Induces Erk5 Phosphorylation in HaCaT Cells
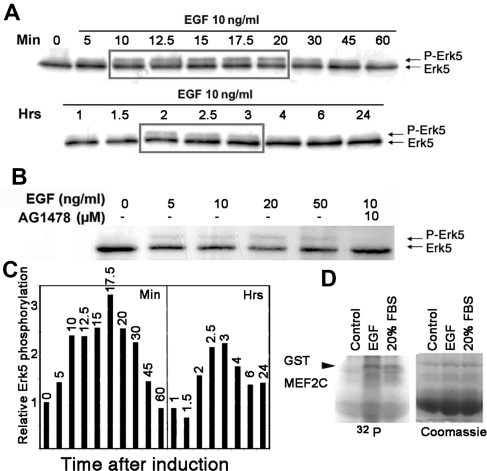
Given the potential link between EGF, Erk5 and Slug expression, we wanted to confirm that that EGF activates the Erk5 pathway in our HaCaT cell model. Mek5 activates Erk5 via phosphorylation on threonine and tyrosine in the TEY motif located in the activation loop, which leads to a second Erk5 band of slowed mobility in SDS-PAGE (Kato et al., 1998). We prepared lysates from HaCaT cells at various times after a 10 ng/ml EGF treatment and visualized Erk5 by Western blotting. A slower Erk5 band appeared 10 min after induction and remained visible for another 20 min, after which it diminished (Figure 5, A and C). Erk1/2 showed a similar pattern of activation (data not shown). Notably, the slower Erk5 band was again visible between 2 and 3 h after treatment, suggesting that two peaks of Erk5 activation are induced by EGF (Figure 5C). This band showed a slight dose dependence, although it was already visible with 5 ng/ml EGF and was inhibited by 10 μM AG1478 (Figure 5B). The biphasic response to EGF might explain the biphasic activation pattern of Slug RNA and protein observed on Figure 1. Unfortunately, immunoblotting with antisera from various suppliers that should specifically detect P-Thr/Glu/P-Tyr-ERK5 proved highly irreproducible and therefore was abandoned. To show that EGF-induced Erk5 phosphorylation reflected its activation, we performed a kinase assay using ERK5 immunoprecipitated from lysates of untreated, EGF-treated or FBS-treated HaCaT cells. Recombinant MEF2C, a specific substrate for Erk5, was phosphorylated by ERK5 isolated from EGF- or FBS-treated cells (Figure 5C), thus confirming that the upper ERK5 band reflects its activation.
Figure 5.
EGF leads to a biphasic induction of Erk5-activating phosphorylation via the EGFR. (A) Serum-starved HaCaT cells were stimulated with 10 ng/ml EGF for the indicated times before extracting protein. Protein extracts (40 μg) were separated on a 7.5% SDS-PAGE and transferred to nitrocellulose, and Erk5 detected by immunoblotting. (B) Serum-starved HaCaT cells were pretreated for 1 h with AG 1478 and then induced for 15 min with increasing amounts of EGF. Erk5 was visualized from protein extracts as described in A. (C) Quantification of Erk5 phosphorylated band from A. The panel presents the time points from A. Results obtained after 1-h or more treatment were standardized to the 60-min time point to maintain proportionality between the two gels in A. (D) Kinase assay. EGF activates ERK5 kinase activity in HaCaT cells. Starved HaCaT cells were induced for 30 min with 10 ng/ml EGF (EGF) or 20% fetal bovine serum (serum). ERK5 was immunoprecipitated from cell lysates and incubated with GST-MEF2C in the presence of [γ-32P]ATP. Proteins were separated by SDS-PAGE and visualized by staining with Coomassie brilliant blue (coom; bottom panel), followed by phosphorimagery of the dried gel (32P; top panel). Both images have been trimmed and are representative of several independent experiments.
Erk1/2 and Erk5 Play Distinct Roles in EGF-driven Slug Activation and Keratinocyte Reepithelialization
To determine the relative roles of the endogenous Erk1/2 and Erk5 pathways in our system, we used PD184352, a Mek inhibitor that selectively blocks the Erk1/2 cascade at low concentrations and both pathways at higher concentrations. Serum-starved HaCaT cells were pretreated with increasing concentrations of PD184352 for 1 h and then stimulated with EGF for 15 min (Figure 6A). Erk1/2 activation was visualized on Western blots using an antibody specific for PThr180/Tyr182Erk1/2. Activation of both Erk1 and Erk2 was progressively inhibited as the concentration of PD184352 rose to 10 μM (Figure 6A, bottom panel). In contrast, this inhibitor, even at 10 μM, did not reduce the band reflecting Erk5-activating phosphorylation (Figure 6D).
Because Slug expression is necessary for keratinocyte reepithelialization (Savagner et al., 2005), we tested the effect of Erk1/2 inhibition in our wound-healing assay. Without further treatment, even though EGF hastens the wound-healing process (data not shown), the latter is complete after 52 h with or without EGF treatment (Figure 6C). PD184352, 10 μM, blocked reepithelialization in the absence of EGF but interestingly did not affect wound healing when EGF was added (Figure 6C). Thus, reepithelialization in the presence of EGF occurs independently of Erk1/2 activation. Because Slug is required for this process, another pathway mediates EGF-induced Slug activation in HaCaT cells.
To identify the relative contribution of the Erk1/2 and Erk5 cascades involvement, we used higher concentrations of PD184352 that also inhibit Mek5 (Figure 6D). Fifty or 100 μM PD184352 clearly inhibited cell spreading and migration in EGF-treated HaCaT cells (Figure 6E), where the wound edge remained unmodified after 48-h culture (arrows). In contrast, cells treated with a lower concentration of inhibitor migrated into the wound like untreated cells. This was not an indirect effect due to general toxicity, because cells pretreated with 50 μM PD spread and migrated upon addition of calf serum (Supplemental Figure S2). These results indicate that the Erk1/2 and Erk5 cascades play distinct roles during EGF-induced reepithelialization by HaCaT cells. The Erk5 pathway seems to regulate cell spreading and migration, which correlates with Slug activation.
The Erk5 Cascade Potentiates EGF-driven Slug Expression and In Vitro Reepithelialization
To further investigate the role of the Erk5 cascade in our system, we generated pools of HaCaT cells that stably overexpress wt Erk5 or the kinase-dead mutant Erk5-AEF. We then measured Slug RNA induction by EGF in the two cell populations. In cells overexpressing Erk5-AEF, the level of Slug mRNA induction by EGF was essentially the same as in control, untransfected cells, namely two- to threefold (Figure 7A, compare with Figures 1 and 2). In contrast, Slug mRNA levels increased sevenfold after EGF treatment of Erk5-overexpressing cells (Figure 7A).
Several clones were established from the Erk5 stable transfectants to study the functional implication of its overexpression. We concentrated on clone Erk5.3, which shows a 3.5-fold protein increase in Erk5 relative to control cells (Supplemental Figure S3) that corresponds to increased spreading (data not shown). Interestingly, in the wound-healing assay, Erk5.3 cells showed a visible increase in the speed of migration (data not shown) and significantly enhanced reepithelialization (Figure 7B). Cells that did not express Erk5 over the endogenous level failed to show a significant change in reepithelialization rate (data not shown).
To clarify the role of Slug after EGF/induction, we analyzed HacaT cells stably overexpressing slug, obtained as described previously (Savagner et al., 2005). Two clones, Slug 1 and Slug 2, were compared with a control clone generated with the vector alone. Cells located at the edge of aggregates of Slug-overexpressing clones expanded further from the cell mass as individual cells (arrow). Some cells detached and migrated alone (Figure 7C). Keratin organization was looser and less coordinated between neighboring cells in Slug-overexpressing cells (arrowhead), indicating a reorganization and partial dissociation from desmosomal structures that was confirmed by desmoplakin immunolocalization (data not shown). Wound-healing assays showed a faster migration rate in slug-overexpressing clones treated with EGF compared with control clone (Figure 7D).
Interfering with Erk5 Pathway Results in Altered Cytoskeleton Organization and In Vitro Reepithelialization
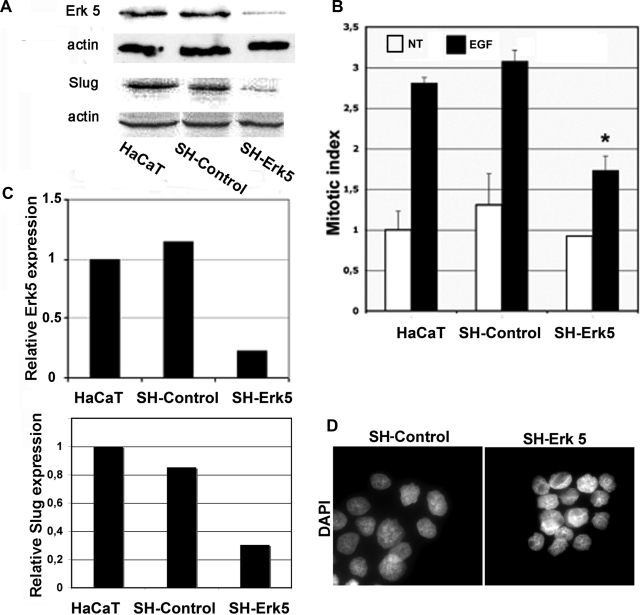
To confirm the role of the Erk5 cascade in enhancing EGF induction of Slug expression and reepithelialization, we used RNA interference to diminish the level of endogenous Erk5. siRNA-mediated knockdown of Erk5 has proven irreproducible in HaCaT cells; therefore we used a lentiviral vector encoding a shRNA specific for Erk5. Infected cells were selected using puromycin and showed an 80% reduction in Erk5 protein levels relative to those present in control cells, either untreated or infected with lentivirus expressing a control shRNA (Figure 8, A and C). The latter showed a normal mitotic response to EGF, whereas that of the SH-Erk5 cell population was reduced nearly twofold (Figure 8B). Moreover, slug protein expression was markedly reduced in SH-Erk5 cells, compared with untransfected or control cells (Figure 8C). The increase normally seen upon EGF treatment was also notably reduced in the SH-Erk5 cells (data not shown). These observations indicate an important role for the Erk5 cascade in EGF-driven Slug mRNA induction and mitogenic signaling in HaCaT cells.
Figure 8.
Erk5 knockdown using a shRNA-expressing vector in HaCaT cells reduces Slug RNA and mitogenesis induction by EGF. (A) Protein extracts from normal HaCaT cells or cells stably expressing a control or Erk5-specific shRNA were immunoblotted for Erk and β-actin. Protein levels were quantified as described in the other figure legends. (B) The same three cell populations were scored for the number of mitotic figures in noninduced cells or cells treated for 48 h with 10 ng/ml EGF. (C) Normal HaCaT cell or cells stably expressing a control or Erk5-specific shRNA were serum-starved and then induced for 1 h with 10 ng/ml EGF. RNAs were extracted. Erk5 and Slug RNA levels determined by quantitative RT-PCR as described in the legend to Figure 3. (D) HaCaT cells stably expressing the shControl or SH-Erk5 RNAs were cultured on glass coverslips and then fixed and stained with DAPI to visualize nuclei. *Significantly different from the SH-Control (Student's test, p < 0.001).
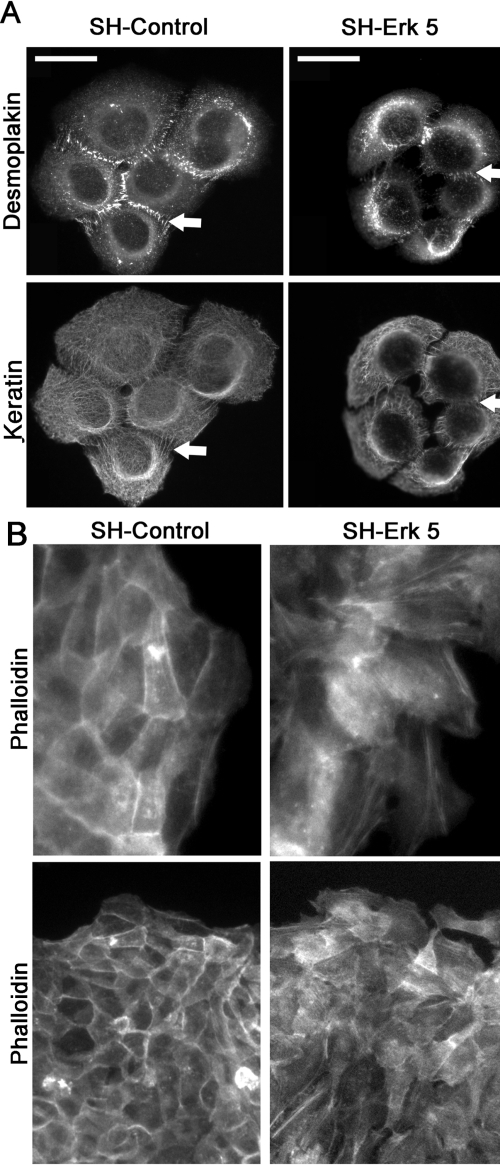
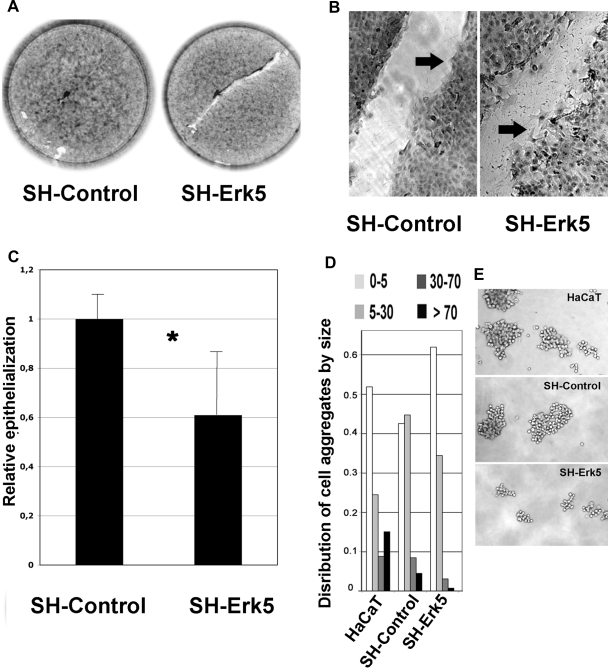
The SH-Erk5-HaCaT cell population showed altered morphology that reflected more compact cell aggregates, as observed by DAPI staining (Figure 8D). Notably, this corresponded to 30% decrease in the average distance between the nuclei of contiguous cells relative to that found with untransfected cells (Table 2), a highly significant difference (p < 0.01 in a Student's test). This overall decrease in cell–substrate contact area was not found in untreated cells and suggested SH-Erk5-HaCaT cells displayed an altered cytoskeleton, leading us to visualize actin and keratin filament mesh organization, as well as cell–cell adhesion structures, in both cell populations (Figure 9). The actin mesh was visualized by phalloidin staining. SHErk5-HaCaT cells displayed an actin mesh found predominantly in regions subcortical to cell–cell contacts (arrow on Figure 9B) that colocalized with E-cadherin (data not shown). No significant protruding cytoplasmic extensions could be detected between these cells. In contrast, it was difficult to distinguish between neighboring cells in the SH-Control because they showed a more diffuse staining that reflected protruding cytoplasmic processes between cells (Figure 9B). E-cadherin staining revealed a similar adherens junction organization (data not shown). We also examined desmosomal organization that was visualized by keratin/desmoplakin double immunolocalization. The extent of desmosomal junctions was dramatically reduced in SH-Erk5-HaCaT but was not suppressed in correspondence with a more confined keratin mesh (Figure 9). This confinement reflected the decrease in cytoplasmic extensions in SH-Erk5-HaCaT cells confirmed by microscopy (Figure 9). Most of the desmoplakin immunoreactivity was confined to cytoplasmic inclusions rather than desmosomal structures. Because desmosomal organization was perturbed, we investigated the functional impact of Erk5 depletion on cell–cell adherence. Isolated SH-control cells grown in suspension clustered into large aggregates involving more than 70 cells. Such large aggregates were rare in SH-Erk5-HaCaT cells (Figure 10, D and E), which instead formed smaller aggregates involving <30 cells, suggesting a defect in cell–cell aggregation mechanisms. These represented more than 90% of cell aggregates formed by SH-Erk5-HaCaT cells (Figure 10D).
Table 2.
Sh-Erk5 HaCat cells grow more compacted on the culture dish
| Average cell–cell distance (relative to untreated HaCaT cells) |
||
|---|---|---|
| NT | EGF | |
| HaCaT | 1.00 ± 0.09a | 1.31 ± 0.43 |
| SH-control | 0.92 ± 0.17a | 1.25 ± 0.22 |
| SH-Erk5 | 0.70 ± 0.08 | 0.93 ± 0.23 |
Average cell–cell distance was estimated by calculating nuclei–nuclei distance between cohesive cells. More than 100 measures were performed on distinct microscopic fields. Experiment was repeated more than three times with similar results.
a Statistically significant difference with untreated SH-Erk5 cells, as found by Student's test (p < 0.05).
Figure 9.
Erk5 knockdown in HaCaT cells induces a destabilization of keratin and actin cytoskeleton. HaCaT cells stably expressing the SH-Control or SH-Erk5 RNAs were cultured on glass coverslips and then fixed and immunostained for desmoplakins, keratins (A) or incubated with fluorescein-labeled phalloidin (B), displayed at low (bottom panel) or high (top panel) magnification. Representative cell aggregates are shown here, based on three independent experiments. The arrows indicate desmosomes (Desmoplakin, keratin) or subcortical actin involved in adherens junctions (Phalloidin). Scale bar, 25 μm.
Figure 10.
Erk5 knockdown in HaCaT cells hampers postwound reepithelialization and reduces cell–cell adherence. SH-Control and SH-Erk5 HaCaT cells were grown to confluency then wounded as described in Material and Methods. Wound area was measured by phase microscopy immediately after the wound (T0) and after 24-h reepithelialization (T24) (A) Low-magnification picture shows a delay in reepithelialization pattern in SH-Erk5 cells. (B) High-magnification picture shows a poor cohesiveness between front migrating SH-Erk5 cells, when SH-Control migrate as a more cohesive sheet (arrows). High-magnification picture from SH-Erk5 cells was taken after about 15-h reepithelialization, when the wound was not fully covered, as seen on A. (C) Relative reepithelialization was computed as T0T24, related to SH-Control. Each value is the average of five distinct wells for each condition. Error bars, SD (population). Statistical analysis using Student's test; *significant difference (p = 0.03) between SH-control and SH-Erk5 cells. (D) Cell–cell adherence was estimated by growing isolated cells in suspension for 120 min on a rotary shaker, to favor aggregation, according to a published method. Cell aggregates were counted and sorted into four size classes, based on the number of cells included. Percentage of cells involved in each class was calculated and reported in D. Examples of larger aggregates are shown in E.
Finally, we looked at cell response to EGF treatment. After EGF treatment, SH-Erk5HaCaT cells extruded fewer cytoplasmic extensions than parental cells and spread less in culture, as quantified by measuring the distance between nuclei of cells in contact compared with untransfected or SH-Control cells (Table 2; p < 0.05 in a Student's test). We performed in vitro wound-healing experiments to monitor cell motility. As expected, SH-Erk 5-HaCaT showed a significant migration defect after wounding (Figure 10, A–C). SH-Erk5 cells displayed a partially dissociated migrating front, compared with the more cohesive SH-Control or nontransfected HaCaT cells (Figure 10B). Experiments were performed both with or without EGF added to the culture medium. EGF had no significant effect on SH-Erk5 relative reepithelialization in these conditions (data not shown). We also found that proliferation was apparently not essential for reepithelialization because mitomycin pretreatment blocked cell proliferation, but did not alter the migration pattern (data not shown). Finally, we checked for vimentin expression. Vimentin was not found in untreated cells. It was slightly induced by EGF in parental HaCaT cells as well as transfected cells, SH-controls, and SH-Erk5 cells (Supplemental Figure S4). We found no difference in vimentin expression, with or without added EGF, between HaCaT and transfected cells.
Given these observations, we analyzed the morphology of mouse embryonic fibroblasts (MEFs) lacking either Erk5 and Mek5. Deletion of either gene is lethal after 10–11 d of embryonic development (Sohn et al., 2005). Erk5-deficient MEFs cultured in vitro were mostly devoid of the large cell–substrate complexes that accumulate at the edge of cytoplasmic extensions in control MEFs (Supplemental Figure S5). At mid-confluency and high confluency, wild-type MEFs became elongated and multilayered, with the cells aligning and developing prominent actin cables. In contrast, Erk5-or Mek5-deficient MEFs showed a thin actin filament network and lacked these long, thick actin cables. These differences were observed using wild-type MEFs from several mice and from two different backgrounds (Supplemental Figure S4). Overall, these results suggest a role for Erk5 in maintaining and/organizing the cytoskeleton in link with cell motility. On the basis of our results, we suggest this link involves Slug as a direct EGF/Erk5 target gene.
DISCUSSION
Our data identify a new pathway involved in EGF-driven keratinocyte activation and migration during reepithelialization. EGF, a physiological effector of wound repair, leads to loosening of cell–cell adhesion during keratinocyte migration. This motility process is linked to the transcription factor Slug (Savagner et al., 2005), and we find that a reporter gene driven by the Slug promoter is specifically activated upon coexpression with components of the Erk5 pathway. Moreover, EGF treatment of HaCaT cells induced activating phosphorylation of endogenous Erk5 that correlated temporally with Slug mRNA expression. Accordingly, overexpression of Erk5 increased the level of Slug expression and led to faster wound healing in EGF-treated keratinocytes. In keratinocytes where Erk5 expression was diminished using shRNA interference induction of Slug expression by EGF was significantly decreased relative to control cells. Moreover, their ability to migrate in a wound-healing assay was decreased. These cells displayed fewer desmosomes and a less organized keratin mesh. Fibrillar actin was found primarily in the subcortical region, linked to lateral walls and adherens junctions. This contrasted with control cells, where actin was visualized in stress fibers and lamellipodia. To confirm that these effects were due to decreased Erk5, we analyzed embryonic fibroblasts from Erk5- and Mek5-deficient mice. Both Erk5−/− and Mek5−/− MEFs showed a dramatic rearrangement of actin cytoskeleton with a reduction in the number of prominent actin cables extending through confluent cell groups. In isolated cells, focal contacts did not converge in growing cytoplasmic extensions, as commonly observed in MEFs from wild-type mice. These results link the Erk5 pathway to cytoskeletal mesh organization in both epithelial cells and MEFs.
Our observations complement previous observations linking Erk5 signaling to actin cytoskeleton remodeling in mesenchymal cells (Barros and Marshall, 2005). That study implicated Erk5 as a cellular effector of Src, and Src mediates Erk5 activation in other cell types, notably by EGF in a mink lung epithelial cell line (Sun et al., 2003). Our data do not address the role of Src in EGF signaling to Erk5 in HaCaT cells. Nevertheless, we observed changes in actin-based cell–cell junctions and cell motility upon Erk5 knockdown or knockout, functions that are regulated in part by Src family kinases. Moreover, EGF-induced cell motility and wound healing was compromised by reducing Erk5 levels, making it reasonable to implicate Src or a related kinase in the pathway between EGF and Erk5 activation in our system.
The phenotype observed upon Erk5 knockdown resembles that of epithelial cells transfected with antisense Slug, namely an inability to migrate normally in response to growth factors. On the other hand, Slug overexpression in keratinocytes resulted in increased spreading, along with remodeling and partial disassociation of the actin cytoskeleton from cell–cell adhesion structures (Figure 7, C and D, and Savagner et al., 1997). The concordance between these observations and those we present strongly suggests a link between Erk5 and Slug in regulation of the cytoskeleton. Slug transcription is induced by diverse intracellular signaling pathways (Conacci-Sorrell et al., 2003; Romano and Runyan, 2000; Thuault et al., 2006), including β-catenin, TGFβ, Erk1/2, and p38MAPK in our system (data not shown). Interestingly, we found that ectopically activated Erk5 was the most potent activator of a Slug promoter-driven reporter gene in our system.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to P. Wade for providing a human Slug promoter and to C. Tournier (University of Manchester, Manchester, United Kingdom) for providing MEFs from Erk5 and Mek5 knockout mice. We acknowledge the invaluable aid provided by D. Kusewitt and L. Hudson through numerous discussions and data sharing. We thank Annick Causse and especially Hélène Vallès for contributing their technical expertise to these studies. This research was supported by the Ligue Regionale contre le Cancer (Languedoc-Roussillon), Groupement des Entreprises Francaises dans la Lutte contre le Cancer, the Fondation de France, and the Association pour la Recherche sur le Cancer (V.A. and R.A.H.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1078) on August 20, 2008.
REFERENCES
- Andree C., Swain W. F., Page C. P., Macklin M. D., Slama J., Hatzis D., Eriksson E. In vivo transfer and expression of a human epidermal growth factor gene accelerates wound repair. Proc. Natl. Acad. Sci. USA. 1994;91:12188–12192. doi: 10.1073/pnas.91.25.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoux V., Come C., Kusewitt D., Savagner P. Cutaneous wound healing: a partial and reversible EMT. In: Savagner P., editor. Rise and Fall of Epithelial Phenotype: Concepts of Epithelial-Mesenchymal Transition. Austin, TX: Landes Biosciences; 2005. pp. 111–134. [Google Scholar]
- Barros J. C., Marshall C. J. Activation of either ERK1/2 or ERK5 MAP kinase pathways can lead to disruption of the actin cytoskeleton. J. Cell Sci. 2005;118:1663–1671. doi: 10.1242/jcs.02308. [DOI] [PubMed] [Google Scholar]
- Boukamp P., Chen J., Gonzales F., Jones P. A., Fusenig N. E. Progressive stages of “transdifferentiation” from epidermal to mesenchymal phenotype induced by MyoD1 transfection, 5-aza-2′-deoxycytidine treatment, and selection for reduced cell attachment in the human keratinocyte line HaCaT. J. Cell Biol. 1992;116:1257–1271. doi: 10.1083/jcb.116.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. L., Nanney L. B., Griffen J., Cramer A. B., Yancey J. M., Curtsinger L. J., 3rd, Holtzin L., Schultz G. S., Jurkiewicz M. J., Lynch J. B. Enhancement of wound healing by topical treatment with epidermal growth factor. N. Engl. J. Med. 1989;321:76–79. doi: 10.1056/NEJM198907133210203. [DOI] [PubMed] [Google Scholar]
- Brunet A., Pages G., Pouyssegur J. Constitutively active mutants of MAP kinase kinase (MEK1) induce growth factor-relaxation and oncogenicity when expressed in fibroblasts. Oncogene. 1994;9:3379–3387. [PubMed] [Google Scholar]
- Chernoff E. A., Robertson S. Epidermal growth factor and the onset of epithelial epidermal wound healing. Tissue Cell. 1990;22:123–135. doi: 10.1016/0040-8166(90)90016-3. [DOI] [PubMed] [Google Scholar]
- Come C., Magnino F., Bibeau F., De Santa Barbara P., Becker K. F., Theillet C., Savagner P. Snail and slug play distinct roles during breast carcinoma progression. Clin. Cancer Res. 2006;12:5395–5402. doi: 10.1158/1078-0432.CCR-06-0478. [DOI] [PubMed] [Google Scholar]
- Conacci-Sorrell M., Simcha I., Ben-Yedidia T., Blechman J., Savagner P., Ben-Ze'ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J. Cell Biol. 2003;163:847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edme N., Downward J., Thiery J. P., Boyer B. Ras induces NBT-II epithelial cell scattering through the coordinate activities of Rac and MAPK pathways. J. Cell Sci. 2002;115:2591–2601. doi: 10.1242/jcs.115.12.2591. [DOI] [PubMed] [Google Scholar]
- Elloul S., Elstrand M. B., Nesland J. M., Trope C. G., Kvalheim G., Goldberg I., Reich R., Davidson B. Snail, Slug, and Smad-interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer. 2005;103:1631–1643. doi: 10.1002/cncr.20946. [DOI] [PubMed] [Google Scholar]
- Fujita N., Jaye D. L., Kajita M., Geigerman C., Moreno C. S., Wade P. A. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113:207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- Hudson L. G., McCawley L. J. Contributions of the epidermal growth factor receptor to keratinocyte motility. Microsc. Res. Tech. 1998;43:444–455. doi: 10.1002/(SICI)1097-0029(19981201)43:5<444::AID-JEMT10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Hemavathy K., Guru S. C., Harris J., Chen J. D., Ip Y. T. Human Slug is a repressor that localizes to sites of active transcription. Mol. Cell. Biol. 2000;26:5087–5095. doi: 10.1128/mcb.20.14.5087-5095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Lan Y., Norton C. R., Sundberg J. P., Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev. Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- Karihaloo A., O'Rourke D. A., Nickel C., Spokes K., Cantley L. G. Differential MAPK pathways utilized for HGF-and EGF-dependent renal epithelial morphogenesis. J. Biol. Chem. 2001;276:9166–9173. doi: 10.1074/jbc.M009963200. [DOI] [PubMed] [Google Scholar]
- Kato Y., Kravchenko V. V., Tapping R. I., Han J., Ulevitch R. J., Lee J. D. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Tapping R. I., Huang S., Watson M. H., Ulevitch R. J., Lee J. D. Bmk1/Erk5 is required for cell proliferation induced by epidermal growth factor. Nature. 1998;395:713–716. doi: 10.1038/27234. [DOI] [PubMed] [Google Scholar]
- Krawczyk W. S. A pattern of epidermal cell migration during wound healing. J. Cell Sci. 1971;49:247–263. doi: 10.1083/jcb.49.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie J. N., L'Allemain G., Brunet A., Muller R., Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J. Biol. Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- Martin P., Hopkinson-Woolley J., McCluskey J. Growth factors and cutaneous wound repair. Prog. Growth Factor Res. 1992;4:25–44. doi: 10.1016/0955-2235(92)90003-z. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Ebisuya M., Honjoh S., Nishida E. ERK activation propagates in epithelial cell sheets and regulates their migration during wound healing. Curr. Biol. 2004;14:731–735. doi: 10.1016/j.cub.2004.03.060. [DOI] [PubMed] [Google Scholar]
- Mazzalupo S., Wawersik M. J., Coulombe P. A. An ex vivo assay to assess the potential of skin keratinocytes for wound epithelialization. J. Invest. Dermatol. 2002;118:866–870. doi: 10.1046/j.1523-1747.2002.01736.x. [DOI] [PubMed] [Google Scholar]
- Mulloy R., Salinas S., Philips A., Hipskind R. A. Activation of cyclin D1 expression by the ERK5 cascade. Oncogene. 2003;22:5387–5398. doi: 10.1038/sj.onc.1206839. [DOI] [PubMed] [Google Scholar]
- Nieto M. A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Nissen L. J., Gelly J. C., Hipskind R. A. Induction-independent recruitment of CREB-binding protein to the c-fos serum response element through interactions between the bromodomain and Elk-1. J. Biol. Chem. 2001;276:5213–5221. doi: 10.1074/jbc.M007824200. [DOI] [PubMed] [Google Scholar]
- Paladini R. D., Takahashi K., Bravo N. S., Coulombe P. A. Onset of re-epithelialization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: defining a potential role for keratin 16. J. Cell Biol. 1996;132:381–397. doi: 10.1083/jcb.132.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano L. A., Runyan R. B. Slug is an essential target of TGFbeta2 signaling in the developing chicken heart. Dev. Biol. 2000;223:91–102. doi: 10.1006/dbio.2000.9750. [DOI] [PubMed] [Google Scholar]
- Savagner P., Kusewitt D. F., Carver E. A., Magnino F., Choi C., Gridley T., Hudson L. G. Developmental transcription factor slug is required for effective re-epithelialization by adult keratinocytes. J. Cell Physiol. 2005;202:858–866. doi: 10.1002/jcp.20188. [DOI] [PubMed] [Google Scholar]
- Savagner P., Yamada K. M., Thiery J. P. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J. Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried J., Wang X., Kharebava G., Tournier C. A novel mitogen-activated protein kinase docking site in the N terminus of MEK5alpha organizes the components of the extracellular signal-regulated kinase 5 signaling pathway. Mol. Cell. Biol. 2005;25:9820–9828. doi: 10.1128/MCB.25.22.9820-9828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih J. Y., et al. Transcription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin. Cancer Res. 2005;11:8070–8078. doi: 10.1158/1078-0432.CCR-05-0687. [DOI] [PubMed] [Google Scholar]
- Sohn S. J., Li D., Lee L. K., Winoto A. Transcriptional regulation of tissue-specific genes by the ERK5 mitogen-activated protein kinase. Mol. Cell. Biol. 2005;25:8553–8566. doi: 10.1128/MCB.25.19.8553-8566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenn K., Depalma L. Re-epithelialiation. In: Clark R.A.F., Henson P. M., editors. The Molecular and Cellular Biology of Wound Repair. New York: Plenum Press; 1988. pp. 321–325. [Google Scholar]
- Sun W., Wei X., Kesavan K., Garrington T. P., Fan R., Mei J., Anderson S. M., Gelfand E. W., Johnson G. L. MEK kinase 2 and the adaptor protein Lad regulate extracellular signal-regulated kinase 5 activation by epidermal growth factor via Src. Mol. Cell. Biol. 2003;23:2298–2308. doi: 10.1128/MCB.23.7.2298-2308.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. Functional correlation between cell adhesive properties and some cell surface proteins. J. Cell Biol. 1977;75:464–474. doi: 10.1083/jcb.75.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuault S., Valcourt U., Petersen M., Manfioletti G., Heldin C. H., Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J. Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner F. E., Broad S., Khanim F. L., Jeanes A., Talma S., Hughes S., Tselepis C., Hotchin N. A. Slug regulates integrin expression and cell proliferation in human epidermal keratinocytes. J. Biol. Chem. 2006;281:21321–21331. doi: 10.1074/jbc.M509731200. [DOI] [PubMed] [Google Scholar]
- Zhang F., Tom C. C., Kugler M. C., Ching T. T., Kreidberg J. A., Wei Y., Chapman H. A. Distinct ligand binding sites in integrin alpha3beta1 regulate matrix adhesion and cell-cell contact. J. Cell Biol. 2003;163:177–188. doi: 10.1083/jcb.200304065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.