Abstract
On starvation, Dictyostelium cells aggregate to form multicellular fruiting bodies containing spores that germinate when transferred to nutrient-rich medium. This developmental cycle correlates with the extent of actin phosphorylation at Tyr-53 (pY53-actin), which is low in vegetative cells but high in viable mature spores. Here we describe high-resolution crystal structures of pY53-actin and unphosphorylated actin in complexes with gelsolin segment 1 and profilin. In the structure of pY53-actin, the phosphate group on Tyr-53 makes hydrogen-bonding interactions with residues of the DNase I-binding loop (D-loop) of actin, resulting in a more stable conformation of the D-loop than in the unphosphorylated structures. A more rigidly folded D-loop may explain some of the previously described properties of pY53-actin, including its increased critical concentration for polymerization, reduced rates of nucleation and pointed end elongation, and weak affinity for DNase I. We show here that phosphorylation of Tyr-53 inhibits subtilisin cleavage of the D-loop and reduces the rate of nucleotide exchange on actin. The structure of profilin–Dictyostelium-actin is strikingly similar to previously determined structures of profilin–β-actin and profilin–α-actin. By comparing this representative set of profilin–actin structures with other structures of actin, we highlight the effects of profilin on the actin conformation. In the profilin–actin complexes, subdomains 1 and 3 of actin close around profilin, producing a 4.7° rotation of the two major domains of actin relative to each other. As a result, the nucleotide cleft becomes moderately more open in the profilin–actin complex, probably explaining the stimulation of nucleotide exchange on actin by profilin.
Keywords: actin phosphorylation, profilin–actin structure, pY53-actin structure, Dictyostelium discoideum actin, gelsolin–actin structure
Multiple cellular functions, including cell motility, cell division, and endocytosis, involve dynamic remodeling of the actin cytoskeleton (1, 2). Various studies have established a connection between actin phosphorylation and cytoskeleton remodeling. For example, fibroblast stimulation with epidermal growth factors induces actin phosphorylation on serine residues and the formation of membrane ruffles (3). Actin is also one of the proteins found to be tyrosine phosphorylated in fibroblasts expressing constitutively active Src tyrosine kinase and displaying significant cytoskeleton rearrangement (4). In Mimosa pudica, a plant that closes its leaves and droops its petioles when touched, actin is heavily tyrosine phosphorylated, and a decrease in actin phosphorylation correlates with petiole bending (5). In none of these examples, however, is the precise connection between actin phosphorylation and cytoskeleton remodeling well understood. Such a connection is better established in Dictyostelium cells, in which the developmental cycle correlates closely with the extent of actin tyrosine phosphorylation (6–11).
Dictyostelium cells grow and divide as amoebae in nutrient medium, but they aggregate and differentiate into multicellular organisms on starvation, ultimately forming fruiting bodies that contain spores that germinate when conditions become favorable for growth (12). The extent of actin phosphorylation, which is very low in growing vegetative cells, begins to increase 12–24 h into the developmental cycle, reaching ≈50% of the total actin at ≈36 h (9–11). At this high level of actin phosphorylation, the spores of the mature fruiting bodies remain viable for ≈20 days, at which time viability and actin phosphorylation levels both decrease, disappearing entirely by ≈30 days. Increases in actin phosphorylation also occur when vegetative cells are exposed to heat stress, sodium azide, and the phosphotyrosine phosphatase inhibitor phenylarsine oxide (6, 8, 11, 13).
The phosphorylation site on Dictyostelium actin has been mapped to residue Tyr-53 (8, 11). This site is near the DNase I-binding loop (D-loop) of actin (residues 40–50), which is implicated in intersubunit contacts in the filament (14–17). The D-loop is disordered in most crystal structures of actin, including in the two structures of unphosphorylated Dictyostelium actin described here in complexes with profilin and gelsolin segment 1 (G1). In contrast, the D-loop is partially stabilized by hydrogen-bonding contacts with the phosphate group on Tyr-53 in the structure of Tyr-53-phosphorylated actin (pY53-actin), which is also described here in complex with G1. The stabilization of the D-loop is further supported by biochemical characterization of pY53-actin in solution.
Profilin stimulates nucleotide exchange on actin (18, 19). However, the structural bases for this activity are not well understood. The crystal structure of profilin–Dictyostelium-actin described here is strikingly similar to previously determined structures of profilin–β-actin (20) and profilin–α-actin (21). Based on this representative group of profilin–actin structures, we analyze the effects of profilin on the conformation of actin and its role in nucleotide exchange.
Results and Discussion
Structures of pY53-Actin and Unphosphorylated Actin Complexed with G1.
The different biochemical properties of pY53-actin and unphosphorylated actin suggested that their structures may be different (11). To test this possibility, we set out to crystallize pY53-actin and unphosphorylated Dictyostelium actin under identical conditions, so that their structures could be compared directly. Actin's natural tendency to polymerize constitutes an obstacle to crystallization. Different approaches have been used to overcome this problem, including the crystallization of complexes of actin with actin-binding proteins (ABPs) (20, 22–26) and toxins (27), and blocking actin polymerization by mutagenesis (28) or chemical cross-linking (29). We chose to attempt crystallization with profilin, gelsolin, vitamin D-binding protein (DBP), and toxofilin, all proteins that bind to actin on the opposite side from subdomain 2, which is where Tyr-53 is located and phophorylation-dependent structural changes are more likely to occur. Unphosphorylated Dictyostelium actin had already been crystallized in complex with G1 (30), and we obtained crystals under similar conditions of both the unphosphorylated and phosphorylated forms (see Materials and Methods). In addition, we obtained crystals of unphosphorylated actin with profilin and the vasodilator-stimulated phosphoprotein (VASP) polyproline peptide 198GAGGGPPPAPPLAAQ213, which binds to profilin on the opposite side from actin (21). pY53-actin failed to crystallize with profilin under similar conditions (see Conclusions). Toxofilin (26) did not crystallize with either form of actin, and although DBP-actin yielded large crystals, they diffracted x-rays poorly.
The structures of complexes of G1 with unphosphorylated actin and pY53-actin were determined to resolutions of 1.6 Å and 1.7 Å, respectively (Table 1). The two structures are strikingly similar to each other and to prior structures of mammalian skeletal α-actin with G1 (23) and gelsolin segments 1 to 3 (31), and Saccharomyces cerevisiae, Caenorhabditis elegans, and Dictyostelium discoideum actin with G1 (30). In particular, the conformation of actin is nearly identical in all these complexes, with the notable exception of the D-loop (supporting information (SI) Fig. S1). The D-loop is one of the most flexible parts of the actin molecule, and it is disordered in most of the structures. Because of its intrinsic flexibility, we cannot compare the conformation of the D-loop among different structures. Factors such as crystal packing, crystallization conditions, actin isoform, and the identity of the ABP used in the crystallization may influence the conformation of the D-loop. In contrast, in the current study we crystallized unphosphorylated actin and pY53-actin under identical conditions, allowing for a direct comparison of the effect of phosphorylation on the conformation of the D-loop.
Table 1.
Crystallographic data and refinement statistics
| Profilin–actin–VASP202–244 | Gelsolin–actin | Gelsolin–pY53–actin | |
|---|---|---|---|
| Space group | P212121 | C2 | C2 |
| Unit cell a/b/c, Å | 38.04/76.18/180.82 | 178.88/69.17/56.56 | 178.42/69.05/56.54 |
| Unit cell α/β/γ, ° | 90/90/90 | 90/104.36/90 | 90/104.2/90 |
| Resolution, Å | 2.3–50 (2.3–2.38) | 1.6–50 (1.6–1.66) | 1.7–50 (1.7–1.76) |
| Completeness, % | 89.7 (80.9) | 96.8 (78.4) | 94.9(57.9) |
| Multiplicity | 11.0 (11.1) | 7.3 (3.8) | 8.0 (2.1) |
| Rsym, %* | 11.7 (34.9) | 7.1 (52.1) | 8.4 (40.7) |
| I/σ | 20.3 (15.8) | 36.6 (2.1) | 23.1 (2.2) |
| Rfactor, %† | 15.7 | 15.0 | 14.1 |
| Rfree, %‡ | 22.7 | 19.7 | 19.1 |
| rms bonds, Å | 0.015 | 0.011 | 0.012 |
| rms angles, ° | 1.504 | 1.326 | 1.365 |
| B-factor actin/ABP, Å2 | 22.54/20.40 | 24.88/23.21 | 29.40/30.32 |
| B-factor solvent, Å2 | 22.89 | 38.60 | 44.71 |
| Number of aa/waters | 513/318 | 491/461 | 494/460 |
| PDB code | 3CHW | 3CIP | 3CI5 |
Values in parentheses correspond to highest resolution shell.
*Rsym = Σ(I− 〈I〉)/ΣsI; I and 〈I〉, intensity and mean intensity of a reflection.
†Rfactor = Σ|Fo − Fc|/Σ|Fo|; Fo and Fc, observed and calculated structure factors.
‡Rfree; Rfactor of 5% of the reflections that were not used in refinement.
The conformation of the side chain of Tyr-53 is essentially the same in pY53-actin and unphosphorylated actin (Fig. 1 and Movie S1) and similar to other structures of actin, except for the phosphate group on Tyr-53, which is clearly defined in the electron density map of pY53-actin, but absent in unphosphorylated actin. To further validate this observation, we analyzed crystals of the two forms of actin using tandem mass spectrometry (Fig. S2). After separation by metal affinity chromatography of phospho-peptides from tryptic digestions of protein samples from the crystals, peptides containing phosphorylated Tyr-53 were detected only in crystals of pY53-actin. This result confirmed that the crystals of unphosphorylated actin were free of the phosphorylated form. Although we cannot exclude the presence of trace amounts of unphosphorylated actin in crystals of pY53-actin, crystallographic occupancies of the phosphate atoms in this high-resolution structure indicate that phosphorylation is ≈100%.
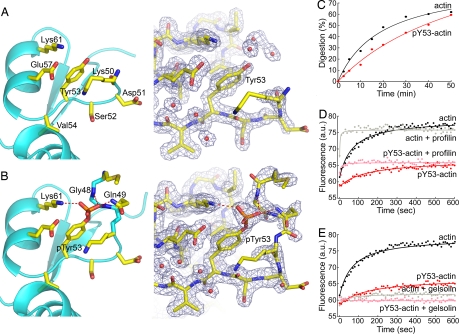
Fig. 1.
Conformational change in actin subdomain 2 on phosphorylation of Tyr-53. (A and B) Close views of subdomain 2 in the structures of unphosphorylated actin and pY53-actin, showing omit electron density maps (contoured at 1 σ) around Tyr-53 (see Figs. S3 and S5 for a full view of the G1-actin structure). The D-loop was not visualized in the unphosphorylated structure. Hydrogen-bonding contacts (red dashed lines) between the oxygen atoms of the phosphate group on Tyr-53 and residues of the D-loop stabilize the conformation of the D-loop in the structure of pY53-actin. This and other figures of the paper were generated with the program PyMOL (http://pymol.sourceforge.net/). (C) Phosphorylation protects the D-loop from subtilisin cleavage, as shown by the ≈50% decrease in the initial rate of digestion. (D and E) Based on the increase in fluorescence as etheno-ATP replaces actin-bound ATP, phosphorylation reduces the rate of nucleotide exchange from 0.011 s−1 for unphosphorylated actin to 0.006 s−1 for pY53-actin, but profilin accelerates and gelsolin inhibits nucleotide exchange to the same extents for both forms of actin. The increase in fluorescence at equilibrium for pY53-actin is only 50% of the increase for unphosphorylated actin. Data were recorded every 10 s.
The phosphate oxygens on Tyr-53 make hydrogen-bonding interactions with the side-chain nitrogen atoms of Lys-61 and Gln-49 and with the main-chain nitrogen atoms of Gln-49 and Gly-48 in the D-loop (Fig. 1B). As a result, the D-loop, which is fully disordered in the unphosphorylated structure (residues 42–49 were not visualized), becomes partially ordered in pY53-actin, where four additional residues of the loop were resolved in the electron density map (Gly-42, Met-47, Gly-48, Gln-49). Although additional density was present, residues 43–46 could still not be unambiguously traced. The average temperature factor for the four additional residues observed in the D-loop is 79.4 Å2, compared to ≈30 Å2 for the rest of the structure (Table 1), suggesting that although more constrained, the D-loop is still quite dynamic in this structure. Other than the changes in the D-loop, the structural differences due to phosphorylation appear minor (Movie S1). However, a symmetry-related molecule in the crystal is located near the D-loop (Fig. S3), and it is possible that the proximity of this crystal contact limited the full extent of the conformational change. Another factor that could have limited the magnitude of the conformational change is the presence of G1 in the structure.
Probing the Structures of pY53-Actin and Unphosphorylated Actin in Solution.
The crystal structures suggest that phosphorylation of Tyr-53 affects the conformation of the D-loop. We used subtilisin cleavage of the D-loop to test this observation in solution. The susceptibility of the D-loop to subtilisin cleavage between residues Met-47 and Gly-48 has been commonly used to monitor conformational changes in the D-loop resulting from factors such as the type of nucleotide and divalent cation bound to actin (32, 33), and the binding of actin-depolymerizing factor (ADF)/cofilin to the filament (34). We found that phosphorylation of Tyr-53 protects the D-loop from subtilisin cleavage, as shown by a ≈50% reduction in the initial rate of cleavage of the D-loop in pY53-actin compared to unphosphorylated actin (Fig. 1C). The protection of the D-loop from subtilisin cleavage is consistent with the more stably folded conformation of the loop observed in the structure of pY53-actin.
Because the exchange of ATP for ADP on actin is thought to alter the conformation of the D-loop (32), we speculated that changes in the conformation of the loop might reciprocally affect nucleotide exchange on actin. Consistent with this prediction, we found that phosphorylation of Tyr-53 reduces the initial rate of nucleotide exchange on actin by ≈45% (as measured by the increase in fluorescence as etheno-ATP replaces bound ATP), doubles the half-time to reach equilibrium, and reduces the fluorescence of etheno-ATP at equilibrium by ≈50% (Fig. 1 D and E). Because the conformational change in the D-loop does not seem to extend to the nucleotide cleft (Movie S1), the structural reasons for the differences in the rate of nucleotide exchange and etheno-ATP equilibrium fluorescence are unclear. Probably, pY53-actin and unphosphorylated actin have different affinities for ATP and etheno-ATP, and the fluorescence of etheno-ATP bound in the nucleotide cleft of pY53-actin may be masked by the conformational change in the D-loop. Finally, it is known that profilin accelerates (18, 19) and G1 inhibits (35) nucleotide exchange on actin. Consistent with these reports, we found that profilin stimulates (Fig. 1D) and G1 inhibits (Fig. 1E) nucleotide exchange on Dictyostelium actin. The effects of these two ABPs are very similar for phosphorylated and unphosphorylated actin.
Structure of Unphosphorylated Dictyostelium Actin Complexed with Profilin.
The structural bases for the stimulation of nucleotide exchange by profilin (18, 19) (Fig. 1D) have remained unclear. Although the original structure of profilin–β-actin revealed a moderately open nucleotide cleft in actin (20), a subsequent structural determination suggested a far more open structure (36). Some biochemical observations have also been interpreted as evidence of a more open cleft in actin than suggested by the majority of the crystal structures (37). However, a wide-open cleft appears to be structurally unstable (38). More important, the wide-open structure of profilin–β-actin (36) was obtained in an unconventional way, by transferring the original crystals (20) into a high-phosphate solution. Thus, opening of the cleft was obtained by crystal manipulation rather than a physiologically relevant factor.
We recently reported two structures of profilin–α-actin (21), determined to resolutions of 1.8 Å and 1.5 Å in the presence of two different fragments of human VASP. Here we describe the structure of profilin–Dictyostelium-actin to 2.3 Å resolution (Fig. 2A and Table 1). The two structures of profilin–α-actin and that of profilin–Dictyostelium-actin are strikingly similar to one another and to the original 2.55 Å structure of profilin–β-actin (20) (Fig. S4A), but different from the wide-open structure of profilin–β-actin (36) (Fig. S4B). Therefore, we now have possession of a representative set of four profilin–actin structures, determined under different crystallization conditions and crystal packing environments, and corresponding to different isoforms and sources of actin.
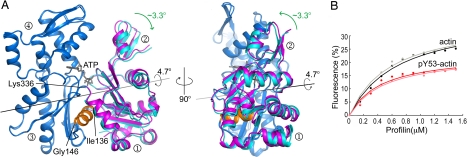
Fig. 2.
Profilin binding causes a moderate opening of the nucleotide cleft in actin. (A) Superimposition of the structures of profilin–Dictyostelium-actin (blue and cyan) and uncomplexed monomeric actin (28) (blue and magenta). Two orientations are shown, rotated by 90°. The latter structure was obtained by mutagenesis in subdomain 4 and is thought to be free of perturbations resulting from the binding of an ABP or chemical cross-linking. For clarity, profilin is not shown in this figure (see Figs. S5 and S6 for a full view of the profilin–actin structure). Subdomains 3 and 4 of the structures were superimposed (blue) to highlight the relative movement of subdomains 1 and 2 (magenta or cyan). Using the classical view of actin as a reference (left view), the 4.7° rotation (calculated with the program DynDom, http://www.sys.uea.ac.uk/dyndom/) between the two major domains of actin can be visualized as two perpendicular rotations of ≈3.3°. The center of this rotation approximately coincides with the junctions between domains, consisting of residue Lys-336 and the helix between residues Ile-136 and Gly-146. Comparison of the profilin–actin structures with any other structure of actin, except for the wide-open structure of profilin–β-actin (36), results in a similar motion of the two major domains (see also Movies S2 and S3). This movement appears less dramatic than previously anticipated (36, 37), but it is probably sufficient to explain the stimulation of nucleotide exchange by profilin. (B) Quenching of tryptophan fluorescence on profilin binding (the results of two identical experiments, with different preparations of both actins, are shown). Profilin binds pY53-actin and unphosphorylated actin with similar affinities (Kd = 0.090 and 0.057 μM, respectively), but the quenching of tryptophan fluorescence is significantly less for profilin–pY53-actin.
Can we identify structural features unique to the profilin–actin complex that would explain the acceleration of nucleotide exchange? To answer this question, we compared the profilin–actin structures to all of the actin structures in the protein data bank. The principal conclusion from this analysis is that compared to any other structure of actin, the nucleotide cleft is moderately more open in the profilin–actin complex (Fig. 2A). The conformational change leading to cleft opening begins with subdomains 1 and 3 of actin closing around the profilin molecule. As a result, the two major domains of actin on each side of the nucleotide cleft rotate by 4.7° relative to each other (Fig. 2A). By using the classical view of actin as a reference (Fig. 1A Left), this rotation can be described as two perpendicular rotations of ≈3.3° (Fig. 2A), roughly corresponding to the propeller-like twisting and scissor-like opening of the domains suggested by normal-mode analysis (39). An important distinction, however, is that rather than a scissor-like motion, we observe a clamp-like motion, i.e., as the target-binding cleft of actin “clamps” on the profilin molecule, the nucleotide cleft opens at the opposite side of the molecule. To better illustrate this movement, we generated a movie of the conformational change by linear interpolation between the atomic coordinates of the profilin–Dictyostelium-actin structure and the ATP-bound structure of uncomplexed monomeric actin (28) (Movie S2). Note that the latter structure was chosen for this comparison because it was obtained by mutagenesis of two residues in subdomain 4 (which prevents polymerization), and it is therefore free of perturbations resulting from the binding of an ABP or chemical cross-linking. To further show that cleft opening is a general feature of the profilin–actin complex, we generated a second interpolation, using as a reference the structure of unphosphorylated Dictyostelium actin in complex with gelsolin (Movie S3).
Although a rotation of the actin domains similar to that described above had already been pointed out when the structure of profilin–β-actin was first reported (20), this effect was attributed to differences between α- and β-actin. In contrast, we find that the profilin-induced rotation is independent of the source of the actin. Moreover, the earlier comparison was made to the only other actin structure available at the time, that of the complex with DNase I (22), which binds atop actin subdomains 2 and 4 and probably limits nucleotide cleft opening.
As mentioned above, we could not obtain crystals of pY53-actin with profilin, which led us to suspect that Tyr-53 phosphorylation affects the interaction with profilin. The binding of profilin to actin is accompanied by a large decrease in the intensity of tryptophan fluorescence, which is thought to result from shielding of actin Trp-356 by profilin in the complex (40). Therefore, we measured the quenching of tryptophan fluorescence to estimate the binding affinities of profilin for pY53-actin and unphosphorylated actin. We found that profilin binds pY53-actin and unphosphorylated actin with similar affinities, although the quenching of tryptophan fluorescence is significantly less for profilin–pY53-actin (Fig. 2B). Because the structure of profilin–pY53-actin could not be obtained, it is unclear what structural differences are responsible for this observation. Interestingly, in the two gelsolin structures, the region around Trp-356 is flexible, with residues Phe-352 to Trp-356 displaying alternative side-chain rotamers (Figs. S1 and S5). In all other structures of actin, Trp-356 presents a single and unique side-chain orientation (Fig. S1), which is also the orientation of Trp-356 in the structure of profilin–Dictyostelium-actin (Fig. S5). If the region around Trp-356 is intrinsically flexible, as suggested by the two gelsolin structures, quenching of tryptophan fluorescence during profilin binding might result from locking Trp-356 into a single conformation that is less exposed to solvent, rather than direct masking of Trp-356 by profilin. Finally, because the binding affinity of profilin is not significantly affected by actin phosphorylation (Fig. 2B), it is likely that our inability to obtain crystals of profilin–pY53-actin resulted from a conflict between the conformational change in the D-loop and crystal contacts. Indeed, analysis of crystal packing contacts reveals a symmetry-related molecule near the D-loop (Fig. S6).
Conclusions
The extent of actin phosphorylation at Tyr-53 varies dramatically during the different stages of the developmental cycle of Dictyostelium cells (9–11). Biochemically, actin Tyr-53 phosphorylation had been shown to increase the critical concentration for polymerization, reduce the rates of nucleation and pointed end elongation, and decrease the affinity for DNase I (11). The proximity of Tyr-53 to the D-loop, which is implicated in intersubunit contacts in the filament (14–17) and mediates the interaction with DNase I (22), was believed to account for most of the biochemical properties of pY53-actin (8, 11). The structures of pY53-actin and unphosphorylated actin seem to confirm this hypothesis. The D-loop is flexible and unresolved in most actin structures. However, in the structure of pY53-actin, hydrogen-bonding contacts between the phosphate oxygens on Tyr-53, and residues of the D-loop help stabilize its conformation (Fig. 1B). Phosphorylation of Tyr-53 protects the D-loop from subtilisin cleavage (Fig. 1C), providing additional evidence for a more stable conformation of the loop in solution. A more stably folded D-loop would probably interfere with the binding of DNase I, and with intersubunit contact in the filament.
Our biochemical analyses suggest that phosphorylation of Tyr-53 also affects other regions of the actin molecule, including the nucleotide cleft and the target-binding cleft between subdomains 1 and 3, where both profilin and gelsolin bind (Fig. S5). Thus, phosphorylation reduces the quenching of tryptophan fluorescence that accompanies the binding of profilin (Fig. 2B), lowers the rate of nucleotide exchange on actin, and reduces the fluorescence of etheno-ATP bound to actin (Fig. 1 D and E). Probably connected with these observations, the hydrolysis of ATP on polymerization was previously shown to be much slower for pY53-actin than for unphosphorylated actin (11). The causes of these biochemical differences are not apparent from analysis of the structures, which show little difference between pY53-actin and unphosphorylated actin other than the conformational change in subdomain 2 (Movie S1). However, there are multiple examples of long-range allosteric interactions in actin, including intramolecular coupling between the target-binding cleft and subdomain 2, which occur both in the monomer and in the filament (41). The causes of these allosteric effects are not always clear from comparisons of the structures. Phosphorylation of Tyr-53 appears to be yet another example of a modification in subdomain 2 that alters the dynamic equilibrium between different regions of the actin molecule, from the D-loop, through the nucleotide cleft, and down to the target-binding cleft.
We find that profilin–actin complexes present a moderately more open nucleotide cleft than other actin structures. Opening of the nucleotide cleft results from a relatively small rotation of 4.7° of the two major domains of actin relative to each other (Fig. 2A and Movies S2 and S3). This movement is less dramatic than previously anticipated (36, 37), but it is probably sufficient to explain the stimulation of nucleotide exchange produced by profilin. Indeed, the most important factor in determining nucleotide exchange might not necessarily be the degree of cleft opening, but rather changes in the intricate network of hydrogen bonding interactions that coordinate the nucleotide in the catalytic cleft. Opening of the cleft is accompanied by slight changes in this network and a small twist of the nucleotide (Movies S2 and S3). Whereas the moderately open cleft stabilized by profilin may explain the stimulation of nucleotide exchange on actin, the short-lived state when actin releases its nucleotide may be characterized by an even more open cleft, probably analogous to that of the wide-open structure of profilin–β-actin (36) (Fig. S4B). Finally, in the actin filament, the nucleotide cleft appears to be more closed than in any of the existing structures of the monomer (17), suggesting that addition of profilin–actin at the barbed end of growing filaments results in closure of the nucleotide cleft, which might then lower the affinity for profilin and stimulate its rapid release.
The conformational changes associated with Tyr-53 phosphorylation and profilin binding described here are less extensive than previously anticipated, but are associated with long-range allosteric effects throughout the actin molecule. Large motions such as the acto-myosin power stroke are visually arresting, but conformational changes in proteins are usually less dramatic and result from changes in the equilibrium between different states in a landscape of nearly isoenergetic conformations (42).
Materials and Methods
Preparation of Proteins and Peptide.
Phosphorylated and unphosphorylated Dictyostelium actin were purified as described (11). Human profilin-I and the VASP peptide 198–213 (corresponding to the last polyPro region of human VASP) were prepared as described (21). The cDNA encoding for human gelsolin was purchased from ATCC (catalog number MGC-39262). Gelsolin segment 1 or G1 (residues Met-52-Phe-176) was amplified by PCR and cloned between the NdeI and XhoI sites of vector pTYB12 (New England BioLabs). This vector comprises a chitin-binding domain (for affinity purification) and an intein (for self-cleavage after purification). BL21(DE3) competent cells (Invitrogen) were transformed with this constructs and grown in LB medium at 37°C until the OD600 reached a value of 0.5. Expression was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and carried out overnight at 20°C. Cells were harvested by centrifugation, resuspended in chitin-affinity-column equilibration buffer (20 mM Tris·HCl, pH 7.5; 0.5 M NaCl; 0.1% Triton X-100; 1 mM EDTA) and lysed using a microfluidizer apparatus (MicroFluidics). Affinity purification on the chitin column was done according to the manufacturer's protocol (New England Biolabs). G1 was eluted from the column after self-cleavage of the intein, which was induced with 50 mM DTT for 2 days at 4°C. The protein was then dialyzed against 10 mM Tris·HCl, pH 8.0; 40 mM NaCl; 0.2 mM EDTA; and 1 mM NaN3 and further purified on a superose12 column (Amersham Pharmacia).
Crystallization.
Phosphorylated and unphosphorylated actin at ≈20 μM concentration in G-buffer (2 mM Tris, pH 7.5; 0.2 mM CaCl2; 0.2 mM ATP; 1 mM NaN3) were mixed with profilin and G1 at a 1:1.2 molar ratio. The complexes were purified on a superose12 column preequilibrated with 2 mM Tris·HCl, pH 8.0; 2 mM ATP; 4 mM MgCl2; 0.5 mM EDTA; and 1 mM NaN3. The profilin–actin and G1–actin complexes were concentrated to ≈5 mg/ml and ≈13 mg/ml, respectively by using Vivaspin centrifugal devices (Sartorius). Crystals of G1–actin were obtained under similar conditions for the unphosphorylated and phosphorylated forms: 100 mM Hepes, pH 7.5; 1.7 M Li2SO4; 2 mM ATP; 1 mM EDTA; and 10% glycerol at 20°C in 4-μl hanging drops. Except for the use of glycerol in this crystallization, these conditions are similar to those published before (30). The crystals were flash-frozen in liquid nitrogen by using 20% glycerol as a cryoprotectant and stored for data collection. Crystals of the profilin–actin complex were obtained only for the unphosphorylated form. This complex was crystallized with a VASP polyproline peptide (VASP198–213), which binds profilin and facilitates crystallization. The crystallization conditions were similar to those described by us for profilin–α-actin (21): 0.1 M Bis-Tris, pH 6.5; 25% (wt/vol) PEG 3350. The crystals were flash-frozen in liquid nitrogen for data collection with Paratone-N as a cryoprotectant.
Data Collection and Structure Determination.
An x-ray dataset was collected to the resolution of 2.3 Å from a profilin–actin crystal by using the beamline F2 of the Cornell High Energy Synchrotron Source (Ithaca, NY). Datasets were collected from crystals of G1 complexes with unphosphorylated and phosphorylated actin to the resolutions of 1.6 Å and 1.7 Å, respectively, using the 17-BM beamline of the IMCA-CAT facility at the Advance Photon Source (Argonne, IL). All of the datasets were indexed and scaled with the program HKL2000 (HKL Research). The structures were determined by molecular replacement using the CCP4 (43) program Phaser and the structures of profilin–α-actin (2PAV) or G1-actin (1NM1) as search models. Model building and refinement were performed with the CCP4 programs Coot and Refmac (Table 1).
Subtilisin Cleavage of the D-Loop.
Actin, 13 μM in G-buffer, was cleaved with subtilisin at a molar ratio of actin:subtilisin of 8,000:1. Similar results were obtained at a ratio of 5,000:1. The percentage digestion for each reaction was determined as the ratio of the intensities of the digested fraction (lower band, ≈38,000 Da, on SDS PAGE) to the sum of the intensities of the digested and undigested actin fractions.
Etheno-ATP Exchange.
The fluorescence of etheno-ATP increases when it replaces the ATP bound to actin. ATP-exchange experiments were carried out in 4 mM Tris, pH 7.5; 1 mM DTT; 0.1 mM CaCl2; 0.01% NaN3; and 10 μM ATP, with addition of 50 μM etheno-ATP at time 0. The experiments were performed with 1 μM actin in the presence of either 0.5 μM profilin, which acts catalytically (19), or 1.2 μM gelsolin.
Quenching of Tryptophan Fluorescence.
Quenching of tryptophan fluorescence by profilin was done with 0.6 μM actin in 4 mM Tris pH 7.5, 1 mM DTT, 0.1 mM CaCl2, 0.01% NaN3 and 0.2 mM ATP. Kaleida graph software was used to fit the data and determine Kd values.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grant GM073791 and the Division of Intramural Research, National Heart, Lung, and Blood Institute. Data collection at the Cornell High Energy Synchrotron Source and Macromolecular Diffraction facilities was supported by National Science Foundation Grant DMR-0225180 and National Institutes of Health Grant RR-01646. Use of IMCA-CAT beamline 17-BM was supported by the Industrial Macromolecular Crystallography Association through a contract with the University of Chicago. The Advanced Photon Source was supported by Department of Energy Contract W-31-109-Eng-38.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3CHW, 3CIP, 3CI5).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805852105/DCSupplemental.
References
- 1.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 2.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: From yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 3.van Delft S, Verkleij AJ, Boonstra J, van Bergen en Henegouwen PM. Epidermal growth factor induces serine phosphorylation of actin. FEBS Lett. 1995;357:251–254. doi: 10.1016/0014-5793(94)01359-9. [DOI] [PubMed] [Google Scholar]
- 4.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 5.Kameyama K, et al. Tyrosine phosphorylation in plant bending. Nature. 2000;407:37. doi: 10.1038/35024149. [DOI] [PubMed] [Google Scholar]
- 6.Schweiger A, Mihalache O, Ecke M, Gerisch G. Stage-specific tyrosine phosphorylation of actin in Dictyostelium discoideum cells. J Cell Sci. 1992;102:601–609. doi: 10.1242/jcs.102.3.601. [DOI] [PubMed] [Google Scholar]
- 7.Howard PK, Sefton BM, Firtel RA. Tyrosine phosphorylation of actin in Dictyostelium associated with cell-shape changes. Science. 1993;259:241–244. doi: 10.1126/science.7678470. [DOI] [PubMed] [Google Scholar]
- 8.Jungbluth A, et al. Stress-induced tyrosine phosphorylation of actin in Dictyostelium cells and localization of the phosphorylation site to tyrosine-53 adjacent to the DNase I binding loop. FEBS Lett. 1995;375:87–90. doi: 10.1016/0014-5793(95)01165-b. [DOI] [PubMed] [Google Scholar]
- 9.Gauthier ML, Lydan MA, O'Day D, Cotter AD. Endogenous autoinhibitors regulate changes in actin tyrosine phosphorylation during Dictyostelium spore germination. Cell Signal. 1997;9:79–83. doi: 10.1016/s0898-6568(96)00113-1. [DOI] [PubMed] [Google Scholar]
- 10.Kishi Y, Clements C, Mahadeo DC, Cotter DA, Sameshima M. High levels of actin tyrosine phosphorylation: Correlation with the dormant state of Dictyostelium spores. J Cell Sci. 1998;111:2923–2932. doi: 10.1242/jcs.111.19.2923. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Shu S, Hong MS, Levine RL, Korn ED. Phosphorylation of actin Tyr-53 inhibits filament nucleation and elongation and destabilizes filaments. Proc Natl Acad Sci USA. 2006;103:13694–13699. doi: 10.1073/pnas.0606321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams HP, Harwood AJ. Cell polarity and Dictyostelium development. Curr Opin Microbiol. 2003;6:621–627. doi: 10.1016/j.mib.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Jungbluth A, et al. Strong increase in the tyrosine phosphorylation of actin upon inhibition of oxidative phosphorylation: correlation with reversible rearrangements in the actin skeleton of Dictyostelium cells. J Cell Sci. 1994;107:117–125. doi: 10.1242/jcs.107.1.117. [DOI] [PubMed] [Google Scholar]
- 14.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 15.Hegyi G, et al. Intrastrand cross-linked actin between Gln-41 and Cys-374. I. Mapping of sites cross-linked in F-actin by N-(4-azido-2-nitrophenyl) putrescine. Biochemistry. 1998;37:17784–17792. doi: 10.1021/bi981285j. [DOI] [PubMed] [Google Scholar]
- 16.Khaitlina SY, Strzelecka-Golaszewska H. Role of the DNase-I-binding loop in dynamic properties of actin filament. Biophys J. 2002;82:321–334. doi: 10.1016/S0006-3495(02)75397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oda T, Stegmann H, Schroder RR, Namba K, Maeda Y. Modeling of the F-actin structure. Adv Exp Med Biol. 2007;592:385–401. doi: 10.1007/978-4-431-38453-3_32. [DOI] [PubMed] [Google Scholar]
- 18.Mockrin SC, Korn ED. Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry. 1980;19:5359–5362. doi: 10.1021/bi00564a033. [DOI] [PubMed] [Google Scholar]
- 19.Goldschmidt-Clermont PJ, Machesky LM, Doberstein SK, Pollard TD. Mechanism of the interaction of human platelet profilin with actin. J Cell Biol. 1991;113:1081–1089. doi: 10.1083/jcb.113.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 21.Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin–actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26:4597–4606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 23.McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993;364:685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 24.Otterbein LR, Cosio C, Graceffa P, Dominguez R. Crystal structures of the vitamin D-binding protein and its complex with actin: Structural basis of the actin-scavenger system. Proc Natl Acad Sci USA. 2002;99:8003–8008. doi: 10.1073/pnas.122126299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chereau D, et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SH, Hayes DB, Rebowski G, Tardieux I, Dominguez R. Toxofilin from Toxoplasma gondii forms a ternary complex with an antiparallel actin dimer. Proc Natl Acad Sci USA. 2007;104:16122–16127. doi: 10.1073/pnas.0705794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klenchin VA, et al. Trisoxazole macrolide toxins mimic the binding of actin-capping proteins to actin. Nat Struct Biol. 2003;10:1058–1063. doi: 10.1038/nsb1006. [DOI] [PubMed] [Google Scholar]
- 28.Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J Biol Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- 29.Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 30.Vorobiev S, et al. The structure of nonvertebrate actin: Implications for the ATP hydrolytic mechanism. Proc Natl Acad Sci USA. 2003;100:5760–5765. doi: 10.1073/pnas.0832273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burtnick LD, Urosev D, Irobi E, Narayan K, Robinson RC. Structure of the N-terminal half of gelsolin bound to actin: Roles in severing, apoptosis and FAF. EMBO J. 2004;23:2713–2722. doi: 10.1038/sj.emboj.7600280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strzelecka-Golaszewska H, Moraczewska J, Khaitlina SY, Mossakowska M. Localization of the tightly bound divalent-cation-dependent and nucleotide-dependent conformation changes in G-actin using limited proteolytic digestion. Eur J Biochem. 1993;211:731–742. doi: 10.1111/j.1432-1033.1993.tb17603.x. [DOI] [PubMed] [Google Scholar]
- 33.Strzelecka-Golaszewska H, Wozniak A, Hult T, Lindberg U. Effects of the type of divalent cation, Ca2+ or Mg2+, bound at the high-affinity site and of the ionic composition of the solution on the structure of F-actin. Biochem J. 1996;316:713–721. doi: 10.1042/bj3160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muhlrad A, et al. Cofilin induced conformational changes in F-actin expose subdomain 2 to proteolysis. J Mol Biol. 2004;342:1559–1567. doi: 10.1016/j.jmb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Bryan J. Gelsolin has three actin-binding sites. J Cell Biol. 1988;106:1553–1562. doi: 10.1083/jcb.106.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chik JK, Lindberg U, Schutt CE. The structure of an open state of beta-actin at 2.65 A resolution. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- 37.Schuler H. ATPase activity and conformational changes in the regulation of actin. Biochim Biophys Acta. 2001;1549:137–147. doi: 10.1016/s0167-4838(01)00255-2. [DOI] [PubMed] [Google Scholar]
- 38.Minehardt TJ, Kollman PA, Cooke R, Pate E. The open nucleotide pocket of the profilin/actin x-ray structure is unstable and closes in the absence of profilin. Biophys J. 2006;90:2445–2449. doi: 10.1529/biophysj.105.072900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tirion MM, ben-Avraham D. Normal mode analysis of G-actin. J Mol Biol. 1993;230:186–195. doi: 10.1006/jmbi.1993.1135. [DOI] [PubMed] [Google Scholar]
- 40.Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- 41.Egelman EH. Actin allostery again? Nat Struct Biol. 2001;8:735–736. doi: 10.1038/nsb0901-735. [DOI] [PubMed] [Google Scholar]
- 42.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 43.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.