Abstract
Apoptosis is a complex process essential for normal tissue development and cellular homeostasis. While biochemical events that occur late in the apoptotic process are better characterized, early physiological changes that initiate the progression of cell death remain poorly understood. Previously, we observed that lymphocytes, undergoing apoptosis in response to growth factor withdrawal, experienced a rapid and transient rise in cytosolic pH. We found that the protein responsible was the pH-regulating, plasma membrane protein Na+/H+ exchanger isoform 1 (NHE1), and that its activity was impeded by inhibition of the stress-activated kinase, p38 MAP kinase. In the current study, we examined how NHE1 is activated during apoptosis. We identified the phosphorylation sites on NHE1 that regulate its alkalinizing activity in response to a cell death stimulus. Performing targeted mutagenesis, we observed that substitution of Ser726 and Ser729 for alanines produced a mutant form of NHE1 that did not alkalinize in response to an apoptotic stimulus, and expression of which protected cells from serum withdrawal- induced death. In contrast, substitution of Ser726 and Ser729 for glutamic acids raised the basal pH and induced susceptibility to death. Analysis of serine phosphorylation showed that phosphorylation of NHE1 during apoptosis decreased upon mutation of Ser726 and Ser729. Our findings thus confirm a necessary function for NHE1 during apoptosis and reveal the critical regulatory sites that when phosphorylated mediate the alkalinizing activity of NHE1 in the early stages of a cell death response.
Keywords: pH, sodium hydrogen exchanger, mitogen-activated protein kinase
regulation of intracellular pH is essential for many cellular processes (38). Most cells maintain a cytosolic pH of 7.1–7.2; however, as a result of metabolic activity, the resting membrane potential, and the passive distribution of proteins, intracellular pH, unless corrected, tends to be more acidic than the extracellular environment. To maintain pH homeostasis, cells express a number of cotransporters, proton pumps, proton channels, or ion exchangers that drive H+ or HCO3− ions in and out of the cell. Of these, one of the most ubiquitous is the Na+/H+ exchanger (NHE) family of integral membrane proteins. Nine mammalian NHE isoforms have been cloned. The ubiquitously expressed isoform, NHE1, the focus of the present study, uses the energy of sodium gradients to catalyze the electroneutral exchange of one Na+ ion for one H+ ion—moving protons out of cells across the plasma membrane. NHE1 is normally activated by acidic conditions or osmotic cell shrinkage; hence its main functions are the maintenance of steady-state pH and cell volume. NHE1 can also control cell migration, cell morphology, proliferation, and apoptosis (6, 39). These diverse functions of NHE1 are regulated through mechanisms still poorly understood.
NHE1 contains 815 amino acids arranged in two domains: an NH2-terminal integral membrane-binding domain and a COOH-terminal regulatory cytoplasmic tail. The activity of NHE1 is regulated through the large cytosolic domain (amino acids 500–815). This region forms a compact β-structure (30) containing domains for the binding of regulatory factors as well as target sites for phosphorylation. Several protein kinases phosphorylate and regulate the activity of NHE1, including Nck-interacting kinase (49), p160ROCK (45), and p90rsk, a downstream substrate of ERK1/2. p90rsk has been shown to phosphorylate NHE1 at Ser703 (44), mutation of which prevented the growth factor-induced activation of NHE1, perhaps because phosphorylation of this site recruited 14-3-3 proteins, which modulate NHE1 activity (27). More recent studies, however, revealed that phosphorylation of Ser770 and Ser771 was responsible for the ERK-mediated activation of NHE1 during acidosis (32). The regulatory proteins that bind to NHE1 include a number of calcium-binding proteins such as calcineurin homologous protein (CHP) (1, 37) and Ca2+-calmodulin (5), the actin-binding proteins ezrin, radixin, and moesin (ERM) (9) and tescalcin (30). Like CHP, tescalcin binds Ca2+ but interacts with the cytoplasmic tail of NHE1 at an unknown site (30). Carbonic anhydrase II also binds to NHE1 and may regulate its activity in response to phosphorylation (28).
In addition to the maintenance of cytosolic pH, mediated by the kinases and regulatory proteins described above, in our previous studies we proposed that NHE1 has a necessary, but previously unrecognized, function during apoptosis that is not related to its housekeeping or growth-promoting activities. We found that, in response to a death stimulus, the activity of the stress kinase p38 MAP kinase (MAPK) was upregulated and that this kinase targeted NHE1 for phosphorylation at four potential sites—Thr718, Ser723, Ser726, and Ser729 (human NHE1 sequence). We identified these sites by in vitro kinase assays and confirmed by mass spectrometry analysis (19). In response to phosphorylation by p38 MAPK, we reported that NHE1 induced a transient alkalinization of the cytosol that triggered the activation of a death protein, BAX (18), and the inhibition of mitochondrial ADP transport (20), and others have observed the deamidation of BCL-XL (51). Our initial findings with cytokine-dependent lymphocytes were supported by multiple studies reporting intracellular alkalinization in response to different apoptotic stimuli such as gamma radiation (8), ceramide treatment (4), and drugs, including proteasome inhibitors (21), staurosporine (STS) (12, 43), or polycyclic aromatic hydrocarbons (16) (reviewed in Ref. 24). In these studies, NHE1 (16, 24, 41), chloride channels (43), and Cl−/HCO3− anion exchangers (12, 21) were all implicated as possible mediators of apoptotic alkalinization. Hence the principal mechanism by which apoptotic alkalinizing activity is induced remains enigmatic. To examine this, we have studied the induction of apoptotic alkalinization in diverse cells types and, using the NHE1-deficient AP1 cell line, identified two sites on NHE1 that are targeted for phosphorylation during apoptosis. Our findings demonstrate that NHE1 is an essential mediator of apoptotic alkalinization whose activity is triggered by phosphorylation of novel sites located at the distal portion of its cytoplasmic domain.
MATERIALS AND METHODS
Cell lines and reagents.
NHE1-deficient Chinese hamster ovary cell line (AP1) (40) was cultured in minimum essential alpha medium (GIBCO) supplemented with 10% fetal bovine serum (FBS) (Hyclone), 50 U/ml penicillin/streptomycin, and 25 mM HEPES. AP1 cells cotransfected with wild-type NHE1 or mutant NHE1 constructs and destabilized yellow fluorescent protein (YFP) were cultured in complete Minimum Essential Medium Alpha Medium supplemented with 500 μg/ml geneticin (Invitrogen). The IL-3-dependent murine pro-B cell line, FL5.12A (a kind gift from James A. McCubrey, East Carolina University, Greenville, SC), was maintained in complete RPMI-1640 supplemented with 10% FBS, 2-β-mercaptoethanol (1,000×, Life Technologies), 50 U/ml penicillin/streptomycin, and 0.2 ng/ml recombinant mouse IL-3 (PeproTech). The IL-7-dependent pro-T cell line D1 was established from the CD4−-CD8− subset sorted from p53−/− mouse thymocytes initially propagated in IL-7 and stem cell factor (22). D1 cells were maintained in 50 ng/ml of recombinant mouse IL-7 (PeproTech) in complete RPMI medium as previously described (15). The IL-2-dependent T cell line, CTLL-2 (no. TIB-214, American Type Culture Collection), was maintained in complete RPMI supplemented with 10 ng/ml recombinant mouse IL-2, and the nerve growth factor (NGF)-dependent cell line, PC-12 (no. CRL-1721, American Type Culture Collection), was maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 50 U/ml penicillin/streptomycin, 15% horse serum, and 5% FBS. To differentiate PC-12 cells, 50 ng/ml of NGF (Sigma) was added to the complete DMEM.
The NHE1 inhibitor dimethyl ameloride (DMA; Sigma), the p38 MAPK inhibitor PD-169316, and STS (Calbiochem) were resuspended as stock solutions in DMSO or water. DMSO alone was used as a mock control. Concentrations used in experiments are indicated in figure legends.
Plasmids and site-directed mutagenesis.
Site-directed mutagenesis was accomplished using the pYN4+ plasmid, which contains the cDNA encoding the human NHE1 isoform with a COOH-terminal hemagglutinin (HA) tag. To construct NHE1 mutant plasmids, PCR site-directed mutagenesis was performed targeting residues T718, S723, S726, and S729 (human NHE1 sequence), substituting serine or threonine for alanine or glutamic acid, and a new restriction digestion site, ApaI, was added to facilitate subcloning into the pYN4+ vector. The PCR primers used are shown in Table 1. Standard conditions for the PCR reactions, using pfx DNA Polymerase (Invitrogen), were used with the following amplification cycles: denaturation at 94°C for 3 min; 25 cycles at 94°C for 30 s, 65°C for 30 s, and 72°C for 30 s; and an extension cycle at 72°C for 7 min. To facilitate restriction enzyme digestion, the PCR inserts were initially inserted into the Zero Blunt TOPO vector (Invitrogen), following manufacturer's protocols. The PCR insert was then digested from the Zero Blunt TOPO vector and inserted into the pYN4+ vector using the ApaI restriction sites. Presence of the PCR insert and orientation were determined by restriction enzyme digest. All plasmids were confirmed by sequencing.
Table 1.
Primers for introducing point mutations in NHE1
| Name | Mutation | Sequence |
|---|---|---|
| F4MUT | T718A, S723A, S726A, S729A | 5′-CgggcccGCATCGGCTCAGACCCACTGGCCTATGAGCCGAAGGAGGACCTGCCTGTCATCgccATCGACCCGGCTgccCCGCAGgcaCCCGAGgctGTGGACC-3′ |
| 3SMUT | S723A, S726A, S729A | 5′-CgggcccGCATCGGCTCAGACCCACTGGCCTATGAGCCGAAGGAGGACCTGCCTGTCATCACCATCGACCCGGCTgccCCGCAGgcaCCCGAGgctGTGGACC-3′ |
| 2SMUT | S726A, S729A | 5′-CgggcccGCATCGGCTCAGACCCACTGGCCTATGAGCCGAAGGAGGACCTGCCTGTCATCACCATCGACCCGGCTTCCCCGCAGgcaCCCGAGgctGTGGACC-3′ |
| 2EMUT | S726E, S729E | 5′-CgggcccGCATCGGCTCAGACCCACTGGCCTATGAGCCGAAGGAGGACCTGCCTGTCATCACCATCGACCCGGCTTCCCCGCAGgaaCCCGAGgaaGTGGACC-3′ |
| S729A | S729A | 5′-CgggcccGCATCGGCTCAGACCCACTGGCCTATGAGCCGAAGGAGGACCTGCCTGTCATCACCATCGACCCGGCTTCCCCGCAGTCACCCGAGgctGTGGACC-3′ |
| S723A | S723A | 5′-CgggcccGCATCGGCTCAGACCCACTGGCCTATGAGCCGAAGGAGGACCTGCCTGTCATCACCATCGACCCGGCTgccCCGCA-3′ |
| S726A | S726A | 5′-CgggcccGCATCGGCTCAGACCCACTGGCCTATGAGCCGAAGGAGGACCTGCCTGTCATCACCATCGACCCGGCTTCCCCGCAGgcaCCCG-3′ |
| T718A | T718A | 5′-CgggcccGCATCGGCTCAGACCCACTGGCCTATGAGCCGAAGGAGGACCTGCCTGTCATCgccATCG-3′ |
| RAp1NHE | Reverse primer with AP1 site | 5′-CTAACCCACGgggcccGTGGCTAT-3′ |
All primers start at the 5′-end. Lowercase residues indicate mutations; underlined residues indicate ApaI restriction sites. NHE1, Na+/H+ exchanger isoform 1.
Transfection of AP1 with NHE1 constructs and YFP.
To establish stable NHE1-expressing AP1 cell lines that coexpressed YFP, Lipofectamine 2000 (Invitrogen) was used to transfect AP1 cells, following the manufacturer's protocol. Twenty-four hours after transfection, cells were resuspended in complete medium with 500 μg/ml geneticin. For the first 3 days, medium was changed daily using complete medium supplemented with 500 μg/ml geneticin. Afterward, medium was replenished every 2 days until cell cultures reached confluency. To isolate cells expressing optimal levels of NHE1 and YFP, high-speed cell sorting was performed. Cells were sorted on the basis of YFP fluorescence following standard protocols using a FACSAria cell sorter (BD Biosciences) equipped with the 488-nm argon laser and FL1-H filter (green fluorescence). Sorted cells were maintained in complete media containing 500 μg/ml geneticin as described above.
Detection of NHE1 and phosphoserines by immunoblot.
To assay NHE1 expression in transfected AP1 cells, whole cell lysates were made using a modified RIPA buffer as previously described (14). Briefly, RIPA buffer [50 mM Tris, 150 mM NaCl, 1% (wt/vol) NP40, 0.5% (wt/vol) sodium deoxycholate, 0.1% (wt/vol) Triton X-100, 0.5 M EGTA, 0.1 M PMSF, 0.1 M benzamidine, 25 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 80 mM NaF, and 1 complete mini EDTA-free protease inhibitor cocktail tablet (Roche)] was prepared. Approximately 1 × 107 cells were resuspended in 100 μl RIPA buffer on ice for 5 min, and lysates were centrifuged at 12,000 g at 4°C for 10 min. SDS-PAGE was performed by loading ∼50 μg protein/lane onto 8% Tris-glycine SDS gels (Hoefer). Protein was transferred to nitrocellulose membranes following standard protocols. Membranes were probed with a mouse monoclonal NHE-1 antibody (Chemicon) (diluted 1:1,000), a mouse monoclonal HA antibody (Covance) (diluted 1:1,000), or a rabbit polyclonal phosphoserine antibody (BD Biosciences) (1:500) overnight at 4°C. Membranes were then incubated with appropriate secondary antibodies conjugated to horseradish peroxidase (HRP; Santa Cruz) (diluted 1:2,000). Membranes were developed using enhanced chemiluminescence (Pierce), following the manufacturer's protocol. Equal loading was verified by probing membranes for GAPDH using an anti-GAPDH mouse antibody (Ambion) (diluted 1:1,000) followed by detection using secondary antibody conjugated to HRP. For immunoprecipitation of serine-phosphorylated proteins, whole cell lysates, prepared as described above, were precleared with protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology) and then immunoprecipitated overnight with antiphosphoserine antibody and protein A/G PLUS-Agarose beads. After stringent washes in RIPA buffer, immunoprecipitates were analyzed by SDS-PAGE and immunoblotted for NHE1 as described above.
Detection of NHE1 by confocal microscopy.
Surface expression of NHE1 and NHE1 mutants was assessed by confocal microscopy. Briefly, glass coverslips were treated with 200 μg/ml of poly-l-lysine, and cells were cultured at a density of 0.5 × 106/ml overnight. Cells grown on coverslips were rinsed with PBS and fixed and permeabilized with ice-cold methanol for 10 min at −20°C. Cells were blocked with 1% BSA in PBS for 1 h and stained with primary antibody anti-HA (F7, Santa Cruz) overnight, followed by secondary antibody, anti-mouse Cy5 (Jackson Labs), for 30 min. After a final rinse in PBS, coverslips were mounted with Gel/Mount (Biomeda). The images were acquired by LSM 510 NLO (Zeiss) at ×40 and ×100 magnification.
Measurement of intracellular pH with BCECF-AM.
Cytokine-dependent cells (FL5.12A, D1, CTLL-2, and PC-12) (0.5 × 106) were resuspended in HEPES pH buffer (25 mM HEPES, 140 mM NaCl, 5 mM KCl, 0.8 mM MgCl2, and 5.0 mM glucose, pH 7.2) supplemented with 1 μM BCECF-AM (Molecular Probes), and pH-dependent changes in fluorescence were measured by flow cytometry, using established methods (11, 19). Briefly, cells were loaded with dye by incubating for 20 min in BCECF-AM- containing buffers. To establish a pH calibration curve, 1 × 106 cells were resuspended in a high-potassium HEPES buffer (25 mM HEPES, 145 mM KCl, 0.8 mM MgCl2, and 5.5 mM glucose) at defined pH standards (6.8, 7.2, and 7.8) and supplemented with the exchange ionophore, nigericin (5 μM), for 20 min. To inhibit NHE1 or p38 MAPK activity, cells were pretreated with DMA (200 μM) or with the p38 MAPK inhibitor PD-169316 (20 μM) for 2 h before apoptosis was induced by 2–8 h of cytokine withdrawal. Cells were immediately assayed by flow cytometry. Data were acquired on a FACSAria flow cytometer excited with a 488-nm argon laser, and emissions were filtered through 525 nm and 610 nm. Dead cells were excluded by forward and side scatter gating. pH values were determined by measuring the absorbance ratio between 525 nm (green fluorescence) and 610 nm (red fluorescence), which compensates for dye concentration, volume, and cell size (34). Actual pH values were determined by correlating the ratio between 525 nm and 610 nm of experiment samples to a pH value on a preestablished calibration curve established as described above. Data were acquired for 20,000 events.
Measurement of intracellular pH using YFP.
AP1 cell lines, stably expressing the NHE1 constructs, were cotransfected with YFP to measure intracellular pH without the need of stressing cells through additional loading of BCECF-AM. The green fluorescent protein (GFP) mutant, YFP, exhibits pH-dependent fluorescence in the pH range 6–8, and fluorescence can be measured by flow cytometry, which eliminates any problems with photobleaching. The fluorescence emission (527-nm peak) and excitation (514-nm peak) of YFP increases with increasing pH (31). Calibration curves were established by incubating cells in a high-potassium HEPES buffer fixed at defined pH values and supplemented with 5 μM nigericin as described above. Experimental samples were resuspended in HEPES buffer (1× PBS, 1% FBS, and 25 mM HEPES). For inhibition of p38 MAPK or NHE1, AP1 cells were pretreated for 2 h with 20 μM PD-169316 or 200 μM DMA before induction of apoptosis. To induce apoptosis, AP1 cells were treated with 1 μM STS (apoptotic stimulus) for 0.5–1 h or serum-withdrawn for 4–5 h. After incubation, cells were immediately analyzed for pH-responsive changes in fluorescence using a FACSCalibur flow cytometer (BD Biosciences) with a 488-nm argon laser; excitation was set at 488 nM, and emissions were filtered through 525-nm band pass (green fluorescence). Dead cells were excluded by forward and side scatter gating. Data were acquired for 20,000 events.
Measurement of NHE1 activity.
Acute acid load was performed by resuspending cells in 50 mM NH4Cl buffer (base buffer: 20 mM HEPES, 120 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 5 mM glucose) for 1 h, followed by a rapid incubation in Na+-free buffer, followed by recovery in the Na+-containing base buffer lacking NH4Cl. Recovery of intracellular pH was examined every 5 min by flow cytometry as described above until intracellular pH levels had returned to original baseline values (∼1 h).
Detection of the mitochondrial translocation of BAX.
Apoptosis was induced in D1 cells by deprivation of IL-7. D1 cells were cultured with or without IL-7 in the presence or absence of DMA (200 μM), SITS, and DIDS (inhibitors of anion exchanger) or a caspase inhibitor (1–100 μM), zVAD-FMK. Mitochondrial and cytosolic protein lysates were made using a mitochondrial extraction kit with minor modifications (Pierce). Briefly, 1 × 107 cells were lysed using the mild detergent method. This method is highly reproducible, but it does not induce detergent-mediated conformational changes that lead to the mitochondrial translocation of BAX (15). The final mitochondrial pellet was washed in 1 M NaCl to remove any proteins peripherally associated with mitochondria (35). Protein lysates were loaded on 12% SDS-PAGE gels (Invitrogen), transferred to polyvinylidene difluoride membranes and immunoblotted. For detection of BAX, an anti-BAX rabbit monoclonal antibody was used (N20, Santa Cruz). For assessment of mitochondrial loading, the control protein, prohibitin, was detected (Ab-2, Fitzgerald). Secondary rabbit antibodies cross-linked to HRP (Cell Signaling) were used for detection. Enhanced chemiluminescence (Pierce) was used for visualization following the manufacturer's protocol.
Detection of apoptosis.
Apoptosis was induced in AP1 cells expressing NHE1 or the NHE1 mutants by withdrawing serum for 24 h. Dead/live cell ratio was determined using the MultiTox-fluor Multiplex Cytotoxicity assay kit (Promega) following manufacturer's protocol. Briefly, 1 × 104 cells were plated per triplicate well in an opaque-walled 96-well flat bottom tissue culture plate supplemented with or without FBS. After 24 h, equal volume of the assay reagent was added. After incubation, viability (live cells) was determined at 400 nm (excitation)/505 nm (emission), and cytotoxicity (dead cells) was determined at 485 nm (excitation)/520 nm (emission). Readings were taken using a Synergy 2 plate reader (Biotek). Cell death was also microscopically determined by examining the number of cells that died as indicated by loss of YFP fluorescence.
Statistical analysis.
Statistical analysis was performed to examine the differences among sample sets. For each sample set, a sample size between 6 and 11 was generated. This sample size does not provide sufficient evidence that the population distributions of the variables of interest are a normal distribution. Therefore, use of the usual t-test would not be appropriate. Data analysis relied on the comparison among the empirical distribution functions and the sample medians. The empirical distribution function of each sample is an estimation of the population distribution; thus a comparison among the empirical distribution functions helps to visualize the overall differences among the sample sets of interests. The sample median m̂ is the middle value of the sample in the sense that 50% of the observations in the sample are larger than m̂ and 50% are less than m̂. The sample median is an estimation of the population median m, which is the middle value of the population distribution. When the sample size is small, it is accepted that the statistical analysis based on sample median is more reliable than that based on sample mean. For testing the difference between the medians, the permutation test (10) was used. The permutation test is nonparametric; that is, this method does not require the underlying population distributions to be normal distributions. In all tests, the usual statistical significant level 0.05 was applied; that is, if P < 0.05, the null hypothesis was rejected in favor of the alternative hypothesis as specified.
RESULTS
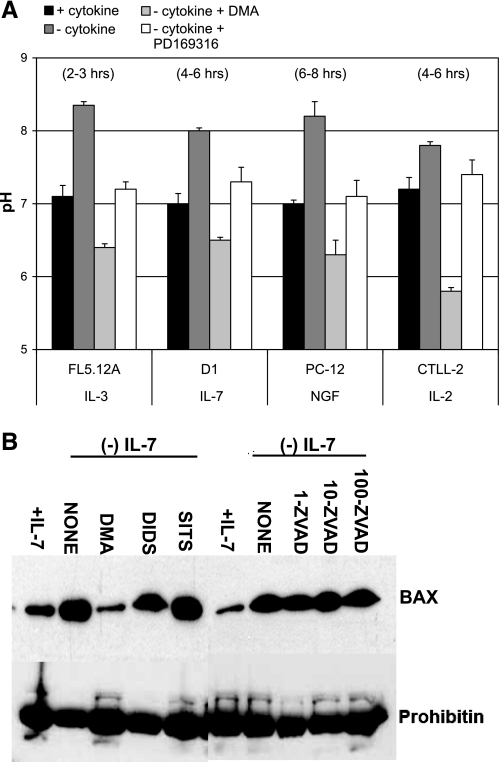
Alkalinization of the cytosol, as we initially reported using cytokine-dependent lymphocytes (19), occurs in response to diverse apoptotic stimuli. We recapitulated these results in Fig. 1, in which four different cell lines (FL5.12A, D1, CTLL-2, and PC-12) were deprived of their dependent growth factors or cytokines (IL-3, IL-7, IL-2, and NGF, respectively), and the subsequent changes in cytosolic pH were measured. BCECF-AM was chosen as an indicator of pH changes because these cells have very low transfection efficiencies, and stable cell lines expressing YFP as a pH indicator, as is later done with AP1 cells, could not be generated. Optimal time points were previously determined by performing time courses that ranged from 2 h to 24 h (19). Results shown in Fig. 1A demonstrated that a rise in cytosolic pH of approximately 0.5–1 pH unit occurred 2–8 h after cytokine withdrawal, the extent of alkalinization and the timing being cell type specific. We also observed that alkalinization occurred upon serum withdrawal in PC-12 cells (not shown). The rise in pH was transient, commonly lasting no more than 2 h. If cells were not rescued from growth factor withdrawal by readdition of the cytokines or serum, cell death ensued with acidification of the cytosol being a later event that coincided with DNA fragmentation (data not shown) (18). Hence our previous and current results indicated that alkalinization is an early physiological event that occurs during apoptosis.
Fig. 1.
Alkalinization is a general response to apoptotic stimuli that activates death factors such as BAX and is prevented by inhibition of Na+/H+ exchanger isoform 1 (NHE1) and p38 MAPK. A: cytosolic pH changes were examined in four cell lines deprived of their respective growth factors. IL-3-dependent FL5.12A pro-B cells, IL-7-dependent D1 pro-T cells, IL-2-dependent CTLL-2 T cells, and NGF-dependent PC-12 cells were cultured in the presence or absence of their respective growth factors for 2–8 h. Inhibition of the cytokine-withdrawal-induced rise in pH was achieved with an NHE1 inhibitor [dimethyl ameloride (DMA); 200 μM] or a p38 MAPK inhibitor (PD-169316, 20 μM). Hours shown at top indicate peak time of transient alkalinization for each cell line. Cytosolic pH was measured with the pH-sensitive probe BCECF-AM. A pH calibration curve was established using the K+ ionophore nigericin and high-K+ buffers at pH 6.8, 7.2, 7.8, and 8.0 (data not shown). BCECF fluorescence was acquired on a FACSAria flow cytometer (BD Biosciences). Actual pH values were determined by correlating the ratio between 525 nm and 610 nm of experimental samples to a pH value on the preestablished calibration curve. Results shown are means ± SD of at least three experiments. B: Western blot analysis of the mitochondrial translocation of BAX in D1 cells in response to IL-7 deprivation. D1 cells were cultured with or without IL-7 for 6 h in the presence or absence of inhibitors of pH-regulatory proteins DMA (NHE1 inhibitor) (200 μM), SITS, and DIDS (inhibitors of anion exchanger) (200 μM) or a caspase inhibitor, zVAD-FMK (1–100 μM). Mitochondrial protein lysates were isolated, and the presence of BAX was determined by immunoblotting. Prohibitin was blotted as a loading control for mitochondrial content. Shown is a representative experiment of three that were performed.
Our previous studies suggested that the mediator of apoptotic alkalinization was NHE1 (19). Inhibiting the activity of NHE1 with the inhibitor DMA during the first 2–8 h after cytokine withdrawal could prevent alkalinization (Fig. 1A). Short-term treatment with DMA in the presence of cytokine caused little or no effect, as was shown in our previous study using FL5.12A and D1 cells. In the same study we found that treatment with DMA for more than 24 h was toxic due to loss of the essential cell activity of NHE1 (19). To determine whether NHE1-induced alkalinization had physiological consequences that contributed to the apoptotic process, we examined the mitochondrial translocation of an apoptotic protein, BAX (14), in IL-7-dependent D1 cells. The mitochondrial translocation of BAX heralds its apoptotic activity and normally precedes mitochondrial disruption, cytochrome c release, the activation of caspases, and cell death (17, 18). We observed that in D1 cells the movement of BAX to mitochondria in response to IL-7 withdrawal coincided with intracellular alkalinization and that this process was blocked by the inhibition of NHE1 with DMA (Fig. 1B). Hence, NHE1-induced alkalinization was involved in the activation of this lethal protein (4, 18, 43). In contrast, inhibition of other pH regulating proteins, such as anion exchangers, or caspases did not impede alkalinization (19) or the activation and movement of BAX to mitochondria (Fig. 1B).
We had further determined that NHE1 was phosphorylated by p38 MAPK during apoptosis (19) and that inhibition of p38 MAPK in parallel with cytokine withdrawal also prevented apoptotic alkalinization (Fig. 1A). As was mentioned, with DMA, in our previous study, treatment with PD-169316 in the presence of cytokine did not induce acidification, although extended treatment for more than 24 h was toxic due to loss of p38 MAPK (19). Performing p38 MAPK in vitro kinase assays using glutathione S-transferase-NHE1 COOH-terminal fusion proteins and mass spectrometry analysis, we had identified four sites on NHE1 (T717, S722, S725, and S728, rabbit NHE1 sequence) that could be phosphorylated in vitro by p38 MAPK (19) (findings summarized in Fig. 2A). The goal of the current study was thus to identify the specific sites by which the alkalinizing activity of NHE1 was regulated and to determine their physiological relevance during apoptosis.
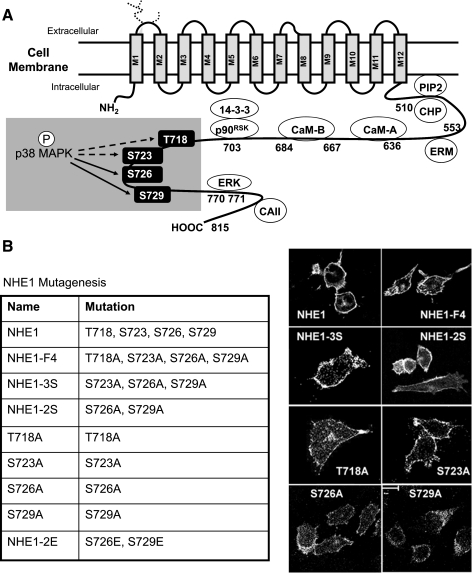
Fig. 2.
Four potential p38 MAPK phosphorylation sites on NHE1 are targets for mutagenesis. A: model of NHE1 displaying the cytosolic domain and sites for interaction of known regulatory binding proteins and kinases. Highlighted are the four residues, T718, S723, S726, and S729 (human NHE1 sequence), previously identified as potential sites for phosphorylation (P) by p38 MAPK. Solid arrows indicate target residues that may regulate the alkalinizing activity of NHE1 during apoptosis. B: list of the NHE1 mutants generated for study. Membrane surface expression of NHE1 and NHE1 mutants in AP1 cells stably cotransfected with cDNA for yellow fluorescent protein (YFP) was demonstrated by confocal microscopy. Cells were fixed and stained for hemagglutinin (HA)-tagged NHE1 proteins using an anti-HA antibody followed by an anti-Cy5 secondary antibody. Images shown were acquired at ×100 magnification. Multiple cells were visualized, and representative cells are shown. ERM, ezrin/radixin/moesin; PIP2, phosphatidylinositol 4,5-bisphosphate; CHP, calcineurin homologous protein; CAII, carbonic anhydrase II.
Our previous studies, establishing NHE1 as the mediator of apoptotic alkalinization (19), were primarily performed using pharmacological inhibitors of NHE1. To study the alkalinizing activity of NHE1 during apoptosis, we constitutively expressed HA-tagged NHE1 (wild-type) in the NHE1-deficient cell line, AP1 (40), producing stable cell lines. In addition, we generated a panel of HA-tagged NHE1 mutants for stable expression in AP1 cells, targeting the four proposed p38 MAPK phosphorylation sites on NHE1: T718, S723, S726, and S729 (human NHE1 sequence). The NHE1 mutants that were generated are summarized in Fig. 2B. To demonstrate that the NHE1 mutants expressed in AP1 cells were exported to the plasma membrane, we assessed surface expression by confocal microscopy, using an anti-HA-antibody. As shown in Fig. 2B, surface expression of NHE1 or the mutant NHE1 proteins were detected, indicating that the targeted mutations did not interfere with the transport of NHE1 to the plasma membrane.
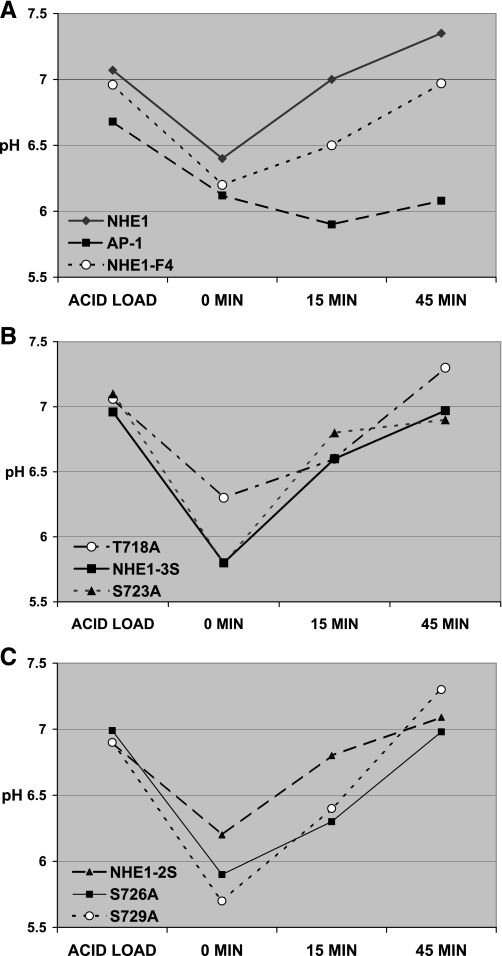
Next, to assess the functionality of NHE1 and the NHE1 mutants proteins expressed in AP1 cells, we performed an acid load test. Results of this test are shown in Fig. 3. We observed that AP1 cells could not recover from an acid load unless these expressed NHE1 (Fig. 3A). AP1 cells expressing the NHE1-F4 mutant (in which T718, S723, S726, and S729 were mutated to alanines) could also recover from an acid load, although to a slight lesser degree than NHE1 (Fig. 3A). Following this pattern, we confirmed that single, double, or triple mutations of the proposed p38 MAPK target sites did not significantly affect the ability of the cells expressing the NHE1 mutant proteins to recover from acid load (Fig. 3, B and C). These results indicated that the targeted mutations of NHE1 did not significantly interfere with the normal housekeeping function of the exchanger.
Fig. 3.
Mutation of T718, S723, S726, and S729 does not impair NHE1-mediated recovery from acid load. NHE1 activity in response to an acid load (described in materials and methods) was assessed in AP1 cells stably transfected with cDNA for YFP or YFP and NHE1 or NHE1-F4 (A), T718A, NHE1-3S, S723A (B), and NHE1-2S, S726A, S729A (C). Recovery of intracellular pH was examined for 0–45 min by detecting pH-sensitive changes in YFP fluorescence by flow cytometry. Representative time points of 0, 15, and 45 min are plotted and are typical of four experiments that were performed.
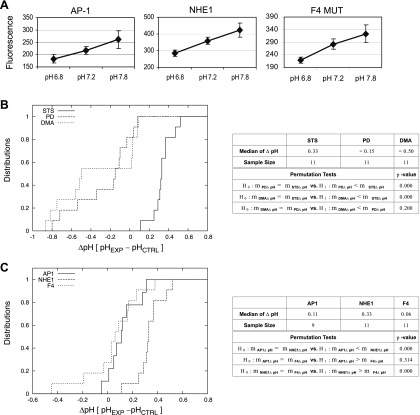
Next we examined how AP1 cells expressing NHE1 or the NHE1 mutant proteins responded to an apoptotic insult. As the apoptotic insult, we treated cells with the drug STS, a potent kinase inhibitor that we previously found strongly activated p38 MAPK at the 1 μM dose used (data not shown). Cells were treated with STS for 30 min to 1 h. This time range was experimentally determined in previous studies in which we observed that STS induced cell death in AP1 cells after 2 h. Intracellular pH was determined by coexpression of the pH-sensitive GFP variant protein YFP. We used YFP as a pH indicator instead of BCECF to directly assess pH changes in response to the apoptotic insult independent of handling stress as occurs when loading a cell with BCECF. We further chose YFP as a pH indicator because of its sensitivity for alkaline pH changes. This is shown in the calibration curves for YFP fluorescence at defined pH values for AP1 cells, and AP1 cells expressing NHE1 or the NHE1-F4 mutant (Fig. 4A). Similar calibration curves were established for AP1 cells expressing each of the NHE1 mutant proteins (data not shown).
Fig. 4.
NHE1 mediates apoptotic alkalinization. A: calibration curves of pH-sensitive changes in YFP fluorescence for AP1 cells and AP1 cells expressing NHE1 or NHE1-F4 mutant (MUT). To establish a pH calibration curve for YFP fluorescence, cells were resuspended in a high-potassium HEPES buffer (as described in materials and methods) at defined pH standards (6.8, 7.2, and 7.8) and supplemented with nigericin. pH-sensitive changes in YFP fluorescence were detected by flow cytometry. Similar calibration curves were established for each of the NHE1 mutant proteins listed in Fig. 2B. B: AP1 cells stably expressing YFP and NHE1 were induced to undergo apoptosis with STS (1 μM), and changes in intracellular pH were detected by flow cytometry. Briefly, cells were treated with STS for 30 min to 1 h, and pH-sensitive changes in YFP fluorescence were detected by flow cytometry as described in materials and methods. To confirm that NHE1 was the mediator of alkalinization, cells were pretreated for 2 h with DMA (200 μM) followed by STS treatment. To confirm that p38 MAPK was required for apoptotic alkalinization, cells were pretreated for 2 h with a p38 MAPK inhibitor [PD-169316 (PD), 20 μM] followed by STS treatment. Experimental pH values were determined from the previously established calibration curve as described in A. ΔpH values were derived from difference between the pH of STS-treated cells (pHEXP) and the pH of mock (DMSO) control cells (pHCTRL). The median ΔpH values were determined as described in materials and methods. The empirical distribution functions of the median ΔpH values are shown in the figure. P values were calculated by performing permutation tests. For determining significance, the threshold P value was 0.05. These results, along with sample sizes, are summarized in the table. C: AP1 cells stably expressing YFP alone or YFP and NHE1 or NHE1-F4 were induced to undergo apoptosis with STS (1 μM), and changes in intracellular pH were detected by flow cytometry as described in B. Median ΔpH values were determined as described in B. The empirical distribution functions of the median ΔpH values are shown in the figure. Sample sizes, the median ΔpH values, and P values (threshold P value was 0.05), calculated by performing permutation tests, can be found in the table.
In Fig. 4B, we displayed the empirical distribution function of the ΔpH values [pH (STS-treated cells) − pH (mock control cells)] for three sample sets represented by NHE1-expressing AP1 cells treated with STS (STS), PD-169316 and STS (PD), and DMA and STS (DMA). Figure 4B clearly shows that the empirical distribution function of the ΔpH values for STS are in the positive range, indicative of STS-induced alkalinization, while the empirical distribution function of the ΔpH values for PD and DMA are close and in the negative range, indicating that treatment with STS in the presence of an NHE1 inhibitor, DMA, or a p38 MAPK inhibitor, PD-169316, did not induce alkalinization and in some instances led to acidification. These observations are supported by the permutation tests summarized in the table adjacent to Fig. 4B, which concluded that the median ΔpH of STS (0.33) was significantly larger than those of PD (−0.15) and DMA (−0.5), and that there was no significant difference between the median ΔpH values of PD and DMA. Note that a median ΔpH of 0.3 units for NHE1-expressing cells represented the difference between an average pH for the mock control cells of 7.2 and an average pH for STS-treated cells of 7.5. Therefore, these results show that NHE1 mediates apoptotic alkalinization, which was prevented by DMA treatment, through a pathway that is dependent on the activity of p38 MAPK.
Having established that NHE1 mediates alkalinization in response to apoptotic insult, we examined the effect of STS treatment in AP1 cells expressing NHE1-F4 mutant to determine whether the potential p38 MAPK phosphorylation sites were necessary for the alkalinizing activity of NHE1. Recall that mutation of these sites did not impair the ability of cells to recover from acid load (Fig. 3A). NHE1-F4-expressing cells were treated with STS as described above and assessed for pH changes by detecting changes in YFP fluorescence. In Fig. 4C, we displayed the empirical distribution functions of the ΔpH values [pH (STS-treated cells) − pH (mock control cells)] data sets represented by AP1 cells (AP1), AP1 cells expressing NHE1 (NHE1), and AP1 cells expressing the NHE1-F4 mutant (F4). These results demonstrated that the empirical distribution functions of AP1 and F4 were close, suggesting that cells lacking NHE1 or expressing the NHE1-F4 mutant could not alkalinize in response to STS treatment (Fig. 4C). In contrast, the empirical distribution function of NHE1 reflected positive median ΔpH values as discussed above. These findings are supported by our permutation tests summarized in the table adjacent to Fig. 4C, which concluded that the median ΔpH of NHE1 (0.33) was significantly larger than those of AP1 (0.11) and NHE1-F4 (0.06), while there was no significant difference between the median ΔpH of AP1 and NHE1-F4. Our experimental results indicated that one or more of the proposed p38 MAPK target sites on NHE1 were required for the exchanger to alkalinize the cytosol of an apoptotic cell.
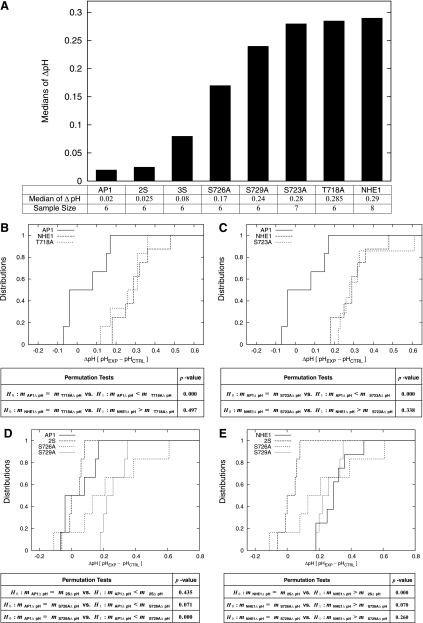
Our next goal was to determine which of the four sites was required for apoptotic alkalinization. To do this, we generated multiple NHE1 mutant constructs: NHE1-2S (S726A and S729A) and NHE1-3S (S723A, S726A, and S729A) as well as single mutations of the four sites, T718A, S723A, S726A, and S729A. Surface expression (Fig. 2B) and functionality (Fig. 3) were previously established for AP1 cells expressing each of these mutants. To examine apoptotic alkalinization, AP1 cells coexpressing the multiple or single NHE1 mutants and YFP were treated with STS and pH changes measured by YFP fluorescence. In Fig. 5A, we summarized the results of these experiments, displaying sample medians of ΔpH, sample sizes, and the bar chart of sample medians in ascending order. We concluded from these results that both S726 and S729 were required for NHE1-mediated apoptotic alkalinization; cells expressing the NHE1-3S mutant and cells expressing the NHE1-2S had low median ΔpH values (<0.08 pH units), indicating minimal alkalization in response to STS treatment. In contrast, cells expressing single NHE1 mutants had larger ΔpH values (>0.24). The exception to this was the single mutation of S726A, discussion of which is forthcoming.
Fig. 5.
Mutation of S726 and S729 inhibits NHE1-mediated apoptotic alkalinization. AP1 cells stably coexpressing YFP and NHE1 or the NHE1 mutants were induced to undergo apoptosis with STS (1 μM) for 30 min to 1 h, and pH-sensitive changes in YFP fluorescence were detected by flow cytometry. pH values determined from previously established calibration curves and ΔpH values were determined from the difference in the pH of STS-treated cells and mock (DMSO) control cells. A: summary of the median ΔpH and sample sizes of each sample set, displaying the median ΔpH in ascending order. B: comparison of the empirical distribution functions of the median ΔpH for AP1 cells expressing YFP alone and AP1 cells coexpressing YFP and NHE1 or T718A mutant. P values were calculated by performing two permutation tests as described in materials and methods and are shown in the table. For determining significance, the threshold P value was 0.05. C: comparison of the empirical distribution functions of the median ΔpH for AP1 cells expressing YFP alone and AP1 cells coexpressing YFP and NHE1 or S723A mutant. P values were calculated by performing two permutation tests as described in materials and methods and are shown in the table. For determining significance, the threshold P value was 0.05. D: empirical distribution functions of the median ΔpH for AP1 cells expressing YFP alone in comparison with AP1 cells coexpressing YFP and NHE1-2S, S726A, or S729A mutants. P values were calculated by performing three permutation tests as described in materials and methods and are shown in the table. For determining significance, the threshold P value was 0.05. E: empirical distribution functions of the median ΔpH for AP1 cells coexpressing YFP and NHE1 in comparison with AP1 cells coexpressing YFP and NHE1-2S, S726A, or S729A mutants. P values were calculated by performing three permutation tests as described in materials and methods and are shown in the table. For determining significance, the threshold P value was 0.05.
These results were then subjected to rigorous statistical analysis. Figure 5B displays the empirical distribution functions of the median ΔpH values of the data sets for AP1 cells, NHE1-expressing cells, and T718A-expressing cells. These results showed that the empirical distributions of the median ΔpH of NHE1 and the single mutant, T718A, were larger and both distant from the empirical distribution of the median ΔpH of AP1. The table adjacent to Fig. 5B summarizes the P values of two permutation tests performed on the median ΔpH values. From these results, we concluded that the median ΔpH of AP1 was significantly smaller (0.02) than that of T718A (0.285), but that there were no significant differences between the medians of NHE1 (0.29) and T718A (0.285). A similar result was observed for the single mutation of S723, shown in Fig. 5C, in which the empirical distribution function of the median ΔpH of S723A-expressing cells was close to that of NHE1-expressing cells but not AP1 cells. This finding was supported by the results of the permutation tests summarized in the table adjacent to Fig. 5C. Thus neither T718 nor S723 were involved in regulating the apoptotic alkalinizing activity of NHE1.
We next compared that data sets derived from mutations of S726 and S729. Shown in Fig. 5D are the empirical distribution functions of the median ΔpH values for NHE1-2S-expressing cells and the single mutants, S726A and S729-expressing cells, in comparison to AP1 cells. Figure 5D shows that the empirical distribution function of AP1 cells is in the low range of median ΔpH and distant from that of cells expressing S729A, suggesting overall smaller median ΔpH values for AP1 than those of S729A. Figure 5D also shows that the empirical distribution functions of the median ΔpH of AP1 cells and NHE1-2S-expressing cells were close, and that the empirical distribution functions of the median ΔpH of AP1 cells and S726A-expressing cells had some overlap portion in the low range of median ΔpH. These results indicated that while mutation of both S726 and S729 impeded apoptotic alkalinization mediated by NHE1, S726 is a site of importance in this process. The table adjacent to Fig. 5D summarizes the P values of three permutation tests on the median ΔpH values. The permutation tests supported the conclusion that the median ΔpH of AP1 was significantly smaller than that of S729A, but that there was no significant difference between the median ΔpH of AP1 and NHE1-2S with a large P value of 0.435, and there was also no significant difference between the medians of AP1 and S726A with a relatively smaller P value of 0.071. When a similar comparison was performed between the empirical distribution functions of the median ΔpH values for NHE1-2S-expressing cells, S726A- and S729-expressing cells in comparison to NHE1-expressing cells, we observed that the median ΔpH for NHE1 was larger than those of NHE1-2S (Fig. 5E). The same figure also showed that the empirical distribution functions of the median ΔpH for NHE1 and S729A were close, and that the empirical distribution functions of the median ΔpH of NHE1 and S726A cross at certain points. These are supported by the three permutation tests in the table adjacent to Fig. 5D, which concluded that the median ΔpH of NHE1 is significantly larger than that of NHE1-2S, but there was no significant difference between the median ΔpH of NHE1 and S729A with a P value of 0.260, and that there was no significant difference between the median ΔpH of NHE1 and S726A with a relatively small P value of 0.070. Therefore, these findings resulted in the identification of both S726 and S729 as being the sites in the COOH-terminal domain of NHE1 that are required for the regulation of the exchanger's apoptotic alkalinizing activity.
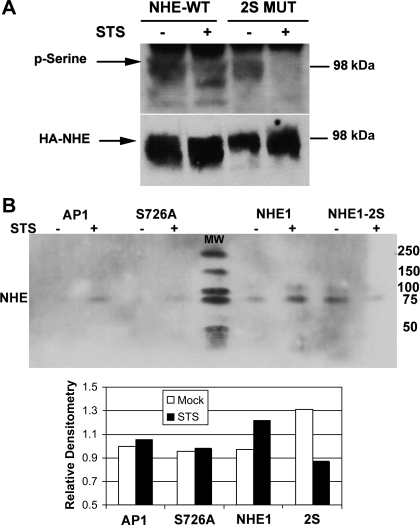
NHE1 can be phosphorylated by multiple kinases, such as ERK (13) or p38 MAPK (19), as discussed in the introduction. Our results suggest that S726 and S729 are phosphorylated by p38 MAPK during apoptosis. We thus examined the phosphorylation status of NHE1 expressed in AP1 cells treated with or without STS (30 min) by immunoblotting for phosphorylated serines. As shown in Fig. 6A, using an anti-HA antibody, we detected a band corresponding to HA-tagged NHE1 around the 98-kDa marker band (Fig. 6A). This HA-specific band was not detected in untransfected AP1 cells (42) (data not shown). Note that the multiple HA-tagged bands observed represent the different glycosylated forms of NHE1. We then detected serine-phosphorylated proteins at the same 98-kDa level, suggesting that this was NHE1 (Fig. 6A). Hence serine-phosphorylated HA-tagged NHE1 protein could be detected in mock and STS-treated cells, suggesting that NHE1 activity was regulated by serine phosphorylation. In contrast, although HA-tagged NHE1-2S could be readily detected in nonapoptotic and apoptotic AP1 cells, only in nonapoptotic cells could serine phosphorylation of NHE1-2S be observed (Fig. 6A). We did not detect serine phosphorylation in NHE1-2S-expressing AP1 cells that were treated with STS.
Fig. 6.
During apoptosis, mutation of S726 and S729 impairs serine phosphorylation of NHE1. A: Western blot shows the serine phosphorylation of NHE1. Analysis of serine phosphorylation was performed by preparing whole cell lysates from AP1 cells stably expressing YFP and NHE1 or NHE1-2S mutant, which were treated with STS (1 μM) for 30 min. Lysates were immunoblotted for serine-phosphorylated proteins and reblotted for HA expression on HA-tagged NHE1 proteins. A molecular weight marker (98-kDa) is indicated. Arrows point to the specific protein bands of the different glycosylated forms of NHE1. Shown are representative results of three experiments performed. WT, wild type. B: Western blot shows results from immunoprecipitation of whole cell lysates from AP1 cells expressing NHE1, S726A, and NHE1-2S muteins for serine-phosphorylated proteins. Phosphoserine-immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblotted for NHE1. Densitometry measurements were acquired relative to the signal from AP1 cells. A representative result from three experiments performed is shown. In-blot molecular weight (MW) markers are indicated.
To confirm that indeed we were detecting serine-phosphorylated NHE1, we next immunoprecipitated phosphoserine-containing proteins with a phosphoserine-specific antibody and blotted the immunoprecipitated proteins for NHE1. This approach was chosen because in our previous studies we observed that the process of metabolic labeling with the millicurie amounts of radioactive phosphate caused stressful conditions that could activate p38 MAPK in nonapoptotic cells. Shown in Fig. 6B are the results of a representative experiment of three performed in which we detected increased serine phosphorylation of NHE1-expressing cells upon STS treatment (30 min), while minimal increases in serine phosphorylation were detected in AP1 cells (note a small amount of nonfunctional NHE1 protein is detectable in AP1). As shown above (Fig. 6A), mock-treated NHE1-2S expressing cells had elevated levels of basal serine-phosphorylated NHE1, which decreased upon STS treatment. Interestingly, the single mutation of S726A also did not result in increased serine phosphorylation of NHE1 during STS treatment, although the effect was not as striking as the double mutation of both S726 and S729. These results are highlighted in the densitometry plot below Fig. 6B. Together with the ΔpH results shown in Fig. 5E, our findings suggest that phosphorylation of S726 is likely critical to the apoptotic alkalinizing activity of NHE1. We therefore conclude that S726 and S729 are phosphorylated during apoptosis and that the responsible kinase activity is induced during apoptosis as would occur with p38 MAPK. This conclusion is supported by our previously published studies in which NHE1 was phosphorylated by ERK in the presence of a growth stimulus (19) or stimulated by intracellular acidosis (32) and by p38 MAPK in the absence of a growth stimulus (19), indicating that p38 MAPK-mediated phosphorylation of S726 and S729 induces the alkalinizing activity of NHE1 only during the early stages of the apoptotic process.
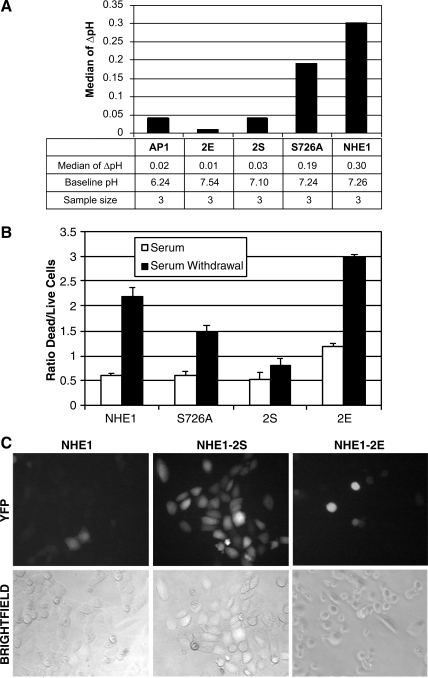
It follows that loss of the alkalinizing activity of NHE1, through mutagenesis of S726 and S729, should be protective during apoptosis. To test this, we treated AP1 cells expressing NHE1, the single mutation, S726A, and NHE1-2S to apoptotic insult. In addition, we generated a new NHE1 mutant in which S726 and S729 were mutated to glutamic acid (Fig. 2B). The substitution of Glu for Ser would mimic phosphorylation, creating a constitutively active form of NHE1. Shown in Fig. 7A is a bar chart of the median ΔpH of AP1 cells expressing NHE1, S726A, NHE1-2S, and the new NHE1-2E mutant. Results for AP1 cells expressing YFP and NHE1, S726A, or NHE1-2S upon STS treatment were comparable to those previously shown in Fig. 5A. Results of AP1 cells expressing the NHE1-2E mutant revealed that STS treatment did not induce any further increase in intracellular pH. However, this occurred because the basal pH of the NHE1-2E-expressing cells was already 0.3–0.4 pH units higher than NHE1- or NHE1-2S-expressing cells, indicating that in NHE1-2E-expressing cells, the alkalinizing activity of NHE1 was being induced independently of any apoptotic stimuli.
Fig. 7.
S726 and S729 on NHE1 regulate cell viability during serum withdrawal-induced death. A: change in pH (ΔpH) for AP1 cells expressing YFP and NHE1, S726A, NHE1-2S, and the constitutively active NHE1-2E was measured by YFP fluorescence detected by flow cytometry. Bar chart summarizes the median ΔpH, baseline pH, and sample sizes of each sample set. B: ratio of dead (fluorescence at 400 nm) versus live cells (fluorescence at 485 nm) was determined using the MultiTox-Fluor cytotoxicity assay (Promega). AP1 cells expressing NHE1, S726A, NHE1-2S, or NHE1-2E were cultured for 24 h with or without serum. Dead and live cells were determined as described in materials and methods. Fluorescence was measured with a Synergy 2 plate reader (Biotek). A representative experiment of three performed is shown. C: viability of AP1 cells expressing YFP and NHE1, NHE1-2S, or NHE1-2E was visualized by fluorescence and brightfield microscopy. Images were acquired with a ×40 objective lens.
Using serum withdrawal as a way to induce a physiologically relevant form of apoptosis, we next examined cell death in AP1 cells expressing NHE1 and the mutant NHE1 proteins. The results displayed in Fig. 7B show that after 24 h of serum withdrawal, there was a fourfold increase in the ratio of dead to live cells expressing NHE1 and a threefold increase in cell death of cells expressing the S726A mutant, indicating that the mutation of S726 alone was not completely protective. We also measured intracellular pH and noted that these cells alkalinized 4–5 h after serum withdrawal (data not shown). Significant induction of apoptosis did not occur in cells expressing NHE1-2S, although the live cell number was reduced because of growth arrest caused by serum withdrawal. These results suggest that mutation of both S726 and S729 was required to prevent apoptotic alkalinization and the resulting cell death. In contrast, cells expressing NHE1-2E, which we observed had a more alkaline basal pH (Fig. 7A), were less viable in serum-containing medium (twofold more dead cells in comparison to NHE1-2S) and exhibited greatly increased cell death during serum withdrawal (threefold more dead cells in comparison to NHE1-2S). These results are visualized in Fig. 7C, in which cell death is correlated with a decreased number of YFP-expressing cells. Under conditions of serum withdrawal, YFP-expressing cells could only be observed with NHE1-2S expressing cells but not with NHE1-expressing or NHE1-2E-expressing cells. We therefore conclude that the alkalinizing activity of NHE1 promotes cell death in response to an apoptotic insult and that the critical mediator of this activity is the phosphorylation of the S726 and S729. Apoptotic alkalinization is, therefore, mediated by NHE1 as an essential part of death induced by loss of a growth or trophic factor, which is a mitochondrial-mediated cell death process.
DISCUSSION
Intracellular alkalinization, as described in our study, is an early event occurring in response to apoptosis. We found that cells lacking a functional NHE1 could not alkalinize in response to STS and that this activity was restored upon expression of wild-type NHE1, indicating that NHE1 is a mediator of apoptotic alkalinization. The activity of NHE1 was triggered by phosphorylation of two sites in the protein's COOH terminus: Ser726 and Ser729. An NHE1 mutant (2S) lacking phosphorylation sites was unable to alkalinize upon STS treatment or serum withdrawal but could alkalinize in response to an acid load. In contrast, an NHE1 mutant (2E), which mimicked constitutive phosphorylation, maintained a more alkaline basal pH and promoted cell death. These results suggest that the alkalinizing activity of NHE1, normally required for maintenance of volume or pH homeostasis, can be subverted during apoptosis to initiate the cell death process. To regulate these two activities of NHE1—maintenance of intracellular pH versus apoptotic alkalinization—the protein is phosphorylated by diverse enzymes targeting different sites. We propose that, during apoptosis, NHE1 is phosphorylated by p38 MAPK at Ser726 and Ser729 and that this differs significantly from how the protein is phosphorylated and its activity is regulated in response to a proliferative or mitogenic signal.
The alkalinizing activity of NHE1 markedly increases in response to acidosis (26). Acidosis in turns activates the kinases ERK and p90rsk (13, 32). These kinases are proposed to phosphorylate NHE1 within the COOH terminus, at serines 770 and 771 (32), and to regulate the exchanger's activity (7, 36, 44), suggesting one pathway by which NHE1 maintains intracellular pH during cell growth. NHE1 can also function as a structural anchor by binding the actin-binding proteins ezrin, radixin, and moesin (ERM) (9) and serve as a scaffold for the assembly of signaling complexes (i.e., kinases, CHP, CaM, 14-3-3, ERM). (3). The NHE1-ERM interaction may also promote survival signaling by recruiting phosphoinositide 3-kinase and its substrate Akt (46). Hence NHE1 is essential for cell growth and survival by maintaining intracellular pH and recruiting survival signaling factors. It follows that loss of NHE1 function would have detrimental consequences in a normal, nonapoptotic cell.
In an apoptotic cell, however, our results suggest that NHE1 has a different function from the survival and growth activities described above. We propose that NHE1, in fact, modulates the cell death process in a number of different cell types. In our studies, we found that NHE1 promoted apoptosis through an active process of intracellular alkalinization, triggering downstream death effectors such as BAX (18) or inhibiting mitochondrial ADP import (20). Other have reported that BCL-XL deamidation is a consequence of NHE1-induced alkalinization and that preventing the antiapoptotic activity of this protein promoted cell death (51). Unlike inhibition of NHE1 in a nonapoptotic cell, inhibition of NHE1-induced alkalinization in apoptotic cells could be protective, as we observed in serum-starved cells expressing the NHE1-2S mutant. However, assessing the positive effects of inhibiting NHE1 activity during different forms of apoptosis can be difficult. For example, in some apoptotic studies where NHE1 inhibition was not protective, cell death ensued because the housekeeping activity of NHE1 was needed to oppose acidification (that activates DNAses) and reverse the volume loss induced by Fas-receptor engagement or long-term STS treatment (25, 47). Hence, inhibition of NHE1 may not necessarily be protective under all conditions that induce apoptosis. It is likely, from our results, that the apoptotic alkalinizing activity of NHE1 is most physiologically relevant during induction of cell death through loss of a growth stimulus. Since death induced upon loss of a cytokine or growth factor involves a mitochondrial-dependent mechanism, we surmise that the apoptotic alkalinizing activity of NHE1, though triggered by diverse forms of stress or lethal stimuli, is most important in the form of mitochondrial-induced cell death that ensues when cells die through neglect or lack of the environmental signals that maintain their survival.
Many regulators of NHE1 activity have been described; however, most of these enzymes, such as ERK (19, 32, 48), are inactive during apoptosis and could not be responsible for inducing the alkalinizing activity of NHE1. In contrast, stress or apoptotic insults upregulate the kinase activity of p38 MAPK (50) that we found responsible for activating NHE1 (19). Oxidative stress is, in fact, a consequence of apoptosis and a well-known stimulus of p38 MAPK activity (23). Such results suggest that during apoptosis, oxidative stress could activate p38 MAPK, phosphorylating NHE1 at S726 and S729.
The requirement for phosphorylation at closely located, dual sites is not unique to NHE1. Other membrane proteins, such as the cystic fibrosis transmembrane regulator (2) and the voltage-dependent calcium channel (33), are examples of how multiple phosphorylation sites are required to optimally regulate physiological activity. The need for multiple phosphorylations may thus be a way to tightly control biological function. Phosphorylation of NHE1 at S726 could enable the subsequent phosphorylation of S729, which in turn may induce alkalinizing activity by providing a binding site for another regulatory protein. As an example, carbonic anhydrase II binding to NHE1 is known to be greatly increased upon phosphorylation of the exchanger's COOH terminus (29). Phosphorylation of S726 and S729 could also change the conformation of COOH terminus, altering the pH sensitivity of the exchanger much in the same way that is shown to occur in the presence of calcium (30).
In summary, our results show that the phosphorylation of NHE1 at two COOH-terminal serine residues is an early apoptotic event that produces a significant physiological change—intracellular alkalinization—that is responsible for inducing multiple apoptotic activities, eventually priming the cell for death.
GRANTS
This work was supported in part by National Institutes of Health (NIH)/National Cancer Institute K22 career transition award CA-097984 (to A. R. Khaled), NIH/National Institute of General Medical Sciences Grant GM-083324 (to A. R. Khaled), and National Science Foundation Grant DMS-0604488 (to J.-J. Ren).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Ammar YB, Takeda S, Hisamitsu T, Mori H, Wakabayashi S. Crystal structure of CHP2 complexed with NHE1-cytosolic region and an implication for pH regulation. EMBO J 25: 2315–2325, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldursson O, Berger HA, Welsh MJ. Contribution of R domain phosphoserines to the function of CFTR studied in Fischer rat thyroid epithelia. Am J Physiol Lung Cell Mol Physiol 279: L835–L841, 2000. [DOI] [PubMed] [Google Scholar]
- 3.Baumgartner M, Patel H, Barber DL. Na+/H+ exchanger NHE1 as plasma membrane scaffold in the assembly of signaling complexes. Am J Physiol Cell Physiol 287: C844–C850, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Belaud-Rotureau MA, Leducq N, Macouillard PdG, Diolez P, Lacoste L, Lacombe F, Bernard P, Belloc F. Early transitory rise in intracellular pH leads to Bax conformation change during ceramide-induced apoptosis. Apoptosis 5: 551–560, 2000. [DOI] [PubMed] [Google Scholar]
- 5.Bertrand B, Wakabayashi S, Ikeda T, Pouyssegur J, Shigekawa M. The Na+/H+ exchanger isoform 1 (NHE1) is a novel member of the calmodulin-binding proteins. Identification and characterization of calmodulin-binding sites. J Biol Chem 269: 13703–13709, 1994. [PubMed] [Google Scholar]
- 6.Besson P, Fernandez-Rachubinski F, Yang W, Fliegel L. Regulation of Na+/H+ exchanger gene expression: mitogenic stimulation increases NHE1 promoter activity. Am J Physiol Cell Physiol 274: C831–C839, 1998. [DOI] [PubMed] [Google Scholar]
- 7.Bianchini L, L'Allemain G, Pouyssegur J. The p42/p44 mitogen-activated protein kinase cascade is determinant in mediating activation of the Na+/H+ exchanger (NHE1 isoform) in response to growth factors. J Biol Chem 272: 271–279, 1997. [DOI] [PubMed] [Google Scholar]
- 8.Dai HY, Tsao N, Leung WC, Lei HY. Increase of intracellular pH in p53-dependent apoptosis of thymocytes induced by gamma radiation. Radiat Res 150: 183–189, 1998. [PubMed] [Google Scholar]
- 9.Denker SP, Huang DC, Orlowski J, Furthmayr H, Barber DL. Direct binding of the Na–H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H+ translocation. Mol Cell 6: 1425–1436, 2000. [DOI] [PubMed] [Google Scholar]
- 10.Efron B, Tibshirani RJ. Permutation tests. In: An Introduction to the Bootstrap, New York: Chapman & Hall/CRC, 1993, p. 207–208.
- 11.Franck P, Petitipain N, Cherlet M, Dardennes M, Maachi F, Schutz B, Poisson L, Nabet P. Measurement of intracellular pH in cultured cells by flow cytometry with BCECF-AM. J Biotechnol 46: 187–195, 1996. [DOI] [PubMed] [Google Scholar]
- 12.Fujita H, Ishizaki Y, Yanagisawa A, Morita I, Murota SI, Ishikawa K. Possible involvement of a chloride-bicarbonate exchanger in apoptosis of endothelial cells and cardiomyocytes. Cell Biol Int 23: 241–249, 1999. [DOI] [PubMed] [Google Scholar]
- 13.Haworth RS, McCann C, Snabaitis AK, Roberts NA, Avkiran M. Stimulation of the plasma membrane Na+/H+ exchanger NHE1 by sustained intracellular acidosis. Evidence for a novel mechanism mediated by the ERK pathway. J Biol Chem 278: 31676–31684, 2003. [DOI] [PubMed] [Google Scholar]
- 14.Hsu YT, Wolter KG, Youle RJ. Cytosol-to-membrane redistribution of Bax and Bcl-X(L) during apoptosis. Proc Natl Acad Sci USA 94: 3668–3672, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu YT, Youle RJ. Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem 272: 13829–13834, 1997. [DOI] [PubMed] [Google Scholar]
- 16.Huc L, Sparfel L, Rissel M, Dimanche-Boitrel MT, Guillouzo A, Fardel O, Lagadic-Gossmann D. Identification of Na+/H+ exchange as a new target for toxic polycyclic aromatic hydrocarbons. FASEB J 18: 344–346, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Khaled AR, Durum S. From cytosol to mitochondria: the Bax translocation story. J Biochem Mol Biol 34: 391–394, 2001. [Google Scholar]
- 18.Khaled AR, Kim K, Hofmeister R, Muegge K, Durum SK. Withdrawal of IL-7 induces Bax translocation from cytosol to mitochondria through a rise in intracellular pH. Proc Natl Acad Sci USA 96: 14476–14481, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khaled AR, Moor AN, Li A, Kim K, Ferris DK, Muegge K, Fisher RJ, Fliegel L, Durum SK. Trophic factor withdrawal: p38 mitogen-activated protein kinase activates NHE1, which induces intracellular alkalinization. Mol Cell Biol 21: 7545–7557, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khaled AR, Reynolds DA, Young HA, Thompson CB, Muegge K, Durum SK. Interleukin-3 withdrawal induces an early increase in mitochondrial membrane potential unrelated to the Bcl-2 family. Roles of intracellular pH, ADP transport, and F0F1-ATPase. J Biol Chem 276: 6453–6462, 2001. [DOI] [PubMed] [Google Scholar]
- 21.Kim JM, Bae HR, Park BS, Lee JM, Ahn HB, Rho JH, Yoo KW, Park WC, Rho SH, Yoon HS, Yoo YH. Early mitochondrial hyperpolarization and intracellular alkalinization in lactacystin-induced apoptosis of retinal pigment epithelial cells. J Pharmacol Exp Ther 305: 474–481, 2003. [DOI] [PubMed] [Google Scholar]
- 22.Kim K, Khaled AR, Reynolds D, Young HA, Lee CK, Durum SK. Characterization of an interleukin-7-dependent thymic cell line derived from a p53−/− mouse. J Immunol Methods 274: 177–184, 2003. [DOI] [PubMed] [Google Scholar]
- 23.Kurata S Selective activation of p38 MAPK cascade and mitotic arrest caused by low level oxidative stress. J Biol Chem 275: 23413–23416, 2000. [DOI] [PubMed] [Google Scholar]
- 24.Lagadic-Gossmann D, Huc L, Lecureur V. Alterations of intracellular pH homeostasis in apoptosis: origins and roles. Cell Death Differ 11: 953–961, 2004. [DOI] [PubMed] [Google Scholar]
- 25.Lang F, Madlung J, Bock J, Lukewille U, Kaltenbach S, Lang KS, Belka C, Wagner CA, Lang HJ, Gulbins E, Lepple-Wienhues A. Inhibition of Jurkat-T-lymphocyte Na+/H+-exchanger by CD95(Fas/Apo-1)-receptor stimulation. Pflügers Arch 440: 902–907, 2000. [DOI] [PubMed] [Google Scholar]
- 26.Leem CH, Lagadic-Gossmann D, Vaughan-Jones RD. Characterization of intracellular pH regulation in the guinea-pig ventricular myocyte. J Physiol 517: 159–180, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehoux S, Abe J, Florian JA, Berk BC. 14-3-3 Binding to Na+/H+ exchanger isoform-1 is associated with serum-dependent activation of Na+/H+ exchange. J Biol Chem 276: 15794–15800, 2001. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Alvarez B, Casey JR, Reithmeier RA, Fliegel L. Carbonic anhydrase II binds to and enhances activity of the Na+/H+ exchanger. J Biol Chem 277: 36085–36091, 2002. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Liu Y, Alvarez BV, Casey JR, Fliegel L. A novel carbonic anhydrase II binding site regulates NHE1 activity. Biochemistry 45: 2414–2424, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Liu Y, Kay CM, Muller-Esterl W, Fliegel L. The Na+/H+ exchanger cytoplasmic tail: structure, function, and interactions with tescalcin. Biochemistry 42: 7448–7456, 2003. [DOI] [PubMed] [Google Scholar]
- 31.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci USA 95: 6803–6808, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malo ME, Li L, Fliegel L. Mitogen-activated protein kinase-dependent activation of the Na+/H+ exchanger is mediated through phosphorylation of amino acids Ser770 and Ser771. J Biol Chem 282: 6292–6299, 2007. [DOI] [PubMed] [Google Scholar]
- 33.Martin SW, Butcher AJ, Berrow NS, Richards MW, Paddon RE, Turner DJ, Dolphin AC, Sihra TS, Fitzgerald EM. Phosphorylation sites on calcium channel alpha1 and beta subunits regulate ERK-dependent modulation of neuronal N-type calcium channels. Cell Calcium 39: 275–292, 2006. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy DA Fluorescence and fluorochromes. In: Cytometric Analysis of Cell Phenotype, edited by McCarthy DA and Macey MG. new York: Cambridge University Press, 2001, p. 68–69.
- 35.Meisinger C, Ryan MT, Hill K, Model K, Lim JH, Sickmann A, Muller H, Meyer HE, Wagner R, Pfanner N. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol Cell Biol 21: 2337–2348, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moor AN, Fliegel L. Protein kinase-mediated regulation of the Na+/H+ exchanger in the rat myocardium by mitogen-activated protein kinase-dependent pathways. J Biol Chem 274: 22985–22992, 1999. [DOI] [PubMed] [Google Scholar]
- 37.Pang T, Su X, Wakabayashi S, Shigekawa M. Calcineurin homologous protein as an essential cofactor for Na+/H+ exchangers. J Biol Chem 276: 17367–17372, 2001. [DOI] [PubMed] [Google Scholar]
- 38.Puceat M pHi regulatory ion transporters: an update on structure, regulation and cell function. Cell Mol Life Sci 55: 1216–1229, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Putney LK, Barber DL. Na-H exchange-dependent increase in intracellular pH times G2/M entry and transition. J Biol Chem 278: 44645–44649, 2003. [DOI] [PubMed] [Google Scholar]
- 40.Rotin D, Steele-Norwood D, Grinstein S, Tannock I. Requirement of the Na+/H+ exchanger for tumor growth. Cancer Res 49: 205–211, 1989. [PubMed] [Google Scholar]
- 41.Shibanuma M, Kuroki T, Nose K. Superoxide as a signal for increase in intracellular pH. J Cell Physiol 136: 379–383, 1988. [DOI] [PubMed] [Google Scholar]
- 42.Slepkov E, Ding J, Han J, Fliegel L. Mutational analysis of potential pore-lining amino acids in TM IV of the Na+/H+ exchanger. Biochim Biophys Acta 1768: 2882–2889, 2007. [DOI] [PubMed] [Google Scholar]
- 43.Tafani M, Cohn JA, Karpinich NO, Rothman RJ, Russo MA, Farber JL. Regulation of intracellular pH mediates Bax activation in HeLa cells treated with staurosporine or tumor necrosis factor-alpha. J Biol Chem 277: 49569–49576, 2002. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi E, Abe J, Gallis B, Aebersold R, Spring DJ, Krebs EG, Berk BC. p90RSK is a serum-stimulated Na+/H+ exchanger isoform-1 kinase. Regulatory phosphorylation of serine 703 of Na+/H+ exchanger isoform-1. J Biol Chem 274: 20206–20214, 1999. [DOI] [PubMed] [Google Scholar]
- 45.Tominaga T, Ishizaki T, Narumiya S, Barber DL. p160ROCK mediates RhoA activation of Na-H exchange. EMBO J 17: 4712–4722, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu KL, Khan S, Lakhe-Reddy S, Jarad G, Mukherjee A, Obejero-Paz CA, Konieczkowski M, Sedor JR, Schelling JR. The NHE1 Na+/H+ exchanger recruits ezrin/radixin/moesin proteins to regulate Akt-dependent cell survival. J Biol Chem 279: 26280–26286, 2004. [DOI] [PubMed] [Google Scholar]
- 47.Wu KL, Khan S, Lakhe-Reddy S, Wang L, Jarad G, Miller RT, Konieczkowski M, Brown AM, Sedor JR, Schelling JR. Renal tubular epithelial cell apoptosis is associated with caspase cleavage of the NHE1 Na+/H+ exchanger. Am J Physiol Renal Physiol 284: F829–F839, 2003. [DOI] [PubMed] [Google Scholar]
- 48.Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 270: 1326–1331, 1995. [DOI] [PubMed] [Google Scholar]
- 49.Yan W, Nehrke K, Choi J, Barber DL. The Nck-interacting kinase (NIK) phosphorylates the Na+-H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J Biol Chem 276: 31349–31356, 2001. [DOI] [PubMed] [Google Scholar]
- 50.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res 15: 11–18, 2005. [DOI] [PubMed] [Google Scholar]
- 51.Zhao R, Oxley D, Smith TS, Follows GA, Green AR, Alexander DR. DNA damage-induced Bcl-xL deamidation is mediated by NHE-1 antiport regulated intracellular pH. PLoS Biol 5: e1, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]