Abstract
Protein kinases targeted by small-molecule inhibitors develop resistance through mutation of the ‘gatekeeper’ threonine residue of the active site. Here we show that the gatekeeper mutation in the cellular forms of c-ABL, c-SRC, platelet-derived growth factor receptor-α and -β, and epidermal growth factor receptor activates the kinase and promotes malignant transformation of BaF3 cells. Structural analysis reveals that a network of hydrophobic interactions—the hydrophobic spine—characteristic of the active kinase conformation is stabilized by the gatekeeper substitution. Substitution of glycine for the residues constituting the spine disrupts the hydrophobic connectivity and inactivates the kinase. Furthermore, a small-molecule inhibitor that maximizes complementarity with the dismantled spine (compound 14) inhibits the gatekeeper mutation of BCR-ABL-T315I. These results demonstrate that mutation of the gatekeeper threonine is a common mechanism of activation for tyrosine kinases and provide structural insights to guide the development of next-generation inhibitors.
Deregulated protein kinases have been linked to numerous diseases including cancer and diabetes as well as inflammation. The targeted inhibition of protein tyrosine kinases is now well established as an effective therapeutic regimen for chronic myeloid leukemia (CML) and several solid tumors1-4. Many small-molecule kinase inhibitors have exploited a conserved threonine residue within the ATP binding site for binding specificity5. This threonine controls access of the inhibitors to a hydrophobic pocket deep in the active site that is not contacted by ATP, hence leading to its designation as a ‘gatekeeper’ residue6. Substitution of the gatekeeper threonine residue with bulky side chains is a common mechanism of resistance to pharmacological ATP-competitive kinase inhibitors7,8.
Imatinib has been used successfully to inhibit BCR-ABL in CML9, c-KIT in gastrointestinal stromal tumor (GIST)10 and platelet-derived growth factor receptor-α (PDGFRA) in hypereosinophilic syndrome (HES)11,12. The first imatinib-resistant mutation described in CML patients was an isoleucine substitution at the gatekeeper residue Thr315 (numbered according to the sequence for the type Ia isoform of c-ABL)13. The T315I mutation has been detected in imatinib-naïve CML patients and accounts for ∼20% of the total burden of clinical resistance14. Mutation at the analogous position to Thr315 in other imatinib targets such as c-KIT (Thr670) and PDGFRA (Thr674) have been linked to imatinib resistance in patients with GIST and HES, respectively15,16. Similarly, the gatekeeper mutation T790M in EGFR causes resistance to gefitinib and erlotinib, and has been detected in lung cancer patients before drug treatment and in the germ line of a family pedigree with several cases of lung cancer17,18. The association of gatekeeper-residue mutations with malignancy even in the absence of treatment with kinase inhibitors implies a role in activation of the transforming function of several protein tyrosine kinases. However, the mechanism for this oncogenic activation has not been defined.
Recent inferences drawn from crystallographic and chemical genetic studies suggest that ABL and SRC are regulated in a similar fashion and have highly conserved tertiary structures19-26. Interestingly, mutation of the gatekeeper residue has been noted in the sequence of v-SRC from several independent strains of avian Rous sarcoma virus (RSV), but the mechanism responsible for cellular transformation was not clearly defined27-29. On the basis of the strong correlation between substitution of the gatekeeper threonine and oncogenic activation of v-SRC, and on structural considerations gleaned from our previous studies with dual SRC-ABL kinase inhibitors30, we reasoned that substitution of the gatekeeper threonine residue with bulkier residues would be a common mechanism of activation of tyrosine kinases.
In this study, we show that the substitution of a bulkier hydro-phobic residue for the gatekeeper threonine in the human tyrosine kinases activates tyrosine phosphorylation. In silico modeling of the native and gatekeeper variants of different kinases and crystallographic analysis of SRC-T341I suggest that an isoleucine substitution for the gatekeeper threonine stabilizes a ‘hydrophobic spine’31 that is a characteristic feature of the active state of several kinases. In support of this hypothesis, we show that disruption of hydrophobic connectivity either pharmacologically or by mutagenesis of residues constituting the spine can effectively inhibit the kinase activity of the gatekeeper variants of c-ABL and BCR-ABL.
RESULTS
Gatekeeper mutations in c-SRC and c-ABL activate the kinase
The gatekeeper residue threonine is found in many tyrosine kinases (Fig. 1a). It lies in the hinge region between the N and C lobes of the kinase (Fig. 1b), where it controls access to a hydrophobic pocket that helps anchor kinase inhibitors to the active site (Fig. 1c). Because v-SRC and BCR-ABL are already deregulated, enzymatically activated protein kinases, we examined the effect of mutation of the gatekeeper residue on the kinase function of c-SRC and c-ABL isoform1b (the gatekeeper threonine is residue 334).
Figure 1.

Sequence conservation and structural features of the gatekeeper residue threonine in tyrosine kinases and activation of kinase activity by gatekeeper residue mutation. (a) Sequence alignment of the kinase domain hinge region and conservation of the gatekeeper residue in v-SRC, c-SRC, c-ABL and several receptor tyrosine kinases. (b) Structural alignment of inactive c-SRC (cyan; PDB 1Y57) and c-ABL (gray; PDB 2G2I) showing the conformational similarities; the kinase hinge region is marked by the arrow. (c) An enlarged view of the ABL active site bound to PD166326 (above; PDB 1OPK) and an ATP analog (below; PDB 2G2F), showing the specific interaction of PD166326 but not ATP with the gatekeeper threonine. (d) Immunoblot analysis of HEK293T cells expressing BCR-ABL and different variants of c-ABL. Above, total cell lysates probed with anti-phosphotyrosine antibody (anti-PY). The blot was stripped and reprobed with anti-ABL. Below, immunoblots of Ni-NTA–purified histidine-tagged c-ABL proteins probed with anti-PY, followed by stripping and reprobing with anti-ABL antibody. (e) Immunoblot analysis of HEK293T cells expressing different c-SRC kinase variants. Above, total cell lysates probed with anti-PY. The blots were stripped and reprobed with anti-SRC, anti-phosphotyrosine-416 (anti-SRC-PY416) and anti-phosphotyrosine-527 (anti-SRC-PY527) antibodies. (f) Immunoblot of Ni-NTA–purified c-SRC kinase variants probed with anti-PY followed by stripping and reprobing as described in e. MW, molecular weight.
Transient expression in human embryonic kidney cells (HEK293T) of C-terminally His-tagged T334I, T334A and G2A c-ABL mutants revealed elevated kinase activity as compared to native c-ABL (Fig. 1d). The myristoylation-defective G2A mutant of c-ABL, which activates the c-ABL kinase23, was included as a positive control for kinase activation. The T334I variant induced equivalent levels of total cellular phosphotyrosine in cell lysates as BCR-ABL, whereas T334A and G2A substitutions had weaker effects (Fig. 1d). After purification of c-ABL and its variants by nickel-resin, we likewise observed increased autophosphorylation of ABL variants, with T334I showing the greatest degree of phosphotyrosine (Fig. 1d, below).
Similarly, we found that substitution of isoleucine and methionine at the homologous gatekeeper threonine residue in c-SRC (T341) induced high levels of total cellular phosphotyrosine and increased tyrosine autophosphorylation; methionine substitution, which likewise introduces hydophobicity, has a more dramatic effect than isoleucine substitution (Fig. 1e,f). To further explore the structural consequences of amino acid substitution at the gatekeeper threonine, we created the T341K and T341P mutations. Lysine, which introduces a strong positive charge into the enzymatic active site, caused reduced kinase activation relative to isoleucine and methionine. Proline, which is expected to kink the polypeptide backbone at hinge region, inactivated the enzyme (Fig. 1e,f). These observations demonstrate the sensitivity of the enzyme to amino acid substitutions at the threonine residue of the hinge region. Immuno-blotting with phosphospecific antibodies against Tyr416 of c-SRC, which is phosphorylated in the enzymatically active state, and Tyr527, which is phosphorylated in the inactive state, revealed that several gatekeeper variants of SRC are expressed as catalytically active enzymes.
Gatekeeper variants of c-ABL and c-SRC transform BaF3 cells
Next, we sought to determine whether kinase-activated variants of c-ABL and c-SRC would show similar patterns of autophosphorylation and induce leukemic transformation in BaF3 cells, a cytokine-dependent hematopoietic progenitor line that has been widely used in assays of oncogenicity by activated tyrosine kinases32,33. We expressed gatekeeper mutants and other variants of c-ABL and c-SRC in BaF3 cells by retroviral transduction and puromycin selection. Immuno-blotting of total cell protein lysates and purified c-ABL and c-SRC showed a comparable pattern of kinase activation as HEK293T cells for all variants except c-SRC-G2A, which showed partial kinase activation in HEK293T cells but not in BaF3 cells (Fig. 2a-c).
Figure 2.
Kinase activation and BAF3 cellular transformation by gatekeeper mutants of SRC and ABL. (a) Immunoblot analysis of BaF3 cells expressing BCR-ABL and gatekeeper mutants of c-ABL. Above, total cell lysates probed with anti-phosphotyrosine antibody (anti-PY). Blots were stripped and reprobed with anti-ABL. Below, immunoblots of Ni-NTA–purified histidine-tagged c-ABL proteins probed with anti-PY, followed by stripping and reprobing with anti-ABL antibody. MW, molecular weight. (b) Immunoblot analysis of BaF3 cells expressing different c-SRC kinase variants. Above, total cell lysates probed with anti-PY. Blots were stripped and reprobed as described in Figure 1e. (c) Immunoblot of Ni-NTA–purified histidine-tagged c-SRC proteins probed with anti-PY. Blots were stripped and reprobed as in Figure 1e. (d) Cell-proliferation assay of BaF3 cells expressing BCR-ABL and different constructs of ABL variants. Cells were plated in quadruplicate in 96-well plates at a density of 5,000 cells per well in the absence of IL-3, and scored when the wells became confluent. (e) Cell-proliferation assay of BaF3 cells expressing c-SRC kinase variants. (f) Survival of mice injected with BaF3 cells expressing BCR-ABL and gatekeeper variants of c-SRC and c-ABL. Ten mice were injected for each construct.
Expression of BCR-ABL and activated PDGFRB, PDGFRA, FMS-like tyrosine kinase-3 (FLT3), EGFR and Janus kinase-2 (JAK2) induces interleukin-3 (IL-3)-independent proliferation of BaF3 cells, which correlates tightly but not absolutely with malignant transformation32,34-37. We therefore tested BaF3 cells expressing c-ABL and c-SRC variants for their capacity to survive and proliferate when cultured in the absence of IL-3. Like BCR-ABL, c-ABL-T334I supported robust survival in short-term assays (Supplementary Fig. 1 online) and rapid IL-3–independent proliferation of BaF3 cells in prolonged cell culture (Fig. 2d). In contrast, cells expressing the weakly activated c-ABL variants T334A and G2A or overexpressing native c-ABL showed no detectable enhanced survival or proliferation (Fig. 2 and Supplementary Fig. 1). Cells expressing the activated c-SRC variants T341I, T341M, T341K, T381K and Y530F demonstrated only modest degrees of survival (Supplementary Fig. 1) and delayed IL-3–independent cell growth after 10 d of culture (Fig. 2e). The c-SRC-G2A and c-SRC-T341A cells showed neither survival nor proliferation (Fig. 2e and Supplementary Fig. 1). Taken together with the autophosphorylation data, these results demonstrate a correlation between the level of kinase activation and cellular transformation for c-ABL and further suggest that c-ABL is more effective at transforming this hematopoietic cell line than activated variants of SRC. The longer latency of transformation by SRC kinase variants may be due to the fact that activated SRC kinase alone is not sufficient for transformation and may require additional epigenetic or genetic alterations to transform the BaF3 cells.
To determine whether the in vitro assays of IL-3–independent cell survival and proliferation correlated with the development of leukemia in vivo, we injected BaF3 cells expressing the c-ABL and c-SRC variants into syngenic Balb/c mice and observed for leukemia induction, with BCR-ABL–expressing BaF3 cells serving as a control. Mice injected with c-ABL-T334I or BCR-ABL–transformed BaF3 cells died within 4 weeks with peripheral blood leukocytosis and massive splenomegaly. In contrast, none of the other variants of c-ABL or c-SRC developed leukemia (Fig. 2f). These data demonstrate that mutation at the gatekeeper threonine can activate the intrinsic tyrosine kinase activity of both c-ABL and c-SRC, but only activated variants of c-ABL induce leukemia in BaF3 cells, consistent with a long-standing observation that activated SRC is ineffective at transforming BaF3 cells38.
Gatekeeper mutation in receptor tyrosine kinases is activating
Transient expression of the receptor tyrosine kinase PDGFRB and its gatekeeper mutation variants (T681A, T681I and T681M) in HEK293T cells showed enhanced kinase autophosphorylation and increased tyrosine phosphorylation of total cellular protein. Like the c-SRC kinase, introduction of the methionine residue (PDGFRB-T681M) generated the most active kinase (Supplementary Fig. 2a online). For PDGFRA, only the T674M variant was associated with kinase activation, as demonstrated by autophosphorylation of purified protein (Supplementary Fig. 2b). For unknown reasons, we were unable to achieve adequate expression of EGFR in HEK293T cells.
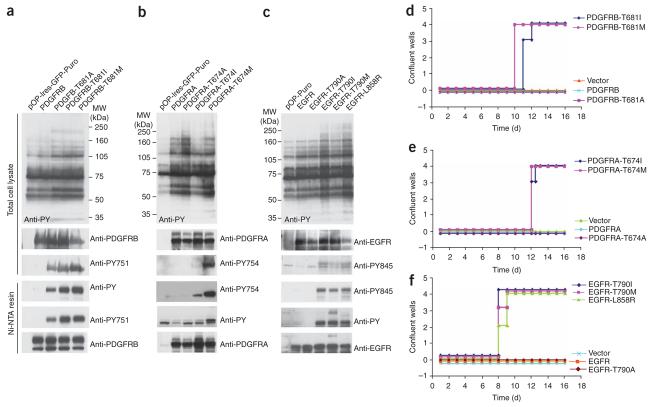
We next expressed these receptor tyrosine kinase mutants in BaF3 cells. Immunoblotting of total cell lysates and partially purified proteins showed kinase activation for each of the gatekeeper variants of PDGFRB (T681A, T681I and T681M; Fig. 3a), PDGFRA (T674I and T674M; Fig. 3b) and EGFR (T790I, T790M and L858R; Fig. 3c). For these experiments, EGFR-L858R served as a control, because EGFR activation and BaF3 cell transformation has been described previously for this mutation36. The results of these gatekeeper mutagenesis studies revealed that, like c-ABL and c-SRC, PDGFRB is maximally activated by the substitution with bulkier residues (isoleucine and methionine) and more modestly activated by alanine substitution (Fig. 3a), whereas PDGFRA and EGFR can be activated only by the substitution with isoleucine or methionine but are tolerant of substitution with smaller side chain substitutions such as alanine (Fig. 3b,c). Unlike BCR-ABL or c-ABL-T334I, none of the receptors bearing gatekeeper mutations supported short term IL-3–free survival in BaF3 cells (Supplementary Fig. 3 online). Like c-SRC-T341M/I, BaF3 cells carrying receptors bearing the gatekeeper variants PDGFRB-T681I, PDGFRB-T681M; PDGFRA-T674I, PDGFRA-T674M; EGFR-T790I, EGFR-T790M and EGFR-L858R showed delayed but robust growth of BaF3 cells after 9–10 d (Fig. 3d-f). The longer latency of transformation by these receptor kinase variants may be due to the fact that the activated kinase alone is not sufficient for transformation and may require additional epigenetic or genetic alterations to transform the BaF3 cells. These data demonstrate that the gatekeeper mutations activate the kinase activity in receptor tyrosine kinases and can transform the cytokine-dependent cell line BaF3, although with longer latency than for activated variants of c-ABL.
Figure 3.
Kinase activation and BAF3 cellular transformation by gatekeeper residue mutation of receptor tyrosine kinases. (a) Immunoblot analysis of BaF3 cells expressing gatekeeper mutants of PDGFRB. Above, total cell lysates probed with anti-phosphotyrosine antibody (anti-PY). Blots were stripped and reprobed with anti-PDGFRB antibody and anti–PDGFRB-phosphotyrosine-751 (anti-PY751). Below, immunoblots of Ni-NTA–purified histidine-tagged PDGFRB proteins probed with anti-PY, followed by stripping and reprobing with anti-PDGFRB antibody and anti-PY751. (b) Immunoblot analysis of BaF3 cells expressing gatekeeper mutants of PDGFRA. Above, total cell lysates probed with anti-PY. Blots were stripped and reprobed with anti-PDGFRA antibody and anti–PDGFRA-phosphotyrosine-754 (anti-PY754). Below, immunoblots of Ni-NTA–purified histidine-tagged PDGFRA proteins probed with anti-PY, followed by stripping and reprobing with anti-PDGFRA antibody and anti-PY754. (c) Immunoblot analysis of BaF3 cells expressing gatekeeper mutants of EGFR. Above, total cell lysates probed with anti-PY. Blots were stripped and reprobed with anti-EGFR antibody and anti–EGFR-phosphotyrosine-845 (anti-PY845). Below, immunoblots of Ni-NTA–purified histidine-tagged EGFR proteins probed with anti-PY, followed by stripping and reprobing with anti-EGFR antibody and anti-PY845. (d–f) Cell-proliferation assays of BaF3 cells expressing wild-type and gatekeeper mutants of PDGFRB (d), PDGFRA (e) and EGFR (f). Cells were plated in quadruplicate in 96-well plates at a density of 5,000 cells per well in the absence of IL-3 and scored when the wells became confluent.
A hydrophobic spine is assembled during kinase activation
Crystal structures of protein kinases in their active and inactive conformations have provided a general understanding of their regulation via intramolecular autoregulatory interactions39. Typically, activation involves changes in the position and orientation of the activation loop and the catalytic c-helix. A surface comparison of active kinase structures has revealed a network of highly conserved hydrophobic or amino-aromatic interactions called the hydrophobic spine, which stabilizes the active kinase conformation31.
In ABL kinase, the spine comprises four residues, Leu320, Met309, Phe401 and His380, which form a chain of hydrophobic interactions from the N lobe via the active site to the Tyr412 of the activation loop (Fig. 4a). These interactions are disrupted in the inactive state (Fig. 4b). The gatekeeper residue Thr334 is situated near the tip of the hydrophobic spine. Our modeling analysis suggests that substitution of a bulkier hydrophobic residue at this position would stabilize the active state by strengthening the spine. In our model, Leu403 serves as a bipositional switch that positions Asp400 of the DFG motif in the correct orientation to enable catalysis (Supplementary Fig. 4a,b online). In the active state the backbone of Leu403 coordinates Arg381 of the catalytic HRD motif, whereas in the inactive state the sidechain of Leu403 stabilizes Phe401 in the catalytically quiescent ‘DFG-out’ conformation. Furthermore, computational modeling of the published structures of SRC and EGFR revealed similar hydrophobic interactions in SRC (Supplementary Fig. 5 online) and EGFR (Supplementary Fig. 6 online), where the gatekeeper residue sits at the top of the spine, stabilizing the active state. The active SRC and EGFR kinases maintain a hydrophobic spine from the gatekeeper threonine to Tyr419 and Tyr869 of the activation loop, respectively (Supplementary Figs. 5b and 6b). These interactions are disrupted in the inactive state (Supplementary Figs. 5d and 6d). As in ABL, Leu410 of SRC and Leu858 of EGFR serve as a bipositional switch for kinase regulation40,41. Altogether, these in silico analyses suggest that gatekeeper mutants stabilize the active state of the kinase by contributing to the hydrophobic spine.
Figure 4.

The active conformation of ABL is stabilized by a hydrophobic spine linking the gatekeeper threonine to the activation loop. (a) The surface projections of the hydrophobic spine that assembled during ABL kinase activation in ABL-ADP conformation (shown in blue; PDB 2G2I). The gatekeeper residue, Thr334, is shown as an orange surface. (b) The surface projections of the dismantled hydrophobic spine in the inactive kinase conformation of ABL–imatinib (PDB 1OPJ). Imatinib binds to the inactive kinase which is stabilized by the DFG-out conformation caused by dismantling of the hydrophobic spine.
SRC-T341I adopts an active conformation
We crystallized the isoleucine substitution of the gatekeeper residue of the chicken c-SRC kinase domain (residues 251–533) in complex with ATPγS and determined the structure to a resolution of 2.0 Å. The four molecules present in the asymmetric unit are in virtually identical conformations; further discussion focuses on one of these (Fig. 5a). There is well-defined electron density for the adenine, ribose and the α-phosphate groups of ATPγS and less well-defined density for the β- and γ-phosphates. The activation loop is fully ordered and Tyr419 is phosphorylated. The overall conformation of the c-Src-T341I mutant resembles that of the activation loop–phosphorylated Lck (PDB 1QPC), where Phe408 of the DFG motif forms contacts with residues His387 and Met317 of the hydrophobic spine, such that the hydrophobic spine is assembled (Fig. 5a). However, in the inactive state of the SRC kinase conformation, the hydrophobic spine is crippled and the activation loop is not phosphorylated and extended (Fig. 5b).
Figure 5.

The hydrophobic spine in active and inactive SRC kinases. (a) Crystal structure of chicken c-SRC-T338I bound to ATPγS. SRC-T338I is homologous to T334I in human c-SRC. Chicken SRC residues are numbered according to the human c-SRC kinase numbering. The residues Leu328, Met317, Phe408 and His387, which constitute the hydrophobic spine, are shown in blue. The gatekeeper isoleucine residue is shown in orange. The activation loop is shown in red. (b) The inactive conformation of chicken c-SRC (PDB 2SRC), colored as in a. (c) Active site of Lck kinase (PDB 1QPC) in the active state bound with AMP-PNP shown in yellow. Gatekeeper residue Thr316 and the catalytic Lys273 are shown as green surfaces. The water molecules sandwiched between the residues Thr316 and Lys273 are shown as red circles. The interactions of lysine with ANP and the catalytic Glu288 are shown; bond distances are presented in angstroms. (d) Active site of SRC-T341I kinase domain (PDB 3DQW) bound with ATPγS shown in yellow. The surfaces of the side chains for residues Ile341 and Lys298 are shown in green. Interactions of Lys298 with Glu310 and ATPγS are mapped and the bond distances are indicated in angstroms. (e) Active site of insulin receptor kinase (IRK; PDB entry 1GAG) bound with ANP shown in yellow. The surfaces of the side chains for residues Met1076 and Lys1030 are shown in green. Interactions of catalytic Lys1030 with Glu1047 and ANP are mapped, and the bond distances are indicated in angstroms.
Gatekeeper mutation efficiently organizes the active site
We compared the structure of the gatekeeper variant of c-SRC with a high-resolution structure of a wild-type SRC family kinase—Lck bound to the ATP analog AMP-PNP, also in the active conformation (PDB 1QPC). In both structures, an active-site lysine residue (SRC Lys298 and Lck Lys273) forms a hydrogen bond with a glutamate residue in helix αC (SRC Glu310 and Lck Glu288), but the conformation of the lysine residue is different in the two structures (Fig. 5c,d). In the wild-type Lck, two water molecules are interdigitated between Thr316 and Lys273, and the lysine side chain does not form hydrogen bonds with the phosphate groups of the ATP analog (the closest distance between the lysine nitrogen and a phosphate oxygen is 5.5 Å). In the SRC structure, the substitution of isoleucine leads to the exclusion of these water molecules and the rearrangement of the lysine side chain, which packs closely to the isoleucine residue and forms a hydrogen bond to the α-phosphate of the nucleotide (N-O distance of 3.4 Å). The hydrophobic packing of Ile341 and Lys298 seals off the interior of the kinase domain, thereby excluding water and potentiating the hydrophobic spine and interaction of Lys298 with ATP relative to the wild-type structure. A similar orientation of the lysine residue and ATP is seen in the structure of kinases with hydrophobic residues at the gatekeeper position of the insulin receptor kinase (Fig. 5e). On the basis of these observations, we infer that the T341I mutation activates the kinase by optimizing the coordination of ATP and by stabilizing the hydrophobic spine.
Mutagenesis of the hydrophobic spine causes kinase inactivation
Our model predicts that kinase activity is dependent on assembly of the hydrophobic spine. To test this, we mutated all four residues that constitute the spine in the ABL kinase (Leu320, Met309, Phe401 and Thr334) to glycine and analyzed kinase activity in a cell-based assay. M309G, F401G and T334G completely abrogated kinase activity (Fig. 6a), whereas L320G showed a decreased level of phosphorylation relative to ABL-T334I. Molecular modeling of the L320G revealed only a modest effect on spine assembly, consistent with the modestly reduced kinase activation of ABL-T315I (Fig. 6b,c), whereas, M309G and F401G mutations completely disrupted the assembly of the spine, inactivating the kinase (Fig. 6d,e). Similarly, double or triple mutations of the residues that disrupted the spine assembly also resulted in an inactive kinase (Fig. 6a). These mutagenesis data suggest that the hydrophobic spine is a key structural element of the enzymatically active conformation of the ABL kinase.
Figure 6.
Disruption of hydrophobic-spine assembly by mutagenesis inactivates the ABL-T334I kinase. (a) Immunoblot analysis of HEK293T cells expressing BCR-ABL and different variants of ABL-T334I (analogous to the T315I substitution in BCR-ABL). Above, total cell lysates probed with anti-phosphotyrosine antibody (anti-PY) followed by stripping and reprobing with anti-ABL antibody. (b–e) Surface projections of the hydrophobic spine in the active ABL-ANP conformation (PDB 2G2I). Isoleucine substitution enhances spine assembly (b). L320G (c), M309G (d) and F401G (e) substitutions disrupt the spine. WT, wild type.
Compound 14 inhibits the gatekeeper variants of BCR-ABL
Given our hypothesis that gatekeeper mutations stabilize the hydrophobic spine, we reasoned that a small-molecule ATP-competitive inhibitor that can disrupt the hydrophobic spine may achieve the necessary binding affinity to successfully inhibit the kinase activity of gatekeeper mutants. We performed structural alignment of the ABL kinase crystallized with different inhibitors to identify an inhibitor that can dismantle the spine without any physical interaction with the activation loop. By this analysis, we identified compound 14 as being able to efficiently compete for the spine residues without directly affecting the mobility of the activation loop.
Compound 14 was generated by a rational ‘hybrid design’ approach by appending a trifluoromethyl group to the parent inhibitor PD166326 (ref 42; Fig. 7a,b). Compound 14, like imatinib, disrupts the assembly of the hydrophobic spine, thereby locking the kinase in an inactive ‘DFG-out’ conformation42. The shape of compound 14 resembles that of the core of imatinib, with the key difference that the hydrophilic methyl-piperazine group in imatinib is replaced with the more hydrophobic trifluoromethyl moiety.
Figure 7.

The ATP competitive inhibitor compound 14 disrupts the hydrophobic spine and inhibits c-ABL-T334I and BCR-ABL-T315I. (a) Chemical structures of PD166326, compound 14 and imatinib. (b) Superimposed structures of PD166326, compound 14 and imatinib in the active and inactive conformations of ABL kinase, showing the physical interaction with activation loop. (c) The ABL–compound 14 cocrystal structure (PDB 2HIW)42 indicates that compound 14 binds to the inactive DFG-out conformation, and the trifluoromethyl group of compound 14 binds deeply into the hydrophobic pocket partially formed by the dismantled hydrophobic spine. The additional anchorage gained through this interaction may drive conformational adjustments in the presence of the bulkier isoleucine side chain. (d–f) Immunoblot analysis of lysates of BaF3 cells transformed by c-ABL-T334I (d), BCR-ABL-native (e) and BCR-ABL-T315I (f), and exposed to various concentrations of PD166326 (left) and compound 14 (right). Blots were probed with anti–phosphotyrosine-412 antibody, anti-phosphotyrosine antibody and anti-ABL antibody as indicated on the right side of each blot. Concentrations of drugs are given above each lane.
The structure also suggests that the trifluoromethylbenzamide ring of compound 14 competes for Phe401 of the DFG motif without having any direct physical interaction with the activation loop (Fig. 7b,c). Therefore, we reasoned that compound 14 may, at a higher concentration, efficiently inhibit the gatekeeper mutant ABL-T315I by destabilizing the active state. Additionally, the trifluoromethyl group of compound 14 fits into a hydrophobic cavity created by the disruption of the spine residues Phe401 and His387, and this may provide extra anchorage for affinity. We tested compound 14 and the parent inhibitor PD166326 for their ability to inhibit native and gatekeeper variants of ABL and BCR-ABL. Compound 14 inhibited c-ABL-T334I, BCR-ABL and BCR-ABL-T315I variants with a 50% inhibitory concentration (IC50) of 0.25 μM, 0.09 μM and 0.590 μM, respectively (Fig. 7d-f). In contrast, PD166326 barely inhibited c-ABL-T334I, with an IC50 of 8.0 μM, and showed a complete lack of activity up to 10 μM against BCR-ABL-T315I. Similarly, imatinib and nilotinib has been shown to be completely resistant against the gatekeeper mutant7,21,43. These results demonstrate that the gatekeeper mutants stabilize the active state by potentiating the hydrophobic spine, and disruption of the spine by a small-molecule inhibitor can efficiently inhibit the gatekeeper mutant. Although imatinib and nilotinib disrupt the hydrophobic spine, the fact that they fail to show any activity against the gatekeeper mutant may be due to the requirement of higher free energy to destabilize the active state imposed by their physical interactions with the activation loop.
DISCUSSION
Substitution of the gatekeeper threonine residue by isoleucine in the BCR-ABL oncoprotein (T315I) was the first resistance mutation noted in imatinib-treated patients13, and mutation in the homologous residue has emerged as a common mechanism of resistance for numerous kinases7. Structural studies have shown that this threonine stabilizes imatinib binding through hydrogen-bond interactions and regulates access to a deep hydrophobic pocket in the active site44,thus providing a compelling reason why Rstitution should mediate drug resistance via a steric mechanism alone. However, the identification of T315I in a significant cohort of CML patients with primary imatinib resistance14,45, the association of germline mutations in the gatekeeper residue of EGFR in familial lung cancer18 and the historical identification of gatekeeper residue mutations in transforming isolates of the v-SRC viral oncoprotein29 imply a role for this mutation in promoting malignancy that has hitherto been unexplained.
In this study, we demonstrate that the highly conserved threonine gatekeeper residue that lies within the hinge region of the enzymatic cleft of many kinases represents a crucial structural feature whose mutation promotes kinase activation. Our data demonstrate that threonine mutation is sufficient to activate kinase activity and transforming function for several native nonreceptor and receptor tyrosine kinases. Evidence that the T315I variant of BCR-ABL has higher oncogenic penetrance in in vitro models supports a direct role in disease pathogenesis46,47. However, the enhanced oncogenicity has been attributed to increased tyrosine phosphorylation at Tyr257 of BCR-ABL46, rather than increased kinase activity, which may be difficult to discern within the context of BCR-ABL, because of its high intrinsic kinase activity. Additionally, the increased binding of ATP demonstrated by the gatekeeper mutants of ABL48 and EGFR40,49 may further contribute to increased phosphorylation of cellular subtrates.
Biochemical and structural analyses of protein kinases have revealed that a complex set of conformational mechanisms regulates the catalytically active and quiescent states39. Conformational dynamics of the activation loop and the conserved C helix contribute to formation of an ion pair between a crucial lysine residue within the β3 strand of the N lobe and a crucial glutamate residue within the C helix. Ultimately, protein dynamics ensures that a conserved DFG motif lying at the tip of the activation loop is oriented in a manner that allows coordinated binding of magnesium and ATP39. In adopting the enzymatically competent ‘DFG in’ conformation, the N lobe and C lobe move toward one another to accommodate these structural changes. Our study supports a model whereby the hydrophobic spine serves as a structural element linking the N and C lobes. The gatekeeper residue, by virtue of its location, stabilizes the spine in the active conformation, whereas the inactive state is stabilized by autoinhibitory regulatory domain interactions and by association with negative regulatory proteins20,39. Mutations that disrupt this autoinhibition promote enzymatically active kinase conformations that are intrinsically resistant to imatinib binding14,21. Although direct steric hindrance has been invoked to explain the resistance of BCR-ABL-T315I, KIT-T670I, PDGFRA-T674I and EGFR-T790M to kinase inhibitor binding12,13,15,17, our data suggest that threonine mutation acts as well to promote the assembly of an enzymatically active kinase conformation by stabilizing the hydrophobic spine. We anticipate that this regulatory mechanism is conserved across the kinase family.
Substitution of the gatekeeper threonine residue with alanine, a smaller hydrophobic residue, likewise causes kinase activation, but less robustly than substituton with isoleucine or methionine. A recent report using hydrogen-exchange (HX) studies of the native and gatekeeper-mutated ERK2-Q103A kinase demonstrated that substitutions of smaller hydrophobic residues create a cavity within the hydrophobic cluster that destabilizes the autoinhibited state and increases the flexibility of the DFG motif and activation loop, and increased flexibility of the activation segment would be expected to favor autophosphorylation50.
Given our data showing that gatekeeper residue mutations cause kinase activation in several receptor and nonreceptor tyrosine kinases, we speculate that this phenomenon may be a generic feature of kinase regulation that helps to explain why this class of mutation has been observed in drug-naïve patients. In future drug design, structures that maximize complementarity to the dismantled spine should be favored, as these represent an inhibitor class that optimizes specificity by recognizing inactive kinase conformations while gaining sufficient affinity to be active against kinases that carry mutations at the gatekeeper residue.
METHODS
Plasmid constructs
We cloned full-length c-ABL isoform Ib by replacing the BCR portion of BCR-ABL with the N terminus of c-ABL-Ib. A 0.6-kb N-terminal DNA fragment of c-ABL-Ib (starting with initiation codon ATG and ending with the amino acid residue G163 of the SH2 domain) was amplified by PCR using gene-specific primers (Supplementary Table 1 online) and cDNA from HEK293T cells. This 0.6-kb fragment was digested with KpnI and BamHI and ligated to BamHI/KpnI digested pEYKBA21, yielding pEYKABL-Ib. Because pEYK vectors lack a selectable marker, we constructed two different selectable expression vectors (pOP-Ires-GFP-Puro and pOP-Ires-RFP-Puro) for stable expression of transgenes in mammalian cells. To construct pOP-Ires-GFP-Puro, pEYK3.1 was digested with EcoR I and DNA ends were filled in by Klenow polymerase, followed by ligation with DNA fragments containing Ires-GFP (pMSCV-IresGFP was digested with SalI and the ends were filled in by Klenow polymerase, followed by DNA purification by agarose gel electrophoresis) and SV40-PURO (pBABE was digested with Sal1 and MluI, followed by DNA purification by agarose gel electrophoresis). To construct pOP-Ires-RFP-puro, pOP-Ires-GFP-Puro was digested with SalI and DNA ends were filled by the Klenow polymerase, followed by digestion with Bam H1. The vector backbone was purified and ligated to a DNA fragment containing Ires-RFP, which was derived from the pIRES2-DsRed (Clontech) vector by digesting it with NotI and BglII. pEYKABL-Ib was digested with EcoRI. The DNA fragment containing ABL-Ib was purified from pEYKABL-Ib by EcoRI digestion and ligated to EcoRI-digested pOP-Ires-GFP-Puro, yielding pOPABL-Ib. A six-histidine tag was attached at the C terminus of pOPABL-Ib by PCR (primers are listed in Supplementary Table 1) using the Quickchange mutagenesis kit (Stratagene). We found no difference in the activity of the histidine-tagged and native versions of ABL and SRC or their gatekeeper variants (data not shown), and therefore the kinase activation observed is unlikely to be influenced by the histidine tag.
SRC, PDGFRA and EGFR were cloned by PCR using Hi-fidelity polymerase (Roche), gene-specific primers (Supplementary Table 1), and cDNA from HEK293T cells. PDGFRB was cloned from total human cDNA (Clontech) by PCR using gene-specific primers (Supplementary Table 1). These PCR products were cloned into TA-cloning vectors pCRXL-TOPO (Invitrogen) and pSCB (Stratagene). c-SRC and PDGFRB were cloned in pCRXL-TOPO vectors to yield pCRXL-SRC and pCRXL-PDGFRB, whereas PDGFRB and EGFR were cloned into the pSC vector to yield pSC-EGFR and pSC-PDGFRA. Four different clones for each gene were sequenced. The clones that were devoid of mutations were used for subcloning into pOP expression vectors. The EcoRI fragment of pCRXL-SRC containing the SRC coding gene was purified and ligated to EcoRI-digested pOP-Ires-GFP-Puro vector, yielding pOP-SRC-Ires-GFP-Puro. Similarly, PDGFRB was cloned into the EcoRI site of the pOP-Ires-GFP-Puro vector to give rise pOP-PDGFRB-Ires-GFP-Puro. The plasmid pSC-PDGFRA was digested with BamHI and AgeI to excise the fragment containing the coding region, followed by cloning in pOP-Ires-RFP-Puro at the BamHI site to yield pOP-PDGFRA-Ires-RFP-Puro. Finally, pSCEGFR was digested with XhoI and PvuI; the DNA fragment was purified and ligated to SalI/XhoI-digested pOP-Ires-GFP-Puro to yield pOP-EGFR-Puro. To construct gatekeeper mutant versions of these kinases, oligonucleotides overlapping the gatekeeper residue were synthesized and used to perform site-directed mutagenesis on the respective plasmid template (Quickchange XL mutagenesis kit; Stratagene). Each mutant was confirmed by sequencing.
Cell culture, transfection and retroviral transduction
We carried out HEK293T cell culture, proviral transfection and viral productions, and transduced BaF3 cells as described previously21. At 24 h after transduction, 2.5 μg ml−1 puromycin was added to the media to select for transduced cells. After 4–6 d, puromycin-resistant cells were counted and plated at 5,000 cells per well in 96-well plates, with and without IL-3, in quadruplicate. Cells were incubated at 37 °C, and the viable cell number was assessed 60 h later using the WST-1 reagent (Roche), according to the manufacturer's specifications. For prolonged IL-3 survival, 5,000 BaF3 cells were plated in quadruplicate in 96-well plates in RPMI without IL3. Cells were incubated at 37 °C for 14 d. Numbers of confluent wells were scored as IL-3–independent growth.
Balb/c mouse injection
We injected 106 BaF3 cells expressing c-SRC, c-ABL and gatekeeper variants intravenously into Balb/c mice by tail vein injection. Ten mice were injected for each construct. Analyses of BaF3-injected mice were performed as described previously51.
Immunoblotting
HEK293T and BaF3 cells were lysed in a buffer containing 50 mM sodium phosphate, pH 7.4, 300 mM NaCl, 1 mM EDTA, 10 mM imidazol, 4 mM sodium vanadate, 2 mM sodium floride, 5 mM benzamidine hydrochloride, 2 mM sodium glycerophosphate and 1 μl ml−1 Triton X-100; 50 μl ml−1 of glycerol protease inhibitors (Roche) and phosphatase inhibitors I and II (Sigma) were added immediately before use. Cells were suspended in lysis buffer and homogenized by 4–6 bursts of sonication. Samples were normalized for total protein content and total cell lysates were clarified by centrifugation for 5 min at 15,000g. For partial purification, 50 μl of Ni-NTA beads were added to each sample and incubated 2–4 h at 4 °C. Afterwards, Ni-beads were washed twice with lysis buffer. Total cell lysates and Ni-bead–purified proteins were suspended in sample buffer and denatured at 70 °C. For drug treatment, 10 mM stocks of PD166326 and compound 14 in DMSO were used to supplement the growth media for the desired concentration, 6 × 106 cells were treated for each drug concentration for 6 h, followed by washing with chilled PBS, and crude extracts were prepared as reported previously21. Proteins were resolved by 7% SDS-PAGE (BIORAD) under denaturing conditions, transferred to nitrocellulose and probed as described21. Antibodies used for immunoblotting were: anti-EGFR, anti–phospho-EGFR-Y1068, anti–phospho-EGFR-Y845, anti–phospho-PDGFRB, anti–phospho-PDGFRA, anti-SRC, anti–phospho-SRC-416, anti–phospho-SRC-527, anti–nonphospho-SRC-416, anti–nonphospho-SRC-527 and anti–phospho-ABL-Y412, from Cell Signaling Technology; anti–phosphotyrosine-PY99, anti-ABL, anti-PDGFRA and anti-PDGFRB from Santa Cruz Biotechnology. Bands were visualized using ECL reagents (Pierce) according to the supplier's instructions.
Protein preparation and crystallization
Wild-type and T338I mutant chicken c-SRC kinase domains (residues 251–533) were expressed in Escherichia coli and purified as described previously52. Crystals were grown at 25 °C by the hanging-drop method, mixing 1 μl of protein at a concentration of 10 mg ml−1 (in 20 mM Tris, pH 8.0, 100 mM NaCl, 5% (v/v) glycerol, 0.5 mM (v/v) Tris-(2-carboxyethyl)phosphine (TCEP), 1 mM ATPγS) with 1 μl well solution (10% (v/v) PEG4000, 50 mM ammonium acetate, 100 mM (v/v) Bis-Tris, pH 5.5, 5% (v/v) glycerol). Plate-shaped crystals of approximate dimensions 200 μm × 200 μm × 50μm grew overnight. Crystals were cryoprotected by serial soaks in well solution with increasing glycerol concentration up to 20% (v/v) and snap frozen and stored in liquid nitrogen.
Data collection and structure determination
Diffraction data were collected at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory beamlines 8.3.1 and 8.2.1, and processed to a resolution of 2.0 Å (SRC-T338I) and 2.3 Å (SRC wild type) with Denzo/Scalepack53. SRC-T338I crystallizes in space group P21 with unit cell dimensions of a = 82.4 Å, b = 103.7 Å, c = 83.7 Å, α = γ = 90°, β = 103.9° and four kinase molecules in the asymmetric unit (Table 1). Wild-type SRC crystallizes in space group P21 with unit cell dimensions of a = 42.5 Å, b = 119.2 Å, c = 63.7 Å, α = γ = 90°, β = 90.4° and two kinase molecules in the asymmetric unit (Table 1). The structures were solved by molecular replacement using the kinase domain of chicken c-SRC residues 250–533 (PDB 2SRC) 24 with the C helix and the activation loop deleted as a search model in the program Phaser54. The models were build in Coot55. SRC-T338I was refined with CNS 1.2 (ref. 56) and Phenix57. The Rwork and Rfree values are 0.205 and 0.247, respectively. Wild-type SRC was refined in REFMAC 5.2 (ref. 57) with TLS refinement with an Rwork and Rfree of 19.7 and 0.245, respectively.
Table 1.
Data collection and refinement statistics
| Chicken c-SRC-T338I, ATPγS (PDB 3DQW) |
Chicken c-SRC-WT, ATPγS (PDB 3DQX) |
|
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 82.4, 103.7, 83.7 | 42.5, 119.1, 63.7 |
| α, β, γ (°) | 90, 103.9, 90 | 90, 90.4, 90 |
| Resolution (Å) | 50–2.01 (2.08–2.01) | 50–2.3 (2.38–2.3) |
| Rsym | 6.6% (35.9%) | 6.2% (20.5%) |
| I / σI | 17 (2.7) | 18 (4.4) |
| Completeness (%) | 97.5 (94.4) | 96.6 (81.4) |
| Refinement | ||
| Resolution (Å) | 50–2.01 | 35.3–2.3 |
| No. reflections | 87,293 | 25,776 |
| Rwork / Rfree | 20.5% / 24.7% | 19.7% / 24.5% |
| No. atoms | 9,718 | 4,550 |
| Protein | 8,961 | 4,299 |
| Ligand/ion | 128 | 46 |
| Water | 629 | 205 |
| Average B-factors | ||
| Protein | 46.0 | 54.8* |
| Ligand/ion | 47.8 | 78.5* |
| Water | 39.2 | 54.7* |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.01 |
| Bond angles (°) | 1.0 | 1.2 |
Data were refined with TLS refinement and isotropic B-factors. Values in parentheses are for highest-resolution shell.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the US National Institutes of Health (NIH), the NIH Director's Pioneer Award of the NIH Roadmap for Medical Research, the Leukemia and Lymphoma Society, and by private funds from the Thomas Anthony Pappas Charitable Foundation. G.Q.D. is a recipient of the Burroughs Wellcome Fund Clinical Scientist Award in Translational Research. M.A.S. was supported by a Johnson & Johnson fellowship of the Life Science Research Foundation, Baltimore, and by NIH K99GM08009.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Accession codes. Protein Data Bank: Coordinates for the ATPγS-bound structures of wild-type c-SRC and c-SRC-T341I have been deposited with accession codes 3DQX and 3DQW, respectively.
References
- 1.Cohen P. Protein kinases—the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 2.Dibb NJ, Dilworth SM, Mol CD. Switching on kinases: oncogenic activation of BRAF and the PDGFR family. Nat. Rev. Cancer. 2004;4:718–727. doi: 10.1038/nrc1434. [DOI] [PubMed] [Google Scholar]
- 3.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–297. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 4.Sawyers CL. Opportunities and challenges in the development of kinase inhibitor therapy for cancer. Genes Dev. 2003;17:2998–3010. doi: 10.1101/gad.1152403. [DOI] [PubMed] [Google Scholar]
- 5.Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Shah K, Yang F, Witucki L, Shokat KM. A molecular gate which controls unnatural ATP analogue recognition by the tyrosine kinase v-Src. Bioorg. Med. Chem. 1998;6:1219–1226. doi: 10.1016/s0968-0896(98)00099-6. [DOI] [PubMed] [Google Scholar]
- 7.Azam M, Daley GQ. Anticipating clinical resistance to target-directed agents: the BCR-ABL paradigm. Mol. Diagn. Ther. 2006;10:67–76. doi: 10.1007/BF03256446. [DOI] [PubMed] [Google Scholar]
- 8.Daub H, Specht K, Ullrich A. Strategies to overcome resistance to targeted protein kinase inhibitors. Nat. Rev. Drug Discov. 2004;3:1001–1010. doi: 10.1038/nrd1579. [DOI] [PubMed] [Google Scholar]
- 9.Druker BJ, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 10.Heinrich MC, Blanke CD, Druker BJ, Corless CL. Inhibition of KIT tyrosine kinase activity: a novel molecular approach to the treatment of KIT-positive malignancies. J. Clin. Oncol. 2002;20:1692–1703. doi: 10.1200/JCO.2002.20.6.1692. [DOI] [PubMed] [Google Scholar]
- 11.Buchdunger E, et al. Abl protein-tyrosine kinase inhibitor STI571 inhibits in vitro signal transduction mediated by c-kit and platelet-derived growth factor receptors. J. Pharmacol. Exp. Ther. 2000;295:139–145. [PubMed] [Google Scholar]
- 12.Cools J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N. Engl. J. Med. 2003;348:1201–1214. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 13.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 14.Shah NP, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 15.Tamborini E, et al. A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology. 2004;127:294–299. doi: 10.1053/j.gastro.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 16.Cools J, et al. PKC412 overcomes resistance to imatinib in a murine model of FIP1L1-PDGFRaα-induced myeloproliferative disease. Cancer Cell. 2003;3:459–469. doi: 10.1016/s1535-6108(03)00108-9. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 18.Bell DW, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat. Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 19.Levinson NM, et al. A Src-like inactive conformation in the Abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison SC. Variation on an Src-like theme. Cell. 2003;112:737–740. doi: 10.1016/s0092-8674(03)00196-x. [DOI] [PubMed] [Google Scholar]
- 21.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/ imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 22.Nagar B, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 23.Hantschel O, et al. A myristoyl/phosphotyrosine switch regulates c-Abl. cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 24.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 25.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 26.Moarefi I, et al. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 27.Takeya T, Hanafusa H. Structure and sequence of the cellular gene homologous to the RSV src gene and the mechanism for generating the transforming virus. Cell. 1983;32:881–890. doi: 10.1016/0092-8674(83)90073-9. [DOI] [PubMed] [Google Scholar]
- 28.Hunter T. A tail of two src's: Mutatis mutandis. cell. 1987;49:1–4. doi: 10.1016/0092-8674(87)90745-8. [DOI] [PubMed] [Google Scholar]
- 29.Kato JY, et al. Amino acid substitutions sufficient to convert the non-transforming p60c-src protein to a transforming protein. Mol. Cell. Biol. 1986;6:4155–4160. doi: 10.1128/mcb.6.12.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azam M, et al. Activity of dual SRC-ABL inhibitors highlights the role of BCR/ABL kinase dynamics in drug resistance. Proc. Natl. Acad. Sci. USA. 2006;103:9244–9249. doi: 10.1073/pnas.0600001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daley GQ, Baltimore D. Transformation of an interleukin 3-dependent hematopoietic cell line by the chronic myelogenous leukemia-specific P210bcr/abl protein. Proc. Natl. Acad. Sci. USA. 1988;85:9312–9316. doi: 10.1073/pnas.85.23.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koh EY, Chen T, Daley GQ. Genetic complementation of cytokine signaling identifies central role of kinases in hematopoietic cell proliferation. Oncogene. 2004;23:1214–1220. doi: 10.1038/sj.onc.1207209. [DOI] [PubMed] [Google Scholar]
- 34.Carroll M, Tomasson MH, Barker GF, Golub TR, Gilliland DG. The TEL/platelet-derived growth factor β receptor (PDGFβR) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGFβR kinase-dependent signaling pathways. Proc. Natl. Acad. Sci. USA. 1996;93:14845–14850. doi: 10.1073/pnas.93.25.14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuki M, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96:3907–3914. [PubMed] [Google Scholar]
- 36.Jiang J, et al. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 2005;65:8968–8974. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- 37.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 38.Mathey-Prevot B, Nabel G, Palacios R, Baltimore D. Abelson virus abrogation of interleukin-3 dependence in a lymphoid cell line. Mol. Cell. Biol. 1986;6:4133–4135. doi: 10.1128/mcb.6.11.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huse M, Kuriyan J. The conformational plasticity of protein kinases. cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 40.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 42.Okram B, et al. A general strategy for creating “inactive-conformation” Abl inhibitors. Chem. Biol. 2006;13:779–786. doi: 10.1016/j.chembiol.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 43.O'Hare T, et al. In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res. 2005;65:4500–4505. doi: 10.1158/0008-5472.CAN-05-0259. [DOI] [PubMed] [Google Scholar]
- 44.Schindler T, et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 45.Pfeifer H, et al. Kinase domain mutations of BCR-ABL frequently precede imatinib-based therapy and give rise to relapse in patients with de novo Philadelphia-positive acute lymphoblastic leukemia (Ph+ ALL) Blood. 2007;110:727–734. doi: 10.1182/blood-2006-11-052373. [DOI] [PubMed] [Google Scholar]
- 46.Skaggs BJ, et al. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc. Natl. Acad. Sci. USA. 2006;103:19466–19471. doi: 10.1073/pnas.0609239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griswold IJ, et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to imatinib. Mol. Cell. Biol. 2006;26:6082–6093. doi: 10.1128/MCB.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corbin AS, Buchdunger E, Pascal F, Druker BJ. Analysis of the structural basis of specificity of inhibition of the Abl kinase by STI571. J. Biol. Chem. 2002;277:32214–32219. doi: 10.1074/jbc.M111525200. [DOI] [PubMed] [Google Scholar]
- 49.Yun CH, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emrick MA, et al. The gatekeeper residue controls autoactivation of ERK2 via a pathway of intramolecular connectivity. Proc. Natl. Acad. Sci. USA. 2006;103:18101–18106. doi: 10.1073/pnas.0608849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J. Exp. Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seeliger MA, et al. c-Src binds to the cancer drug imatinib with an inactive Abl/c-Kit conformation and a distributed thermodynamic penalty. Structure. 2007;15:299–311. doi: 10.1016/j.str.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 54.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr. D Biol. Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.