Abstract
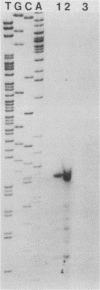
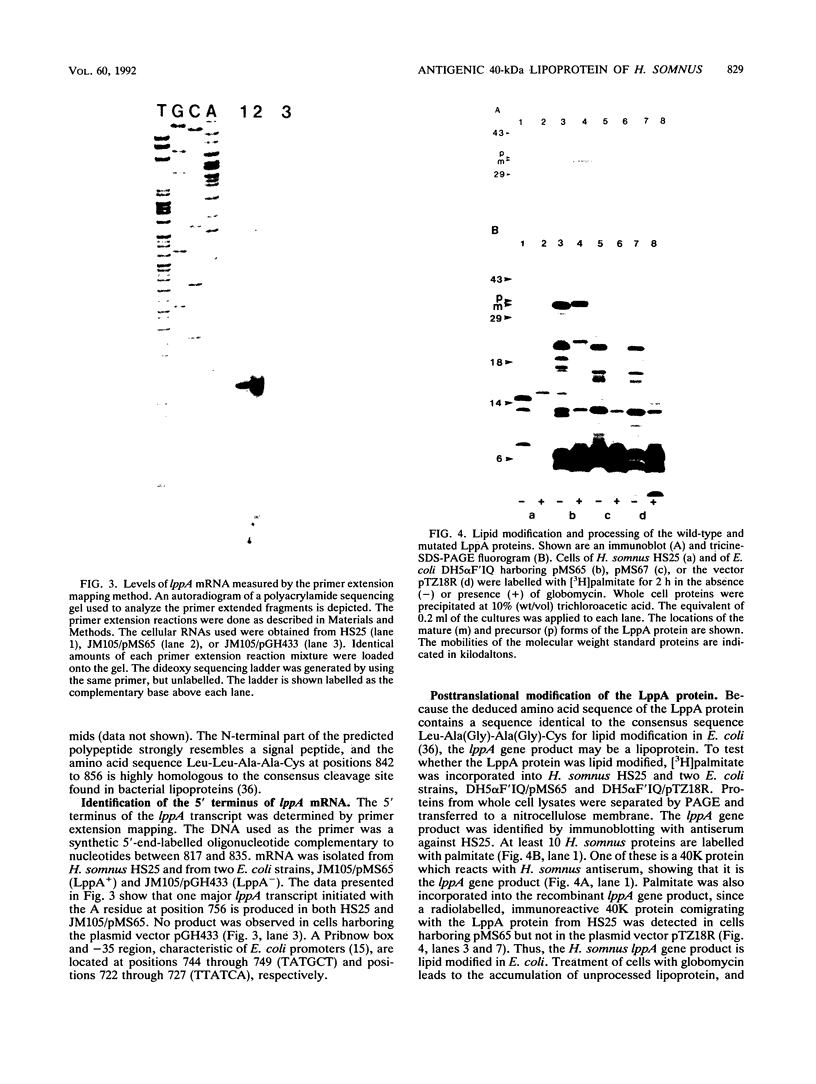
A gene of Haemophilus somnus encoding the major 40,000-molecular-weight antigen (LppA) was cloned on a 2-kb Sau3AI fragment. The nucleotide sequence of the entire DNA insert was determined. One open reading frame, encoding a 247-residue polypeptide with a calculated molecular weight of 27,072, was identified. This reading frame was confirmed by sequencing the fusion joint of two independent IppA::TnphoA gene fusions. The 21 amino-terminal amino acids of the deduced polypeptide showed strong sequence homology to the signal peptide of secreted proteins, and the sequence Leu-Leu-Ala-Ala-Cys at the putative cleavage site is identical to the consensus cleavage sequence of lipoproteins from gram-negative bacteria. The presence of the lipid moiety on the protein was shown by incorporation of radioactive palmitic acid into the natural H. somnus protein. Palmitic acid could also be incorporated into the recombinant protein in Escherichia coli. Synthesis of the mature LppA lipoprotein was inhibited by globomycin, showing that cleavage of the signal peptide is mediated by signal peptidase II in both organisms. By using site-directed mutagenesis, the cysteine residue at the cleavage site was changed to glycine. Radiolabelled palmitate was not incorporated into the mutated protein, showing that lipid modification occurs at the Cys-22 residue.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson B. E., Baumstark B. R., Bellini W. J. Expression of the gene encoding the 17-kilodalton antigen from Rickettsia rickettsii: transcription and posttranslational modification. J Bacteriol. 1988 Oct;170(10):4493–4500. doi: 10.1128/jb.170.10.4493-4500.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessler W. G., Cox M., Lex A., Suhr B., Wiesmüller K. H., Jung G. Synthetic lipopeptide analogs of bacterial lipoprotein are potent polyclonal activators for murine B lymphocytes. J Immunol. 1985 Sep;135(3):1900–1905. [PubMed] [Google Scholar]
- Bessler W. G., Ottenbreit B. P. Studies on the mitogenic principle of the lipoprotein from the outer membrane of Escherichia coli. Biochem Biophys Res Commun. 1976 May 23;76(2):239–246. doi: 10.1016/0006-291x(77)90717-3. [DOI] [PubMed] [Google Scholar]
- Bessler W., Resch K., Hancock E., Hantke K. Induction of lymphocyte proliferation and membrane changes by lipopeptide derivatives of the lipoprotein from the outer membrane of Escherichia coli. Z Immunitatsforsch Immunobiol. 1977 Apr;153(1):11–22. [PubMed] [Google Scholar]
- Bricker T. M., Boyer M. J., Keith J., Watson-McKown R., Wise K. S. Association of lipids with integral membrane surface proteins of Mycoplasma hyorhinis. Infect Immun. 1988 Feb;56(2):295–301. doi: 10.1128/iai.56.2.295-301.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Henning U. Nucleotide sequence of the gene for the peptidoglycan-associated lipoprotein of Escherichia coli K12. Eur J Biochem. 1987 Feb 16;163(1):73–77. doi: 10.1111/j.1432-1033.1987.tb10738.x. [DOI] [PubMed] [Google Scholar]
- Corbeil L. B., Arthur J. E., Widders P. R., Smith J. W., Barbet A. F. Antigenic specificity of convalescent serum from cattle with haemophilus somnus-induced experimental abortion. Infect Immun. 1987 Jun;55(6):1381–1386. doi: 10.1128/iai.55.6.1381-1386.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbeil L. B. Molecular aspects of some virulence factors of Haemophilus somnus. Can J Vet Res. 1990 Apr;54 (Suppl):S57–S62. [PubMed] [Google Scholar]
- Dev I. K., Harvey R. J., Ray P. H. Inhibition of prolipoprotein signal peptidase by globomycin. J Biol Chem. 1985 May 25;260(10):5891–5894. [PubMed] [Google Scholar]
- French B. T., Maul H. M., Maul G. G. Screening cDNA expression libraries with monoclonal and polyclonal antibodies using an amplified biotin-avidin-peroxidase technique. Anal Biochem. 1986 Aug 1;156(2):417–423. doi: 10.1016/0003-2697(86)90275-7. [DOI] [PubMed] [Google Scholar]
- Gogolewski R. P., Kania S. A., Liggitt H. D., Corbeil L. B. Protective ability of antibodies against 78- and 40-kilodalton outer membrane antigens of Haemophilus somnus. Infect Immun. 1988 Sep;56(9):2307–2316. doi: 10.1128/iai.56.9.2307-2316.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C., Barondess J., Manoil C., Beckwith J. The use of transposon TnphoA to detect genes for cell envelope proteins subject to a common regulatory stimulus. Analysis of osmotically regulated genes in Escherichia coli. J Mol Biol. 1987 May 20;195(2):289–297. doi: 10.1016/0022-2836(87)90650-4. [DOI] [PubMed] [Google Scholar]
- Hanson M. S., Hansen E. J. Molecular cloning, partial purification, and characterization of a haemin-binding lipoprotein from Haemophilus influenzae type b. Mol Microbiol. 1991 Feb;5(2):267–278. doi: 10.1111/j.1365-2958.1991.tb02107.x. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris F. W., Janzen E. D. The Haemophilus somnus disease complex (Hemophilosis): A review. Can Vet J. 1989 Oct;30(10):816–822. [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Wright A. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5107–5111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard C. L., Gherardini F. C., Bassford P. J., Jr, Stamm L. V. Molecular cloning and characterization of a 35.5-kilodalton lipoprotein of Treponema pallidum. Infect Immun. 1991 Apr;59(4):1521–1528. doi: 10.1128/iai.59.4.1521-1528.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. U., Gutierrez C., Villarejo M. R. Sequence of an osmotically inducible lipoprotein gene. J Bacteriol. 1989 Jan;171(1):511–520. doi: 10.1128/jb.171.1.511-520.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986 Sep 26;233(4771):1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers F., Braun V., Galanos C. The lipoprotein of the outer membrane of Escherichia coli: a B-lymphocyte mitogen. J Exp Med. 1975 Aug 1;142(2):473–482. doi: 10.1084/jem.142.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nelson M. B., Apicella M. A., Murphy T. F., Vankeulen H., Spotila L. D., Rekosh D. Cloning and sequencing of Haemophilus influenzae outer membrane protein P6. Infect Immun. 1988 Jan;56(1):128–134. doi: 10.1128/iai.56.1.128-134.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase I., Ishino F., Wachi M., Kamata H., Doi M., Asoh S., Matsuzawa H., Ohta T., Matsuhashi M. Genes encoding two lipoproteins in the leuS-dacA region of the Escherichia coli chromosome. J Bacteriol. 1987 Dec;169(12):5692–5699. doi: 10.1128/jb.169.12.5692-5699.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen M., Potter A. A. Cloning, sequencing, expression, and functional studies of a 15,000-molecular-weight Haemophilus somnus antigen similar to Escherichia coli ribosomal protein S9. J Bacteriol. 1992 Jan;174(1):17–23. doi: 10.1128/jb.174.1.17-23.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirkell D., Myles A. D., Russell W. C. Palmitoylated proteins in Ureaplasma urealyticum. Infect Immun. 1991 Mar;59(3):781–784. doi: 10.1128/iai.59.3.781-784.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. C., Tokunaga M. Biogenesis of lipoproteins in bacteria. Curr Top Microbiol Immunol. 1986;125:127–157. doi: 10.1007/978-3-642-71251-7_9. [DOI] [PubMed] [Google Scholar]
- von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983 Feb;80(3):653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]