Abstract
Humans and most other animals use 2 different genetic codes to translate their hereditary information: the standard code for nuclear-encoded proteins and a modern variant of this code in mitochondria. Despite the pivotal role of the genetic code for cell biology, the functional significance of the deviant mitochondrial code has remained enigmatic since its first description in 1979. Here, we show that profound and functionally beneficial alterations on the encoded protein level were causative for the AUA codon reassignment from isoleucine to methionine observed in most mitochondrial lineages. We demonstrate that this codon reassignment leads to a massive accumulation of the easily oxidized amino acid methionine in the highly oxidative inner mitochondrial membrane. This apparently paradoxical outcome can yet be smoothly settled if the antioxidant surface chemistry of methionine is taken into account, and we present direct experimental evidence that intramembrane accumulation of methionine exhibits antioxidant and cytoprotective properties in living cells. Our results unveil that methionine is an evolutionarily selected antioxidant building block of respiratory chain complexes. Collective protein alterations can thus constitute the selective advantage behind codon reassignments, which authenticates the “ambiguous decoding” hypothesis of genetic code evolution. Oxidative stress has shaped the mitochondrial genetic code.
Keywords: evolution, methionine sulfoxide, nonstandard genetic code, protein oxidation, oxidative stress
After the first demonstration that the genetic code was not universal (1), various hypotheses have been elaborated to explain how the coordinate alteration of the major components carrying the genetic code, i.e., tRNAs, aminoacyl-tRNA-synthetases, and polypeptide chain release factors, might have been accomplished during evolution (2, 3). Because it has been deemed impossible that the collective alteration of all amino acid positions in a given proteome, which are encoded by a certain codon, might ever be of evolutionary benefit for an organism (4, 5), neutral mechanisms of evolution have arisen rapidly, and they have largely dominated the field except until recently (2–4, 6–9). Still, any adaptive nature of the later changes in the genetic code has continued to be doubted, because there was apparently little to be gained from code alterations in modern organisms (9). The main tenet and raison d'être of all neutral mechanisms of codon reassignment is that collective amino acid alterations would ineluctably be detrimental, why changes in the genetic code would not change protein primary structure (4, 10, 11). This postulate can be achieved by assuming a complex multistep process of codon reassignment that involves alternating evolutionary pressures acting on the DNA level to influence GC contents and thus codon usage (3). This concept is known as the “codon capture” hypothesis. The codon capture hypothesis has been cited as particularly expedient for the explanation of the unusual encoding of methionine instead of isoleucine by the codon AUA in different lineages of mitochondria (2, 4, 12). The latter codon reassignment has occurred independently at least 5 times during evolution, making it the most frequently evolved sense-to-sense genetic code reassignment known (8, 9).
Methionine is a sulfur-containing proteinogenic amino acid that is readily oxidized by reactive oxygen species (ROS) (13). The primary products of methionine oxidation are R- and S-methionine sulfoxide, which can be rereduced by 2 classes of stereospecific methionine sulfoxide reductases (MSRs) (14, 15) at the low metabolic cost of 1 molecule of NADPH/H+. Both types of MSRs have been shown to be vital for a variety of functions in bacteria, fungi and animals (14–16). In particular, knockout mice for MSRA have been shown to suffer from a decreased lifespan and several additional pathologies (16, 17). Moreover, mitochondrial dysfunction and decreased cell viability have been observed in human lens cells after MSRA silencing (18). Levine et al. (19) have proposed that the collective functional property of methionine in proteins was competitive antioxidant protection, i.e., the rapid scavenging of ambient ROS before they may attack other oxidant-labile sites, e.g., cofactors, within the carrier protein or closely adjacent macromolecular structures. Such a strategy might be of particular advantage if the methionine-protected structures were either more difficult or costly to repair than oxidized methionine itself, or if unrepaired methionine oxidation was less detrimental to the cell than the otherwise occurring oxidation reactions. Considering the straightforwardness of the MSR system (14, 17) and the observed tolerance of model proteins to surface methionine oxidation (19), this may be viewed to be a rather plausible assumption.
Nevertheless, the mere essentiality of the MSR system in various biological settings does not prove that the biochemical value of these enzymes extends beyond mere methionine maintenance. If, however, methionine was of additional antioxidant value to the cell in terms of the protection of adjacent and more critical biochemical sites, the seemingly, but not in truth paradoxical, situation might be encountered that this oxidant-labile amino acid was enriched in cellular compartments characterized by high ROS load. In contrast, if methionine was just an unusually oxidant-labile amino acid and the MSR enzymes just designed for the either complete or partial repair of inadvertently forming methionine sulfoxide, then methionine would be expected to be either unaltered or even depleted in cellular proteins subjected to high ROS fluxes.
Mitochondria are generally thought to constitute the major source and target of ROS in most cell types (20–22). In particular, the inner mitochondrial membrane displays a variety of peculiar adaptations and responses to oxidative stress (22–24). Hence, we have used a comparative genomics approach and analyzed an extensive set of mitochondrial and nuclear genomes for the encoded methionine contents. Thereby, we have (i) investigated our hypothesis that methionine usage in proteins exposed to particularly high ROS fluxes may show characteristic idiosyncrasies and (ii) tested the main tenet of the codon capture hypothesis, i.e., that encoded protein structures must not be altered by the use of a nonstandard genetic code. We have found that methionine is strikingly enriched in many mitochondrially encoded proteomes, especially in animals with high aerobic metabolic rate, and that the enrichment depended almost exclusively on the use of a second codon (AUA) in these mitochondria to encode the amino acid methionine.
Results and Discussion
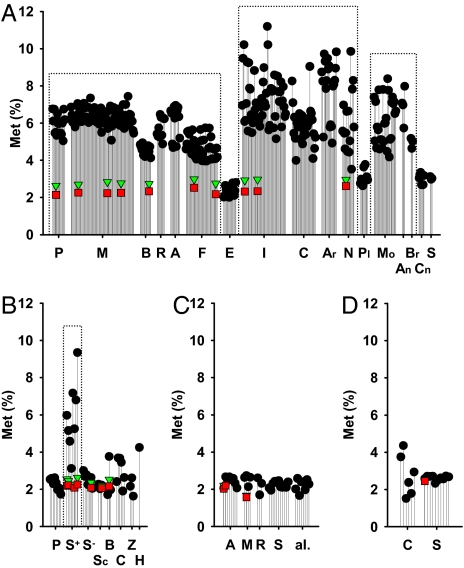
We have determined methionine content in the mitochondrially encoded respiratory chain proteins of 361 neutrally selected animals and various other eukaryotes, and we have compared the obtained methionine contents in mitochondria with baseline methionine usage in nuclear-encoded proteomes of a variety of genomically sequenced species. The results in Fig. 1 and supporting information (SI) Table S1 demonstrate that methionine use in nuclear-encoded proteins is very uniform in all eukaryotes (animals: 2.32% ± 0.14%; fungi: 2.15% ± 0.07%; other eukaryotes: 2.06% ± 0.32%, Arabidopsis thaliana: 2.45%). Mitochondrially encoded methionine contents in animals using the standard coding for this amino acid, i.e., echinoderms, platyhelminthes, cnidarians, and sponges, were quite similar to these values (overall average: 2.74% ± 0.48%). Moreover, also fungi, plants, and a collection of other eukaryotes all sharing the use of a single codon (AUG) for mitochondrial methionine did not differ from the rule that proteomic methionine usage generally amounts to 2–3% (fungi: 2.53% ± 0.64%; plants: 2,67% ± 0.63%; other eukaryotes: 2.42% ± 0.46%). In contrast, animals that also used a second codon for methionine in mitochondria considerably accumulated this amino acid in the encoded respiratory chain proteins (overall average: 6.17% ± 1.33%), with several insects reaching contents of 10% and more (Fig. 1A). Using 3 different approaches, the latter association was found to be statistically significant: regarding animal species as independent entities, the level of significance was P = 2.1×10−41 (n = 361); regarding only phyla as independent entities, significance was P = 5.7×10−5 (n = 10); phylogenetically independent contrast analysis on clades sharing the same AUA codon assignment returned a significance level of P = 0.003 (n = 5).
Fig. 1.
Mitochondrially encoded methionine contents in 361 animal species, 39 fungi, 34 unicellular eukaryotes, and 16 plants, compared with a reference selection of nuclear-encoded, proteomic methionine contents. (A) Methionine contents of mitochondrially encoded proteomes of 361 animal species (black circles). P, primates; M, other mammals; B, birds; R, reptiles; A, amphibians; F, fish; E, echinoderms; I, insects; C, crustaceans; Ar, arachnids; N, nematodes; Pl, platyhelminthes; Mo, molluscs; An, annelids; Br, brachiopods; Cn, cnidarians; S, sponges. Genomically encoded methionine contents of 10 reference species are shown for comparison (red squares). Adjustment of these nuclear-encoded proteomes to the higher transmembrane domain contents of the belonging mitochondrially encoded proteomes led to only marginally higher reference values (green triangles). The 10 reference species were (from left to right): Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus, Gallus gallus, Danio rerio, Tetraodon nigroviridis, Anopheles gambiae, Drosophila melanogaster, Caenorhabditis elegans. Clades, which use 2 codons (AUG and AUA) to encode mitochondrial methionine are marked by dotted frames. (B) Mitochondrially encoded methionine contents in 39 fungi. Symbols and frames are used as in A. P, pezizomycotina; S+, saccharomycotina, which decode AUA as methionine; S−, saccharomycotina, which decode AUA as isoleucine; Sc, schizosaccharomycetes; B, basidiomycota; C, chytridiomycota; Z, zygomycota; H, Hyaloraphidium curvatum. The 7 reference species were: Ashbya gossypii, Candida glabrata, Saccharomyces cerevisiae, Yarrowia lipolytica, Kluyveromyces lactis, Schizosaccharomyces pombe, Cryptococcus neoformans. (C) Mitochondrially encoded methionine contents in 34 unicellular eukaryotes. Symbols are used as in A. A, alveolata; M, mycetozoa; R, rhodophyta; S, stramenopiles; al., other eukaryotes. The 3 reference species were: Paramecium tetraurelia, Plasmodium falciparum, Dictyostelium discoideum. (D) Mitochondrially encoded methionine contents in 16 plants. Symbols are used as in A. C, chlorophyta; S, streptophyta. The reference species was Arabidopsis thaliana. All species names and calculated numeric values pertaining to this figure are given in Table S1.
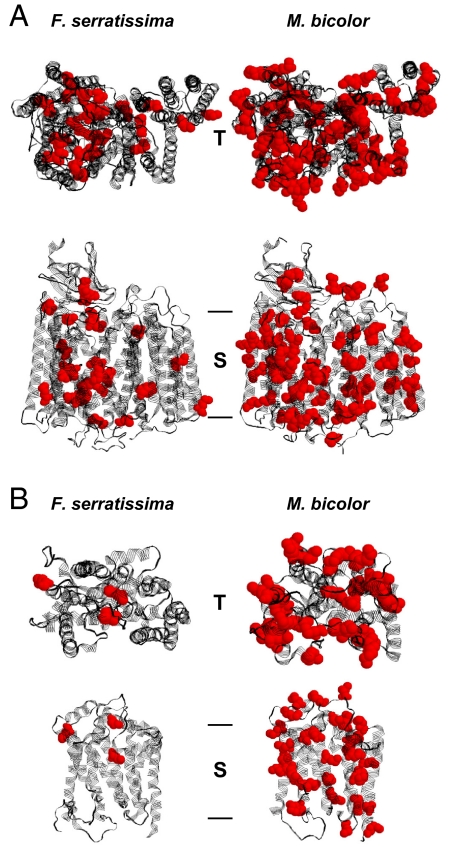
We have investigated the protein structural consequences of the resulting methionine accumulation for 2 of the encoded respiratory chain complexes, the core subunits of cytochrome c oxidase (COX), and cytochrome b, the central subunit of ubiquinone-cytochrome c oxidoreductase (Fig. 2 and Table 1). A comparison of the modeled structures of Florometra serratissima, an echinoderm using the standard code (2.0% methionine), and Melipona bicolor, an insect using the nonstandard code (11.2% methionine), indicates that methionine accumulation in the insect is massive, particularly pronounced in transmembrane domains, and primarily affects protein surfaces. A quantification of these observations is given in Table 1, for which the absolute surface exposure of all methionine residues was analyzed. Respecting COX, the average M. bicolor methionine is ≈2.5-fold more surface-exposed than the average F. serratissima methionine, which adds to the effect that the M. bicolor enzyme contains ≈3 times more methionine than its echinoderm counterpart. Ultimately, methionine builds 10.8% of the insect enzyme's surface, as opposed to 1.7% in the echinoderm.
Fig. 2.
Structural models of mitochondrially encoded respiratory chain proteins from the feather star Florometra serratissima and the stingless bee Melipona bicolor. (A) COX core, consisting of COX subunits I, II, and III. COX core is the central part of respiratory chain complex IV. (B) Cytochrome b, the mitochondrially encoded core peptide of ubiquinone-cytochrome c oxidoreductase (respiratory chain complex III). The top view representations (T) approximate the perspective from the mitochondrial intermembrane space. The side view representations (S) show the identical structures as beheld from within the inner mitochondrial membrane, placing the intermembrane space on top. Methionine residues are shown in red. Bars indicate the approximate membrane boundaries. A distinct accumulation of methionine can be discerned in the insect enzymes. The displayed structures correspond to the following methionine contents: COX core: 2.9% in F. serratissima, 7.8% in M. bicolor; cytochrome b: 1.1% in F. serratissima, 9.5% in M. bicolor. Mitochondrially encoded NADH dehydrogenase subunits of M. bicolor exhibited an even more pronounced methionine accumulation, reaching approximately double the level (13.5%) of the displayed COX core structure.
Table 1.
Surface exposure of methionine residues in respiratory chain proteins
| n(Met) | Average Met surface, Å2 | Total Met surface, Å2 | Total protein surface, Å2 | Met surface fraction, % | |
|---|---|---|---|---|---|
| COX core | |||||
| F. serratissima | 26 | 7.75 | 202 | 12,138 | 1.7 |
| M. bicolor | 73 | 18.08* | 1320 | 12,267 | 10.8 |
| B. taurus | 58 | 13.76† | 758 | 11,391 | 7.0 |
| Cytochrome b | |||||
| F. serratissima | 3 | 14.89 | 45 | 6,626 | 0.7 |
| M. bicolor | 33 | 27.73 | 915 | 6,116 | 15.0 |
| B. taurus | 15 | 20.92 | 314 | 6,172 | 5.1 |
Structural models of COX core and cytochrome b from F. serratissima, M. bicolor, and B. taurus were analyzed for methionine content (column 1), average solvent-accessible surface area per methionine residue (column 2), total methionine-covered protein surface area (column 3), and total protein surface area (column 4). Column 5 gives the percentage of the protein surface that is made up by methionine. Symbols indicate significantly increased surface exposure of the average methionine residue as compared with F. serratissima: *P = 0.003; †P = 0.037. Statistical analysis of both proteins combined yielded: M. bicolor vs. F. serratissima, P = 9×10−5; B. taurus vs. F. serratissima, P = 0.02.
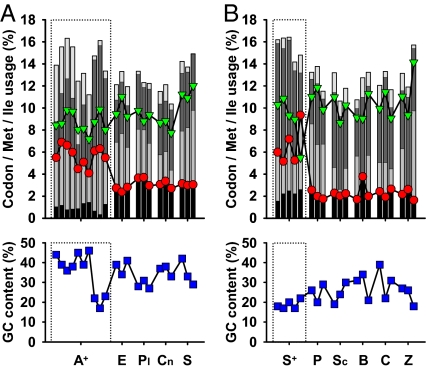
In summary, the recent evolutionary change of the genetic code in different lineages of mitochondria has resulted in an extraordinary increase in the use of methionine. Consequently, the codon capture hypothesis is inappropriate to account for the basic outcome of the AUA codon reassignment in mitochondria, namely methionine accumulation in the encoded proteins. To investigate whether this conclusion was also borne by the underlying coding characteristics, we have performed codon usage analyses on a variety of animals and fungi (Fig. 3). Although methionine was clearly increased in AUA-reassigning species, isoleucine was only mildly decreased due to an increased use of isoleucine codons (AUU and AUC) and an increased overall usage of the AUX family box. GC content was unrelated to the codon reassignment. Significantly, the AUA codon is not used less frequently in species that have undergone the reassignment than in species that have not; rather, animals make less use of the AUG codon if they can decode methionine by AUA. These observations diametrically contradict the postulates of the codon capture hypothesis (8), whereas they lend support to the hypothesis of an adaptive nature of the AUA codon reassignment.
Fig. 3.
Animal and fungal codon usage in relation to the mitochondrial AUA codon reassignment. (A) Methionine content (red circles), isoleucine content (green triangles), codon usage (bars, from bottom to top: AUG, AUA, AUU, AUC), and GC content (blue squares, Lower) in a representative selection of animals, which decode AUA as methionine (the 10 reference species from Fig. 1A, marked by dotted frames), vs. 3 representatives from each phylum decoding AUA as isoleucine. These representatives were selected based on the total number of protein sequences known for each species. A+, animals, which decode AUA as methionine; E, echinoderms; Pl, platyhelminthes; Cn, cnidarians; S, sponges. (B) The same data as in A, pertaining to fungi. Four reference species from Fig. 1B (A. gossypii, C. glabrata, S. cerevisiae, Y. lipolytica) plus Saccharomyces castellii, all decoding AUA as methionine and marked by dotted frames, compared with 3 representatives from each fungal clade decoding AUA as isoleucine. The latter representatives were selected on the same grounds as in A. S+, saccharomycotina, which decode AUA as methionine; P, pezizomycotina; Sc, schizosaccharomycetes; B, basidiomycota; C, chytridiomycota; Z, zygomycota.
Which other theory of genetic code evolution might have predicted the results observed? Only 1 of the alternative concepts allows and explicitly considers protein alterations as evolutionary driving force of codon reassignments, the “ambiguous decoding” hypothesis (3, 5, 7, 10, 11). According to this model, simple mutations in tRNAs or aminoacyl-tRNA-synthetases would more or less gradually lead to the loading of a tRNA with a “new” amino acid, resulting in the insertion of this new amino acid at a site in the protein that would still be encoded by the “old” codon. Thus, genomic factors such as GC pressure are irrelevant in this scenario, whereas the whole reassignment process would be determined by the outcome on the protein level. Specifically, the ambiguous decoding hypothesis predicts that “immediately” after the codon reassignment, the same protein sites formerly occupied by the old amino acid would now be occupied by the new amino acid. This prediction seems to be surprisingly fulfilled by the AUA codon reassignment, because the new methionines cluster on the protein surfaces of the encoded membrane proteins (Table 1), a topological position that would typically be expected to be rich in isoleucine (encoded by any codon), but not methionine. In animals, the old AUG-encoded internal methionines would even be partially lost (Fig. 3), whereas pressures from the fewer isoleucine-encoding codons (2 instead of 3) would be counterbalanced by an increased frequency of the remaining codons and the complete family box.
What might have been the selective advantage of the observed accumulation of methionine in inner mitochondrial membrane respiratory chain complexes? Two biochemically fundamental premises point toward a specific solution: first, methionine is a very easily oxidized amino acid, and it may be the most readily oxidized residue in certain biochemical settings (13). Despite efficient reduction enzymes, steady-state levels of methionine sulfoxide amount to 5–10% of all protein methionine residues in the majority of tissues (25). Second, the mitochondrial compartment, particularly the inner mitochondrial membrane, is characterized by exceptionally oxidative conditions (20, 22). Thus, pronounced methionine oxidation in mitochondria is inevitable, such that the observed methionine accumulation would be clearly counterproductive for at least 2 reasons if there were no compensatory beneficial mechanisms of exceeding selective advantage: first, the inescapable structural alterations imposed by the mere exchange of amino acids (4, 5), even if those alterations may be uncommonly mild in the present example (Table 2), and second, the need to permanently repair or replace the newly introduced unstable methionine residues, which are henceforth used instead of stable isoleucine residues.
Table 2.
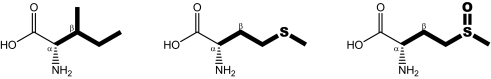
Chemical structures and physicochemical properties of isoleucine, methionine, and methionine sulfoxide
 |
|||
|---|---|---|---|
| LogP | 2.38 | 1.63 | −0.50 |
| p [D] | 0.01 | 1.56 | 3.79 |
| A [Å2] | 117.7 | 122.0 | 125.0 |
| Ap [Å2] | 0.0 | 0.0 | 22.3 |
The following properties were calculated for the displayed amino acid side chains depicted in bold: LogP, octanol-water partition coefficient; p, dipole moment; A, Connolly molecular surface area; Ap, polar surface area. Transitions from isoleucine to methionine are accompanied by a loss in sterical demand in the β position, whereas the overall elongated and hydrophobic character of the side chain is largely maintained.
The most parsimonious explanation for this conundrum would postulate that the redox properties of methionine themselves constituted the exceeding selective advantage, because most other physicochemical properties of isoleucine and methionine are not greatly dissimilar (Table 2). In fact, several biochemical studies have described protein methionine as an intrinsic antioxidant shield of proteins against the oxidative destruction of other, potentially more critical biochemical sites within the same or in neighboring proteins (15, 17, 19). Thus, targeted antioxidant activities of methionine might provide a plausible origin of significant vantages of methionine accumulation as effected by the AUA codon reassignment. This concept is supported by the following observations: first, the structural models of Fig. 2 evidence that methionine is selectively enriched on the protein surface or the lipid bilayer interface. At these sites, methionine residues are especially reactive (17, 19), whereas their oxidation to sulfoxides does not entail major structural distortion of the carrier protein (19). Second, methionine can be economically repaired by MSRs, which are known to be expressed in mitochondria and whose silencing has been shown to compromise mitochondrial membrane potential in cultivated cells (18). We have examined all genomically sequenced, methionine-accumulating animals and fungi for sequences homologous to Escherichia coli MSR-A and -B, and we have found corresponding sequences in all cases (data not shown), indicating that this potentially mandatory condition for the extensive use of methionine as antioxidant was fulfilled. In contrast, various unicellular organisms reading AUA for isoleucine, particularly anaerobes, did not reveal MSR-homologous sequences. It remains to be determined, though, how far MSRs are capable of repairing intramembrane sulfoxides, or whether those methionine residues are rather installed for “single use.” Third, mitochondrial methionine in animals apparently runs parallel to aerobic metabolic capacity. The anaerobic platyhelminthes of Fig. 1A and slowly respiring sponges and echinoderms do not accumulate methionine and have refrained from altering their genetic code, whereas insects, arachnids, and mammals synthesize particularly methionine-rich respiratory chain complexes (Fig. 1).
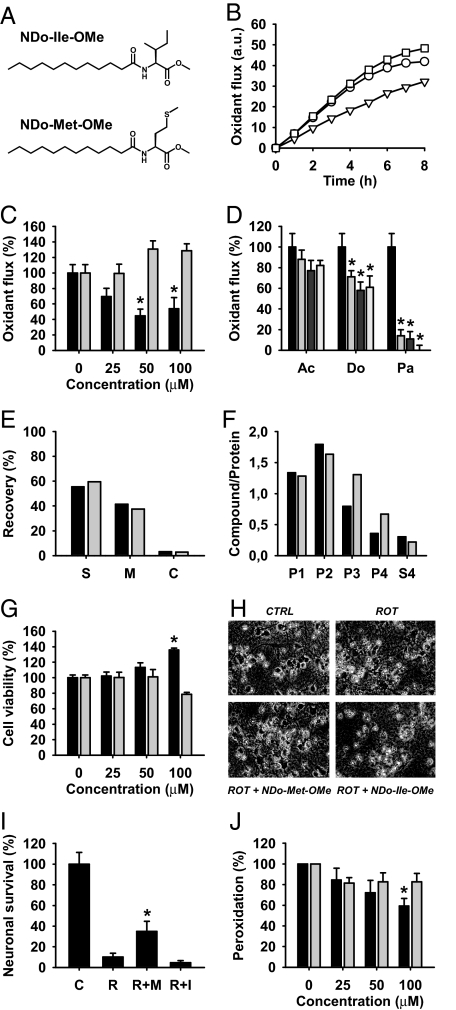
We sought to experimentally confirm our biochemical and evolutionary reasoning. All proteins affected by the AUA codon reassignment are integral membrane proteins, whose overall transmembrane domain content amounts to some exceptional 55%. In addition, they seem to expose the majority of their newly gained methionines to the lipid bilayer (Fig. 2; Table 1). Thus, we have synthesized lipophilic methionine derivatives (Fig. 4A) to mimic the biochemical result of the evolutionary AUA codon reassignment from isoleucine to methionine and have investigated these compounds for their potential to decrease oxidative stress in intact eukaryotic cells and isolated mitochondria. The results in Fig. 4 show that intramembrane methionine enrichment in fact suppresses the measurable flux of oxidants in respiring tumor cells (Fig. 4B). A similar result was found after the exogenous addition of the physiological peroxide H2O2, which can emerge from numerous metabolic processes and mediates signaling and toxic responses (26, 27). A comparable reduction of cellular ROS was not observed with isoleucine enrichment of the cells (Fig. 4C). In addition, the reduction of ROS by methionine derivatives depended on the intramembrane localization of the applied thioether moiety (of methionine), because more hydrophilic but cell-permeable compounds, such as acetyl methionine methyl ester, were hardly effective (Fig. 4D).
Fig. 4.
Antioxidant and cytoprotective effects of intramembrane methionyl accumulation. (A) Chemical structures of 2 model compounds used to mimic the cellular effect of the AUA codon reassignment in mitochondria. The 2 structures are N-dodecanoyl methionine methyl ester (NDo-Met-OMe) and N-dodecanoyl isoleucine methyl ester (NDo-Ile-OMe). (B) Metabolic oxidant flux in respiring SH-SY5Y neuroblastoma cells as measured by the temporal increase in cellular DCF fluorescence. Treatments were vehicle (circles), 50 μM NDo-Ile-OMe (squares), and 50 μM NDo-Met-OMe (triangles). (C) Induced oxidant flux in SH-SY5Y cells after addition of hydrogen peroxide (H2O2; 500 μM). Shown are normalized increases in cellular DCFA fluorescence 1 h after application of the peroxide. Black bars represent NDo-Met-OMe, gray bars represent NDo-Ile-OMe. (D) Comparison of N-acylated methionine derivatives as oxidant quenchers in SH-SY5Y cells. Experiments were done as in C; measurements were done after 3 h of incubation with H2O2. Ac is N-acetyl methionine methyl ester, Do is N-dodecanoyl methionine methyl ester, Pa is N-palmitoyl methionine. Each stack of bars represents concentrations of 0, 25, 50, 100 μM of the corresponding compound from left to right. (E) Cell permeability and membrane accumulation of NDo-Met-OMe (black bars) and NDo-Ile-OMe (gray bars), analyzed by HPLC with fluorescence detection. SH-SY5Y cells were treated with both compounds (100 μM) as in C, after which the different compartments were separated by centrifugation. (S) supernatant (incubation medium); M, membrane fraction; C, cytosolic fraction. Data are presented as relative recovery of each applied compound. (F) Subcellular distribution of NDo-Met-OMe (black bars) and NDo-Ile-OMe (gray bars) in SH-SY5Y cells. Organelle-enriched fractions were prepared after 1-h incubation (100 μM concentration) as in C. P1, nuclear fraction; P2, mitochondrial fraction; P3, Golgi fraction; P4, endoplasmic reticulum fraction; S4, cytosolic fraction. (G) Primary mesencephalic cell culture survival under hyperoxic culture conditions (20% oxygen). Metabolic MTT reduction was measured in mature cultures after a differential incubation with NDo-Met-OMe vs. NDo-Ile-OMe for 3 d. (H) Micrographs of primary mesencephalic cells incubated with rotenone (50 nM) and the indicated amino acid derivatives (100 μM) for 3 d. (I) The same experiment as in H, quantified by microscopic evaluation of neuronal survival. (J) Lipid peroxidation in rat liver mitochondria (1 mg/ml protein), induced by 10-min incubation with 200 μM Fe2+/100 μM H2O2. Mitochondria were preincubated with the compounds to be tested for 30 min. TBARS formation was measured by fluorimetric assay. Black bars represent NDo-Met-OMe, gray bars represent NDo-Ile-OMe. Numeric results are given as mean ± standard deviation. Asterisks denote significantly decreased oxidant flux (C and D), significantly increased cellular survival (G and I), or significantly decreased lipid peroxidation (J) as compared with the control (n ≥ 3; P < 0.01 by ANOVA with Student–Newman–Keul's test).
The cell penetration and intramembrane accumulation of both model compounds of Fig. 4A were verified by means of an HPLC-based analytical protocol. The results in Fig. 4E indicate that, under the authentic conditions of the antioxidative activity assay in Fig. 4C, both compounds achieved approximately the same cell penetration (NDo-Met-OMe: 44%; NDo-Ile-OMe: 40%). In addition, both compounds elicited considerable accumulation in membranes as expected: the membrane-to-cytosol distribution factor was 13.4 for NDo-Met-OMe and 13.5 for NDo-Ile-OMe. Tracking the organelle distribution of the applied compounds by subcellular fractionation (Fig. 4F), it was found that both compounds attained the highest concentrations in the mitochondrial fraction (P2), followed by the nuclear fraction (P1). Hence, both compounds reached their assumed site of action to a similar degree, which rules out differential distribution as an explanation for their disparate antioxidant activities.
In primary mesencephalic neurons, which were used for their high sensitivity to oxidants of mitochondrial origin (28), intramembrane methionine enrichment resulted in increased survival under the physiologically hyperoxic conditions of the incubator (20% O2) (Fig. 4G). Moreover, treatment with 100 μM NDo-Met-OMe was able to partially prevent cell death from enhanced mitochondrial oxidant production (Fig. 4 H and I) as induced by the mitochondrial toxin rotenone (29). Rotenone is a well-established inhibitor of mitochondrial complex I that has been demonstrated to induce mitochondrial oxidative stress in vitro, resulting in a syndrome resembling Parkinsons's disease in vivo (29).
Finally, we aspired to obtain a quantitative comparison between the degree of methionine accumulation effected by the mitochondrial codon reassignment and the pharmacological application of our model compounds. Rat liver mitochondria arguably constitute the best-characterized mitochondrial species. Using literature values on the frequency of each respiratory chain complex per mitochondrion (30) and data from the National Center for Biotechnology Information (NCBI) Genome Database, we were able to calculate the approximate increase in protein methionine effected by the codon reassignment (Table S2). In rat liver mitochondria, a “sudden loss” of the codon reassignment would decrease the total protein-bound methionine concentration by ≈8 mM. Similarly, a difference of 11.5 mM methionine can be calculated for M. bicolor vs. F. serratissima mitochondria, if similar respiratory chain complex copy numbers as in rat liver mitochondria are assumed (Table S2). In return, freshly prepared rat liver mitochondria were significantly protected from iron/hydrogen peroxide toxicity by 100 μM NDo-Met-OMe (Fig. 4J). Under the conditions of the experiment, 42% of the applied substance entered the mitochondria (data not shown). Using published values (30) for rat liver mitochondrial volume (2.7×10−16 L) and protein content (1.1×10−13 g), these figures translate into a mitochondrial NDo-Met-OMe concentration of ≈17 mM. Hence, the mitochondrial genetic code reassignment and the pharmacological application of NDo-Met-OMe appear to match on a topological, qualitative, and even quantitative level. Thus, intramembrane methionine enrichment seems to provide significant antioxidant protection to eukaryotic cells, illustrating why mitochondrial inner membranes as prime targets of ROS have been selected by evolution to acquire and sustain unrivalled contents of methionine.
In summary, mitochondrially encoded proteomes of most animals contain conspicuously higher contents of the easily oxidized amino acid methionine than nuclear-encoded proteomes. This increase is due to the use of a second codon for methionine as part of a nonstandard genetic code, which could be installed against strong evolutionary pressures of conservation, because isoleucine and methionine were of similar size and shape, minimizing protein structural distortion that may have been caused by their exchange, whereas methionine provided an immediate selective advantage as antioxidant on the formerly isoleucine-occupied hydrophobic surfaces. Our results offer a specific and functional explanation for the modern evolution of a nonstandard genetic code as a unique adaptation of mitochondria to oxidative stress.
Materials and Methods
Data Sources and Calculations.
Sequences for mitochondrial genome and proteome analysis were obtained at the indicated time points from the NCBI genome database (www.ncbi.nlm.nih.gov/genomes/static/euk_o.html). Nuclear-encoded proteomes were from the European Bioinformatics Institute (EBI) Integr8 database (www.ebi.ac.uk/integr8). Proteomic amino acid contents were quantified by using customized Perl scripts. Protein transmembrane domains were identified by means of a hidden Markov model algorithm (TMHMM) (31).
Inclusion Criteria for the Analyzed Species.
All metazoan phyla with 3 or more fully sequenced mitochondrial genomes were considered for analysis. Phyla with <50 sequenced representatives were analyzed in full (sponges, cnidarians, brachiopods, and annelids were processed in April 2006; mollusks, platyhelminthes, nematodes, and echinoderms were processed in November 2005). Phyla with >50 sequenced mitochondrial genomes were partially considered: of the arthropods, all insects, crustaceans, and arachnids were analyzed in November 2005; of the chordates, all species listed in an arbitrarily chosen reference book (32) were sampled in August 2005. Pertaining to fungi, plants, and other eukaryotes, all totally sequenced mitochondria were analyzed in April 2006. All nuclear proteomes available from the EBI Integr8 database were analyzed in November 2005.
Molecular Modeling.
3D models of respiratory chain proteins were generated by alignment with the experimental crystal structures of corresponding bovine sequences [COX: Protein Data Bank (PDB) 1v54; cytochrome b: PDB 1bgy], which were obtained from the PDB (www.rcsb.org). Structural calculations were performed on the Geno3D server of the Pôle Bioinformatique Lyonnais (33) (http://geno3d-pbil.ibcp.fr). Surface exposure data were calculated on the WHAT IF server (34) (http://swift.cmbi.ru.nl/whatif).
Biochemical and Cell Culture Experiments.
All experimental procedures were conducted according to established protocols published elsewhere; customized chemicals were synthesized as described (35) from commercially available acyl halides (Sigma–Aldrich) and amino acid esters (Bachem) and were analyzed by TLC, mass spectrometry, and 1H-NMR. Clonal and primary cell culture were essentially done as described (36, 37); rat primary mesencephalic cells were cultivated in neurobasal medium (Invitrogen) supplemented with B27 without antioxidants. Primary cells were used for experiments after 9 days of differentiation in vitro.
Cellular oxidative flux was quantified on stringently washed SH-SY5Y neuroblastoma cells in suspension (106 cells per ml) loaded with the redox-sensitive fluorescent dye 2′,7′-dichlorofluorescin diacetate (DCFA) (38) (5 μM). Cell viability analyses were either done by the metabolic 3-(4,5-dimethylthiazol-2-yl-)-2,5-diphenyltetrazolium bromide (MTT) reduction method or by microscopic examination for morphologically intact neurons and counting of the resulting cell numbers (38).
Subcellular fractionation of neuroblastoma cells was done by stepwise differential centrifugation (39). Cell membranes were prepared by sonication of the cells in authentic homogenization buffer (39), followed by ultracentrifugation at 90,000 ×g for 40 min.
NDo-Met-OMe and NDo-Ile-OMe were analyzed by means of a fluorescence derivatization RP-HPLC routine. In brief, cellular fractions were supplemented with 50 nmol of the internal standards (N-dodecanoyl valine ethyl ester, NDo-Val-OEt, and heptadecanoic acid), after which they were extracted with chloroform-methanol as described (40). The resulting lipids were dried and hydrolyzed in 0.5 M methanolic KOH (80 °C, 1 h). After acidification with HCl, the hydrolysate was extracted with hexane, dried, redissolved in acetonitrile, and derivatized with the carboxylic acid-reactive fluorescent label 4-bromomethyl-7-methoxycoumarin as published (41). The obtained fluorescently labeled fatty acids and dodecanoyl amino acids were quantified by reversed-phase HPLC at 325-nm excitation/398-nm emission, using a 35-min acetonitrile-water gradient (60–100% acetonitrile).
Rat liver mitochondria were prepared following standard protocols (30, 42). Thiobarbituric acid reactive substances (TBARS) were measured after 10-min induction of lipid peroxidation with 200 μM Fe2+/100 μM H2O2 as free radical initiation system (42).
Statistical Analyses.
Experimentally generated data were tested for statistical significance by 1-way ANOVA followed by Student–Newman–Keul's multiple comparisons test. Mitochondrially encoded methionine contents and surface exposures were analyzed with the same algorithm. Evolutionary trees and phylogenetically independent contrasts were generated as described (24). The obtained independent contrasts were evaluated by correlation analysis.
Supplementary Material
Acknowledgments.
We thank P. Dehghanpoor for developing software tools, A. Hohberger for assisting with the analysis of mitochondrial genomes, A. Krogh for providing transmembrane domain prediction software (TMHMM), T. Garland for contributing phylogenetic analysis software (PDAP), J. Mocko for sharing primary mesencephalic cultures, and C. Behl for stimulating discussions. This work was supported by the Hans-Gottschalk-Stiftung.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802779105/DCSupplemental.
References
- 1.Barrell BG, Bankier AT, Drouin J. A different genetic code in human mitochondria. Nature. 1979;282:189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- 2.Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: Evolvability of the genetic code. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 3.Santos MA, Moura G, Massey SE, Tuite MF. Driving change: The evolution of alternative genetic codes. Trends Genet. 2004;20:95–102. doi: 10.1016/j.tig.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Osawa S, Jukes TH, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crick FH. The origin of the genetic code. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 6.Pezo V, et al. Artificially ambiguous genetic code confers growth yield advantage. Proc Natl Acad Sci USA. 2004;101:8593–9597. doi: 10.1073/pnas.0402893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultz DW, Yarus M. On malleability in the genetic code. J Mol Evol. 1996;42:597–601. doi: 10.1007/BF02352290. [DOI] [PubMed] [Google Scholar]
- 8.Swire J, Judson OP, Burt A. Mitochondrial genetic codes evolve to match amino acid requirements of proteins. J Mol Evol. 2005;60:128–139. doi: 10.1007/s00239-004-0077-9. [DOI] [PubMed] [Google Scholar]
- 9.Sengupta S, Yang X, Higgs PG. The mechanisms of codon reassignments in mitochondrial genetic codes. J Mol Evol. 2007;64:662–688. doi: 10.1007/s00239-006-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jukes TH, Osawa S. Further comments on codon reassignment. J Mol Evol. 1997;45:1–3. doi: 10.1007/pl00006192. [DOI] [PubMed] [Google Scholar]
- 11.Yarus M, Schultz DW. Further comments on codon reassignment. Response. J Mol Evol. 1997;45:3–6. doi: 10.1007/pl00006171. [DOI] [PubMed] [Google Scholar]
- 12.Castresana J, Feldmaier-Fuchs G, Pääbo S. Codon reassignment and amino acid composition in hemichordate mitochondria. Proc Natl Acad Sci USA. 1998;95:3703–3707. doi: 10.1073/pnas.95.7.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogt W. Oxidation of methionyl residues in proteins: Tools, targets, and reversal. Free Radic Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 14.Weissbach H, et al. Peptide methionine sulfoxide reductase: Structure, mechanism of action, and biological function. Arch Biochem Biophys. 2002;397:172–178. doi: 10.1006/abbi.2001.2664. [DOI] [PubMed] [Google Scholar]
- 15.Moskovitz J. Methionine sulfoxide reductases: Ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim Biophys Acta. 2005;1703:213–219. doi: 10.1016/j.bbapap.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Moskovitz J, et al. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci USA. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stadtman ER, Moskovitz J, Berlett BS, Levine RL. Cyclic oxidation and reduction of protein methionine residues is an important antioxidant mechanism. Mol Cell Biochem. 2002;234–235:3–9. [PubMed] [Google Scholar]
- 18.Marchetti MA, et al. Silencing of the methionine sulfoxide reductase A gene results in loss of mitochondrial membrane potential and increased ROS production in human lens cells. Exp Eye Res. 2006;83:1281–1286. doi: 10.1016/j.exer.2006.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 21.Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowaltowski AJ, Vercesi AE. Mitochondrial damage induced by conditions of oxidative stress. Free Radic Biol Med. 1999;26:463–471. doi: 10.1016/s0891-5849(98)00216-0. [DOI] [PubMed] [Google Scholar]
- 23.Kowaltowski AJ, Castilho RF, Vercesi AE. Mitochondrial permeability transition and oxidative stress. FEBS Lett. 2001;495:12–15. doi: 10.1016/s0014-5793(01)02316-x. [DOI] [PubMed] [Google Scholar]
- 24.Moosmann B, Behl C. Mitochondrially encoded cysteine predicts animal lifespan. Aging Cell. 2008;7:32–46. doi: 10.1111/j.1474-9726.2007.00349.x. [DOI] [PubMed] [Google Scholar]
- 25.Stadtman ER, Van Remmen H, Richardson A, Wehr NB, Levine RL. Methionine oxidation and aging. Biochim Biophys Acta. 2005;1703:135–140. doi: 10.1016/j.bbapap.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Behl C, Davis JB, Lesley R, Schubert D. Hydrogen peroxide mediates amyloid beta protein toxicity. Cell. 1994;77:817–827. doi: 10.1016/0092-8674(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 27.Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen peroxide: A metabolic by-product or a common mediator of ageing signals? Nat Rev Mol Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 28.Moon Y, Lee KH, Park JH, Geum D, Kim K. Mitochondrial membrane depolarization and the selective death of dopaminergic neurons by rotenone: Protective effect of coenzyme Q10. J Neurochem. 2005;93:1199–1208. doi: 10.1111/j.1471-4159.2005.03112.x. [DOI] [PubMed] [Google Scholar]
- 29.Sherer TB, et al. Mechanism of toxicity in rotenone models of Parkinson's disease. J Neurosci. 2003;23:10756–10764. doi: 10.1523/JNEUROSCI.23-34-10756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwerzmann K, Cruz-Orive LM, Eggman R, Sänger A, Weibel ER. Molecular architecture of the inner membrane of mitochondria from rat liver: A combined biochemical and stereological study. J Cell Biol. 1986;102:97–103. doi: 10.1083/jcb.102.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 32.Carey JR, Judge DS. Life Spans of Mammals, Birds, Amphibians, Reptiles, and Fish. Monographs on Population Aging. No. 8. Odense, Denmark: Odense Univ Press; 2000. [Google Scholar]
- 33.Combet C, Jambon M, Deléage G, Geourjon C. Geno3D: Automatic comparative molecular modelling of protein. Bioinformatics. 2002;18:213–214. doi: 10.1093/bioinformatics/18.1.213. [DOI] [PubMed] [Google Scholar]
- 34.Vriend G. WHAT IF: A molecular modeling and drug design program. J Mol Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 35.Fincher TK, Yoo SD, Player MR, Sowell JW, Michniak BB. In vitro evaluation of a series of N-dodecanoyl-L-amino acid methyl esters as dermal penetration enhancers. J Pharmacol Sci. 1996;85:920–923. doi: 10.1021/js9600787. [DOI] [PubMed] [Google Scholar]
- 36.Moosmann B, Behl C. The antioxidant neuroprotective effects of estrogens and phenolic compounds are independent from their estrogenic properties. Proc Natl Acad Sci USA. 1999;96:8867–8872. doi: 10.1073/pnas.96.16.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bayatti N, Zschocke J, Behl C. Brain region-specific neuroprotective action and signaling of corticotropin-releasing hormone in primary neurons. Endocrinology. 2003;144:4051–4060. doi: 10.1210/en.2003-0168. [DOI] [PubMed] [Google Scholar]
- 38.Moosmann B, Skutella T, Beyer K, Behl C. Protective activity of aromatic amines and imines against oxidative nerve cell death. Biol Chem. 2001;382:1601–1612. doi: 10.1515/BC.2001.195. [DOI] [PubMed] [Google Scholar]
- 39.Petit N, et al. Selenoprotein N: An endoplasmic reticulum glycoprotein with an early developmental expression pattern. Hum Mol Genet. 2003;12:1045–1053. doi: 10.1093/hmg/ddg115. [DOI] [PubMed] [Google Scholar]
- 40.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 41.Wolf JH, Korf J. Automated solid-phase catalyzed pre-column derivatization of fatty acids for reversed-phase high-performance liquid chromatographic analysis with fluorescence detection. J Chromatogr. 1988;436:437–445. doi: 10.1016/s0021-9673(00)94603-x. [DOI] [PubMed] [Google Scholar]
- 42.Nagababu E, Lakshmaiah N. Inhibitory effect of eugenol on non-enzymatic lipid peroxidation in rat liver mitochondria. Biochem Pharmacol. 1992;43:2393–2400. doi: 10.1016/0006-2952(92)90318-d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.