Abstract
Transition path sampling (TPS) has been applied to the chemical step of human purine nucleoside phosphorylase (PNP). The transition path ensemble provides insight into the detailed mechanistic dynamics and atomic motion involved in transition state passage. The reaction mechanism involves early loss of the ribosidic bond to form a transition state with substantial ribooxacarbenium ion character, followed by dynamic motion from the enzyme and a conformational change in the ribosyl group leading to migration of the anomeric carbon toward phosphate, to form the product ribose 1-phosphate. Calculations of the commitment probability along reactive paths demonstrated the presence of a broad energy barrier at the transition state. TPS identified (i) compression of the O4′···O5′ vibrational motion, (ii) optimized leaving group interactions, and (iii) activation of the phosphate nucleophile as the reaction proceeds through the transition state region. Dynamic motions on the femtosecond timescale provide the simultaneous optimization of these effects and coincide with transition state formation.
Keywords: dynamics in catalysis, promoting vibrations, purine nucleoside phosphorylase, transition path sampling, transition state lifetime
Protein motions are widely accepted to be intimately connected to the catalytic function of enzymes. Important motions on different timescales have been experimentally and computationally observed, and a direct link between microsecond to millisecond domain motions and enzymatic function has been established (1–3). Although gating modes have been identified that link dynamics of the protein environment to hydrogen transfer (4) the role of dynamics on the timescale of bond vibrations (femtoseconds to picoseconds) in enzyme catalytic efficiency is less well understood. We have proposed that these sub-ps motions, which we termed “protein promoting vibrations,” may be coupled to the reaction coordinate and influence catalysis (5–8).
Molecular dynamics (MD) simulations can provide details at an atomistic level, however, the accessible simulation time is usually in the picosecond to nanosecond timescale. It is currently not possible for a conventional MD simulation to study a rare chemical event, such as an enzymatic reaction, whose overall turnover rate is on the millisecond timescale. Techniques such as umbrella sampling, which impose a bias potential on a guess for the reaction coordinates, have been widely used in studies of enzymatic reaction mechanisms. Chandler and coworkers developed the Transition Path Sampling (TPS) method to overcome both the long timescale problem and the lack of knowledge about reaction mechanisms (9, 10). This method performs Monte Carlo (MC) walks through either trajectory space (MD) or stochastic (MC) space and is used in conjunction with either MD or MC. The principal advantage of TPS is to allow the study of reaction mechanisms in atomic detail, without prior knowledge of the reaction coordinate and transition state.
The TPS algorithm has been applied in studies of lactase dehydrogenase (LDH) (11). The results suggested that hydride transfer is facilitated by linked motions of residues originating far from the active site (11). A subsequent study (12) determined the residue motions that define the reaction coordinate. The transition state was tightly coupled to these protein motions.
Here, we explore the connection between enzyme dynamics and transition state formation in purine nucleoside phosphorylase (PNP). PNP is the only enzyme in humans that catalyzes phosphorolysis of 6-oxypurine (deoxy)nucleosides to generate the corresponding purine bases and (deoxy)ribose 1-phosphate. The reaction involves phosphorolysis at C1′ of ribose with inversion of stereochemistry (β to α) at this position (Fig. 1). The structure of PNP is trimeric with the catalytic sites near the subunit interfaces (Fig. 2). Previous studies suggested that compression of the O4′···O5′ ribosyl oxygens is linked to catalysis (13, 14). The full protein dynamic architecture is proposed to be linked to transition state formation because amino acid mutations far from the active site can enhance the catalytic efficiency and can alter transition state structure (15). Remote mutations caused stronger coupling of catalytically favorable compression motions in the mutant enzyme than in the native PNP (14). Moreover, KIE measurements in the mutant enzyme revealed altered transition state structures compared with the native enzyme (16). These computational and experimental findings indicate that transition state structure is altered by mutations far from the catalytic site, because these mutations alter dynamic coupling of protein motion to the reaction coordinate. Here, we employ TPS to link dynamic motion with formation of the transition state. Protein dynamics on the femtosecond to picosecond timescale are linked to enzymatic function. We show at the atomic bond vibrational level exactly how the chemical reaction is accomplished by PNP.
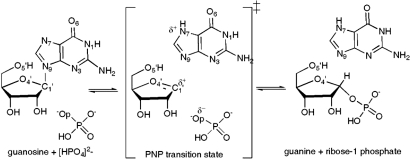
Fig. 1.
Phosphorolysis of guanosine catalyzed by PNP. A schematic rendering of reactants, ribooxacarbenium ion transition state, and products.
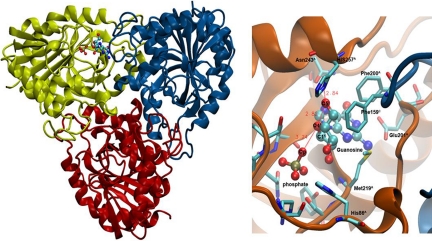
Fig. 2.
Structure of PNP trimer with substrates guanosine and phosphate bound in a catalytic site near the trimer interface.
Results and Discussion
Transient Time.
The output of the TPS algorithm is an ensemble of unbiased trajectories that connect reactants and products, to provide the dynamic motions linked to the reaction. Atomic details of the protein and reactant motion are revealed as they progress through the transition state. A total of 220 reactive trajectories were generated and analyzed to find common features of bond excursions linked to transition state formation in the phosphorolysis reaction catalyzed by PNP (Fig. 1).
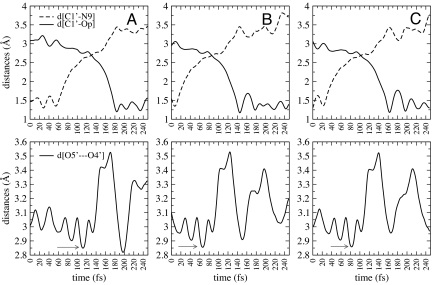
Bond-breaking (C1′–N9) and bond-forming (C1′–OP) distances as the reaction proceeds from reactant to product are shown for 2 trajectories that belong to statistically distinct parts of phase space of the transition path ensemble (Fig. 3). The reactive trajectories generated from TPS of 250 fs in length are long enough to capture transition state excursions. A more extended reactive trajectory (20 ps) revealed no recrossing. The transient time (Tts) was defined as the time a trajectory spent after leaving the reactant state but before it entered the product state. Specifically, it is the time spent when the distances d(C1′−N9) ≥ 1.7 Å and d(C1′−Op) ≥ 1.8 Å. Thus, the transient time is the typical time for a trajectory to cross the transition state barrier and commit to 1 of the 2 stable states (10). The transient time was ≈100 fs from the collected transition path ensemble. This time is long compared with the Tts = 10 fs observed in a TPS study of hydride transfer in LDH (11, 12), consistent with the more complex dynamic motion required in the transition state for PNP than for hydride transfer in LDH. These motions will be examined in detail in the following section. A similar range of Tts was observed (85 fs) for the isomerization of butane by Zuckerman and Woolf (17) and (≥130 fs) for the Claisen rearrangement catalyzed by chorismate mutase (18). In addition, the frequency factory was computed from the transition path ensemble and plateaued at 150 femtoseconds, which verified that 250 femtoseconds was an appropriate path length [see supporting information (SI) Fig. S1] (19).
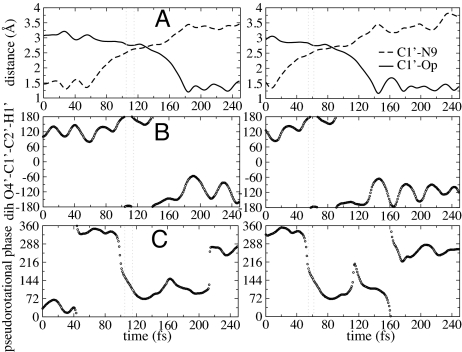
Fig. 3.
Geometry changes associated with the transition state excursion of PNP. Plot of bond-breaking (C1′−N9) and bond-forming (C1′−Op) distances (A); dihedral angle of O4′–C1′–C2′–H1′representing a hybridization orbital of the C1′reactive center (B); and the pseudorotational phase of the ribose ring (C) as a function of time, as the chemical reaction proceeds in the direction from reactants (guanosine and phosphate) to products (guanine and ribose-1 phosphate). Two examples of trajectories selected from the transition path ensemble are shown. Transition state regions were found from commitment probabilities and are located at 105–115 fs (Left) and at 57–62 fs (Right), indicated by the vertical lines.
Location of Transition State.
The transition state structures were identified based on the statistical definition of the stochastic seperatrix (20). In a complex system like PNP, the transition state structures do not coincide with a single saddle point, but are identified by the equal probability to commit to reactants and products. Fig. 4 shows the commitment probabilities of a given configuration along 2 reactive trajectories from the TPS ensemble, each from a different part of phase space. In the reactant region, PP = 0, and as the reaction progresses, its value increases to 1 in the product region. We identify the transition state as the configurations for which the value of Pp was in the range 0.4–0.6. There are ≈10 configurations that satisfy this requirement, located at 105–115 fs in Fig. 4 Upper and at 57–62 fs in the Fig. 4 Lower. This wide range (10 structures; see Fig. S2) of transition states suggests a broad activation energy barrier and indicates a lifetime for transition states of ≈10 fs. In the transition state region, these structures all have oxacarbenium ion character with the leaving group departing before participation from the phosphate nucleophile (Fig. 3). The change of the orbital hybridization of the C1′ reactive center from sp3 to sp2 as indicated by the dihedral angle of O4′–C1′–C2′–H1′ becoming planar (180°) identifies the transition state region. The ribocationic charge of the transition state is delocalized over the anomeric C1′ reactive center. Transition state configurations show development of a C2′-endo conformation of the ribose ring as indicated by the pseudorotational phase defined by Altona and Sundaralingam (21). Transition state lifetime in PNP is longer than that characterized for LDH, where a hydride is transferred in less then 5 fs and passes through a single TS structure in each reactive trajectory. A single transition state per reactive path was also reported in the TPS study of a Claisen rearrangement reaction catalyzed by chorismate mutase (18). In addition to the variety of transition states found within a given path, the range of transition state structures extends to the transition state ensemble. Four groups of transition states were identified from 4 unique parts of phase space (see Tables S1–S4). Among the transition states identified, there was a distribution of distances of the C1′–N9, C1′–Op, and O4′–O5′atom pairs. Also, the reactant and product configurations showed variation as well. The lack of a single reactant, transition state and product exemplifies the variety of ways that the enzyme can accomplish this transition.
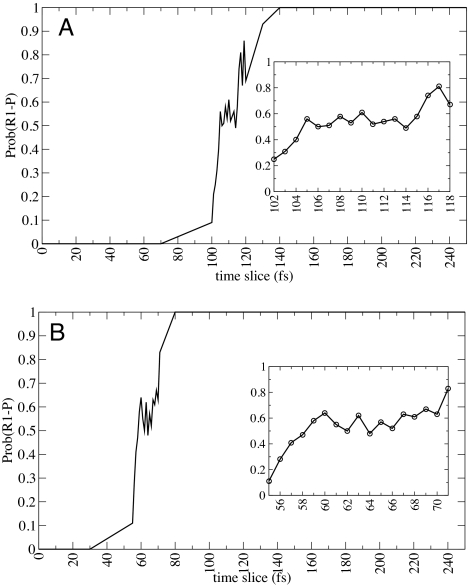
Fig. 4.
The commitment probabilities for product formation were calculated along sample trajectories from the transition path ensemble. The configuration at which the values of the probabilities committed to reactant and product are equal (0.5), is defined as the transition state of the path. The transition state structures were located at 105–115 fs in A and 57–62 fs in B.
Reaction Mechanism Revealed by TPS.
Most N-ribosyltransferases (including PNP) catalyze an inverting nucleophilic displacement reaction. The usual chemical view of nucleophilic displacement has a nucleophile with sufficient kinetic energy to approach an electrophilic site. However, this is not the case for PNP, since the phosphate is immobilized by forming multiple H-bonds with active site residues that hold the phosphate nucleophilic oxygen >3 Å away from the reaction center in the Michaelis complex. From structural and transition state considerations, the reaction mechanism for PNP was proposed to be “nucleophilic displacement by electrophile migration” were the atomic excursions involve migration of the anomeric ribosyl carbocation between purine base and the phosphate nucleophile (22). The TPS study is fully consistent with this mechanism.
The first step in the PNP reaction is loss of the N-ribosidic bond at the transition state, generating substantial ribooxacarbenium ion character (Fig. 3 and Fig. 5). Leaving group departure is facilitated by (i) compression of 3 oxygen atoms (O5′, O4′and Op) to push electrons from the ribosyl group into the C1′–N9 bond, and to the purine base and (ii) electron delocalization to the protonated-N7 and H-bonded O6 and N1 positions, which are stabilized by active site residues Asn-243 and Glu-201.
Fig. 5.
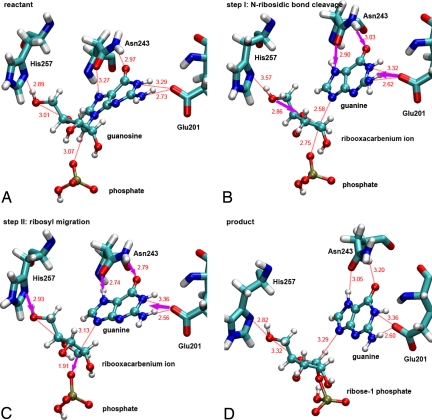
Snapshots taken from a reactive trajectory as the reaction progresses from reactant (A), passing through the transition state region during the cleavage of the C1′–N9 ribosidic bond (B). After the leaving group is fully dissociated, conformational changes and migration of the ribosyl group occur (C), to form ribose 1-phosphate as a product (D). A movie of the trajectory is provided in Movie S1.
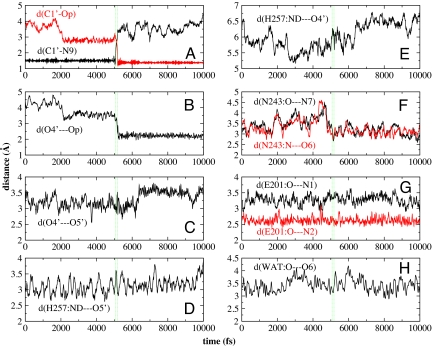
Relevant bond distances for ribosyl and leaving group activation are shown just before, during and after the C1′–N9 bond breaks (the green line at 5050 fs) (Fig. 6). The motion of His-257 contributes to compression of the 3 oxygen stack to cause an electron 'push' from the ribosyl group, leading to loss of the C1′–N9 ribosidic bond. Simultaneously, bond distances shorten between the guanine base and Asn-243, Glu-201, and a catalytic site water molecule(WAT). The C1′–N9 bond loss occurs, indicating that these bond changes play an important role in facilitating departure of the guanine base. At the transition state, the His257—O5′ distance increases causing maximum compression of O4′···O5′ to form the oxacarbenium ion transition state.
Fig. 6.
Distances of guanosine with the important active site residues Asn-243, Glu-201 and His-257, as the reaction progresses. The transition state regions are located at 5105–5115 fs, (vertical lines).
Formation of the ribooxacarbenium transition state is followed by conformational changes in the ribosyl group leading to migration of the anomeric carbon of the ribosyl group toward phosphate, forming ribose 1-phosphate. The geometry of the ribose ring adapts the C1′-exo conformation (Fig. 3). This lengthens the His-257 and O4′distances and the protein facilitates the reaction by His-257 impinging on the O5′-hydroxyl to complete the ribosyl migration process.
Migration of the C1′-ribosyl group after passing the TS takes place in ≈60 fs and occupies ≈60% of the total transient time. Motion of the C1′–H1′bond within this time scale maintains the character of a sp2 bond with a C–H out-of-plane bending mode, which is relevant to the experimentally observed motion of 40 fs (900 cm−1). This mode gives rise to the large normal kinetic isotope effect from [1′-3H] inosine reported for human PNP (23). The ensemble of TPS trajectories all include the excursion of the ribose ring toward phosphate with an almost 2 Å displacement of the C1′reactive center. This result corresponds to the crystallographic structural data comparing the crystal structures of PNP in complex with reactant and product, PDB entries 1A9S and 1A9T, respectively (22, 24). Analysis of trajectories in the TPS ensemble correlate with a role of His-257 in facilitating ribosyl migration by pushing the 5′-hydroxyl into a configuration assisting the anomeric carbon (C1′) to move toward phosphate and formation of ribose 1-phosphate (Figs. 5 and 6).
Enzyme Dynamics on the Transition Event Timescale.
Since TPS generates unbiased reactive trajectories of the transition event, protein motions on the timescale of barrier crossing are observed. In this TPS study, we used the crystal structure of PNP complexed with a picomolar transition state analogue inhibitor that provides a stable and well-ordered complex as the starting configuration (25). The transition state analogue was replaced with guanosine before analysis. Fluctuation of distances between substrate and active site residues in the reactant or product state occurs within a narrow geometry (Fig. 6). The TPS trajectory reveals short distances before the start of the transition event (region between the green dashed lines in Fig. 6). These femtosecond-to-picosecond motions of the active site residues initiate the C1′–N9 bond loss that dominates this reaction coordinate.
Crystal structures of PNP bound to transition state analogue inhibitors reveal an unusual geometric arrangement of the atoms O5′, O4′, and Op, in van der Waals contact in a unique oxygen stack. This arrangement destabilizes ribosyl electrons for loss into the purine leaving group. Protein vibrational modes cause these 3 oxygen atoms to squeeze together, pushing electrons toward the N-ribosidic bond, making the purine a better leaving group, thus facilitating the reaction (14, 26).
The TPS result presented here resolves the importance of compression of the O5′···O4′···Op vibrational motion on the time scale of single bond vibrations as the reaction passes the transition state barrier. His-257 initiates compression of the oxygens (O5′ and Op) surrounding the ribosyl ring oxygen (O4′), destabilizing bonding electrons (see Fig. 6 B–E). During the transition state period, O5′···O4′ became more compressed, stabilizing the oxacarbenium ion as determined by the commitment probability criterion (marked by the arrows in Fig. 7).
Fig. 7.
Plot of the reaction coordinate distances (Upper) and the O4′···O5′ distance (Lower) as the reaction proceeds through the transition state (arrow in the Lower). The transition state corresponds to maximum compression of the O4′···O5′ (Lower). Three exemplary reactive trajectories are shown from different regions of phase space in the transition path ensemble.
After transition state formation, the O5···O4′distance increases as H257:ND···O5′distance decreases and the migration of the anomeric carbon occurs. Ribosyl migration is completed with the help of His-257 holding O5′ in place to promote the ribosyl group to undergo the conformational change, causing migration of the anomeric carbon toward phosphate. Recent KIE studies with PNP mutations His257Asp, Gly, and Phe showed that His-257 is critical in polarizing the 5′-hydroxyl for transition state formation. Native and His257Asp exhibit normal [5′-3H] KIEs, whereas those incapable of H-bonding (His257Gly and His257Phe) resulted in inverse [5′-3H]KIEs (27).
Motions contributed from the PNP protein to the purine base are also important. As the C1′–N9 bond breaks, electron withdrawal into the purine is facilitated by H-bonds from Asn-243, Glu-201 and the water immobilized in the catalytic site (Fig. 6 F–H). In particular, the d(N243:O···N7) and d(N243:N···O6) reach their minimum during the transition state to generate fully dissociative leaving group. This explains the necessity of the protonation at N7 before chemical reaction.
Conclusions
Work reported here provides an advance in the increasing body of data supporting the concept that protein dynamic motion couples directly to transition state formation in enzymatically catalyzed reactions and are an integral part of the reaction coordinate. Slower protein dynamic motion also influences the heights of barriers in enzymatic reactions (28). The present work departs from earlier studies by demonstrating that motions on the femtosecond-to-picosecond timescale are a critical part of transition state formation. That rapid motions influence catalysis, even in reactions that occur on the slower millisecond timescale, is expected. In all condensed-phase reactions, individual bond vibrations contribute to even the slowest reactions. The statistical concept of an ensemble rate should not be confused with individual passages over a barrier. Slower processes also contribute to the rate of enzymatically-catalyzed reactions, and it is often the case that the overall turnover is limited by product release. The presence of the slower steps does not diminish the critical involvement of rapid motions at the chemical step. Enzymatic catalysis viewed in this fashion is a collection of stochastic processes operating on a wide time scale. The dynamic motions reported here reveal that motions on the timescale of individual bond vibrations contribute critically to transition state formation. Only when all dynamic excursions occur in precisely the correct fashion does catalysis occur. Thus, enzymatic catalysis, must be studied on timescales ranging from femtoseconds to milliseconds to capture the atomic vibrational events leading to transition state formation and slow protein conformational changes.
Methods
System Preparation.
We used the crystal structure of the trimeric PNP in a complex with the transition state analogue, Immucillin-H, and phosphate at 2.5 Å resolution (PDB entry 1RR6) as a starting structure (Fig. 2). Hydrogen atoms were added and assigned protonation states for all ionizable residues according to their solution pKa value assuming a pH of 7. The Biopolymer module of InsightII was used to make these assignments. The catalytic site residue Glu-201 was deprotonated at neutral pH to stabilize the purine base. His-257 was also modeled as the neutral form with a proton at NE leaving ND to interact with 5′-hydroxyl of ribose ring. We first modified the TS inhibitor to the guanosine substrate by replacing atoms at N4′ with O4′ and C9 with N9, to make the TS inhibitor and the substrate isosteric. We also protonated N7 of the purine base, because transition state analysis has established N7 protonation to N7H before reaching the transition state (23). Classical MD simulation in explicit water was performed for 400 ps. The details of this methodology is given in the SI.
Quantum Mechanical and Molecular Mechanical MD Simulation (QM/MM).
The structure obtained from the classical MD simulation was then used for the QM/MM MD simulations. It is CPU-expensive to do a QM/MM MD simulation on the fully solvated trimeric PNP system, and so we performed QM/MM MD without solvating water molecules, but all crystallographic waters were retained. All QM/MM simulations were performed on a Linux Cluster, using the CHARMM (29)/MOPAC (30) interface. The QM region comprised the protonated N7 guanosine and HPO42− molecules, which contain a total of 40 atoms (drawn in ball and stick model in Fig. 2). This region was treated with the PM3 semiempirical method (31). We have chosen the PM3 method because it has been used extensively to successfully model phosphorus and phosphate groups in enzymatic reaction studies (32). The remaining atoms of the protein and crystal waters were in the MM region and described with the CHARMM27 all-atom force field for the protein and the TIP3P model for water. None of the protein was treated quantum mechanically, so there were no covalent bonds between the atoms of the QM and the MM regions. The interaction between the QM and MM parts is well-treated with an effective Hamiltonian that includes the electrostatic and van der Waals interactions between electrons and nuclei of QM atoms with the MM atoms (point charges). Minimization, heating, equilibration, and dynamics were performed included the full QM/MM potential assignment. The MM-MM nonbonding cutoffs were 14 Å, and there were no cutoffs for QM-MM nonbonding interactions. For the MM atoms, the SHAKE algorithm was used to constrain all bonds involving hydrogen atoms. The time step for integration was 1 fs. The system was minimized using 500 steps steepest descent and 5,000 steps adopted-basis Newton–Raphson algorithm. The temperature of the system was slowly increased over the first 10 ps to 300 K, and this temperature was held for the rest of the simulations. After the heating phase, the system was equilibrated with velocities assigned from a Gaussian distribution every 100 steps at 300 K for another 20 ps and then dynamically equilibrated for a final 70 ps. The final structure was further used as a starting configuration for a series of QM/MM bias simulations to generate a free energy profile as described in the following section.
Transition Path Sampling
Definition of the Order Parameter.
The TPS algorithm requires an order parameter that divides the phase space into 3 nonoverlapping regions, (reactants, products, and intermediate). We used the bond-breaking (C1′–N9) and bond-forming (C1′–OP) distances as the order parameter. The reactant (product) region included all configurations that had a bond length of C1′–N9 shorter (longer) than 1.7 Å and a bond length of C1′–Op longer (shorter) than 1.8 Å.
Finding an Initial Reactive Trajectory.
A challenge to application of the TPS algorithm is that it requires one starting reactive trajectory to generate the ensemble of reactive paths. Fortunately, this starting trajectory need not be a true dynamical trajectory, but it can be any trajectory that connects reactants and products (33). We used the umbrella sampling technique as a guideline to obtain approximate transition state configurations. To this end, a series of QM/MM MD simulations were performed with the bias potential applied on a simple approximate reaction coordinate defined as the difference between the bond-breaking (C1′–N9) and bond-forming distances, d(C1′−N9) − d(C1′−Op). Each simulation consisted of 10 ps of equilibration followed by 20 ps of sampling dynamics. Each subsequent equilibration was started from the 1 ps slice of the previous adjacent equilibration run. The weighted histogram analysis method (WHAM) was used to combine all 22 windows to construct a potential of mean force (PMF). Several configurations from the top of the activation barrier exhibited ribooxacarbenium ion character and were further used in an attempt to generate an initial reactive trajectory. Simulations were initiated both forward and backward in time until 1 trajectory was generated that began in the reactant region and finished in the product region. The length of this trajectory was 250 fs, long enough to capture the transition event.
Generating a Transition Path Ensemble.
We employ the TPS algorithm described in ref. 10 and generated an ensemble of reactive trajectories. The protocol of the TPS algorithm was implemented within CHARMM C29b2 (11). The 250-fs initial reactive trajectory, described above, was used as a seed to start an iterative process of generating transition paths. To generate a new reactive trajectory, a shooting algorithm was used. The momenta of the previous reactive trajectory were slightly perturbed, then rescaled to conserve linear and angular momentum and energy, as described in detail in ref. 11. Trajectories were then run forward and backward in time to complete a 250-fs trajectory. Using the definition of the order parameter, it was checked whether the initial and final configurations of the newly generated trajectory were in the reactant or product states. If the newly generated trajectory was reactive (i.e., it connected reactant and product), then it was accepted as an additional member of the TPS ensemble of reactive trajectories, and used as a seed to initiate a new shooting move. If the new trajectory was not reactive, then a different time slice from the prior reactive trajectory was randomly chosen for a new shooting move, until a new reactive trajectory was generated. A total of 1,000 shooting attempts were made, resulting in a total of 220 reactive trajectories. The reactive trajectories decorrelated after ≈40 successful shooting trials.
Location of Transition States.
The probability of a configuration to relax into reactant (PR) or product (PP) state is called the “commitment probability” or “committor” (34). A transition state is defined as a configuration that has an equal probability to commit to reactant or product. To find the transition state we calculated the commitment probability of configurations along a reactive trajectory in the TPS ensemble, by initiating multiple trajectories with random initial velocities chosen from a Boltzmann distribution. For each trajectory, dynamics was run for 150 fs, sufficient time to reach a stable state. In practice, it is very CPU-expensive if trajectories are begun from every configuration along a reactive trajectory. Instead, we first created 10 trajectories, if all of them went to the same state, either reactant or product, then we assumed the commitment probability to be close to 0 or 1. Otherwise, we shot more trajectories until we obtain an error of ±0.05. More than 100 trajectories were run for configurations where the PR and PP were both in the range of 0.4–0.6, which we defined as the transition state.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grant GM068036.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808413105/DCSupplemental.
References
- 1.Henzler-Wildman KA, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 2.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme dynamics during catalysis. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 3.Hammes-Schiffer S, Benkovic S. Relating motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 4.Knapp J, Klinman J. Temperature-dependent isotope effects in soybean lipoxygenase-1: Correlating hydrogen tunneling with protein dynamics. J Am Chem Soc. 2002;124:3865–3874. doi: 10.1021/ja012205t. [DOI] [PubMed] [Google Scholar]
- 5.Antoniou D, Schwartz SD. Internal enzyme motions as a source of catalytic activity: Rate promoting vibrations and hydrogen tunneling. J Phys Chem B. 2001;105:5553–5558. [Google Scholar]
- 6.Antoniou D, Basner J, Núñez S, Schwartz SD. Computational and theoretical methods to explore the relation between enzyme dynamics and catalysis. Chem Rev. 2006;106:3170–3187. doi: 10.1021/cr0503052. [DOI] [PubMed] [Google Scholar]
- 7.Schramm V. Enzymatic transition states: Thermodynamics, dynamics and analogue design. Arch Biochem Biophys. 2005;433:13–26. doi: 10.1016/j.abb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 8.Cui Q, Karplus M. Promoting modes and demoting modes in enzyme-catalyzed proton transfer reactions: A study of models and realistic systems. J Phys Chem B. 2002;106:7927–7947. [Google Scholar]
- 9.Bolhuis P, Chandler D, Dellago C, Geissler P. Transition path sampling: Throwing ropes over mountain passes, in the dark. Annu Rev Phys Chem. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 10.Dellago C, Bolhuis P, Geissler P. Transition path sampling. Adv Chem Phys. 2001;123:1–86. [Google Scholar]
- 11.Basner JE, Schwartz SD. How enzyme dynamics helps catalyze a reaction, in atomic detail: A transition path sampling study. J Am Chem Soc. 2005;127:13822–13831. doi: 10.1021/ja043320h. [DOI] [PubMed] [Google Scholar]
- 12.Quaytman SL, Schwartz SD. Reaction coordinate of an enzymatic reaction revealed by transition path sampling. Proc Natl Acad Sci USA. 2007;104:12253–12258. doi: 10.1073/pnas.0704304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nuñez S, Antoniou D, Schramm VL, Schwartz SD. Promoting vibrations in human PNP: A molecular dynamics and hybrid quantum mechanical/molecular mechanical study. J Am Chem Soc. 2004;126:15720–15729. doi: 10.1021/ja0457563. [DOI] [PubMed] [Google Scholar]
- 14.Saen-oon S, Ghanem M, Schramm VL, Schwartz SD. Remote mutations and active site dynamics correlate with catalytic properties of purine nucleoside phosphorylase. Biophys J. 2008;94:4078–4088. doi: 10.1529/biophysj.107.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghanem M, Li L, Wing C, Schramm VL. Altered thermodynamics from remote mutations altering human toward bovine purine nucleoside phosphorylase. Biochemistry. 2008;47:2559–2564. doi: 10.1021/bi702132e. [DOI] [PubMed] [Google Scholar]
- 16.Luo M, Li L, Schramm VL. Remote mutations alter transition-state structure of human purine nucleoside phosphorylase. Biochemistry. 2008;47:2565–2576. doi: 10.1021/bi702133x. [DOI] [PubMed] [Google Scholar]
- 17.Zuckerman DM, Woolf TB. Transition events in butane simulations: Similarities across models. J Chem Phys. 2002;116:2586–2591. [Google Scholar]
- 18.Crehuet R, Field MJ. A transition path sampling study of the reaction catalyzed by the enzyme chorismate mutase. J Phys Chem B. 2007;111:5708–5718. doi: 10.1021/jp067629u. [DOI] [PubMed] [Google Scholar]
- 19.Dellago C, Bolhuis P, Chandler D. On the calculation of reaction rate constants in the transition path ensemble. J Chem Phys. 1999;110:6617–6625. [Google Scholar]
- 20.Bolhuis P, Dellago C, Chandler D. Reaction coordinates of biomolecular isomerization. Proc Natl Acad Sci USA. 2000;97:5877–5882. doi: 10.1073/pnas.100127697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altona C, Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. New description using the concept of pseudorotation. J Am Chem Soc. 1972;94:8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 22.Schramm VL, Shi W. Atomic motion in enzymatic reaction coordinates. Curr Opin Chem Biol. 2001;11:657–665. doi: 10.1016/s0959-440x(01)00269-x. [DOI] [PubMed] [Google Scholar]
- 23.Lewandowicz A, Schramm VL. Transition state analysis for human and plasmodium falciparum purine nucleoside phosphorylases. Biochemistry. 2004;43:1458–1468. doi: 10.1021/bi0359123. [DOI] [PubMed] [Google Scholar]
- 24.Fedorov A, et al. Transition state structure of PNP and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- 25.Schramm VL. Enzymatic transition states and transition state analogues. Curr Opin Struct Biol. 2005;15:604–613. doi: 10.1016/j.sbi.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 26.Nunez S, Antoniou D, Schramm VL, Schwartz SD. Promoting vibrations in human purine nucleoside phosphorylase. a molecular dynamics and hybrid quantum mechanical/molecular mechanical study. J Am Chem Soc. 2004;126:15720–9. doi: 10.1021/ja0457563. [DOI] [PubMed] [Google Scholar]
- 27.Murkin AS, et al. Neighboring group participation in the transition state of human purinenucleoside phosphorylase. Biochemistry. 2007;46:5038–5049. doi: 10.1021/bi700147b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frauenfelder H, McMahon BH, Austin RH, Chu K, Groves JT. The role of structure, energy landscape, dynamics, and allostery in the enzymatic function of myoglobin. Proc Natl Acad Sci USA. 2001;98:2370–2374. doi: 10.1073/pnas.041614298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 30.Field M, Bash P, Karplus M. A combined quantum mechanical and molecular mechanical potential for molecular dynamics simulations. J Comp Chem. 1990;11:700–733. [Google Scholar]
- 31.Stewart JJP. Optimization of parameters for semiempirical methods I. Method. J Comp Chem. 1989;10:209–220. [Google Scholar]
- 32.Hart JC, Burton NA, Hillier IH, Harrison MJ, Jewsbury P. Prediction of transition state structure in protein tyrosine phosphatase catalysis using a hybrid QM/MM potential. Chem Commun. 1997:1431–1432. [Google Scholar]
- 33.Dellago C, Bolhuis P. Transition path sampling simulations of biological systems. Top Curr Chem. 2007;268:291–317. [Google Scholar]
- 34.Bolhuis P, Dellago C, Chandler D. Sampling ensembles of deterministic transition pathways. Faraday Discuss. 1998;110:421–436. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.