Abstract
Base excision repair (BER) is essential for maintaining genome stability both to counter the accumulation of unusual bases and to protect from base loss in the DNA. Herpes simplex virus 1 (HSV-1) is a large dsDNA virus that encodes its own DNA replication machinery, including enzymes involved in nucleotide metabolism. We report on a replicative family B and a herpesvirus-encoded DNA Pol that possesses DNA lyase activity. We have discovered that the catalytic subunit of the HSV-1 DNA polymerase (Pol) (UL30) exhibits apurinic/apyrimidinic (AP) and 5′-deoxyribose phosphate (dRP) lyase activities. These activities are integral to BER and lead to DNA cleavage on the 3′ side of abasic sites and 5′-dRP residues that remain after cleavage by 5′-AP endonuclease. The UL30-catalyzed reaction occurs independently of divalent cation and proceeds via a Schiff base intermediate, indicating that it occurs via a lyase mechanism. Partial proteolysis of the Schiff base shows that the DNA lyase activity resides in the Pol domain of UL30. These observations together with the presence of a virus-encoded uracil DNA glycosylase indicates that HSV-1 has the capacity to perform critical steps in BER. These findings have implications on the role of BER in viral genome maintenance during lytic replication and reactivation from latency.
Keywords: base excision repair, abasic DNA, uracil DNA glycosylase
Herpes simplex virus 1 (HSV-1) is a dsDNA virus with a genome of ≈152 kbp (1). HSV-1 switches between lytic replication in epithelial cells and a state of latency in sensory neurons during which there is no detectable DNA replication (1). Replication of the genome is mediated by seven essential virus-encoded factors with the following functions: a DNA polymerase (Pol) catalytic subunit (UL30), its associated processivity factor (UL42), a ssDNA binding protein (UL29), a heterotrimeric helicase-primase (UL5, UL8, and UL52) and an initiator protein (UL9) that binds to and unwinds the viral replication origins (1–4). HSV-1 also encodes several other enzymes that are dispensable for replication in cell culture, which are involved in nucleotide metabolism and perform other important roles in maintaining the viral genome including a thymidine kinase (UL23), a ribonucleotide reductase (UL39 and UL40), a dUTPase (UL50), an exonuclease (UL12), and notably a uracil DNA glycosylase (UDG) (UL2) (4).
The presence of a virus-encoded UDG suggests that excision of uracil may be important during viral replication. Hence, it has been shown that uracil substitutions in the viral origins of replication alters their recognition by the viral initiator protein (5). Moreover, whereas UL2 may be dispensable for viral replication in fibroblast (6), UL2 mutants exhibit reduced neurovirulence and a decreased frequency of reactivation from latency (7). Thus, UDG action in HSV-1 may be important for viral reactivation after quiescence in neuronal cells during which the genome may accumulate uracil as a result of spontaneous deamination of cytosine. In cytomegalovirus, the viral UDG was shown to be required for the transition to late-phase DNA replication (8, 9).
The removal of unusual bases, including uracil, from DNA by the action of DNA glycosylases together with spontaneous base loss are two processes that contribute to the formation of apurinic/apyrimidinic (AP) sites. In the case of HSV-1, a recent study showed that DNA isolated from HSV-1-infected cultured fibroblasts contains a steady state of 2.8–5.9 AP sites per viral genome equivalent (10). Because AP sites are noninstructional lesions, they may either induce mutagenesis or cause replisome stalling. Indeed, the HSV-1 Pol (UL30) cannot replicate beyond a model AP site (tetrahydrofuran residue) (10), indicating the necessity to repair this type of lesion. In mammalian single-nucleotide base excision repair (BER) initiated by monofunctional DNA glycosylases, AP sites are incised hydrolytically at the 5′ side by AP endonuclease (APE) followed by one nucleotide addition by Pol β to generate a 5′deoxyribose phosphate (dRP) flap (11). The 5′ dRP residue is subsequently removed by the 5′ dRP lyase activity of Pol β to leave a nick that is ligated by DNA ligase I or III (12, 13). Certain enzymes, including Pol β, also exhibit AP lyase activity that cleaves 3′ to AP sites, independently of preincision by APE, leaving a 5′ phosphate and an unsaturated sugar-phosphate at the 3′ end (13, 14).
In this study, we explored the existence of additional HSV-1-encoded BER activities. We report on a replicative family B and a herpesvirus Pol that exhibits DNA lyase activity. We show that the HSV-1 Pol (UL30) exhibits AP and 5′ dRP lyase activities. These findings have implications for virus-mediated BER and the emergence of the viral genome after quiescence in neuronal cells.
Results
Cleavage of AP DNA by UL30.
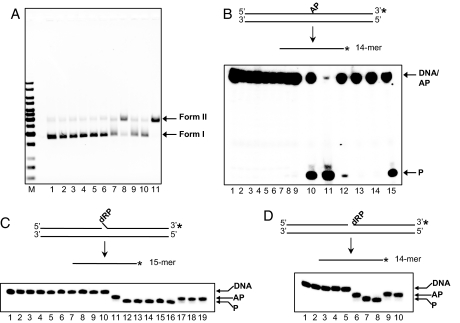
We investigated whether UL30 was capable of specifically cleaving abasic DNA. We found that UL30 exhibited nicking activity with supercoiled DNA that had been heated at 70°C for 90 min at pH 5.5 to induce base loss (data not shown). Consequently, we systematically examined whether UL30 was able to cleave abasic DNA. Fig. 1A shows the results of an experiment in which UL30 was tested for its ability to nick supercoiled plasmid DNA that contained abasic sites prepared by pretreating uracil-containing pSV011 with UDG. Consistent with our preliminary findings, UL30 was able to convert supercoiled (form I) DNA into form II DNA (Fig. 1A, lane 8). This activity resembles that of Pol β, which is known to cleave AP DNA (Fig. 1A, lane 11) (14). The conversion of supercoiled abasic plasmid to form II DNA depended on the native conformation of UL30 because neither heat nor chemically denatured UL30 showed activity above that observed with buffer alone (Fig. 1A, compare lanes 9 and 10 with lane 7). Importantly, no nicking by UL30 or Pol β was observed with intact, uracil-containing DNA that had not been pretreated with UDG, indicating that the cleavage is specific for abasic DNA (Fig. 1A, lanes 3 and 6). It is also noteworthy that the cleavage of abasic DNA by UL30 occurred in the presence of EDTA and is independent of Mg2+.
Fig. 1.
UL30 exhibits AP and 5′ dRP cleavage activities. (A) Cleavage of supercoiled AP DNA. Lane 1, pSV011 form I; lanes 2–6, reactions with intact substrate; lanes 7–11, reactions with AP substrate. Lanes 2 and 7, buffer; lanes 3 and 8, 100 nM UL30; lanes 4 and 9, 100 nM heat denatured UL30 (UL30*); lanes 5 and 10, 100 nM heat and SDS denatured UL30 (UL30*SDS); lanes 6 and 11, 100 nM Pol β. (B) Cleavage of duplex AP substrate. Lane 1, substrate; lanes 2–8, reactions with intact substrate; lanes 9–15, reactions with AP substrate. Lanes 2 and 9, buffer; lanes 3 and 10, 2 mM spermidine; lanes 4 and 11, 0.2 M NaOH; lanes 5 and 12, 450 nM UL30; lanes 6 and 13, 450 nM UL30*; lanes 7 and 14, 450 nM UL30*SDS; lanes 8 and 15, 400 nM Pol β. (C) 5′ dRP cleavage with the flap substrate. Lane 1, substrate; lanes 2–10, reactions with intact substrate; lanes 11–19, reactions with AP substrate. Lanes 2 and 11, buffer; lanes 3 and 12, 2 mM spermidine; lanes 4 and 13, 2 mM spermidine and 1% SDS; lanes 5 and 14, 0.2 M NaOH; lanes 6 and 15, 400 nM Pol β; lanes 7 and 16, 450 nM UL30; lanes 8 and 17, 450 nM UL30*; lanes 9 and 18, 450 nM UL30*SDS; lanes 10 and 19, 750 nM BSA. (D) 5′ dRP cleavage with the nicked substrate. Lanes 1–5, reactions with intact substrate; lanes 6–10, reactions with AP substrate. Lanes 1 and 6, buffer; lanes 2 and 7, 2 mM spermidine; lanes 3 and 8, 450 nM UL30; lanes 4 and 9, 450 nM UL30*; lanes 5 and 10, 450 nM UL30*SDS. The asterisk indicates a 3′-32P label. The positions of form I, form II, markers (M, 1-kb DNA Ladder; Fermentas), intact DNA (DNA), AP-DNA (AP), and product (P) are as indicated.
To support the above finding, we examined the ability of UL30 to cleave a linear 31-mer duplex oligonucleotide substrate in which the 3′-32P-labeled top strand contains an abasic site at position 17 (Fig. 1B). Cleavage 3′ to the AP site would generate a 3′-32P-labeled 14-mer. Treatment of this substrate with spermidine or NaOH, reagents that are know to cleave abasic DNA by β or β/δ elimination (15), resulted in 14-mer product formation (Fig. 1B, lanes 10 and 11). Consistent with its ability to cleave AP DNA (14), Pol β gave rise to 14-mer product (Fig. 1B, lane 15). Importantly, 14-mer product was also formed after incubation with UL30 but not with heat or chemically denatured UL30 (Fig. 1B, compare lane 12 with lanes 13 and 14). As was observed with the assay using abasic plasmid DNA, no cleavage occurred on intact DNA with spermidine/NaOH or UL30/Pol β, indicating that the reaction is specific for abasic DNA (Fig. 1B, lanes 3–8). Moreover, cleavage of the AP oligonucleotide substrate occurred in the presence of EDTA. Under these reaction conditions, cleavage by Pol β was fairly robust (≈40% product formation after 40 min). In contrast, UL30 was ≈10-fold less active. This observation is attributed to a Km effect (see below). Similar results were obtained with 5′-32P labeled substrate (data not shown). Thus far, the results indicate that UL30 cleaves DNA 3′ to abasic sites independently of Mg2+.
Cleavage of 5′ dRP by UL30.
In single-nucleotide BER, abasic sites are incised at the 5′ side by APE followed by one nucleotide addition by Pol β to generate a 5′ dRP flap (11). The 5′ dRP residue is subsequently removed by the 5′ dRP lyase activity of Pol β (11). To examine whether UL30 also cleaves 3′ to dRP residues, we constructed a substrate that mimics a preincised DNA duplex with a 5′ dRP flap (“flap” substrate; Fig. 1C). In this case, the substrate contains a 3′-32P-labeled 16-mer that possesses a 5′ U flap, which upon pretreatment with UDG is converted to an oligonucleotide with a 5′ dRP flap that has the electrophoretic mobility of a 15.5-mer. Removal of the 5′ dRP would generate a 3′-32P-labeled 15-mer. Treatment of this substrate with spermidine or NaOH resulted in 15-mer product formation (Fig. 1C, lanes 12–14). Consistent with its ability to remove 5′ dRP residues (14), Pol β gave rise to 15-mer product (Fig. 1C, lane 15). UL30, but not heat or chemically denatured UL30, was able to convert the 15.5-mer into 15-mer, indicating removal of the 5′ dRP residue (Fig. 1C, compare lanes 16 and 17 with lane 18). No product formation was observed with DNA substrate that possessed a 5′ U flap, indicating that it is specific for the removal of 5′ dRP (Fig. 1C, lanes 3–10). 5′ dRP removal by UL30 occurred in the presence of EDTA.
The 5′ dRP cleavage activity of UL30 was further examined with a substrate that mimics a preincised DNA duplex with a 5′ dRP at the site of the nick (“nicked” substrate; Fig. 1D). In this case, the substrate contains a 3′-32P-labeled 15-mer that possesses a 5′ terminal U, which upon pretreatment with UDG is converted to an oligonucleotide with a 5′ dRP that has the electrophoretic mobility of a 14.5-mer. Removal of the 5′ dRP would generate a 3′-32P-labeled 14-mer. Treatment of this substrate with spermidine resulted in 14-mer product formation (Fig. 1D, lane 7). Consistent with its activity on the flap DNA substrate, UL30 converted the 14.5-mer into 14-mer product in the presence of EDTA (Fig. 1D, lane 8). However, neither the heat nor chemically denatured UL30 showed product formation (Fig. 1D, lanes 9 and 10). Consistent with the previous reactions, no product formation was observed with DNA substrate that possessed a 5′ U (Fig. 1D, lanes 2–5). Collectively, these results demonstrate that UL30 cleaves 3′ to AP sites and is capable of removing 5′ dRP residues at sites that mimic preincision with APE, in a manner that is independent of Mg2+. Moreover, it should be noted that the activity of UL30 on the preincised flap and nicked substrates (Fig. 1 C and D) was subsgtantially greater (i.e., complete substrate conversion) than that observed on AP DNA (Fig. 1B). This finding is similar to what has been found for Pol β, whose kcat for 5′ dRP lyase activity is ≈200-fold higher than that for AP lyase activity (14).
Kinetics of 5′ dRP Cleavage.
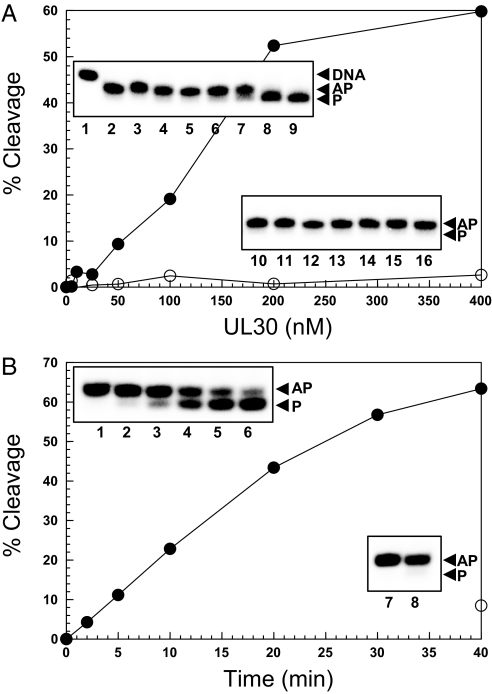
Fig. 2A demonstrates the dose dependency of 5′ dRP cleavage by UL30. Under the specified reaction conditions, 5′ dRP removal was linear up to 200 nM UL30, with maximum activity (≈60% product formation) observed at 400 nM UL30. Over the same concentration range, heat-inactivated UL30 only exhibited background activity (<5% product formation) (Fig. 2A). The time course of 5′ dRP cleavage by UL30 is shown in Fig. 2B. The reaction is linear for the first 20 min and approaches saturation after ≈40 min with >60% product formation.
Fig. 2.
5′ dRP cleavage activity of UL30. Reactions were performed as a function of UL30 concentration (A) or time (B). (A) UL30 (●) or heat and SDS-denatured UL30 (○) was incubated with the flap substrate. (Insets) The corresponding storage phosphor images for this experiment. Lane 1, substrate; lanes 2–9, 0, 5, 10, 25, 50, 100, 200, and 400 nM UL30, respectively; lanes 10–16, 5, 10, 25, 50, 100, 200, and 400 nM heat and SDS-denatured UL30, respectively. (B) UL30 (100 nM) (●) was incubated with the nicked substrate for the times indicated. ○ represents the background activity after incubation with buffer. (Insets) The corresponding storage phosphor images for this experiment. Lanes 1–6, 2, 5, 10, 20, 30, and 40 min with 100 nM UL30, respectively; lanes 7 and 8, 0 and 40 min with buffer, respectively. The positions of intact DNA (DNA), AP-DNA (AP), and product (P) are as indicated.
A steady-state kinetic analysis shows that 5′ dRP removal by UL30 follows Michaelis–Menten kinetics, with an apparent Km of 1.8 ± 0.2 μM and a kcat of 0.45 ± 0.03 min−1 [supporting information (SI) Fig. S1]. Whereas the kcat for UL30 is 10-fold lower than that observed for Pol β (4.5 min−1) it is slightly higher than that reported for Pol γ (0.26 min−1) (14, 16). On the other hand, the Km for UL30 is ≈3-fold higher than that for both Pol β and Pol γ (14, 16). Assuming that the Km for AP cleavage is similar to that for 5′ dRP removal, it would explain the relative difference in AP cleavage observed with the plasmid (Fig. 1A) and AP duplex oligonucleotide (Fig. 1B), with the former containing a ≈100-fold higher concentration of AP sites (assuming ≈100 uracil residues per plasmid molecule) than the oligonucleotides-based substrate.
5′ dRP Cleavage Occurs by a Lyase Mechanism.
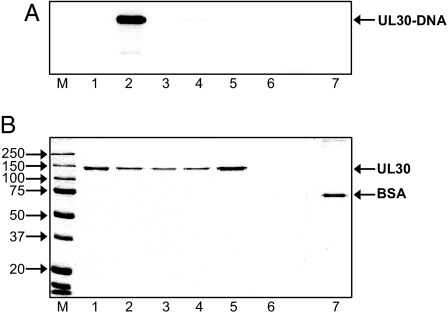
The finding that the AP and 5′ dRP cleavage activities of UL30 occur in the presence of EDTA and are therefore independent of divalent cation suggests that the reaction does not proceed via a hydrolytic mechanism. Bona fide DNA lyases catalyze reactions in which β-elimination proceeds via a Schiff base intermediate between a lysine in the enzyme and the DNA substrate (13). This protein–DNA intermediate can be covalently trapped via reduction with sodium borohydride. Fig. 3 shows the results of the NaBH4 trapping. The covalent UL30–DNA complex was formed only in the presence of UL30 and NaBH4 (Fig. 3A, lane 2). Substitution of NaBH4 with NaCl did not lead to formation of a covalent UL30–DNA complex and, importantly, covalent complex formation was not observed when using BSA as a nonspecific control protein (Fig. 3A, lanes 3 and 7). Moreover, neither heat nor chemically denatured UL30 formed a covalent complex with DNA, indicating that Schiff base formation depends on the native conformation of UL30 (Fig. 3A, lanes 4 and 5). The finding that a covalent UL30–DNA complex can be trapped by reduction with NaBH4 demonstrates that the reaction proceeds by a β-elimination mechanism and that UL30 functions as a bona fide lyase. Furthermore, the finding that UL30 is the species involved in the Schiff base indicates that the activity is not caused by a contaminant in the UL30 preparation.
Fig. 3.
UL30 cleaves 5′ dRP residues via a lyase mechanism that involves a Schiff base. Reactions were performed with 450 nM UL30 and the flap substrate. Storage phosphor (A) and Coomassie blue-stained (B) images of the same gel. Lane 1, UL30 only; lane 2, UL30 with DNA and NaBH4; lane 3, UL30 with DNA and NaCl, lane 4, heat-denatured UL30 with DNA and NaBH4; lane 5, heat- and SDS-denatured UL30 with DNA and NaBH4; lane 6, DNA only; lane 7, 750 nM BSA with DNA and NaBH4. The positions of markers (M, Precision Plus Protein Standards; BioRad), UL30, and BSA are as indicated.
Copurification of 5′ dRP Lyase and DNA Pol Activities.
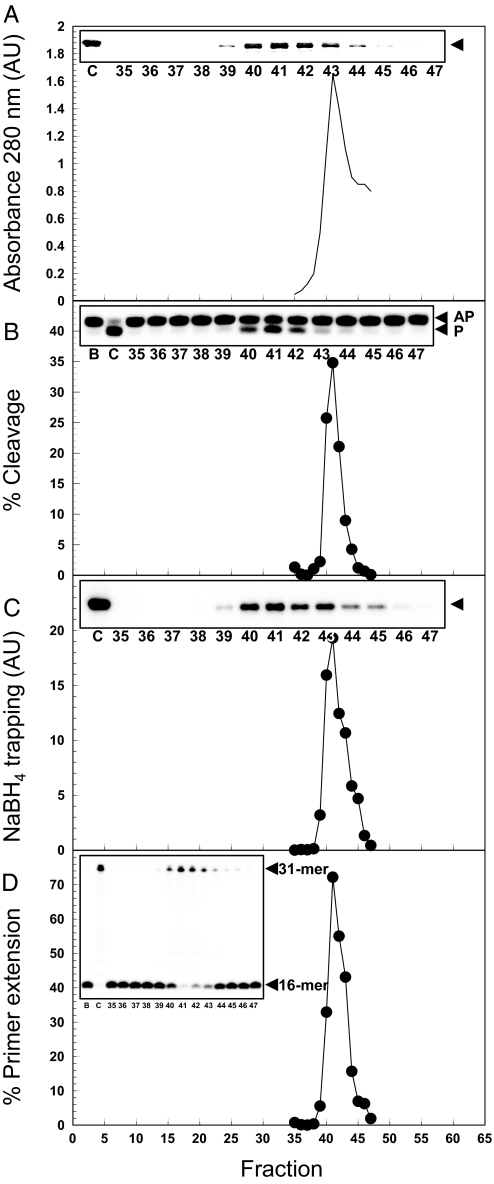
To further demonstrate that the 5′ dRP lyase activity is inherent to UL30 we examined whether it copurifies with the DNA Pol activity of UL30. Thus, purified UL30 was subjected to analytical gel filtration and the peak column fractions were assayed for 5′ dRP cleavage activity, covalent intermediate formation, and DNA Pol activity. Fig. 4 shows that the peak of UL30 protein (fraction 41) is perfectly coincident with the peaks of all three activities. These data indicate that the ability of UL30 to cleave AP DNA and remove 5′ dRP residues is an intrinsic property of UL30 and not caused by a contaminant.
Fig. 4.
Copurification of 5′ dRP lyase activity with DNA Pol activity on gel filtration. (A) Absorbance at 280 nm. (B) 5′ dRP cleavage (25 min reactions) with the nicked substrate. (C) Schiff base trapping performed with the nicked substrate. (D) Primer extension activity. AU, arbitrary units. (Insets) The relevant gel images (Coomassie blue staining, A; storage phosphor, B–D) with the indicated fraction numbers. C, UL30 control; B, buffer control. The arrow heads in A and C indicate the positions of UL30 and the UL30-DNA complex, respectively. The positions of AP-DNA (AP), product (P), 16-mer (primer), and 31-mer (product) are as indicated.
The DNA Lyase Activity Maps to the Pol Domain of UL30.
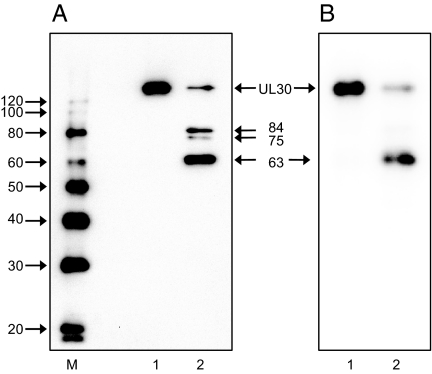
To further eliminate the possibility that the DNA lyase activity is caused by a contaminant of the same mass, we performed partial proteolysis of the Schiff base by using endoproteinase Glu-C. The UL30-specific digestion products were detected by immunoblotting using an anti-UL30 rabbit serum. Fig. 5A shows the formation of three UL30 digestion products with estimated masses of 84, 75, and 63 kDa that are identical to those reported by Weisshart et al. (17). The storage phosphor image of the same membrane shows that, apart from full-length UL30, the 63-kDa species is the one predominantly linked to DNA (65% of all of the digestion species) (Fig. 5B). The finding that the DNA is linked to a protease digestion product that is specifically recognized by antibodies against UL30 demonstrates that the Schiff base involves UL30. The 63-kDa endoproteinase Glu-C polypeptide was previously shown to map to the C terminus of UL30, approximately between residue 650 and the C terminus (17). Based on the structure of UL30 (18), the DNA lyase is situated in the Pol domain of UL30.
Fig. 5.
The DNA lyase of UL30 maps to the C-terminal Pol domain. The Schiff base between UL30 and the nicked substrate was covalently trapped by the addition of 1 mM NaBH4. One-half of the reaction was treated with 7.5 μg/ml endoproteinase Glu-C for 20 min followed by termination with 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin A, 20 mM EDTA, and 1 mM phenylmethylsulfonylfluoride. After SDS/PAGE, protein species were transferred to PVDF and probed with an anti-UL30 rabbit serum (26). (A) Chemiluminescent detection of UL30. Lane 1, UL30; lane 2, endoproteinase Glu-C-digested UL30. (B) Storage phosphor image of the same membrane. Lane 1, UL30; lane 2, endoproteinase Glu-C-digested UL30. The positions of UL30 and the 84-, 75-, and 63-kDa endoproteinase Glu-C digestion products and markers (M, Magicmark; Invitrogen) are as indicated.
Discussion
A recent study in cultured fibroblasts has estimated the steady-state number of AP sites in the HSV-1 genome between 2.8 and 5.9 (10). These may arise through spontaneous base loss and as a result of DNA glycosylase action, presumably that of the virus-encoded UL2. Because AP sites are noninstructional and cause UL30 to stall (10), their occurrence in the viral genome necessitates that they are repaired by BER. Here, we have shown that UL30, a replicative B family Pol and a Pol from the herpesviridae family, possesses both AP and 5′ dRP lyase activities that are integral to BER. We report that such activities are associated with both a replicative B family Pol or with a Pol from the herpesviridae family. In summary, our data demonstrate that UL30 can cleave 3′ to abasic sites in duplex DNA and remove 5′ dRP residues from both nicked and flap structures, that resemble DNA that is either preincised with APE or that which has been preincised with APE and extended by one nucleotide to create a 5′ dRP flap. Furthermore, we showed that this activity occurs via a bona fide lyase mechanism that proceeds via a Schiff base intermediate and not via a hydrolytic reaction. Although the catalytic lysine that is involved in Schiff base formation remains to be identified, our experiments have mapped the activity to the C-terminal Pol domain of UL30.
Our finding that UL30 possesses AP and 5′ dRP lyase activities adds to the list of activities already associated with this enzyme, namely DNA Pol, 3′–5′ proofreading exonuclease and RNaseH activities (4). In this regard, UL30 resembles the mitochondrial replicase Pol γ, which, apart from its Pol activity, also exhibits proofreading exonuclease and 5′ dRP lyase activities (16, 19–21). Although cellular organisms have evolved to encode specialized DNA repair enzymes, there are several examples where multiple tasks are performed by a small set of enzymes. Mitochondrial genome maintenance is one such example. In this case Pol γ, with its class IIa aminoacyl tRNA synthetase-type processivity factor, TWINKLE helicase, and mitochondrial ssDNA binding protein mediate mitochondrial DNA replication (21). However, Pol γ also plays a primary role in mitochondrial genome maintenance via BER, where it is required for repair synthesis, and as a DNA lyase (16, 19). Similarly, in HSV-1, the seven essential replication factors perform DNA replication functions but have also been shown to directly mediate recombination and DNA repair reactions as illustrated by the recombination functions of ICP8 (22, 23) and the DNA lyase activity of UL30 described herein.
Undoubtedly, given the frequency of AP sites in the HSV-1 genome, BER is important to prevent mutagenesis and replisome stalling at abasic sites during viral replication. However, if the requirement for UL2 is taken as an indicator for the necessity of virus-mediated BER, the fact that UL2 is dispensable for HSV-1 replication in epithelial cells (6) suggests that there may be redundancy in BER in these cells, i.e., BER performed by either viral or abundant cellular factors. On the other hand, the reduced neurovirulence and reduced reactivation frequency of UL2 mutants (7) indicates that the HSV-1 UDG, and by extension virus-mediated BER, may play an important role during viral replication in neurons. In this regard, it is easy to envisage that spontaneous deamination of cytosine and depurination/depyrimidation during latency in postmitotic neurons necessitates repair via BER. We propose that virus-mediated BER is necessary to prepare the quiescent genome for replication in neurons where certain cellular BER enzymes (e.g., UDG) are lacking (24, 25). Because uracil substitutions in the viral origins impacts recognition by UL9 protein (5), uracil excision and subsequent repair of AP sites would ensure efficient replication initiation. Similarly, BER would also allow faithful replication fork progression.
In a search for Pol-interacting factors that may orchestrate events at the viral replication fork, we identified the viral UDG (UL2) as a protein that interacts with UL30 (F.B., Ilsa Corrediera, Virneliz Fernandez, Ulrike Satter, Martine Defais, and P.E.B., unpublished work). The association of UL30 with UL2 further supports our notion that UL30 is involved in BER. Thus, a complex of UL30 and UL2 would be able to perform all steps in BER except for incision 5′ to AP sites and ligation, presumably these steps are performed by cellular APE and ligases I or III. In summary, the existence of both virus-encoded UDG (UL2) and AP/5′ dRP lyase (UL30) activities indicates that HSV-1 has the capacity to perform critical steps in BER that may be important for genome surveillance during lytic replication and reactivation from latency.
Materials and Methods
Enzymes and Reagents.
UL30 was expressed in Spodoptera frugiperda cells and purified as described (26). The experiments described here used fraction V. UL30 was inactivated at 75°C for 10 min in the absence or presence of 1% SDS. Calf thymus terminal deoxynucleotidyl transferase (TdT) was purchased from Fermentas Life Sciences. Escherichia. coli UDG, bacteriophage T4 polynucleotide kinase, and BSA were obtained from New England Biolabs. Human DNA Pol β was from Trevigen. Sequencing grade endoproteinase Glu-C was from Promega. Unlabeled deoxyribonucleoside 5′-triphosphates (disodium salts) were purchased from GE Healthcare. [γ-32P]ATP (3,000 Ci/mmol) and [α-32P]cordycepin 5′-triphosphate (5,000 Ci/mmol) were from PerkinElmer. Spermidine and sodium borohydride were from Sigma–Aldrich.
Nucleic Acids.
pSV011 (27) was extracted from E. coli CJ236 (ung-1) grown in the presence of 0.25 μg/ml uridine by using the EndoFree Plasmid Maxi Kit (Qiagen). Form I DNA was isolated after electrophoresis through 1% agarose and further purified by using the Promega Wizard Plus DNA purification system and ethanol precipitation. Oligonucleotides were obtained from Operon Biotechnologies. Their sequences are shown in Table S1. Oligonucleotides were either 3′-32P-labeled with TdT and [α-32P]cordycepin 5′-triphosphate or 5′-32P-labeled with T4 polynucleotide kinase and [γ-32P]ATP followed by removal of unincorporated nucleotides using Microspin G-25 columns (GE Healthcare). DNA substrates were constructed by annealing the labeled oligonucleotides with a 1.25-fold molar excess of unlabeled oligonucleotides in 20 mM Tris·HCl (pH 8.0), 1 mM EDTA, and 0.1 M NaCl by heating to 95°C for 5 min followed by gradual cooling to <30°C. Schematics of the duplex (3′-32P-labeled PBAZ7 annealed to PBAZ3), flap (3′-32P-labeled PBAZ5 annealed to PBAZ4 and PBAZ3), and nicked (3′-32P-labeled PBAZ6 annealed to PBAZ4 and PBAZ3) DNA substrates used in the cleavage assays are shown in Fig. 1 B, C, and D, respectively.
AP and 5′ dRP Cleavage Assays.
Reactions were performed on either intact DNA (mock-treated) or DNA that had been treated with E. coli UDG to remove uracil and generate AP sites. To prepare AP DNA, 1–25 pmol of DNA was treated with 1 unit of E. coli UDG in 20 mM Hepes·NaOH (pH 7.5), 2 mM DTT, 50 mM KCl, and 1 mM EDTA for 10 min at 37°C immediately before use. Unless otherwise stated, reactions (10 μl) were performed in the same buffer with either 2.5 nM plasmid or 5 nM oligonucleotide substrate and the indicated concentration of UL30. Reactions were incubated for 40 min at 37°C (15 min at 75°C for reactions with NaOH) and stopped with 100 mM NaBH4 followed by 10 min at 0°C. Reactions with plasmid DNA were combined with 2 μl of loading buffer [20 mM Tris-acetate (pH 8.0), 150 mM EDTA, 6% SDS, 50% glycerol, 0.05% bromophenol blue, and 0.05% xylene cyanol] and analyzed by electrophoresis through 0.8% agarose-Tris acetate EDTA (pH 7.6) at 8 V/cm for 1 h. Gels were stained with ethidium bromide and analyzed by UV transillumination by using a BioRad VersaDoc 1000 imaging system. Reactions with oligonucleotide substrates were combined with an equal volume of stop buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol) and heated for 3 min at 75°C before electrophoresis through 16% polyacrylamide–8 M urea gels in GTG buffer [89 mM Tris (pH 9.0), 28.5 mM taurine, and 0.5 mM EDTA]. Reaction products were visualized and quantified by storage phosphor analysis with a Molecular Dynamics Storm 820 PhosphorImager using ImageQuant version 5.2. Cleavage was quantified as a percentage of total radioactivity. Steady-state kinetic parameters (kcat and Km) were determined by measuring initial rates of 5′ dRP cleavage using the nicked substrate and 100 nM UL30 and analyzed by nonlinear regression using the Enzfitter (Biosoft) Michaelis–Menten equation.
Schiff Base Trapping.
Reactions were performed as described for 5′ dRP cleavage, using either the flap or nicked substrate, except that NaBH4 was added at a final concentration of 20 mM immediately after the enzyme. After 10 min at 0°C and 20 min at 25°C, reactions were resolved by 10% SDS/PAGE, followed by Coomassie blue staining and storage phosphor analysis.
Primer Extension Assay.
Reactions (10 μl) contained 25 mM Hepes·NaOH (pH 7.5), 50 mM NaCl, 1 mM DTT, 2.5 mM MgCl2, 50 μg/ml BSA, 100 μM dNTP, and 5 nM 5′-32P-labeled PBAZ4 annealed to PBAZ3. Reactions were initiated by the addition of protein, incubated at 37°C for 1 min, and terminated by the addition of 5 μl of stop buffer. Samples were heated for 5 min at 75°C, rapidly cooled to 4°C, and resolved by electrophoresis through 16% polyacrylamide–8 M urea gels in GTG buffer. Reaction products were visualized and quantified by storage phosphor analysis. Primer extension products are expressed as a percentage of total radioactivity.
Gel Filtration.
UL30 (≈0.1 mg) was injected onto a Superdex 200 HR 10/30 column (GE Healthcare) that had been equilibrated with 20 mM Hepes·NaOH (pH 7.5), 0.2 M NaCl, 1 mM EDTA, 2 mM DTT, and 4% glycerol. The column was developed at 0.2 ml/min. After the first 6 ml, 13 ml of column eluate was collected in 0.2-ml fractions.
Supplementary Material
Acknowledgments.
This work was supported by Public Health Service Grant GM62643.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806375105/DCSupplemental.
References
- 1.Boehmer PE, Nimonkar AV. Herpes virus replication. IUBMB Life. 2003;55:13–22. doi: 10.1080/1521654031000070645. [DOI] [PubMed] [Google Scholar]
- 2.Boehmer PE, Villani G. Herpes simplex virus type 1: A model for genome transactions. Prog Nucleic Acid Res Mol Biol. 2003;75:139–171. doi: 10.1016/s0079-6603(03)75005-3. [DOI] [PubMed] [Google Scholar]
- 3.Lehman IR, Boehmer PE. Replication of herpes simplex virus DNA. J Biol Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 4.Boehmer PE, Lehman IR. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 5.Focher F, Verri A, Verzeletti S, Mazzarello P, Spadari S. Uracil in oris of herpes simplex 1 alters its specific recognition by origin binding protein (OBP): Does virus-induced uracil-DNA glycosylase play a key role in viral reactivation and replication? Chromosoma. 1992;102:S67–S71. doi: 10.1007/BF02451788. [DOI] [PubMed] [Google Scholar]
- 6.Mullaney J, Moss HW, McGeoch DJ. Gene UL2 of herpes simplex virus type 1 encodes a uracil-DNA glycosylase. J Gen Virol. 1989;70:449–454. doi: 10.1099/0022-1317-70-2-449. [DOI] [PubMed] [Google Scholar]
- 7.Pyles RB, Thompson RL. Evidence that the herpes simplex virus type 1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J Virol. 1994;68:4963–4972. doi: 10.1128/jvi.68.8.4963-4972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prichard MN, Duke GM, Mocarski ES. Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J Virol. 1996;70:3018–3025. doi: 10.1128/jvi.70.5.3018-3025.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courcelle CT, Courcelle J, Prichard MN, Mocarski ES. Requirement for uracil-DNA glycosylase during the transition to late-phase cytomegalovirus DNA replication. J Virol. 2001;75:7592–7601. doi: 10.1128/JVI.75.16.7592-7601.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y, Song L, Stroud J, Parris DS. Mechanisms by which herpes simplex virus DNA polymerase limits translesion synthesis through abasic sites. DNA Repair. 2008;7:95–107. doi: 10.1016/j.dnarep.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava DK, et al. Mammalian abasic site base excision repair: Identification of the reaction sequence and rate-determining steps. J Biol Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 12.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Gen. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 13.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase β. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 14.Prasad R, Beard WA, Strauss PR, Wilson SH. Human DNA polymerase β deoxyribose phosphate lyase: Substrate specificity and catalytic mechanism. J Biol Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl T, Anderson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972;11:3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- 16.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci USA. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisshart K, Kuo AA, Hwang CBC, Kumura K, Coen DM. Structural and functional organization of herpes simplex virus DNA polymerase investigated by limited proteolysis. J Biol Chem. 1994;269:22788–22796. [PubMed] [Google Scholar]
- 18.Liu S, et al. Crystal structure of the herpes simplex virus 1 DNA polymerase. J Biol Chem. 2006;281:18193–18200. doi: 10.1074/jbc.M602414200. [DOI] [PubMed] [Google Scholar]
- 19.Pinz KG, Bogenhagen DF. Characterization of a catalytically slow AP lyase activity in DNA polymerase γ and other family A DNA polymerases. J Biol Chem. 2000;275:12509–12514. doi: 10.1074/jbc.275.17.12509. [DOI] [PubMed] [Google Scholar]
- 20.Kaguni LS. DNA polymerase γ, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 21.Garesse R, Kaguni LS. A Drosophila model of mitochondrial DNA replication: Proteins, genes, and regulation. IUBMB Life. 2005;57:555–561. doi: 10.1080/15216540500215572. [DOI] [PubMed] [Google Scholar]
- 22.Nimonkar AV, Boehmer PE. On the mechanism of strand assimilation by the herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) Nucleic Acids Res. 2003;31:5275–5281. doi: 10.1093/nar/gkg740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuven NB, Willcox S, Griffith JD, Weller SK. Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: Potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J Mol Biol. 2004;342:57–71. doi: 10.1016/j.jmb.2004.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Focher F, Mazzarello P, Verri A, Hübscher U, Spadari S. Activity profiles of enzymes that control the uracil incorporation into DNA during neuronal development. Mutat Res. 1990;237:65–73. doi: 10.1016/0921-8734(90)90012-g. [DOI] [PubMed] [Google Scholar]
- 25.Wilson DM, 3rd, McNeill DR. Base excision repair and the central nervous system. Neuroscience. 2007;145:1187–1200. doi: 10.1016/j.neuroscience.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 26.Boehmer PE. Expression, purification, and characterization of the herpes simplex virus type 1 DNA polymerase. Methods Enzymol. 1996;275:16–35. doi: 10.1016/s0076-6879(96)75004-8. [DOI] [PubMed] [Google Scholar]
- 27.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase α and Δ during initiation of leading and lagging strand synthesis. J Biol Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.