Abstract
Profilins are actin binding proteins, which also interact with polyphosphoinositides and proline-rich ligands. On the basis of the genome sequence, three diverse profilin homologues (PFN) are predicted to exist in Caenorhabditis elegans. We show that all three isoforms PFN-1, PFN-2, and PFN-3 are expressed in vivo and biochemical studies indicate they bind actin and influence actin dynamics in a similar manner. In addition, they bind poly(l-proline) and phosphatidylinositol 4,5-bisphosphate micelles. PFN-1 is essential whereas PFN-2 and PFN-3 are nonessential. Immunostainings revealed different expression patterns for the profilin isoforms. In embryos, PFN-1 localizes in the cytoplasm and to the cell–cell contacts at the early stages, and in the nerve ring during later stages. During late embryogenesis, expression of PFN-3 was specifically detected in body wall muscle cells. In adult worms, PFN-1 is expressed in the neurons, the vulva, and the somatic gonad, PFN-2 in the intestinal wall, the spermatheca, and the pharynx, and PFN-3 localizes in a striking dot-like fashion in body wall muscle. Thus the model organism Caenorhabditis elegans expresses three profilin isoforms and is the first invertebrate animal with tissue-specific profilin expression.
Keywords: Profilin, Caenorhabditis elegans, actin cytoskeleton, poly(l-proline), polyphospho-inositides
INTRODUCTION
Profilin is a central protein in actin cytoskeletal dynamics that is abundantly expressed in many organisms. Next to the well-characterized mammalian profilins [Di Nardo et al., 2000; Lambrechts et al., 2000a; Witke, 2004], profilins have been studied in lower eukaryotes such as Saccharomyces cerevisiae [Magdolen et al., 1988; Haarer et al., 1990], Schizosaccharomyces pombe [Balasubramanian et al., 1994], Dictyostelium discoideum [Haugwitz et al., 1991], Acanthamoeba castellanii [Ampe et al., 1985, 1988], in nonvertebrates such as Drosophila [Cooley et al., 1992], in plants [Valenta et al., 1991; Staiger et al., 1993], and in Vaccinia virus [Blasco et al., 1991]. In general, profilins vary considerably in their primary structures, with only few residues conserved among all profilins. In spite of this variation, crystal structures show that the overall fold is well conserved [Thorn et al., 1997; Eads et al., 1998].
Accordingly, biochemical studies indicated that all known profilins bind actin and influence actin polymerization [Carlsson et al., 1977; Schluter et al., 1997; Lu and Pollard, 2001] (and references therein). In vitro experiments suggest a dual activity for profilin with respect to actin dynamics. When barbed ends are free, profilins accelerate actin assembly by desequestering actin monomers from the actin–thymosin β4 pool and presenting them to the free barbed ends. On the other hand, profilins behave as sequestering proteins when barbed ends are capped [Pantaloni and Carlier, 1993; Kang et al., 1999]. In addition, profilins promote in vivo nucleotide exchange on actin monomers [Wolven et al., 2000; Lu and Pollard, 2001].
Furthermore, with the exception of Vaccinia profilin and mouse profilin IIb [Machesky et al., 1994; Di Nardo et al., 2000], all profilins bind proline-rich sequences and interactions with enabled/vasodilator-stimulated phosphoprotein (Ena/VASP) proteins, Wiskott–Aldrich syndrome protein (WASP) family members and formins have been described [Reinhard et al., 1995; Imamura et al., 1997; Watanabe et al., 1997; Suetsugu et al., 1998; Lambrechts et al., 2000b]. The proline binding residues [Mahoney et al., 1997] are strongly conserved in all known profilins [Thorn et al., 1997]. Another group of profilin ligands are the polyphospho-inositides such as phosphatidylinositol 4,5-bisphosphate (PI-4,5-P2) and phosphatidylinositol 3,4,5-trisphosphate (PI-3,4,5-P3) [Lassing and Lindberg, 1985; Machesky et al., 1990; Haugwitz et al., 1991; Haarer et al., 1993; Machesky et al., 1994; Lu et al., 1996].
The role of profilin in actin dynamics and the results from gene disruption studies in different organisms suggest an essential role for profilin in the cell. In mouse, gene disruption of profilin I leads to lethality during early embryogenesis [Witke et al., 2001], probably caused by a cell division defect. In addition, gene disruptions of profilin in Dictyostelium and Schizoaccharomyces result in impaired cytokinesis [Balasubramanian et al., 1994; Haugwitz et al., 1994]. In the latter organism profilin localizes specifically to the medial region of dividing cells, where the contractile ring forms [Balasubramanian et al., 1994]. In organisms expressing more than one isoform, profilins may have different biological functions. Acanthamoeba profilins have different subcellular localizations [Haugwitz et al., 1994; Bubb et al., 1998] and mammals also express several isoforms. In both organisms the profilins have distinct biochemical properties [Machesky et al., 1990; Lambrechts et al., 2000a; Hu et al., 2001] and may have different cellular functions [Da Silva et al., 2003; Neuhoff et al., 2005].
Mining the Caenorhabditis elegans (C. elegans) genome using vertebrate or nonvertebrate profilin sequences yields three profilin (PFN) isoforms. PFN-1 is required for assembly of cortical microfilaments [Severson and Bowerman, 2003] and was shown to be involved in cytokinesis and to genetically interact with the formin Cyk-1 [Severson et al., 2002]. The other two isoforms are hypothetical forms in the C. elegans database (www.wormbase.org). Unlike vertebrate profilin I, IIa, and IIb, which show at least 61% similarity [Lambrechts et al., 2000a], the C. elegans profilins show intermediate to low similarity to each other. Potentially, C. elegans would be the first invertebrate animal that expresses three such diverse profilins. We show that all three isoforms PFN-1, PFN-2, and PFN-3 are expressed in vivo and that they behave as classical nonvertebrate profilins with respect to actin sequestering, influence on actin dynamics and poly(l-proline), and PI-4,5-P2 binding [Lassing and Lindberg, 1985; Lambrechts et al., 2002]. Gene knock-out of PFN-2 and PFN-3 suggest these isoforms are not essential. In vivo localization of these three profilins revealed a diverse expression pattern, suggesting different biological functions.
MATERIALS AND METHODS
cDNA Cloning, Profilin Purification, and Expression
EST clones yk531a6 and yk615f11 for PFN-1 and yk124e8 and yk392b9 for PFN-2 were obtained from Dr. Y. Kohara (National Institute of Genetics, Japan). cDNA for PFN-3 was amplified from a C. elegans cDNA library. We amplified these clones using the following primers with the start and stop codons in bold: CTGAACATGCCATGGCCTCGGATGGAATGCC and CGCGGATCCGCGTTAGTATCCAGCATTGTTG for PFN-1, CTGAACATGCCATGGCCTCTGGCTGGGACGACTAC and CGCGGATCCGCGTCACTTAAGAAAAGAATG for PFN-2, and CTGAACATGCCATGGCCTCGTGGTCTGATATTATC and CGCGGATCCGCGTCAGTACTTGATGGACC for PFN-3. We cloned the cDNAs of PFN-1, PFN-2, and PFN-3 in the NcoI/BamHI sites of the pET11d plasmid. Proteins were expressed in E. coli strain MC1061 harboring pT7POL26 [Mertens et al., 1995]. Cell pellets were collected by centrifugation and washed with buffer A (20 mM Tris-HCl (pH 8.1), 1 mM EDTA, and 5 mM dithiotreitol). Cells were resuspended in buffer A, lysed using a French press, and centrifuged at 32,000 rpm for 1 h. The cleared supernatants was loaded on a poly(l-proline)-CNBr Sepharose column. After washing the column with buffer A, profilin was eluted with increasing concentration of urea (3, 5, and 8 M in buffer A). The profilin-containing fractions were dialyzed in a stepwise fashion in decreasing concentration of urea to refold the protein. The protein was further purified using gel filtration [Lambrechts et al., 1995]. We determined protein concentrations by alkaline hydrolysis and ninhydrine reaction.
Biochemical Methods
Interaction With Actin
α-Actin was purified from rabbit skeletal muscle [Spudich and Watt, 1971] with modifications [Van Troys et al., 1996] and pyrene labeled at Cys 375 using the protocol of Brenner and Korn [Brenner and Korn, 1983]. We determined the equilibrium dissociation constant of the profilins for actin using a sequestration assay with gelsolin capped filaments [Pantaloni and Carlier, 1993]. In experiments with uncapped filaments [Pantaloni and Carlier, 1993], the samples were allowed to reach steady state by overnight incubation at room temperature before the relative fluorescence was measured using a Hitachi F-4500 spectrophotometer (excitation, 365 nm; emission, 388 nm).
For time-course experiments, we incubated 10 μM actin (5% pyrene labeled) in G-buffer (5 mM Tris-HCl [pH 7.7], 0.2 mM ATP, 0.2 mM dithiothreitol, 0.1 mM CaCl2) in the presence of different profilin concentrations indicated in the legend of Fig. 2, for 30 min at room temperature. Then MgCl2 and KCl were added (final concentration of 2 and 100 mM, respectively) to induce actin polymerization, which was measured fluorimetrically as a function of time.
Fig. 2.
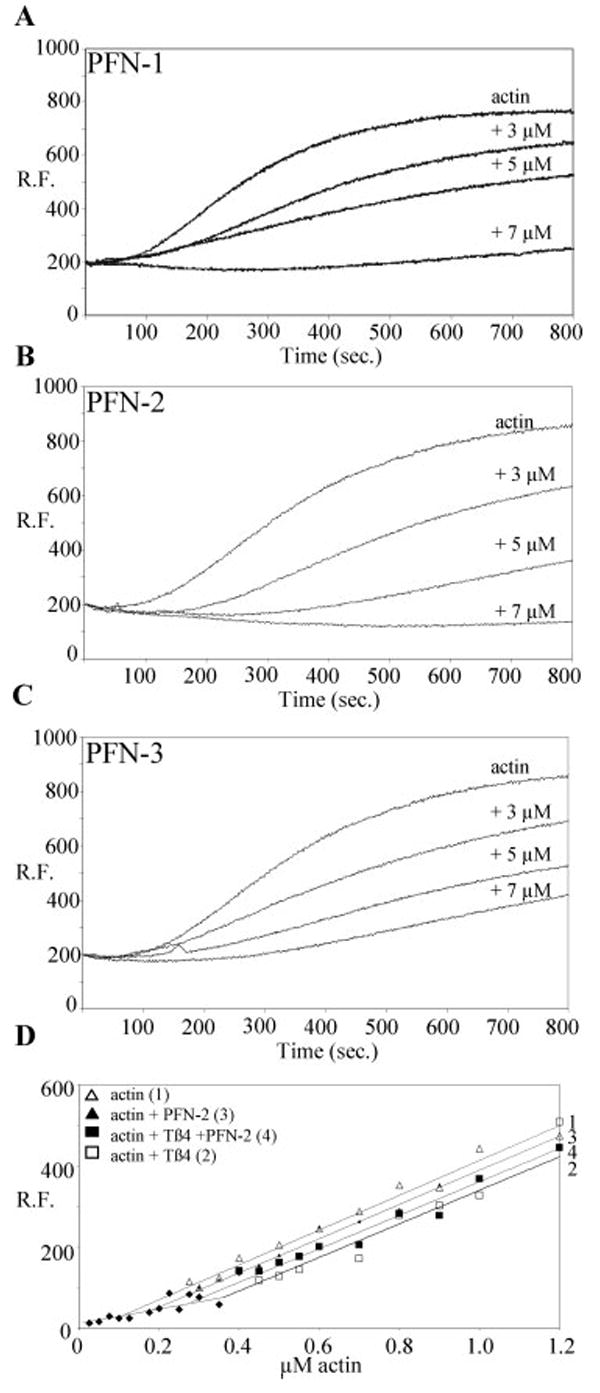
PFN-1, PFN-2, and PFN-3 have characteristic actin binding properties. (A–C) Time-course showing inhibition of actin polymerization activity of C. elegans profilins at the indicated profilin concentrations. The curve labeled actin is the control curve without profilin. The curves show the relative fluorescence (RF), proportional to the amount of F-actin formed, in function of time. (D) PFN-2 promotes actin assembly at uncapped barbed ends in the presence of thymosin β4. Different concentrations of actin filaments alone (curve 1) or incubated with thymosin β4 (5 μM) (curve 2), profilin (5 μM) (curve 3), or thymosin β4 and profilin (curve 4) were allowed to reach steady state before measurements. The symbols represent the actual data points.
PI-4,5-P2 and Poly(l-proline) Binding Assays
For the gel filtration experiment, we incubated profilin (10 μM) for 30 min on ice with different concentrations of PI-4,5-P2 in micelles (see legend Fig. 3A). Next, the samples were separated on a Superdex 75 gel filtration column (Smart, Pharmacia). The apparent Mr of profilin shifts after binding with phospholipid micelles. The height of the peak of free profilin was used to calculate the percentage of bound profilin in each sample [Lambrechts et al., 2002].
Fig. 3.
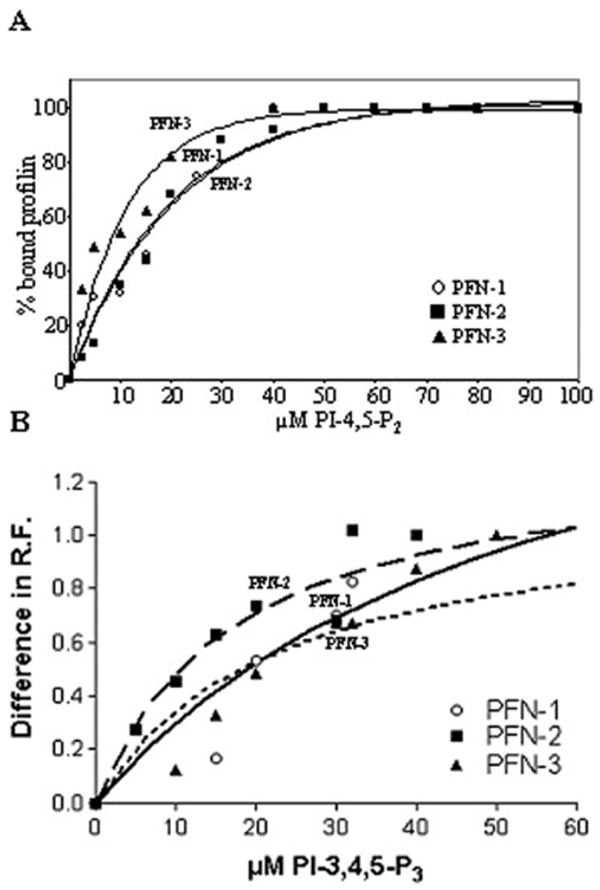
C. elegans profilins bind to poly-phosphoinositides. (A) We examined by gel filtration the interaction between PI-4,5-P2 and PFN-1 (circle), PFN-2 (blocks) or PFN-3 (triangle). The percentage of PI-4,5-P2-bound profilin is plotted vs. PI-4,5-P2 concentration. The curves for PFN-2 and PFN-1 nearly coincide. The concentration of PI-4,5-P2, for which 50% of profilin is bound to PI-4,5-P2 micelles (C50%), is a measure for the affinity and is shown in Table I. The curves, fitted with Graphpad (one phase exponential association, nonlineair regression), had a goodness of fit (R2) of at least 0.96. (B) We examined the interaction between PI-3,4,5-P3 and PFN-1 (circle), PFN-2 (blocks) or PFN-3 (triangle) by measuring the changes in intrinsic relative fluorescence (RF) of Trp in function of total PI-3,4,5-P3 concentration.
Interaction with PI-3,4,5-P3 micelles (Sigma) was monitored with Trp fluorescence. We incubated PFN-1, PFN-2, and PFN-3 (16 μM) for 15 min at room temperature with different concentrations of these micelles and measured changes in intrinsic fluorescence of Trp (excitation, 290 nm; emission, 300–450 nm) using fluorimetry. For each profilin, we determined the wavelength for which the protein alone showed maximal fluorescence and calculated the difference in relative fluorescence for profilin with increasing PI-3,4,5-P3 concentrations at that wavelength (347.6 nm for PFN-1, 334.8 nm for PFN-2, and 338.2 nm for PFN-3).
PI-4,5-P2/poly(l-proline) competition
we preincubated 10 μM profilin with a 6-fold molar excess of PI-4,5-P2 for 30 min on ice and loaded the sample (300 μl) on 250 μl of poly(l-proline) CNBr-Sepharose column equilibrated with buffer A. After washing the column with 500 μl of buffer A, profilins were eluted with 500 μl of 8 M urea in buffer A. In the control experiment, 300 μl of 10 μM profilin without PI-4,5-P2 was loaded on the column, washed with buffer A, and eluted with 8 M urea in buffer A. The presence of profilin in the flow through, in the wash, and in the eluate with 8 M urea was monitored on SDS-PAGE and quantified using density scanning (Total Lab).
Surface plasmon resonance experiment [Jonckheere et al., 1999]
We immobilized chemically synthesized biotinylated Caenorhabditis elegans enabled (Ce-Ena) peptides on an SA streptavidin chip (BIAcore) (for sequences see Table II) and tested binding by sending different concentrations of profilin isoforms over the sensor chip in 0.01 M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% polysorbate. For each concentration, the response values, in arbitrary units, are listed in Table II.
TABLE II.
Interaction of C. elegans Profilins With Ce-Ena Peptides
| PFN-1 | PFN-2 | PFN-3 | Hum prof I | |
|---|---|---|---|---|
| Peptide 1: 218GAPPPPPLPPVGAGAPPPPPPPPPPA243 | ||||
| [max] | 205 | 263 | ND | 337 |
| 65 μM | 118 | 209 | 250 | 159 |
| 50 μM | 98 | 175 | 209 | 125 |
| 25 μM | 53 | 106 | 116 | 76 |
| Peptide 2(A + B): 182SIPHAPPPPVPLTSNIPQAPPAPPPPIG209 | ||||
| [max] | 62 | 148 | ND | 157 |
| 65 μM | 37 | 117 | ND | 79 |
| 50 μM | 27 | 100 | 65 | 66 |
| 25 μM | 19 | 54 | 34 | 32 |
| Peptide 2A: 182 SIPHAPPPPVPLTS195 | ||||
| 50 μM | 5 | 6 | 7 | 7 |
| Peptide 2B: 196 NIPQAPPAPPPPIG209 | ||||
| 50 μM | 9 | 17 | 17 | 17 |
The interaction between the proline-rich peptides and the profilin isoforms is expressed in surface plasmon resonance response units. The peptides are derived from Ce-Ena (UNC-34, Y50D4C.1a) and are numbered according to their position in the protein sequence.
ND indicates not determined, [max] is the maximal concentration used, 92 μM for PFN-2, 126 μM for PFN-1, and 190 μM for human profilin I, respectively.
Antibody Purification, Immunostaining, and Phalloidin Staining
Antibodies against recombinant PFN-1 (no. G316), PFN-2 (no. G322) and PFN-3 (no. G321) were raised in rabbits (Centre d’economie rurale, laboratoire d’hormonologie animale, Marloie). Antibodies were affinity purified and tested by Western blotting and ELISA on purified recombinant profilins. Anti-PFN-2 and anti-PFN-3 antibodies were further adsorbed with acetone-fixed powder of the pfn-2 and pfn-3-null mutant worms respectively to remove nonspecific reactivity as described by Miller and Shakes [Miller and Shakes, 1995].
We prepared worm lysates that were used for immunoblotting, as described previously [Ono and Ono, 2002]. Briefly, C. elegans of mixed developmental stages were suspended in SDS-lysis buffer (2% SDS, 80 mM Tris-HCl, 5% β-mercaptoethanol, 15% glycerol, 0.05% bromophenol blue, pH 6.8) and lysed by sonication and incubation at 97°C for 2 min.
Immunostaining of worm embryos were performed using two methods. For PFN-1 and PFN-2, embryos were obtained by a hypochlorite/NaOH lysis of adults [Epstein et al., 1993], fixed for 15 min at room temperature with 4% formaldehyde in 1× cytoskeleton buffer (10 mM MES-KOH, 138 mM KCl, 3 mM MgCl2, and 2 mM EGTA, pH 6.1) containing 0.32 M sucrose, permeabilized with methanol for 5 min at −20°C, and stained with antibodies diluted (1/250) in PBS containing 1% bovine serum albumin, 0.5% Triton X-100 and 30 mM glycine. For PFN-3, embryos were obtained by cutting gravid adults on poly-lysine-coated slides, freeze-cracked as described [Epstein et al., 1993], fixed with methanol for 5 min at −20°C, and stained with antibodies diluted (1/250) in PBS containing 1% bovine serum albumin. Immunostaining of adult worms was performed as described by Finney and Ruvkun [1990].
In addition to the anti-profilin antibodies, we used the following mouse monoclonal antibodies: anti-vinculin (MH24), anti-α-actinin (MH40) [Francis and Waterston, 1985] (gifts of Dr. Michelle Hresko, Washington Univ., St. Louis, MO), anti-myoA (clone 5.6) [Miller et al., 1983], (gifts of Dr. Henry Epstein, University of Texas Medical Branch, Galveston, TX), and anti-actin (C4; ICN Biomedicals). Secondary antibodies used were Alexa488-labeled goat anti-mouse IgG (Molecular Probes) and Cy3-labeled goat anti-rabbit IgG (Jackson ImmunoResearch Labratories). To stain DNA, 4′6-dia-midino-2-phenylindole (DAPI) (Sigma-Aldrich) was included in the solution of secondary antibodies at 0.1 μg/ml.
Filamentous actin was visualized by staining worms with tetramethylrhodamine-phalloidin (Sigma-Aldrich), as described previously [Ono, 2001].
Promoter-GFP Fusion Analysis
The pfn-1 promoter sequence (2920 bp) was amplified from wild-type N2 genomic DNA by PCR using Y18D10A-A (5′-CGTTTGCAGCTCCGTTTTA-3′) and Y18D10A-B (5′-AGTCGACCTGCAGGCATGCAAGCTTTTGTGTTTTGGAGGAGGTTG-3′). The green fluorescent protein (GFP) coding sequence was amplified from pPD95-67 (provided by Dr. Andrew Fire, Stanford University, Stanford, CA) and fused to the promotor by fusion PCR, as described by Hobert [Hobert, 2002]. The nested primer for the promoter was Y18D10A-A1 (5′-TCCAAGTTTTCTTTCTTTTTCCC-3′). The fusion construct was co-injected with pCeh-361 (a dpy-5 rescuing plasmid as a transgenic marker) into the dpy-5(e907) mutants, and transgenic animals were isolated as described previously [Zhao et al., 2004].
RNA Interference of Worms
Wild-type strain N2 was obtained from Caenorhabditis Genetics Center (Minneapolis, MN). The pfn-2-null allele F35C8.6(ok458)X was isolated and provided by the C. elegans Gene Knockout Project at Oklahoma Medical Research Foundation, which is part of the International C. elegans Gene Knockout Consortium. The pfn-3-null allele K03E6.6(tm1362)X was isolated and provided by Dr. Shohei Mitani’s National Bioresource Project (Tokyo Women’s Medical University, Tokyo, Japan).
The full-length cDNA of PFN-2 and PFN-3 were cloned in the restricted RNA interference vector L4440 (provided by Dr. Andrew Fire, Stanford University, Stanford, CA) between the two T7 promotors. RNA interference (RNAi) was performed by feeding, using the technique described by Timmons [Timmons et al., 2001] under conditions as described previously [Ono and Ono, 2002].
A motility assay was performed as described previously [Epstein and Thomson, 1974]. Briefly, adult worms were placed in M9 buffer (22 mM KH2PO4, 42 mM Na2HPO4, 85.5 mM NaCl, and 1 mM MgSO4). Then, one beat was counted when a worm swung its head to either left or right. The total number of beats in 30 sec was recorded. This monitoring and the counting of the brood size were measured for worms incubated at different temperatures, as indicated in Table III.
TABLE III.
Brood Size and Motility of the Mutated Worms
| Brood Size (average ± SD, n = 5)
|
Motility (beats/30 sec: average ± SD, n ± 10)
|
|||||
|---|---|---|---|---|---|---|
| 15°C | 20°C | 25°C | 15°C | 20°C | 25°C | |
| WT | 327 ± 17 | 249 ± 16 | 166 ± 34 | 112 ± 1.7 | 104 ± 4.9 | 94.5 ± 4.5 |
| pfn-2(null) | 298 ± 20 | 270 ± 49 | 155 ± 23 | 115 ± 6.8 | 111 ± 8.0 | 89.1 ± 7.2 |
| pfn-3(null) | 226 ± 14 | 285 ± 39 | 144 ± 28 | 110 ± 5.6 | 90 ± 3.8 | 84.1 ± 5.4 |
| WT; control RNAi | 275 ± 58 | 301 ± 22 | 166 ± 34 | 100 ± 6.5 | 118 ± 4.9 | 89.3 ± 4.6 |
| WT; pfn-2(RNAi) | 283 ± 21 | 326 ± 23 | 152 ± 18 | 100 ± 3.5 | 103 ± 3.6 | 88.6 ± 5.3 |
| pfn-3(null); control RNAi | 268 ± 36 | 254 ± 35 | 125 ± 14 | 88.0 ± 7.6 | 96.3 ± 5.3 | 87.1 ± 4.4 |
| pfn-3(null); pfn-2(RNAi) | 276 ± 33 | 285 ± 23 | 117 ± 5.4 | 92.0 ± 5.0 | 106 ± 7.4 | 88.8 ± 6.6 |
These measurments were performed at the indicated temperatures. WT: wild-type C. elegans, pfn-2(null): pfn-2 null mutant, pfn-3(null): pfn-3 null mutant, WT; control RNAi: wild-type worm treated with the RNAi vector with no insert; WT; pfn-2(RNAi): RNAi of pfn-2, pfn-3(null); control RNAi: pfn-3 null mutant treated with the RNAi vector with no insert; and pfn-3(null); pfn-2(RNAi): pfn-3 null mutant with RNAi of pfn-2.
RESULTS
C. elegans Expresses Three Profilin Homologues
Based on the genome sequence, three profilin homologues are predicted in the C. elegans database corresponding to clone numbers Y18D10A.20, F35C8.6, and K03E6.61. Y18D10.20 was previously described as pfn-1 [Severson et al., 2002]. We designate the other profilins as pfn-2 and pfn-3, respectively (Suppl. Fig. 1). pfn-1 is located on chromosome I, while pfn-2 and pfn-3 are linked on the left arm of the X chromosome (Suppl. Fig. 3). Interestingly, the number of exons differs among the pfn genes (Suppl. Fig. 3). We note that the gene sequence of pfn-2 in Wormbase contains two, in frame, ATG codons close to each other, resulting in predicted proteins of ~19 and 14 kDa. We think that the second ATG codon (predicted Met-39) preferentially serves as start codon for translation of PFN-2. This is supported by our immunoblot data (see below) and the sequences of the EST clones, since none of them extends to the predicted first start codon, whereas most of them contain the second ATG.
We aligned the amino acid sequences of the C. elegans profilins to each other and to profilins from other species and performed phylogenetic analysis (Suppl. Fig. 2). PFN-1 and PFN-2 are most similar (58%) whereas the similarity between these two forms and PFN-3 (25 and 28%) is not significantly higher than the similarity with profilins from other species. We note that the similarity between profilins from the same species is usually relatively high (between 40 and 84%) and therefore they cluster together in phylogenetic trees. C. elegans PFN-3 seems to form an exception here. Its primary structure clusters with the one from Entamoeba histolytica profilin. The similarity of all three C. elegans profilins was however sufficiently high to construct 3-D models using the crystal structure of Acanthamoeba castellanii profilin IA as a template [Fedorov et al., 1994]. Despite, the low degree of similarity modeling was straightforward, suggesting that these proteins adopt the profilin fold (Suppl. Fig. 4). Comparison of the sequences and of these structures readily indicated that the potential poly(l-proline) binding residues are conserved [Mahoney et al., 1997; Eads et al., 1998]. Information on the identity of actin binding residues is available from the bovine actin–profilin crystal structure [Schutt et al., 1993] and is mapped for various profilins by mutagenesis studies [Lu and Pollard, 2001] (and references therein). There is, however, little conservation between these residues with those in nonvertebrate profilins (see Suppl. Fig. 1). Also information on the identity of nonvertebrate profilin residues involved in PI-4,5-P2-binding is limited, in part because PI-4,5-P2 interaction seems to involve different residues [Lambrechts et al., 2002; Skare et al., 2002] (see also below). In view of this, and the fact that in recent years a number of other proteins with a profilin-like architecture but without profilin-like function have been identified [Tochio et al., 2001; Qian et al., 2005], it is important to show that these three C. elegans proteins have biochemical properties typical of profilins. Therefore, we biochemically characterized each of them.
We expressed PFN-1, PFN-3, and the profilin-like domain of PFN-2 in E. coli and purified them using poly(l-proline) affinity chromatography [Lambrechts et al., 1995]. PFN-1 was eluted in the 3 M urea fraction, whereas PFN-2 and PFN-3 were eluted with 5 M urea. We affinity purified polyclonal antibodies that displayed no cross-reactivity with the other PFN isoforms on Western blot (Fig. 1A), and used these to probe the presence of the isoforms in C. elegans lysates. Figure 1B shows that C. elegans expresses each of the three profilins. The anti-PFN-2 antibody recognized a single band with the same apparent molecular weight as the recombinantly produced form. In addition its size was similar to those of other profilins, suggesting that the longer form of PFN-2 is rare or does not exist.
Fig. 1.
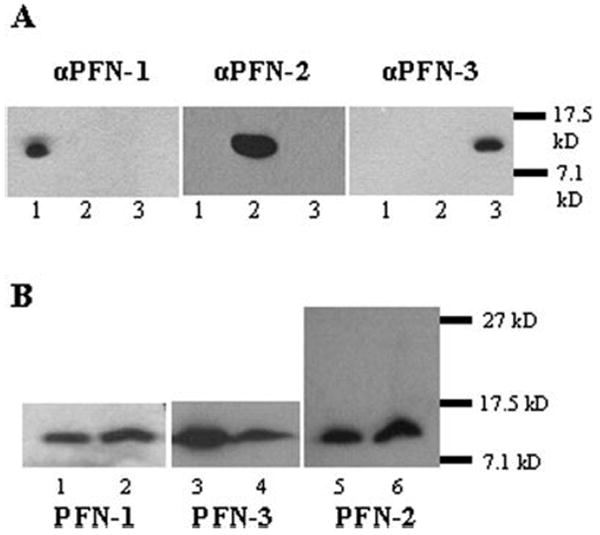
Three profilins are expressed in C. elegans. (A) We purified the PFN antibodies using affinity chromatography and tested their specificity by Western blotting on recombinant PFNs. Blot anti-PFN-1 and anti-PFN-3: 0.25 μg PFN-1, 2, and 3; blot anti-PFN-2: 0.1 μg PFN-1, 2, and 3. (1) PFN-1, (2) PFN-2, (3) PFN-3. The antibodies show no cross-reactivity at the dilution used (1/10 000). (B) We used these purified polyclonal antibodies and Western blotting to probe profilin expression in C. elegans. Note that for PFN-2 the observed molecular weight matches the molecular weight of the recombinant protein representing the profilin only domain, and we could not detect a larger profilin-like protein (translation starting at a prior start codon) in the worm lysate. (1) 100 ng recombinant PFN-1, (2) worm lysates, 50 μg total protein, (3) 200 ng recombinant PFN-3 and (4) worm lysates, 240 μg total protein, (5) 20 ng recombinant PFN-2, (6) worm lysates, 25 μg total protein.
The Three Profilin Homologues Bind Actin and Influence Actin Polymerization in a Similar Manner
To assess actin binding capacity of the profilin homologues, we first probed their influence on salt induced actin polymerization using fluorimetry (Figs. 2A–2C). Actin monomers were incubated without or with profilin at different concentrations. For each of the profilins, we observed an increased lag phase with increasing concentration of profilin, suggesting inhibition of nucleation. In all cases, the amount of F-actin formed decreased with increasing profilin concentration, indicative of sequestering activity.
In addition, we determined the equilibrium dissociation constant for the actin–profilin interaction using an actin sequestration assay with capped filaments [Pantaloni and Carlier, 1993]. Although this is an indirect assay that may underestimate equilibrium dissociation (Kd)-values [Vinson et al., 1998] it shows that the C. elegans profilins have comparable micromolar affinities for G-actin (Table I). We also investigated the effect of the profilins on actin polymerization in the presence of thymosin β4, when barbed ends are free. Similar results were obtained for the three profilin isoforms (see Fig. 2D, for PFN-2 and data not shown). Thymosin β4 and the tested profilins exhibited nonadditive sequestering effects, and the concentration of unpolymerized actin was lower in the presence of both profilin and thymosin β4 (Fig. 2D, curve 4) than that in the presence of thymosin β4 alone (Fig. 2D, curve 2). These results suggest all three C. elegans profilins are not simple actin-sequestering agents, but like other profilins can add actin–profilin complexes to free fast growing ends [Pantaloni and Carlier, 1993; Kang et al., 1999].
TABLE I.
Summary of Binding Data for PFN-1, PFN-2, and PFN-3
| Kd (μM) | C50% (μM) | [Urea] (M) | |
|---|---|---|---|
| PFN-1 | 2 | 14 | 3 |
| PFN-2 | 2.4 | 15 | 5 |
| PFN-3 | 3.4 | 7 | 5 |
| Hum prof I | 0.4a | 44a | 8b |
| Rat prof IIa | 0.4a | 155a | 8b |
| Yeast prof | 2.9c | ND | ND |
Kd indicates the equilibrium dissociation constant for actin; C50%, the concentration of PI-4,5-P2 for which 50% of profilin is bound to PI-4,5-P2 micelles; and [Urea], the concentration of urea required for eluting profilin from the poly(l-proline) sepharose column during the purification procedure. ND indicates that these data are not available.
The C. elegans Profilin Homologues Interact with Medium Affinity to Proline-Rich Peptides
The profilin purification method already revealed the poly(l-proline) binding capacity of the three C. elegans profilins (Table I). To investigate this property further, we used surface plasmon resonance with various chemically synthesized biotinylated proline-rich peptides derived from a potential C. elegans profilin interaction partner: Ce-Ena (UNC-34, (uncoordinated), Y50D4C.1) [Yu et al., 2002] on streptavidin sensor chips. All three profilins interact with medium affinity, with the two longer proline-rich peptides (peptide 1 and peptide 2(A + B)) (Table II). This is comparable to the binding affinity of human profilin I for these peptides. We consistently observed that PFN-1 displayed the lowest binding capacity to these peptides, in agreement with the lower urea concentration needed in the purification protocol. Additionally, the response unit (RU)-values for the shorter peptides 2A and 2B, representing two halves of peptide 2(A + B), were very low and the sum of the RU’s for peptide 2A and peptide 2B was significantly lower than the RU-value measured for binding to peptide 2(A + B). This suggests some co-operativity in binding of C. elegans profilins to the longer peptide 2(A + B). In neither case, the maximum levels of theoretical response (RUmax) were reached, and so we were unable to determine the stoichiometry of profilin-peptide complexes.
The C. elegans Profilin Homologues Interact with PI-4,5-P2
Next, we studied PI-4,5-P2 binding in a gel filtration assay, using a constant amount of each isoform and series of PI-4,5-P2-concentrations (Fig. 3A). In all the cases, the amount of bound profilin increased with increasing PI-4,5-P2-concentration. We calculated that 50% of PFN-1, PFN-2, or PFN-3 is bound to PI-4,5-P2-micelles in the presence of 14, 15, and 7 μM PI-4,5-P2, respectively (Table I), suggesting PFN-3 has a slightly higher affinity for PI-4,5-P2-micelles. In addition, we studied the interaction with PI-3,4,5-P3 using changes in intrinsic Trp-fluorescence (Fig. 3B). Each of the PFN isoforms interact with PI-3,4,5-P3 micelles.
Mutually exclusive binding between poly(l-proline) and PI-4,5-P2 has been described for mammalian profilins using affinity chromatography [Lambrechts et al., 1997]. Here we perform a similar assay. In a control experiment, PFN-1, PFN-2, and PFN-3 without PI-4,5-P2 were loaded on a poly(l-proline) column, and after washing eluted with 8 M urea (Fig. 4). Each of the profilins was fully recovered in the urea eluate. We next preincubated each of the profilins with PI-4,5-P2, choosing a condition where most of the protein molecules are associated with the micelles (i.e. a 6-fold molar excess of PI-4,5-P2 based on the experiment in Fig. 3A), and loaded the sample onto the poly(l-proline) column. The profilins are now recovered in the flow-through and wash fractions. The amount eluted with 8 M urea is significantly reduced compared with that with the control (Fig. 4). These data indicate that binding of PFN-1, PFN-2, or PFN-3 to PI-4,5-P2 (micelles) and poly(l-proline) is mutually exclusive.
Fig. 4.
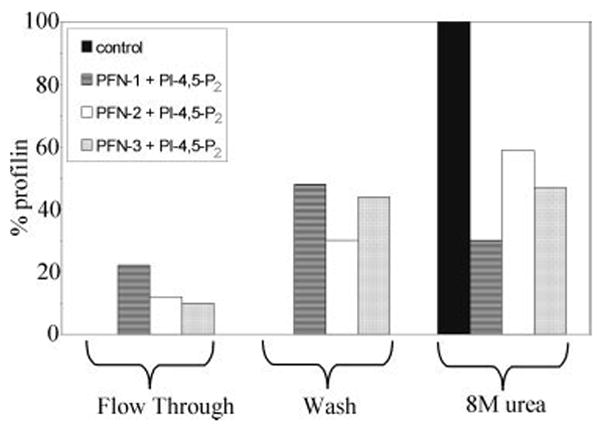
PI-4,5-P2 and poly(l-proline) compete for profilin binding. In the control experiment, we used profilin without PI-4,5-P2 and observed for each isoform no profilin in the flow-trough or in the wash fraction. The total amount of profilin, eluted with 8 M urea, was set at 100%. We incubated PFN-1, PFN-2, or PFN-3 with micellar PI-4,5-P2 and passed the sample over a poly(l-proline) column. PFN-1, PFN-2, and PFN-3 were partly recovered in the flow-through and the wash fraction.
PFN-2 and PFN-3 are not Essential for Viability of C. elegans
PFN-1 plays an essential role in the cell, since RNAi of this isoform results in a strong cytokinesis defect (“Cyk” phenotype) [Severson et al., 2002] (Wormbase: [Kamath et al., 2003]). To examine the biological functions of PFN-2 and PFN-3, we characterized null and RNAi phenotypes of pfn-2 and pfn-3. pfn-2(ok458) has a deletion of 2.2 kb that removes all the exons (Suppl. Fig. 3). A deletion in pfn-3(tm1362) spans 0.8 kb that eliminates part of the promoter region, exon 1, and intron 1 (Suppl. Fig. 3). Homozygous pfn-2(ok458) and pfn-3(tm1362) animals lack the PFN-2 and PFN-3 proteins, respectively, without significantly altering the levels of the other isoforms (Fig. 5A, lanes 2 and 3), and are considered null alleles for each gene. We found that RNAi of pfn-2 effectively knocked down PFN-2 (Fig. 5A, lane 5), while RNAi of pfn-3 did not affect the PFN-3 level (data not shown). We additionally performed pfn-2(RNAi) on the pfn-3(null) background to eliminate two PFN isofoms (Fig. 5A, lane 7). All these mutant worms with or without the RNAi treatments are viable and showed no apparent phenotypes under a dissecting microscope. However, quantitative determination of worm motility and brood size at 15, 20, or 25°C (Table III) revealed that brood size was reduced in the pfn-3(null) worms at 15°C and the pfn-3(null) worms with control RNAi or pfn-2(RNAi) at 25°C and that worm motility was slightly slower in the pfn-3(null) worms at 20 or 25°C. No alteration in the F-actin organization in body wall muscle (Fig. 5B), pharynx, intestine, vulva, gonad, and embryos (data not shown) was detected in these worms at three different temperatures except for the pfn-3(null) worms treated with pfn-2(RNAi) that had subtly altered muscle structure with wider actin striations (Fig. 5Bg). This phenotype was observed in 10–20% of the worms and may explain why no major defects in motility were detected (Table III). In summary, silencing of PFN-1 [Severson et al., 2002] and our results suggest that C. elegans expresses one essential (PFN-1) and two nonessential profilins, PFN-2 and PFN-3.
Fig. 5.
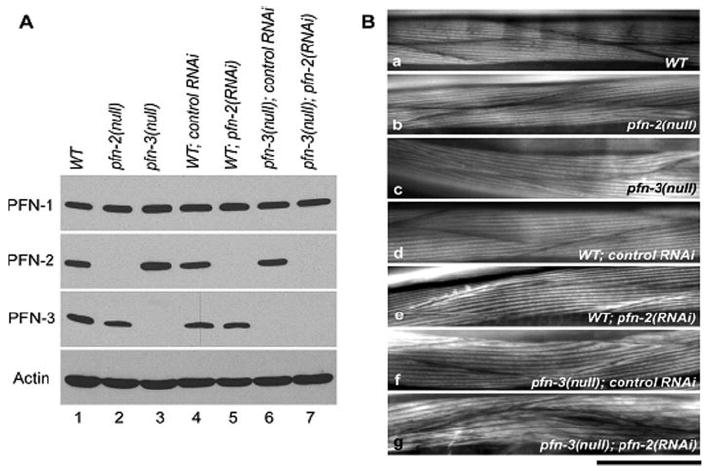
PFN-2 and PFN-3 are not essential for viability of C. elegans. (A) Effects of gene knockout and RNAi treatments on the levels of the profilin proteins. Total worm lysates (20 μg protein) of wild-type (lane 1), pfn-2(null) (lane 2), and pfn-3(null) (lane 3) mutants under standard culture conditions, and wild-type after control RNAi (lane 4) or pfn-2 (RNAi) (lane 5), and pfn-3(null) after control RNAi (lane 6) or pfn-2 (RNAi) (lane 7) were examined by Western blot with antibodies against PFN-1, PFN-2, PFN3, or actin. PFN-2 was not detectable in the pfn-2(null) mutant and pfn-2 (RNAi) worms (lanes 2, 5, and 7), and PFN-3 was absent in the pfn-3(null) mutant (lanes 3, 6, and 7). The levels of PFN-1 and actin were not different among these worms. (B) Organization of the muscle structures in mutant or RNAi-treated worms. Striations of actin in the muscle structures are compared after phalloidin staining of the different worms grown at 20°C. WT: wild-type C. elegans, pfn-2(null): pfn-2 null mutant, pfn-3(null): pfn-3 null mutant, WT; control RNAi: wild-type worm transfected with RNAi vector, WT; pfn-2(RNAi): RNAi of pfn-2; pfn-3(null), control RNAi: pfn-3 null mutant transfected with RNAi vector, and pfn-3(null); pfn-2(RNAi): pfn-3 null mutant with RNAi of pfn-2. Pfn-3 null mutants have wider actin bundles and in combination with RNAi of pfn-2 irregularities in actin striations.
Developmental and Tissue-Specific Expression of the Profilin Isoforms
We probed expression of profilins using the affinity-purified isoform-specific antibodies (see Fig. 1A). Although anti-PFN-2 and anti-PFN-3 antibodies showed specific reactivity for respective isoform on Western blot of total C. elegans lysates, they had some reactivity to null mutants in immunofluorescent staining. Therefore, these antibodies were additionally adsorbed to acetone fixed powders of pfn-2 and pfn-3 null mutant worms respectively to ascertain the specificity of these antibodies. These treatments significantly reduced nonspecific staining of the respective null mutant (Figs. 6 and 7, and data not shown).
Fig. 6.
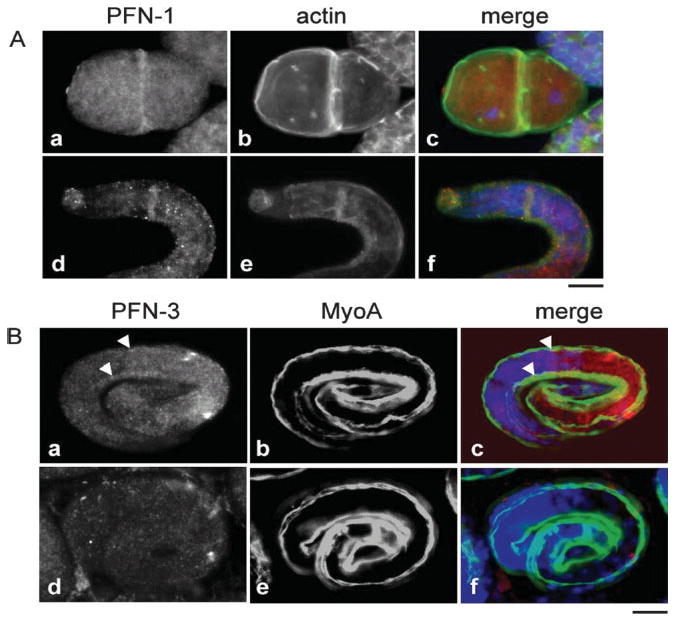
PFN-1 and PFN-3 are expressed at different stages during C. elegans embryonic development. (A) PFN-1: at the two-cell stage PFN-1 (a–c) is expressed diffusely throughout the cytoplasm and is enriched in the cell–cell contacts (a), where it colocalizes with actin (b and c). In contrast to PFN-1, actin is also highly enriched in the membrane region of the cells. In the larval stage (d–f), PFN-1 is expressed in the nerve ring (d) where it also colocalizes with actin (e and f). In the merged pictures (c and f), profilin is red, actin is green, and DAPI stain is blue. (B) PFN-3 expression in body wall muscle starts during embryogenesis, persists through embryonic development and is shown here at the 3-fold stage (a–c). MyoA ((b and e), green in merged pictures (c and f)), a myosin heavy chain, is a marker for the body wall muscle of C. elegans (scale bar, 10 μm). Fig. d–e illustrates the absence of PFN-3 expression (d) in the body wall muscle in pfn-3 null mutants.
Fig. 7.
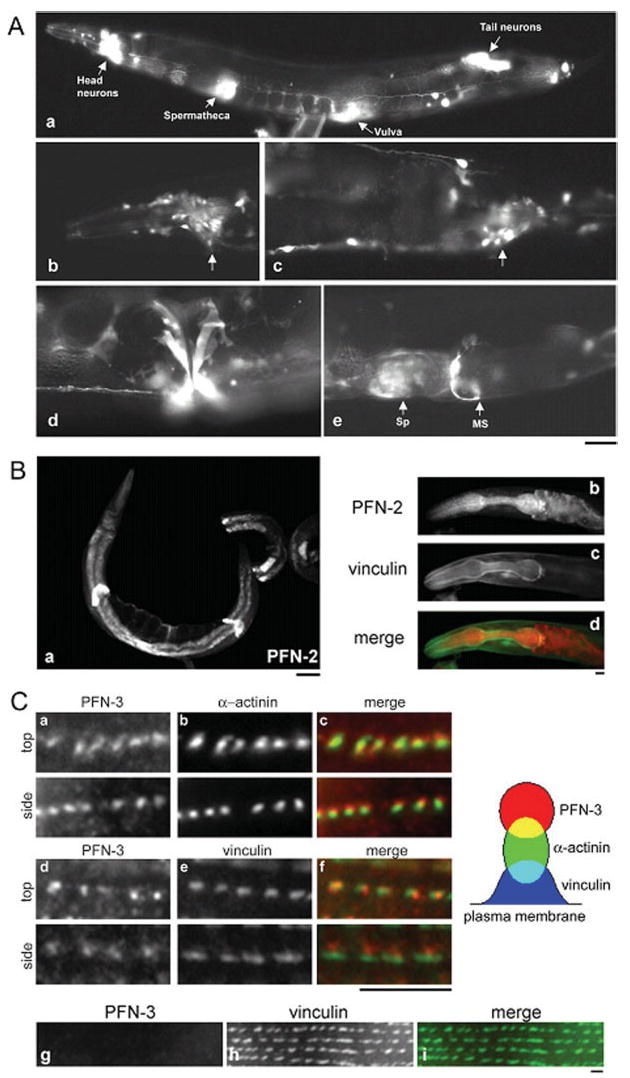
The three C. elegans profilins have different expression patterns in adults. (A) Expression pattern of GFP driven by the pfn-1 promoter. (a) GFP expression was detected in the neurons, spermatheca, vulva, and myoepithelial sheath of the proximal ovary (not shown). (b–e) Micrographs with a higher magnification of neurons (arrows) in the head (b) and the tail (c), vulva (d), and spermatheca (Sp) and myoepithelial sheath (MS) (e). Bar, 40 μm for (a) and 20 μm for (b–e). (B) PFN-2 staining is detected in the intestine, the spermatheca (a), and in the pharynx (b). Vinculin ((c), green in merged picture (d)) was used as a marker to outline the pharynx. (C) PFN-3 (a and d) is found in the body wall muscle. The dot-like structures of PFN-3 partially colocalize with α-actinin ((b), green in merged picture (c)) and localize adjacent to vinculin ((e), green in merged picture (f)). Dense bodies were viewed vertically to the plasma membrane (top) or laterally (side). Schematic representation of relative locations of PFN-3, α-actinin, and vinculin is shown on the right. This dot-like structure is no longer observed in pfn-3 null mutants (g) (scale bars: Fig. 7A, 50 μm; Figs. 7B and 7Cg–Ci, 10 μm; Figs. 7Ca–7Cf, 5 μm). In the merged pictures, PFN is shown in red (Figs. 7Bd, 7Cc, and 7Cf).
In embryos, PFN-1 and PFN-3, but not PFN-2, were detected by immunostaining, and they were expressed in different patterns (Fig. 6). Consistent with a previous report [Severson et al., 2002], we detected expression of PFN-1 from early embryonic stages (Fig. 6A). At the two-cell-stage embryo, PFN-1 showed diffuse staining in the cytoplasm and localized to cell–cell contacts where it colocalized with actin (Fig. 6A). During the late embryonic stage and in the L1 larva, PFN-1 was found in the nerve ring (Fig. 6A). By contrast, staining for PFN-3 was negative in early embryos but detected specifically in body wall muscle cells from the 1.5-fold stage (~350 min after the first cell division) [Epstein et al., 1993] and persisted through embryogenesis (Fig. 6B, upper panel). Note the absence of staining in the pfn-3 null mutants and that, in these embryos, organization of a muscle-specific myosin heavy chain MyoA appears normal (Fig. 6B, lower panel).
All three isoforms are expressed in adult worms. Immunostaining of adult worms with anti-PFN-1 antibody showed diffuse staining throughout the body (data not shown). However, we do not have a pfn-1-null mutant to test the specificity of the antibody, and there are not many alternative protocols for whole-mount staining of adult worms to optimize the staining conditions. Therefore, we additionally tested the activity of the promoter region of the pfn-1 gene using GFP as a reporter (Fig. 7A). The pfn-1 promoter activity was found in many neurons (Figs. 7Aa–7Ac), vulva (Fig. 7Ad), spermatheca and myoepithelial sheath of the proximal ovary (Fig. 7Ae). Immunofluorescent staining of PFN-2 was strong in the intestinal wall, the spermatheca, and the pharynx (Fig. 7B). The pfn-2 promoter-GFP reporter assay confirmed its expression in the intestine (Johnsen et al., unpublished data). In the pharynx, PFN-2 was diffusely localized to the cytoplasm of the pharyngeal cells but did not overlap with vinculin staining, which is generally known to concentrate beneath the plasma membrane. Immunostaining showed PFN-3 was specifically expressed in body wall muscle and localized in a striking dot-like fashion in the dense bodies (Fig. 7C). When the dense bodies were viewed vertically to the plasma membrane (Fig. 7Ca–f, top), PFN-3 colocalized with α-actinin and vinculin. However, when they were observed laterally (Fig. 7Ca–f, side), PFN-3 partially colocalized with α-actinin and localized adjacent to vinculin. Since vinculin is closer to the plasma membrane than that of α-actinin in the dense bodies [Francis and Waterston, 1985], PFN-3 is located at the cytoplasmic tips of the dense bodies (Fig. 7C, schematic representation on the right). As expected, these dot-like staining patterns of PFN-3 were absent in the pfn-3 null mutants (Fig. 7C). Interestingly, the vinculin localization was slightly altered in the mutants, and some neighboring vinculin spots appeared merged (Fig. 7C, lower panels). The pfn-3 promoter∷GFP fusion construct yielded no expression (data not shown), suggesting that an additional enhancer element may be required for its expression or an inhibitory cis-element may be contained in the construct. The patterns of expression and localization of PFN-1 and PFN-3 in the pfn-2 and of PFN-1 and PFN-2 in the pfn-3 null mutants were indistinguishable from those in wild type (data not shown), suggesting that the functions of the three profilins are largely independent of each other. Thus immunolocalization of the profilin proteins revealed diverse expression patterns for the three C. elegans profilin isoforms, implicating different biological functions for the three profilins.
DISCUSSION
In this study, we demonstrate that the three C. elegans profilins PFN-1, PFN-2, and PFN-3 are expressed in vivo and that in spite of the high sequence diversity they are bona fide profilin isoforms. They have conventional profilin-like activities with regard to actin binding and polymerization and they interact with PI-4,5-P2 micelles and proline-rich peptides.
Poly(l-proline) binding of PFN-1, PFN-2, and PFN-3 is consistent with the conserved nature of the residues known to interact with poly(l-proline) [Mahoney et al., 1997] and also with our structural models in which these side chains are positioned at equivalent positions (supplemental data). Severson et al. [2002] demonstrated an yeast two-hybrid interaction between PFN-1 and the proline-rich formin homology 1 (FH1) region of Cyk-1. We extend this observation in two ways. We demonstrate binding to proline-rich peptides derived from C. elegans Ena and that all three isoforms have comparable affinities for proline-rich sequences. This contrasts the situation in mammals, where a 100-fold difference in poly(l-proline) binding affinity between profilins I and IIa is observed [Lambrechts et al., 2000a]. Note that, despite the moderate affinity, our results suggest cooperativity in binding to the proline-rich peptides. Such co-operativity has not been described yet for invertebrate profilins but is similar to observations for mammalian profilin IIa [Jonckheere et al., 1999; Lambrechts et al., 2000a].
Like other profilins, the three C. elegans profilins interact with micelles of PI-4,5-P2 or PI-3,4,5-P3 [Machesky et al., 1994; Lu et al., 1996; Lambrechts et al., 1997], with PFN-3 displaying a slightly higher affinity. In addition we show, for the first time, mutually exclusive interaction of invertebrate profilins with PI-4,5-P2 and poly(l-proline).
C. elegans profilins display diverse expression patterns during embryogenesis and in adults. Such an isoform-specific expression for actin binding proteins in C. elegans is not unprecedented. UNC-60A and UNC-60B are actin depolymerizing factor/cofilin isoforms, of which the latter is expressed in body wall muscle, vulva, and spermatheca while the former is ubiquitously expressed [Ono et al., 2003]. The tissue-specific expression suggests the isoforms are involved in different biological functions. PFN-1 is required for embryonic cytokinesis [Severson et al., 2002] consistent with its localization at the cleavage furrow (Fig. 6A). A role for profilins in cytokinesis may be common in protozoans, yeast, and animals. In Tetrahymena, profilin was also observed at the cleavage furrow [Edamatsu et al., 1992] and in Schizosaccharomyces it was found in the medial region of the cell, where the F-actin contractile ring forms [Balasubramanian et al., 1994; Pelham and Chang, 2002]. Gene disruption of profilin in Dictyostelium, Saccharomyces, Drosophila, and mouse also results in impaired cell division [Haarer et al., 1990; Cooley et al., 1992; Haugwitz et al., 1994; Witke et al., 2001], indicating certain profilin isoforms are essential in this process. Severson et al. [2002] observed two phenotypes upon RNAi inhibition of PFN-1 in embryos. Polarization of the anterior–posterior axis is disrupted and the cells have aberrant furrow ingression. Both processes are microfilament dependent and on the basis of our biochemical data it is tempting to speculate that the promotive effect of PFN-1 on elongation of actin is crucial in forming these filaments. Additionally the polyproline binding capacity of PFN-1 may be important in cytokinesis. Two proteins with such sequences have been shown to be involved in cytokinesis CYK-1 and WSP-1 [Hu et al., 2001; Withee et al., 2004]. However, the fact that in the CYK-1 mutants, used by Severson [Severson et al., 2002], the proline-rich region was still present suggests that the combined actin binding activity of PFN-1 and CYK-1 may prevail in this process [Kovar et al., 2003; Li and Higgs, 2003].
Our experiments indicate that the three C. elegans profilins have similar biochemical properties and one would therefore expect that PFN-2 and PFN-3 should be able to rescue the lethal phenotype. Since we were unable to demonstrate the presence of the two latter isoforms in early embryos, this absence, or the too low expression levels, may simply explain the lack of rescue of silencing PFN-1. Intriguingly, another protein, tetrathymosin β, with profilin-like activity is expressed in the dividing zygote [Van Troys et al., 2004] but neither is it able to overcome the lethal effects of PFN-1 silencing.
Our RNAi experiments and the results from the null mutants suggest that PFN-2 and PFN-3 are not essential. The absence of visible phenotypes for the pfn-2 null mutant may result from redundancy with PFN-1. Indeed PFN-1, which appears diffusely expressed in most of the nonmuscle tissues of the adult worm, may compensate for the function of PFN-2. Alternatively, PFN-2 may have a very specific but nonessential function. PFN-3 has a striking body wall muscle specific staining pattern, which cannot be found for the other isoforms. The disorganization of the muscle actin structure, especially visible in PFN-2 silenced pfn-3 null mutant (Fig. 5B), is possibly linked to altered vinculin organization (Fig. 7C), since vinculin is the major component of the dense bodies [Barstead and Waterston, 1989] that are believed to be the anchoring point of the actin barbed ends. However, these defects do not appear to influence motility to a great extent.
The possible different biological functions may involve interactions with specific ligands. Although our results did not show striking differences in these interactions, we suspect that the profilin isoforms have preferential binding partners but that specificity is in part governed by tissue specific expression. This is similar to the situation in mammals where next to general profilin I expression, certain tissues have one additional isoform for instance, profilin IIa appears to be a neuronal form [Di Nardo et al., 2000; Lambrechts et al., 2000a]. As mentioned above the interaction between CYK-1 and PFN-1 was described [Severson et al., 2002]. It will be interesting to investigate whether CYK-1 also interacts with PFN-2 and PFN-3. Other potential binding proteins with long proline-rich sequences, such as formins, Ce-Ena and WASP-family members exist, or, are predicted to exist in C. elegans. A recent paper showed that UNC-34 and WSP-1 or UNC-34 and WVE-1 play a role in morphogenesis [Withee et al., 2004]. These proteins may be likely partner proteins for PFN-1 at this stage of development. Characterization of their interaction with profilin and their colocalization in cells will shed light on the cellular function and regulation of profilins.
Supplementary Material
The supplemental materials described in this article can be found at http://www.interscience.wiley.com/jpages/0886-1544/suppmat
Acknowledgments
We thank Marc Goethals, Daisy Dewitte, and Veronique Jonckheere for technical assistance. Robert Johnsen for help in the pfn-1∷gfp analysis and Dr. Ann Rose (University of British Columbia, Canada) for providing dpy-5 rescuing construct. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institute of Health Center for Research Resources. A. L. is a recipient of a postdoctoral fellowship of the Fund For Scientific Research Flanders (FWO). D. L. B. is a Canadian Research Chair.
Contract grant sponsor: Geneeskundige Stichting Koningin Elisabeth; Contract grant sponsor: FWO; Contract grant number: G.0007.03; Contract grant sponsor: Concerted Research Actions of the Flemish Community; Contract grant number: GOA 12051401; Contract grant sponsor: National Sciences and Engineering Research Council of Canada; Contract grant sponsor: Genome BC/Canada; Contract grant sponsor: National Institute of Health; Contract grant number: R01 AR48615.
Footnotes
Published online 29 November 2005 in Wiley InterScience (www.interscience.wiley.com).
References
- Ampe C, Vandekerckhove J, Brenner SL, Tobacman L, Korn ED. The amino acid sequence of Acanthamoeba profilin. J Biol Chem. 1985;260(2):834–840. [PubMed] [Google Scholar]
- Ampe C, Sato M, Pollard TD, Vandekerckhove J. The primary structure of the basic isoform of Acanthamoeba profilin. Eur J Biochem. 1988;170(3):597–601. doi: 10.1111/j.1432-1033.1988.tb13739.x. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125(6):1289–1301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989;264(17):10177–10185. [PubMed] [Google Scholar]
- Blasco R, Cole NB, Moss B. Sequence analysis, expression, and deletion of a vaccinia virus gene encoding a homolog of profilin, a eukaryotic actin-binding protein. J Virol. 1991;65(9):4598–4608. doi: 10.1128/jvi.65.9.4598-4608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner SL, Korn ED. On the mechanism of actin monomer-polymer subunit exchange at steady state. J Biol Chem. 1983;258(8):5013–5020. [PubMed] [Google Scholar]
- Bubb MR, Baines IC, Korn ED. Localization of actobindin, profilin I, profilin II, and phosphatidylinositol-4,5-bisphosphate (PIP2) in Acanthamoeba castellanii. Cell Motil Cytoskeleton. 1998;39(2):134–146. doi: 10.1002/(SICI)1097-0169(1998)39:2<134::AID-CM4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Carlsson L, Nystrom LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115(3):465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Cooley L, Verheyen E, Ayers K. Chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell. 1992;69(1):173–184. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- Da Silva JS, Medina M, Zuliani C, Di Nardo A, Witke W, Dotti CG. RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J Cell Biol. 2003;162(7):1267–1279. doi: 10.1083/jcb.200304021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A, Gareus R, Kwiatkowski D, Witke W. Alternative splicing of the mouse profilin II gene generates functionally different profilin isoforms. J Cell Sci. 2000;113(Pt 21):3795–3803. doi: 10.1242/jcs.113.21.3795. [DOI] [PubMed] [Google Scholar]
- Eads JC, Mahoney NM, Vorobiev S, Bresnick AR, Wen KK, Rubenstein PA, Haarer BK, Almo SC. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37(32):11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- Edamatsu M, Hirono M, Watanabe Y. Tetrahymena profilin is localized in the division furrow. J Biochem (Tokyo) 1992;112(5):637–642. doi: 10.1093/oxfordjournals.jbchem.a123952. [DOI] [PubMed] [Google Scholar]
- Epstein HF, Thomson JN. Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature. 1974;250(467):579–580. doi: 10.1038/250579a0. [DOI] [PubMed] [Google Scholar]
- Epstein HF, Casey DL, Ortiz I. Myosin and paramyosin of Caenorhabditis elegans embryos assemble into nascent structures distinct from thick filaments and multi-filament assemblages. J Cell Biol. 1993;122(4):845–858. doi: 10.1083/jcb.122.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov AA, Magnus KA, Graupe MH, Lattman EE, Pollard TD, Almo SC. X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates. Proc Natl Acad Sci USA. 1994;91(18):8636–8640. doi: 10.1073/pnas.91.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63(5):895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101(4):1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer BK, Lillie SH, Adams AE, Magdolen V, Bandlow W, Brown SS. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J Cell Biol. 1990;110(1):105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer BK, Petzold AS, Brown SS. Mutational analysis of yeast profilin. Mol Cell Biol. 1993;13(12):7864–7873. doi: 10.1128/mcb.13.12.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugwitz M, Noegel AA, Rieger D, Lottspeich F, Schleicher M. Dictyostelium discoideum contains two profilin isoforms that differ in structure and function. J Cell Sci. 1991;100(Pt 3):481–489. doi: 10.1242/jcs.100.3.481. [DOI] [PubMed] [Google Scholar]
- Haugwitz M, Noegel AA, Karakesisoglou J, Schleicher M. Dictyostelium amoebae that lack G-actin-sequestering profilins show defects in F-actin content, cytokinesis, and development. Cell. 1994;79(2):303–314. doi: 10.1016/0092-8674(94)90199-6. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32(4):728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hu E, Chen Z, Fredrickson T, Zhu Y. Molecular cloning and characterization of profilin-3: a novel cytoskeleton-associated gene expressed in rat kidney and testes. Exp Nephrol. 2001;9(4):265–274. doi: 10.1159/000052621. [DOI] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16(10):2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere V, Lambrechts A, Vandekerckhove J, Ampe C. Dimerization of profilin II upon binding the (GP5)3 peptide from VASP overcomes the inhibition of actin nucleation by profilin II and thymosin beta4. FEBS Lett. 1999;447(2–3):257–263. doi: 10.1016/s0014-5793(99)00293-8. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kang F, Purich DL, Southwick FS. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J Biol Chem. 1999;274(52):36963–36972. doi: 10.1074/jbc.274.52.36963. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161(5):875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts A, Van Damme J, Goethals M, Vandekerckhove J, Ampe C. Purification and characterization of bovine profilin II. Actin, poly(l-proline) and inositolphospholipid binding. Eur J Biochem. 1995;230(1):281–286. [PubMed] [Google Scholar]
- Lambrechts A, Verschelde JL, Jonckheere V, Goethals M, Vandekerckhove J, Ampe C. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 1997;16(3):484–494. doi: 10.1093/emboj/16.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts A, Braun A, Jonckheere V, Aszodi A, Lanier LM, Robbens J, Van Colen I, Vandekerckhove J, Fassler R, Ampe C. Profilin II is alternatively spliced, resulting in profilin isoforms that are differentially expressed and have distinct biochemical properties. Mol Cell Biol. 2000a;20(21):8209–8219. doi: 10.1128/mcb.20.21.8209-8219.2000. In Process Citation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts A, Kwiatkowski AV, Lanier LM, Bear JE, Vandekerckhove J, Ampe C, Gertler FB. cAMP-dependent protein kinase phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with Actin and SH3 domains. J Biol Chem. 2000b;275(46):36143–36151. doi: 10.1074/jbc.M006274200. [DOI] [PubMed] [Google Scholar]
- Lambrechts A, Jonckheere V, Dewitte D, Vandekerckhove J, Ampe C. Mutational analysis of human profilin I reveals a second PI(4,5)-P2 binding site neighbouring the poly(l-proline) binding site. BMC Biochem. 2002;3(1):12. doi: 10.1186/1471-2091-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature. 1985;314(6010):472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- Li F, Higgs HN. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13(15):1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Lu J, Pollard TD. Profilin binding to poly-l-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12(4):1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Shieh WR, Rhee SG, Yin HL, Chen CS. Lipid products of phosphoinositide 3-kinase bind human profilin with high affinity. Biochemistry. 1996;35(44):14027–14034. doi: 10.1021/bi961878z. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Goldschmidt-Clermont PJ, Pollard TD. The affinities of human platelet and Acanthamoeba profilin isoforms for polyphosphoinositides account for their relative abilities to inhibit phospholipase C. Cell Regul. 1990;1(12):937–950. doi: 10.1091/mbc.1.12.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Cole NB, Moss B, Pollard TD. Vaccinia virus expresses a novel profilin with a higher affinity for polyphosphoinositides than actin. Biochemistry. 1994;33(35):10815–10824. doi: 10.1021/bi00201a032. [DOI] [PubMed] [Google Scholar]
- Magdolen V, Oechsner U, Muller G, Bandlow W. The intron-containing gene for yeast profilin (PFY) encodes a vital function. Mol Cell Biol. 1988;8(12):5108–5115. doi: 10.1128/mcb.8.12.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney NM, Janmey PA, Almo SC. Structure of the profilin-poly-l-proline complex involved in morphogenesis and cytoskeletal regulation. Nat Struct Biol. 1997;4(11):953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- Mertens N, Remaut E, Fiers W. Tight transcriptional control mechanism ensures stable high-level expression from T7 promoter-based expression plasmids. Biotechnology (NY) 1995;13(2):175–179. doi: 10.1038/nbt0295-175. [DOI] [PubMed] [Google Scholar]
- Miller DM, Shakes DC. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–394. [PubMed] [Google Scholar]
- Miller DM, 3rd, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34(2):477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Neuhoff H, Sassoe-Pognetto M, Panzanelli P, Maas C, Witke W, Kneussel M. The actin-binding protein profilin I is localized at synaptic sites in an activity-regulated manner. Eur J Neurosci. 2005;21(1):15–25. doi: 10.1111/j.1460-9568.2004.03814.x. [DOI] [PubMed] [Google Scholar]
- Ono K, Parast M, Alberico C, Benian GM, Ono S. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J Cell Sci. 2003;116(10):2073–2085. doi: 10.1242/jcs.00421. [DOI] [PubMed] [Google Scholar]
- Ono S. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J Cell Biol. 2001;152(6):1313–1319. doi: 10.1083/jcb.152.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156(6):1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D, Carlier MF. How profilin promotes actin filament assembly in the presence of thymosin β4. Cell. 1993;75(5):1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419(6902):82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- Qian C, Zhang Q, Wang X, Zeng L, Farooq A, Zhou MM. Structure of the adaptor protein p14 reveals a profilin-like fold with distinct function. J Mol Biol. 2005;347(2):309–321. doi: 10.1016/j.jmb.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14(8):1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter K, Jockusch BM, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359(2):97–109. doi: 10.1016/s0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365(6449):810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- Severson AF, Bowerman B. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J Cell Biol. 2003;161(1):21–26. doi: 10.1083/jcb.200210171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Baillie DL, Bowerman B. A Formin Homology protein and a profilin are required for cytokinesis and arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol. 2002;12(24):2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- Skare P, Karlsson Evidence for two interaction regions for phosphatidylinositol(4,5)-biphosphate on mammalian profilin I. FEBS Lett. 2002;522(1–3):119–124. doi: 10.1016/s0014-5793(02)02913-7. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246(15):4866–4871. [PubMed] [Google Scholar]
- Staiger CJ, Goodbody KC, Hussey PJ, Valenta R, Drobak BK, Lloyd CW. The profilin multigene family of maize: differential expression of three isoforms. Plant J. 1993;4(4):631–641. doi: 10.1046/j.1365-313x.1993.04040631.x. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T. The essential role of profilin in the assembly of actin for microspike formation. EMBO J. 1998;17(22):6516–6526. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn KS, Christensen HE, Shigeta R, Huddler D, Shalaby L, Lindberg U, Chua NH, Schutt CE. The crystal structure of a major allergen from plants. Structure. 1997;5(1):19–32. doi: 10.1016/s0969-2126(97)00163-9. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263(1–2):103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tochio H, Tsui MM, Banfield DK, Zhang M. An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science. 2001;293(5530):698–702. doi: 10.1126/science.1062950. [DOI] [PubMed] [Google Scholar]
- Valenta R, Duchene M, Pettenburger K, Sillaber C, Valent P, Bettelheim P, Breitenbach M, Rumpold H, Kraft D, Scheiner O. Identification of profilin as a novel pollen allergen; IgE autoreactivity in sensitized individuals. Science. 1991;253(5019):557–560. doi: 10.1126/science.1857985. [DOI] [PubMed] [Google Scholar]
- Van Troys M, Dewitte D, Goethals M, Vandekerckhove J, Ampe C. Evidence for an actin binding helix in gelsolin segment 2; have homologous sequences in segments 1 and 2 of gelsolin evolved to divergent actin binding functions? FEBS Lett. 1996;397(2–3):191–196. doi: 10.1016/s0014-5793(96)01086-1. [DOI] [PubMed] [Google Scholar]
- Van Troys M, Ono K, Dewitte D, Jonckheere V, De Ruyck N, Vandekerckhove J, Ono S, Ampe C. TetraThymosinbeta is required for actin dynamics in C. elegans and acts via functionally different actin binding repeats. Mol Biol Cell. 2004;15(10):4735–4748. doi: 10.1091/mbc.E04-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson VK, De La Cruz EM, Higgs HN, Pollard TD. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry. 1998;37(31):10871–10880. doi: 10.1021/bi980093l. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, A mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16(11):3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withee J, Galligan B, Hawkins N, Garriga G. Caenorhabditis elegans WASP and Ena/VASP proteins play compensatory roles in morphogenesis and neuronal cell migration. Genetics. 2004;167(3):1165–1176. doi: 10.1534/genetics.103.025676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14(8):461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci U S A. 2001;98(7):3832–3836. doi: 10.1073/pnas.051515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolven AK, Belmont LD, Mahoney NM, Almo SC, Drubin DG. In vivo importance of actin nucleotide exchange catalyzed by profilin. J Cell Biol. 2000;150(4):895–904. doi: 10.1083/jcb.150.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Hao JC, Lim W, Tessier-Lavigne M, Bargmann CI. Shared receptors in axon guidance: SAX-3/Robo signals via UNC-34/Enabled and a Netrin-independent UNC-40/DCC function. Nat Neurosci. 2002;5(11):1147–1154. doi: 10.1038/nn956. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Sheps JA, Ling V, Fang LL, Baillie DL. Expression analysis of ABC transporters reveals differential functions of tandemly duplicated genes in Caenorhabditis elegans. J Mol Biol. 2004;344(2):409–417. doi: 10.1016/j.jmb.2004.09.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplemental materials described in this article can be found at http://www.interscience.wiley.com/jpages/0886-1544/suppmat


