Abstract
Previous studies on molecular recognition of uranyl-DCP (dicarboxy-phenanthroline chelator) compound by two distinct monoclonal antibodies (Mabs U04S and U08S) clearly showed the presence of a biphasic shape in Bell-Evans' plots and an accentuated difference in slopes at the high loading rates. To further explore the basis in the slope difference, we have performed complementary experiments using antibody PHE03S, raised against uranyl-DCP but, presenting a strong cross-reactivity toward the DCP chelator. This work allowed us to obtain a reallocation of the respective contributions of the metal ion itself and that of the chelator. Results led us to propose a 2D schematic model representing two energy barriers observed in the systems Mabs U04S- and U08S-[UO2-DCP] where the outer barrier characterizes the interaction between UO2 and Mab whereas the inner barrier characterizes the interaction between DCP and Mab. Using dynamic force spectroscopy, it is thus possible to dissect molecular interactions during the unbinding between proteins and ligands.
From a toxicological point of view, uranium produces both chemical and radiological hazards known to both accumulate in tissues and skeletons and for its nephrotoxicity (1). Although uranium has been widely studied for decades, available data on the chemical toxicity due to long-term ingestions of uranium are insufficient. In aqueous solution, chelated uranyl ion (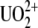 ) is the most common species of uranium (2,3). To study biological targets of
) is the most common species of uranium (2,3). To study biological targets of 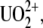 such as proteins or DNA, the approach that at least two teams have adopted was to raise antibodies against the uranyl chelates (4,5) where
such as proteins or DNA, the approach that at least two teams have adopted was to raise antibodies against the uranyl chelates (4,5) where 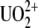 was attached to a dicarboxy-phenanthroline chelator (DCP) coupled to a protein carrier. Using this approach, our group has selected a set of monoclonal antibodies (Mabs) specific for binding with UO2-DCP (5), including U04S, U08S and PHE03S; Mab PHE03S was chosen also for its strong cross-reactivity with the chelator DCP.
was attached to a dicarboxy-phenanthroline chelator (DCP) coupled to a protein carrier. Using this approach, our group has selected a set of monoclonal antibodies (Mabs) specific for binding with UO2-DCP (5), including U04S, U08S and PHE03S; Mab PHE03S was chosen also for its strong cross-reactivity with the chelator DCP.
From our previous dynamic force spectroscopy (DFS) experiments, we have revealed the presence of two unbinding force regimes to unbind Mabs U04S- and U08S-[UO2-DCP] (6,7). The interpretation of DFS measurements has been proposed by Bell (8) and further refined by Evans et al. (9) where the energy landscape of an unbinding process is characterized by two parameters: the width of energy barrier (γ) and the depth of potential that is correlated with the natural off-rate constant (k0) of the association reaction. The two unbinding force regimes thus correspond to at least two energy barriers (10,11); the inner barrier with a narrow width (<1 Å) was measured at high applied loading rates (re > 3000 pN/s) whereas the outer barrier had a larger width (>1 Å) with lower loading rates (re < 3000 pN/s).
To resolve the ambiguity in the correspondence between the chemical bonds and the energy parameters obtained, we have performed new DFS experiments on these systems and that involving Mab PHE03S. We characterized the chemical kinetics of antibody-antigen dissociation and proposed a multiple-step process for the unbinding event.
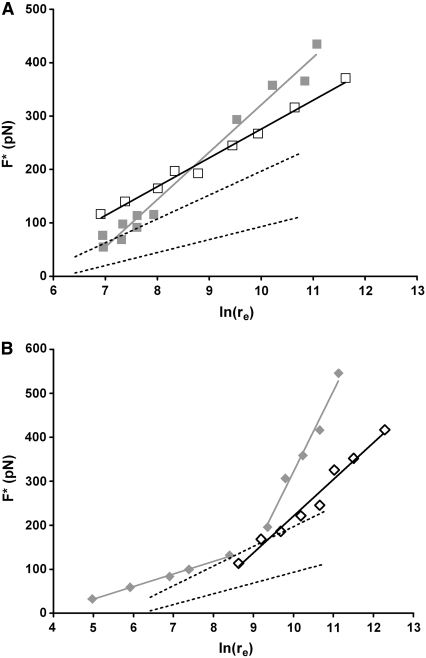
Two systems were established for force-displacement measurements: 1), tip coated with DCP and Mab PHE03S deposited on the gold substrate; and 2), same as 1 except the tip coated with Ni-DCP. The experimental set-ups have been described elsewhere (6,7) and the details are in the Supplementary Material. Using the Yieldfinder software, 1198 and 996 rupture events were collected, representing ∼20% and 15% positive events, for systems 1 and 2, respectively. Fig. 1 displays the Bell-Evans' plots for the two systems. System 3 was set for recording nonspecific interactions: the tip was coated with DCP and an irrelevant Mab anti-GST antibody was deposited on the flat substrate; ∼850 force-displacement curves were recorded for the system 3, representing <1% of the total amount of force-displacement trials. Lower and upper boundaries for nonspecific data are illustrated in Fig. 1, A and B, with black dash lines (complete data are shown in Fig. S1 in the Supplementary Material, Data S1). In systems 1 and 2, the most probable unbinding forces were only measured at high loading rates (re > 1096 pN/s); no experimental condition was obtained for measuring rupture forces at lower loading rates. A set of straight lines in the Bell-Evans' plots represent a single regime in the energy landscape. The presence of multiple parallel bonds in all our study systems have been extensively described previously (6,7,12) and are given in the Supplementary Material.
FIGURE 1.
Bell-Evans' plots of the most probable unbinding forces F*versus the logarithm of the applied loading rate [ln(re)]. (A) Dissociation of Mab PHE03S-DCP (open squares) and Mab PHE03S-[Ni-DCP] (solid gray squares). The dotted lines represent the nonspecific interactions. (B) Unbinding of Mab U08S-[Cu-DCP] (open diamonds) and that for Mab U08S-[UO2-DCP] (solid gray diamonds). A single population of event (pop2 for A and pop1 for B) is retained in the plot for clarity (see the Supplementary Material).
THE URANYL BONDING IS INVOLVED IN A SINGLE UNBINDING FORCE REGIME
The energy landscape for PHE03S interacting with either DCP or Ni-DCP is similar. For both systems, the energy barrier width is very narrow (<1 Å) with an average k0 slightly lower for system 2 than for system 1 (last two lines, Table 1). It reflects that Ni has a marginal effect on the unbinding of DCP from PHE03S; it also implies that Ni has no direct bonding with Mab PHE03S. Thus, the energy landscape we have seen in Fig. 1 A depicts mostly the interaction between PHE03S and DCP. It should be noted that the conformation of DCP in Ni-DCP crystal structure is not symmetrical and suggests that PHE03S likely recognizes a conformationally strained DCP. It should be recalled that Mab PHE03S was obtained from an immunization against UO2-DCP, another conformationally strained DCP. Markedly different from PHE03S, the unbinding events between Mabs U04S and U08S and DCP were rarely observed and were mainly treated as nonspecific interactions (6).
TABLE 1.
Kinetic parameters characterizing the interaction between Mab and ligands
| Inner barrier (high loading rate)
|
Outer barrier (low loading rate)
|
|||
|---|---|---|---|---|
| Mab-metal complex | γ1 (nm) | k0 (s−1) | γ2 (nm) | k0 (s−1) |
| U04S-[UO2-DCP] | 0.04 | 0.9 | 0.28 | 0.1 |
| U08S-[UO2-DCP] | 0.02 | 15.1 | 0.16 | 1.1 |
| U08S-[Cu-DCP] | 0.04 | 8.8 | ||
| PHE03S-DCP | 0.07 | 0.7 | ||
| PHE03S-[Ni-DCP] | 0.04 | 5.9 | ||
In our previous study, two unbinding force regimes were observed in unbinding processes of UO2-DCP from Mabs U04S and U08S (6). To reveal the role of UO2 in the unbinding process, we replaced UO2 with Cu and Ni in DCP of the Mabs U08S and U04S systems. Fig. 1 B shows the Bell-Evans' plot for Cu-DCP dissociated from Mab U08S, and despite a large proportion of nonspecific unbinding ruptures (only 5% of positive events), a single specific unbinding force regime was clearly obtained from 1069 force-displacement curves. Mab U08S does not bind with DCP; thus, the specific unbinding events observed in Fig. 1 B are essentially attributed to the presence of Cu. Consequently, the single regime of unbinding force suggests that Mab U08S recognizes a constrained conformation of DCP due to the presence of Cu. Therefore, the second unbinding force regime observed in the system of Mab U08S and UO2-DCP is thus likely due to the specific interactions between the Mab and UO2.
The weak interaction between Mab U08S and Cu-DCP, as observed using DFS, has also been observed from surface plasmon resonance experiments (5). However, no detectable interaction by surface plasmon resonance was observed for divalent metals complexed with Mab U04S, in agreement with our DFS results that show unbinding events between Mab U04S and Ni-DCP occurring rarely (<0.3% of total events; see Fig. S1 in Data S1).
THE ENERGY LANDSCAPE OF CHELATED-METAL COMPLEXED WITH ANTIBODY
In DFS experiments, the results of PHE03S-DCP, PHE03S-[Ni-DCP] and U08S-[Cu-DCP] have revealed the presence of single regimes of unbinding force, whereas that of U08S-[UO2-DCP] and U04S-[UO2-DCP] showed the presence of two regimes. These facts led us to conclude that UO2 was exclusively responsible for the presence of the second unbinding force regime in the system of Mabs U04S and U08S. Assuming the presence of two energy barriers, the inner one of the unbinding of UO2-DCP from Mabs U04S and U08S was due to the DCP rupture whereas the outer barrier characterized the bond rupture between UO2 and Mabs. The inner energy barrier width was very small (γ1 < 1 Å), a property that could be related to the rigidity of the DCP, although not in a straightforward relationship. The γ-length distance is usually much larger when the ligand is a peptide or a protein (13,14) since they are stretchable over a longer distance. Indeed, the energy barrier width (γ1) is a measure of the extent to which the bonding complex is stretched or deformed before ruptured, also called rupture distance. The rupture distance of the outer barrier was larger (γ2 > 1 Å) for UO2-DCP dissociated from Mabs U04S and U08S, indicating a longer stretching of Mab structures before rupturing the UO2-Mab bonding.
The depth of the energy barrier is related with the natural dissociation rate of the complex. The k0 values corresponding to the inner barrier in Mabs U04S, U08S and PHE03S systems indicated a very low affinity for DCP (k0 > 1 s−1). The average k0 value of the outer barrier, at the best, was 0.06 s−1 for Mabs U04S and U08S systems, highlighting the strong interactions between UO2 and antibody (6).
The very high apparent affinity of Mabs U04S and U08S toward UO2-DCP as measured in bulk solution (5), led us to postulate that such high affinity was gained from an enhanced association rate constant, probably driven by strong Coulombic interactions between uranyl and antibodies.
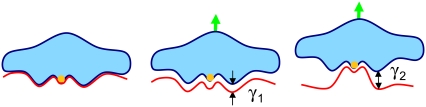
In conclusion, the energy landscape of various designed systems obtained from DFS experiments provides a good understanding of antibody-metal chelate binding. We were able to distinguish the role of individual chemical components in the interactions of the complex; that is Mabs U04S and U08S having a stronger interaction with uranyl than with the DCP chelator. In addition, in the two energy barrier systems observed in Mabs U04S- and U08S-[UO2-DCP] complexes, we attribute the outer barrier to the interaction between UO2 and Mabs. The determination of the natural dissociation rate constant in the binding reaction suggested that the high-affinity of Mab-[UO2-DCP] binding is likely due to an enhanced association rate. Fig. 2 provides a 2D schematic diagram representing the relative locations of the ligand (chelated-metal compound) to the receptor (Mab) in different binding states.
FIGURE 2.
Schematic diagrams of ligand-receptor unbinding process. On the left, the bound state of a metal (orange)-chelated ligand (blue) and a Mab receptor (red) is displayed. The green arrows above indicate the direction of the applied force on the ligand to provoke the rupture between the ligand and the receptor. Using the Bell-Evans' model we determined two energy barriers. The middle diagram describes the inner barrier characterized by a short width (γ1 < 1 Å) attributed to the Mab-DCP rupture, and the right diagram depicts the outer barrier characterized by a longer width (γ2 > 1 Å) attributed to the uranyl-Mab rupture.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank S.-w. W. Chen for editorial assistance of the manuscript. We also acknowledge our collaborators, L. Bellanger, L. Reisser-Rubrecht, and C. Vidaud, for providing Mabs U04S, U08S, and PHE03S, as well as T. Bessou for providing the sequences of the antibody variable regions.
This work has been supported by the program for environmental nuclear toxicology of the Commissariat à l'Énergie Atomique (CEA), France.
Editor: Kathleen B. Hall.
References
- 1.Berradi, H., J. M. Bertho, N. Dudoignon, A. Mazur, L. Grandcolas, C. Baudelin, S. Grison, P. Voisin, P. Gourmelon, and I. Dublineau. 2008. Renal anemia induced by chronic ingestion of depleted uranium in rats. Toxicol. Sci. 103:397–408. [DOI] [PubMed] [Google Scholar]
- 2.Ansoborlo, E., O. Prat, P. Moisy, C. Den Auwer, P. Guilbaud, M. Carriere, B. Gouget, J. Duffield, D. Doizi, T. Vercouter, C. Moulin, and V. Moulin. 2006. Actinide speciation in relation to biological processes. Biochimie. 88:1605–1618. [DOI] [PubMed] [Google Scholar]
- 3.Pible, O., P. Guilbaud, J. L. Pellequer, C. Vidaud, and E. Quemeneur. 2006. Structural insights into protein-uranyl interaction: towards an in silico detection method. Biochimie. 88:1631–1638. [DOI] [PubMed] [Google Scholar]
- 4.Blake, I. R., A. R. Pavlov, M. Khosraviani, H. E. Ensley, G. E. Kiefer, H. Yu, X. Li, and D. A. Blake. 2004. Novel monoclonal antibodies with specificity for chelated uranium(VI): isolation and binding properties. Bioconjug. Chem. 15:1125–1136. [DOI] [PubMed] [Google Scholar]
- 5.Reisser-Rubrecht, L., C. Torne-Celer, W. Renier, O. Averseng, S. Plantevin, E. Quemeneur, L. Bellanger, and C. Vidaud. 2008. High-affinity uranyl-specific antibodies suitable for cellular imaging. Chem. Res. Toxicol. 21:349–357. [DOI] [PubMed] [Google Scholar]
- 6.Odorico, M., J.-M. Teulon, L. Bellanger, C. Vidaud, T. Bessou, S.-w. W. Chen, E. Quéméneur, P. Parot, and J.-L. Pellequer. 2007. Energy landscape of chelated uranyl-antibody interactions by Dynamic Force Spectroscopy. Biophys. J. 93:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teulon, J. M., M. Odorico, S. W. Chen, P. Parot, and J. L. Pellequer. 2007. On molecular recognition of an uranyl chelate by monoclonal antibodies. J. Mol. Recognit. 20:508–515. [DOI] [PubMed] [Google Scholar]
- 8.Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. [DOI] [PubMed] [Google Scholar]
- 9.Evans, E., and K. Ritchie. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 72:1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maki, T., S. Kidoaki, K. Usui, H. Suzuki, M. Ito, F. Ito, Y. Hayashizaki, and T. Matsuda. 2007. Dynamic force spectroscopy of the specific interaction between the PDZ domain and its recognition peptides. Langmuir. 23:2668–2673. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, X., E. Wojcikiewicz, and V. T. Moy. 2002. Force spectroscopy of the leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction. Biophys. J. 83:2270–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odorico, M., J.-M. Teulon, O. Berthoumieu, S.-w. W. Chen, P. Parot, and J.-L. Pellequer. 2007. An integrated methodology for data processing in Dynamic Force Spectroscopy of ligand-receptor binding. Ultramicroscopy. 107:887–894. [DOI] [PubMed] [Google Scholar]
- 13.Schwesinger, F., R. Ros, T. Strunz, D. Anselmetti, H. J. Guntherodt, A. Honegger, L. Jermutus, L. Tiefenauer, and A. Pluckthun. 2000. Unbinding forces of single antibody-antigen complexes correlate with their thermal dissociation rates. Proc. Natl. Acad. Sci. USA. 97:9972–9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morfill, J., K. Blank, C. Zahnd, B. Luginbuhl, F. Kuhner, K. E. Gottschalk, A. Pluckthun, and H. E. Gaub. 2007. Affinity-Matured Recombinant Antibody Fragments Analyzed by Single Molecule Force Spectroscopy. Biophys. J. 93:3583–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.