Abstract
The Group IVA (GIVA) phospholipase A2 associates with natural membranes in response to an increase in intracellular Ca2+ along with increases in certain lipid mediators. This enzyme associates with the membrane surface as well as binding a single phospholipid molecule in the active site for catalysis. Employing deuterium exchange mass spectrometry, we have identified the regions of the protein binding the lipid surface and conformational changes upon a single phospholipid binding in the absence of a lipid surface. Experiments were carried out using natural palmitoyl arachidonyl phosphatidylcholine vesicles with the intact GIVA enzyme as well as the isolated C2 and catalytic domains. Lipid binding produced changes in deuterium exchange in eight different regions of the protein. The regions with decreased exchange included Ca2+ binding loop one, which has been proposed to penetrate the membrane surface, and a charged patch of residues, which may be important in interacting with the polar head groups of phospholipids. The regions with an increase in exchange are all located either in the hydrophobic core underneath the lid region or near the lid and hinge regions from 403 to 457. Using the GIVA phospholipase A2 irreversible inhibitor methyl-arachidonyl fluorophosphonate, we were able to isolate structural changes caused only by pseudo-substrate binding. This produced results that were very similar to natural lipid binding in the presence of a lipid interface with the exception of the C2 domain and region 466-470. This implies that most of the changes seen in the catalytic domain are due to a substrate-mediated, not interface-mediated, lid opening, which exposes the active site to water. Finally experiments carried out with inhibitor plus phospholipid vesicles showed decreases at the C2 domain as well as charged residues on the putative membrane binding surface of the catalytic domain revealing the binding sites of the enzyme to the lipid surface.
The 85-kDa GIVA3 phospholipase A2 (GIVA PLA2) is a member of the superfamily of phospholipase A2 enzymes (1, 2) that cleave fatty acids from the sn-2 position of membrane phospholipids. This enzyme was initially isolated from human platelets, and it is specific for phospholipids containing arachidonic acid in the sn-2 position (3, 4). The release of arachidonic acid is the critical first step in the biosynthesis of eicosanoids, which are potent mediators of inflammation and pain (5). There are a number of enzymes and routes by which arachidonic acid can be released from phospholipids, but experiments with GIVA PLA2 knock-out mice have demonstrated its importance in many inflammatory processes (6-8), thereby confirming the key role of the GIVA enzyme.
The enzyme is composed of two domains, a Ca2+ binding C2 domain and a α/β-hydrolase domain that contains the catalytic site (9). Crystal structures of both the intact enzyme (PDB 1CJY) (10) and the C2 domain alone (11) (PDB 1RLW) have been solved. For the enzyme to be active, it must be sequestered to a phospholipid interface. Binding Ca2+ to the C2 domain accomplishes this as does the binding of two different lipid mediators, e.g. ceramide 1 phosphate (12, 13) and phosphatidyl-inositol-(4,5) bis-phosphate (14, 15).
The Ca2+ binding C2 domain is a conserved domain that is present on many different lipid-binding proteins (16). Ca2+ binding to this domain sequesters the protein to the lipid surface. Extensive studies have been carried out on the C2 domain using a variety of techniques to determine how Ca2+ binding accomplishes lipid surface binding. These studies have included x-ray reflectivity, site-directed mutagenesis, NMR, EPR, and computational methods (17). These studies have shown (18-23) that lipid binding entails the penetration of Ca2+ binding loops one and three, composed of amino acids 35-39 and 96-98, into the interface. However, these studies only deal with C2 binding to the membrane, not the intact cPLA2 enzyme. How the presence of the α/β-hydrolase domain affects surface binding has not been determined in detail due to the difficulties of working with such a large protein.
Unlike the C2 domain, the binding of the catalytic domain of the enzyme to the lipid surface has not been extensively studied. Numerous studies using site-directed mutagenesis of the intact protein have localized amino acids that are important for lipid binding and activation by secondary lipid mediators, as well as activation by phosphorylation of specific residues, especially residue Ser-505 (24, 25). We have also shown that there is an increase in activity upon binding phosphatidyl-inositol-(4,5) bis-phosphate-containing lipids in a specific lysine binding pocket (14, 15). The enzyme has been shown to be minimally active on monomeric lipid substrates but has substantial activation upon binding an interfacial surface (9). The enzyme also contains a lid region spanning amino acids 415-432 that covers the active site. Many different lipase proteins exist in a closed lid conformation, and upon lipid binding, the enzyme shifts to an open lid conformation, causing interfacial activation (26-28). Many of these open lid conformations have been crystallized in the presence of an inhibitor binding in the active site (PDB 1K8Q) (26). Other than the crystal structure of the native GIVA PLA2 with the lid region obstructing the active site, there has been no evidence for this mechanism in the GIVA enzyme. The technique of deuterium exchange mass spectrometry (DXMS) is well equipped to examine the issues of the catalytic domain binding the lipid surface, as well as examining the conformational changes upon the lid region opening. We have recently demonstrated for the first time that DXMS can be used to characterize membrane lipid binding to a PLA2 (29).
Peptide amide hydrogen deuterium exchange analyzed via liquid chromatography/mass spectrometry has been widely used to analyze protein-protein interactions (30, 31), protein conformational changes (32, 33) and protein dynamics (34). We have recently conducted deuterium exchange studies on the GIVA enzyme showing intradomain interactions of the enzyme as well as structural changes caused by Ca2+ binding (35). This technique is an excellent way to probe the effects of lipid binding across the entire enzyme. The present study represents a continuation of our studies of PLA2 binding a lipid surface (29). We have now identified specific regions of the GIVA PLA2 that bind the lipid surface, specifically regions 28-39, 268-279, and 466-470. We now have also demonstrated conformational changes in regions 391-397, 481-495, and 543-553 hypothesized to be due to the conversion from the closed to the open lid conformation of the enzyme induced by natural phospholipid as well as the specific inhibitor MAFP binding to the active site of the enzyme.
MATERIALS AND METHODS
All reagents were analytical reagent grade or better. Protein Expression and Purification—C-terminal His6-tagged GIVA PLA2, and the C2 and catalytic domains were expressed using recombinant baculovirus in a suspension culture of Sf9 insect cells. The cell pellet was lysed in 25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm β-mercaptoethanol, and 2 mm EGTA, and then the insoluble portion was removed by centrifugation at 12,000 × g for 30 min. The supernatant was passed through a column comprised of 6 ml of nickel-nitrilotriacetic acid agarose (Qiagen, Valencia, CA). The protein in the native state was eluted in the “protein buffer” (25 mm Tris-HCl, pH 8.0, 100 mm NaCl, 125 mm imidazole, and 2 mm dithiothreitol). The protein concentration was measured using the Bradford assay (Bio-Rad) to the manufacturer's standards, and the activity was assayed using mixed micelles in a modified Dole assay (36) using the same conditions employed for deuterium exchange experiments. Purified GIVA PLA2 (2 mg/ml) was stored in the protein buffer on ice for DXMS experiments. Experiments were performed immediately after elution from the nickel column.
Preparation of Lipid Vesicles—Lipid vesicles were prepared by evaporating solutions of 1-palmitoyl 2-arachidonic phosphatidyl-choline in chloroform under argon to dryness. The lipid film was resuspended in 100 mm KCl and allowed to sit in a 40 °C water bath for 30 min. The lipid solution was then probe sonicated five times for 30 s each with 30-s breaks in ice between sonications. This solution was then centrifuged at 5,000 × g to remove large lipid aggregates and titanium particles released from the probe sonicator. This solution was then used immediately for deuterium exchange studies. Small unilamellar vesicles were chosen for this study because kinetic studies performed with both small unilamellar vesicles and large unilamellar vesicles with the GIVA PLA2 have shown similar kinetics (25, 37). We have previously used this technique with small unilamellar vesicles on the GIA PLA2 to study lipid binding (29), and we chose to expand on this using GIVA PLA2.
Preparation of Deuterated samples for On-Exchange Experiments—D2O buffer contained 10 mm Tris (pD 7.5), 50 mm NaCl in 98% D20. Hydrogen/deuterium exchange experiments were initiated by mixing 20 μl of GIVA PLA2 or the C2 domain or catalytic domain (containing 40 μg) in protein buffer with 60 μl of D2O buffer to a final concentration of 73% D2O at pH 7.5. In lipid vesicle and MAFP binding experiments, the GIVA PLA2 in protein buffer was preincubated in the presence of 60 μm MAFP added froma 1 mm stock dissolved in ethanol and/or 2 mm PAPC vesicles in a 23 °C water bath for 1 min. The final concentration of ethanol was 0.25% for experiments with MAFP and MAFP controls. MAFP was allowed to incubate with the enzyme for 3 h on ice followed by 10 min at room temperature pre-D2O incubation. The D2O buffer was then added, and the samples were incubated at 23 °C for an additional 10, 30, 100, 300, 1,000, 3,000, or 10,000 s. For experiments with lipid present, time points were only taken while the percentage of hydrolysis of the lipid vesicles was under 10%. The activity of the enzyme against these lipid vesicles was measured using a modified Dole assay that exactly matched deuterium exchange conditions (33). The deuterium exchange was quenched by adding 120 μl of ice-cold quench solution (0.96% formic acid, 1.66 m guanidine hydrochloride (GdHCl)) that acidified the sample to a final pH = 2.5 and concentrations of formic acid of 0.58% and 1 m GdHCl. The samples were placed on ice for 10 min to partially denature the protein and obtain optimal peptide maps. Vials with frozen samples were stored at -80 °C until analysis, usually within 3 days.
Proteolysis-Liquid Chromatography/Mass Spectrometry Analysis of Samples—All steps were performed at 0 °C as described previously (30, 32). The samples were hand-thawed on melting ice and injected onto and passed through a protease column (66-μl bed volume) filled with porcine pepsin (Sigma) and immobilized on Poros 20 AL medium (Applied Biosystems) at 30 mg/ml following the manufacturer's instructions, at a flow rate of 100 μl/min with 0.05% trifluoroacetic acid. The eluate from the pepsin column was directly loaded onto a C18 column (Vydac catalog number 218MS5150). The peptides were eluted at 50 μl/min with a linear gradient of 0.046% trifluoroacetic acid, 6.4% (v/v) acetonitrile to 0.03% trifluoroacetic acid, 38.4% acetonitrile for 30 min. The eluate from the C18 column was directed to a Finnigan Classic LCQ mass spectrometer via its electrospray mass ionization probe operated with a capillary temperature of 200 °C as described previously (30, 32).
Data Processing—SEQUEST software (Thermo Finnigan Inc.) was used to identify the sequence of the peptide ions. DXMS Explorer (Sierra Analytics Inc, Modesto CA) was used for the analysis of the mass spectra as described previously (30, 32). All selected peptides had first passed the quality control threshold of the software and were then manually checked to ensure that the observed mass envelope agreed with the calculated mass envelope. The highest signal/noise ion was picked if multiple ionization charges (1, 2, or 3) of a peptide were detected. Incorporated deuteron number was obtained by measuring the centroid shift between the non-deuterated and the partially deuterated mass envelope.
The deuteration level of each peptide was calculated by the ratio of the incorporated deuteron number to the maximum possible deuteration number. Peptide deuteration levels in replicate samples, measured by our DXMS methods, have been found to vary by less than 10%, and we therefore regard changes greater than 10% as significant (30). All experiments were performed at least twice, and representative data are shown. Trends in the data were similar from experiment to experiment, but total deuterium content varied by roughly 5-10% in similar experiments carried out weeks apart. For all regions that showed a greater then 10% increase or decrease, other peptides that include some or all of the regions of interest are provided in the supplemental data.
RESULTS
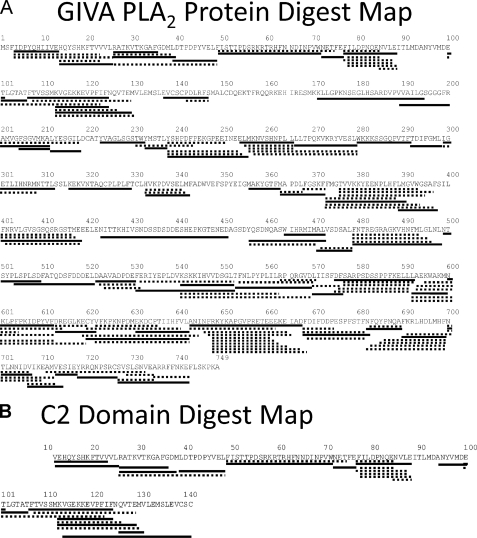
GIVA PLA2 Coverage Map of Pepsin Fragmentation—The protein digestion procedure was optimized to produce a peptide map that yielded the best coverage of GIVA PLA2 as described previously (35). The optimized condition gave 157 distinct peptides that gave 92% coverage of the GIVA PLA2 sequence. The same condition was used to digest the C2 domain, which generated 32 peptides covering 89% of the sequence (Fig. 1).
FIGURE 1.
Pepsin-digested peptide coverage map of GIVA PLA2 and C2 domain. Identified and analyzed pepsin-digested peptides are shown underneath the primary sequence of GIVA PLA2. Only the peptides in bold lines are used in this study.
Sixty-one peptide fragments are shown that cover 82% of the protein, and 17 peptide fragments for the C2 domain, covering 89% of the sequence, were analyzed for the data shown in the figures. These peptides are shown in Fig. 1 as bold lines. In this report, when we use the term “peptide,” we are referring to an actual peptide that was identified in the pepsin digestion. When we use the term “region,” we are referring to a section of the protein for which the deuterium exchange has been calculated, but it may or may not correspond to an actual pepsin peptide.
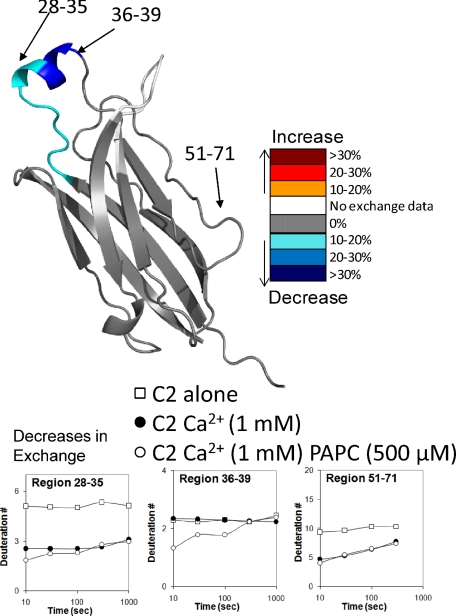
C2 Lipid Binding Experiments—On-Exchange experiments were performed on the isolated C2 domain containing amino acids 12-140. C2 domain lipid binding has been extensively studied by other techniques, and these studies served as an important control to compare this technique against previously published results obtained with other approaches. C2 on-exchange experiments were performed under five different conditions (Fig. 2). The change in exchange upon binding to 500 μm lipid vesicles was tested in the presence of 0, 200, and 1000 μm Ca2+. Significant decreases upon exposure to the lipid surface were only achieved in the presence of 1 mm Ca2+ (Fig. 2) and not with 0 or 200 μm Ca2+ (data not shown). The changes in exchange upon Ca2+ binding were exactly the same as the changes observed with intact full-length cPLA2 (35) and included regions 28-35, 51-71, 101-106, and 125-129 as well as a new region 41-48 (data not shown). This new region 41-48 is part of the C2 domain that shows very slow exchange rates in the full enzyme and faster exchange in the C2-only domain (35). A significant decrease in exchange due to binding of lipid vesicles was seen in the region 36-39, as well as a small change in the region 28-35 in the presence of 1 mm Ca2+ and PAPC lipid vesicles (Fig. 2). These regions include the hydrophobic residues Phe-35, Met-38, and Leu-39 present on the first Ca2+ binding loop. Previous work has also implicated region 96-98 in lipid binding (17-23), but no peptides were identified that contained these amino acids. These results agree with the previous experiments demonstrating lipid binding of the C2 domain alone.
FIGURE 2.
Deuterium exchange of the isolated C2 domain binding the phospholipid membrane in the presence of Ca2+. DXMS was performed on the C2 domain under the following three conditions: 0 and 1 mm Ca2+ without PAPC, as well as 1 mm Ca2+ with PAPC. The numbers of incorporated deuterons for three regions are shown: 28-35, 36-39, and 51-71. Changes in deuteration between the 1 mm Ca2+ and 1 mm Ca2+ + 500 μm PAPC conditions greater then 10% are represented in color on the crystal structure (PDB 1RLW) (see legend).
cPLA2 Lipid Binding Experiments—On-Exchange experiments were performed on the intact cPLA2 enzyme in the presence of a phospholipid membrane to determine how the catalytic domain interacts with the surface and to determine whether it changed membrane binding of the C2 domain. These experiments were carried out in the presence of 500 μm PAPC and 200 μm Ca2+ for time points varying from 10 to 300 s. The literature Kd values of this enzyme interacting with membranes varies from 1 to 90 μm depending on the presentation of the lipid substrate (25, 37). Generation of product interfering with membrane binding was a concern in these experiments. For this reason, these experiments were carried out at shorter time points with low levels of Ca2+ to keep the hydrolysis of phospholipids at the lipid membrane below 10%. Experiments were carried out testing lipid binding in the presence of 1 mm EGTA to test binding without Ca2+. No changes greater then 10% were observed upon lipid binding without Ca2+ present.
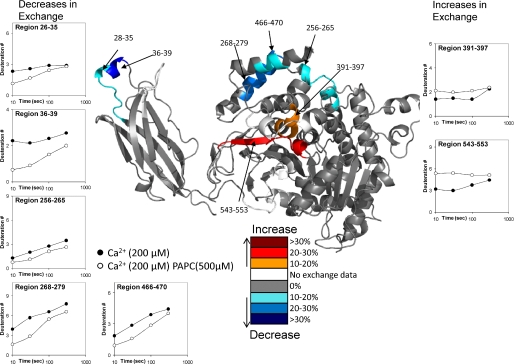
Activity assays were done under the same experimental conditions as the deuterium exchange experiments to measure the levels of arachidonic acid release. In the presence of 200 μm Ca2+, the percentage of hydrolysis at 10 s was ∼0.2%, and the percentage of hydrolysis proceeded to 6% at 300 s. Samples taken at 1000 s showed 16% hydrolysis, and we decided to limit our time course to only 300 s. The specific activity of the enzyme under our conditions was ∼20 nmol min-1 mg-1, which is consistent with literature values under similar protein to lipid ratios (25). Four regions of the protein (28-39, 258-265, 268-279, and 466-470) exhibited decreased exchange in the presence of lipid vesicles (Fig. 3). Multiple additional peptides covered portions of these regions, and peptide data for these regions is shown in supplemental Fig. 1. Region 28-39 exhibited similar changes in deuterium exchange as did the isolated C2 domain. Changes were seen due to membrane surface binding in the intact enzyme in the presence of 200 μm Ca2+ with 500 μm PAPC vesicles that required 1 mm Ca2+ to be seen in the C2 domain alone. It is important to note that there is a large change in exchange induced upon Ca2+ binding in region 28-35 and a much smaller change induced upon lipid binding. Once again, no peptides spanning region 96-98 were identified, so no data on lipid interactions of that region were available. No other regions on the C2 domain showed changes upon lipid binding.
FIGURE 3.
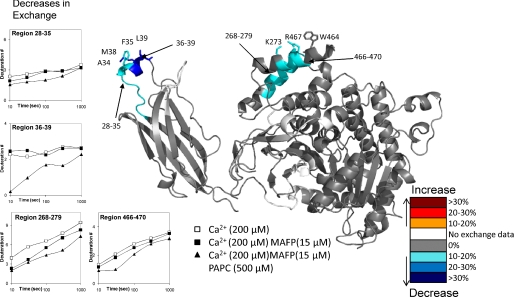
Deuterium exchange of the GIVA PLA2 binding the phospholipid membrane in the presence of Ca2+. Ca2+ was present at 200 μm, and vesicles were present at 500 μm. The numbers of incorporated deuterons at four time points in seven different regions, 28-35, 36-39, 256-265, 268-279, 391-397, 466-470, and 543-553, in GIVA PLA2 are plotted. Decreases or increases in deuteration greater then 10% are represented by the color scheme in the legend.
The region 268-279 showed a 10-15% decrease in exchange upon membrane binding but no change upon Ca2+ binding alone. This region contains a group of charged amino acids, Lys-273, Arg-274, and Glu-277 as well as the polar Gln-270 that are oriented to possibly interact with the zwitterionic head groups of a lipid surface. There are also multiple hydrophobic residues including Val-272, Tyr-275, and Leu-279 pointing toward the catalytic site of the enzyme that may interact with fatty acyl chains of the substrate. The region 466-470 is located on the same face as 268-279 and has a similar decrease in exchange, with a decrease of around 10-15% at all time points. This area contains a charged group at Arg-467 and one hydrophobic residue Ile-469.
There was also a smaller decrease going from 5 to 10% over the time course in the region from 256 to 265 that contains a group of four leucines at 262-265 that have extensive hydrophobic contacts with other non-polar residues on the interior of the protein. There are no specific residues that would obviously interact with the membrane surface, so this decrease could be due to a conformational shift upon the membrane binding to the lipid surface or interactions between the leucines and the fatty acyl chains of the substrate.
There are two regions that show increases in exchange upon lipid binding, and these are 391-397 and 543-553. Region 391-397 has a >10% increase at early time points, with no change in exchange at 300 s. Region 543-553 has a larger increase of ∼25% at early time points with almost no change in exchange seen at 300 s. These rates imply that these regions of the protein are becoming more accessible to solvent. These regions contain hydrophobic residues that are part of a channel that leads to the active site residues Ser-228 and Asp-549. These residues are all in contact with the lid region spanning 415-432. These increases may be caused by a lid opening caused by substrate binding or membrane surface binding. Region 543-553 also contains the active site Asp-549. Experiments were also performed using the isolated catalytic domain in the presence of lipid vesicles, but no changes in deuteration greater than 10% were observed (data not shown).
On-Exchange Results Using MAFP-inhibited GIVA PLA2—MAFP is a potent irreversible inhibitor of the GIVA PLA2 enzyme (39). Inhibitor-bound α/β-hydrolases have been used to crystallize the open lid conformation of various lipases (26). MAFP was used to determine whether substrate binding caused opening of the lid region or whether interfacial binding of the lipid surface caused this change. Using MAFP allowed us to separate substrate binding effects from membrane surface binding effects. MAFP was selected due to its similarity to natural substrate because it contains an arachidonic acid in a similar position to the sn-2 fatty acid chain of the natural substrate, although there is no phospholipid head group, or sn-1 fatty acid tail in MAFP. This acts as an excellent mimic to the lysophospholipase and acyl transferase activity of cPLA2 (40, 41). MAFP binding was tested at seven time points from 10 to 10,000 s at a concentration of 15 μm. There were five regions that saw changes upon MAFP binding. Multiple peptides covered these regions, and additional peptide data for these regions are shown in supplemental Fig. 2.
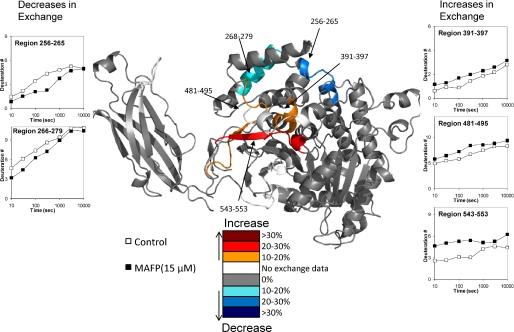
Two regions of the enzyme 256-265 and 268-279 showed decreases in exchange upon MAFP binding (Fig. 4). The region from 268 to 279 showed a decrease in deuteration of >10%, which is less then observed with phospholipid binding experiments. The region from 258 to 265 had a >10% decrease in exchange similar to the phospholipid binding experiments. This indicates either that these regions are binding the arachidonic acid tail of the MAFP molecule or that conformational change that occurs when the lid opens blocks access to these residues or rigidifies the protein structure in this region. Other hydrophobic residues that should bind MAFP in the active site funnel have very low levels of exchange even in the inhibitor free samples, so no changes could be recorded upon inhibitor binding.
FIGURE 4.
Deuterium exchange upon binding of 15μm MAFP. The number of incorporated deuterons at seven time points in five different regions, 258-265, 268-279, 391-397, 481-495, and 543-553, in GIVA PLA2 are plotted. Decreases or increases in deuteration greater then 10% are represented by the color scheme in the legend.
All of the regions that had increases upon lipid binding had increases upon MAFP binding. These increases were of the same magnitude as with lipid binding. Region 391-397 has a constant 5-15% increase in exchange over all time points. Other peptides overlapping this region (supplemental Fig. 2) show that the majority of this increase is localized to region 394-397. The region 543-553 has a 25% increase at early time points with less of an increase at later time points. The region 481-495 also had an increase in exchange upon exposure to MAFP. This region had a 10-20% increase in exchange across all time points. All of these regions are partially located under the lid region spanning 415-432. MAFP binding closely mimics natural phospholipid binding in the increases seen in the catalytic domain of the protein, even without a membrane surface.
On-Exchange Results Using MAFP-inhibited GIVA PLA2 Binding Phospholipid Membrane—On-Exchange experiments were performed with both MAFP and PAPC vesicles in an attempt to identify those changes due to the presence of an interface as opposed to those due to the binding of the pseudo-substrate in the catalytic site. The presence of MAFP in the active site should block any binding of phospholipid in the active site, so the effects should only be from the enzyme binding to the phospholipid surface and not from conformational changes induced by substrate binding. This also acted as an important control to see effects of lipid binding without product generation. The interface experiments were performed for time courses ranging from 10 up to 1,000 s. These experiments were performed in the presence of Ca2+ to activate membrane binding. An experiment was also performed in the presence of EGTA with lipid vesicles to see whether any changes were seen without Ca2+ present. Experiments with EGTA showed no significant changes in deuterium exchange upon exposure to lipid vesicles without Ca2+ present (data not shown).
There were four regions of the protein that showed decreases in exchange upon MAFP-inhibited lipid binding (Fig. 5). Multiple peptides covered these regions, and peptide data for these regions are shown in supplemental Fig. 3.
FIGURE 5.
Deuterium exchange upon Ca2+-mediated PAPC vesicle binding in the presence of 15μm MAFP. Ca2+ was present at 200 μm, and vesicles were present at 500 μm. The numbers of incorporated deuterons at five time points in four different regions, 28-35, 36-39, 268-279, and 466-470, in GIVA PLA2 are plotted. Possible amino acids that may interact with the lipid surface have been shown in stick form. Decreases or increases in deuteration greater then 10% are represented by the color scheme in the legend.
The regions 28-35 and 36-39 are located on the C2 domain. Exchange at peptide 28-35 is decreased significantly by the presence of Ca2+, but even so, the changes induced by lipid were greater then 10%. The greatest change was seen in region 36-39 located on the hydrophobic α helix proposed to penetrate the membrane surface. These peptides showed no changes in the presence of lipid without Ca2+ present, demonstrating that these changes caused by lipid binding are Ca2+-dependent. These changes were almost identical to those seen with uninhibited enzyme mixed with lipid.
The region from 268 to 279 on the catalytic domain also showed a >10% decrease in exchange. This area is also one of the regions that show a significant decrease upon MAFP binding. This region appears to both bind the interfacial surface, as well as bind the single phospholipid molecule in the active site. The region from 466 to 470 that had decreases upon non-inhibited lipid binding shows a similar exchange pattern upon inhibitor-bound lipid binding. These results suggest that the C2 domain, as well as the region of the catalytic domain containing regions 466-470 and 268-279, are important in binding the lipid surface. The regions that increased due to lipid binding in regions 391-397 and 543-553 showed no changes in exchange between the MAFP-inhibited enzyme and the inhibited enzyme incubated with lipid vesicles (supplemental Fig. 3).
DISCUSSION
C2 domain binding to membrane of the GIVA PLA2 has been extensively studied (17-23). However, how the intact enzyme binds a lipid surface and the allosteric changes caused by this binding have been much more difficult to determine. This is the first study to examine the structural basis of the intact enzyme binding to the lipid surface as well as conformational changes upon binding a pseudo-substrate in the active site with deuterium exchange mass spectrometry. Ca2+-mediated C2 binding to the membrane surface has been shown to be mediated by hydrophobic residues in the Ca2+ binding loop one 35-39 and residues in the Ca2+ binding loop two 96-98 (17-23). No peptides existed that spanned region 96-98, so information about how Ca2+ binding loop two penetrates the surface was unavailable. Our results with deuterium exchange of the isolated C2 domain with lipid show decreases in regions 28-35 and 36-39, which are consistent with these regions penetrating the lipid interface. These experiments on the C2 domain alone were used as a control to demonstrate that we could examine membrane surface binding using the DXMS technique.
The results of the intact cPLA2 enzyme binding to the lipid surface showed similar changes to the C2 domain alone, which showed lipid binding at regions 28-35 and 36-39 but with less Ca2+ required to see effects with the intact enzyme in deuterium exchange. This corresponds well with previous work showing an increased residence time of the intact GIVA PLA2 on the membrane surface of the Golgi as compared with the C2 domain alone (38). This implies that the presence of the catalytic domain helps bind the enzyme to the lipid surface or causes changes in the binding of Ca2+ in the intact enzyme. The hydrophobic core of the protein around the active site gave rise to significant increases in exchange upon Ca2+-mediated lipid binding. This region contains many hydrophobic residues that are blocked from solvent exposure in the closed form of the enzyme. When lipid binds in the active site, there must be a change in the structure that causes the lid to reorient in some way that increases solvent accessibility to the hydrophobic core. The other decreases in deuteration in the catalytic domain upon Ca2+-mediated lipid binding were in regions that could be caused by interactions with either the membrane surface or the fatty acyl chains of the substrate.
We used the inhibitor MAFP to differentiate lipid surface binding effects from conformational changes induced by the enzyme binding substrate in the active site. Previous work has used inhibitors to force lipases into an open lid conformation (26). MAFP gave rise to the same increases in the hydrophobic core as seen with phospholipid vesicles (supplemental Fig. 3), except for a slightly greater increase in solvent accessibility in region 481-495 as compared with lipid binding results. The decreases in exchange seen in regions 256-265 and 268-279 of the catalytic domain were very similar to those seen with only lipid binding. MAFP covalently modifies the active site Ser-228 with an arachidonic acid and is very similar to the sn-2 fatty acid of a natural phospholipid substrate. The decreases in exchange in regions 256-265 and 268-279 upon MAFP binding without a lipid surface present suggest that residues are interacting with the arachidonic acid fatty acid. Residues Leu-264, Leu-267, and Tyr-275 could be in the correct orientation to interact with the fatty acid tail of substrate. The other hydrophobic amino acids that would bind to substrate are in the hydrophobic core of the protein that has very low levels of exchange (35), and therefore no changes in exchange can be seen. The increases in exchange seen upon inhibitor binding are almost certainly caused by a shift of the lid region from 415 to 432 as shown in Fig. 6.
FIGURE 6.
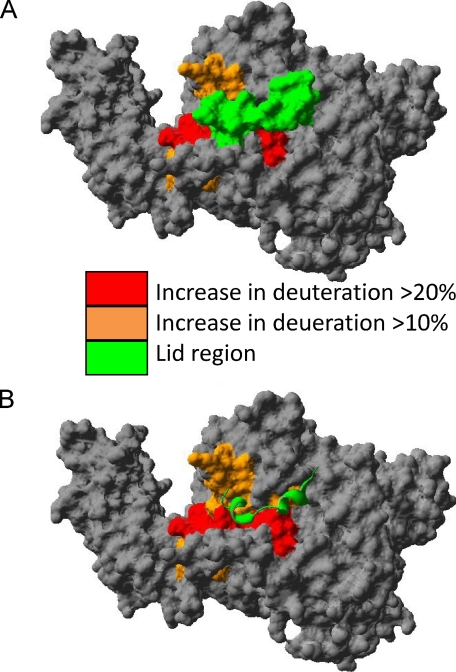
Opening of the lid induced by inhibitor binding. A, the molecular surface of the enzyme with areas that show an increase in exchange upon inhibitor binding is colored red or orange, and the lid region from 415 to 432 is colored green. B, the lid region surface is removed and shown as a ribbon with most areas in red underneath the lid. The deuterium exchange data imply that the lid region from 415 to 432 shifts position to expose the regions underneath. Molecular surface was generated using the Swiss-PdbViewer.
There have been very few studies that have looked at how the intact enzyme binds the surface. The enzyme is assumed to shift to an “open conformation” in which the lid region has moved out of the way of the catalytic site. This hypothesis was suggested due to the inability to otherwise model a phospholipid in the active site (10) but was not based on or verified by experiment. The DXMS results reported here with natural phospholipids and the inhibitor MAFP clearly demonstrate this conformational change.
The lid region of 415-432 has amphipathic character with amino acids involved in hydrophobic interactions with amino acids underneath and many charged amino acids exposed to solvent. There are significant increases in exchange in areas of the protein directly covered by the lid region. There are significant hydrophobic contacts seen in the crystal structure (PDB 1CJY) between the lid region and areas with an increase in exchange upon MAFP binding. There are contacts between four amino hydrophobic amino acids on the lid, Met-417, Leu-421, Ile-424, and Ile-429, and amino acids in the regions with increases in exchange, Ala-396, Phe-397, Ala-486, Val-548, and Leu-552. The breaking of these hydrophobic contacts between the lid region and the regions underneath would explain the increase in deuterium exchange seen in this region. This increase in exchange could be due to the lipid substrate binding to the hydrophobic residues at Phe-397, Ile-399, Leu-400, Val-404, and Leu-405 and causing the lid region to undergo a conformational shift opening up the active site. Previous studies using mutants of the GIVA PLA2, specifically mutants of Ile-399 and Leu-400, showed a vast decrease in activity, membrane affinity, and membrane penetration (24). The proposal was that these amino acids were mediating penetration of the membrane surface. This is unlikely due to the many charged residues that would also need to penetrate the membrane surface to allow Ile-399 and Leu-400 to penetrate. It is possible that these residues are mediating substrate binding, and this substrate binding helps mediate activity and surface binding by moving the lid region. These increases in exchange are seen with natural phospholipid substrate and with the MAFP substrate analog. This implies that the presence of an inhibitor in the active site closely mimics the open form of the enzyme that works on natural phospholipid substrate.
Our results using MAFP-inhibited enzyme binding to the lipid surface were used to show how the enzyme binds to the lipid surface without complicating effects caused by conformational changes upon substrate binding since those changes have already occurred due to MAFP in the active site. Upon the addition of phospholipid to the MAFP-inhibited enzyme, the majority of decreases are seen in the region from 28 to 35 and 36 to 39, which contains the hydrophobic α helix thought to penetrate the membrane surface. However, there is still a significant change in exchange in regions 268-279 and 466-470. These regions contain a group of charged amino acids Lys-271, Lys-273, Arg-274, Arg-467 as well as the polar Gln-270 that could all interact with zwitterionic PC head groups. Lys-27, Arg-274, and Arg-467 are the only residues from the crystal structure (PDB 1CJY) that are oriented in the correct way to interact with PC head groups. However, region 268-279 had significant decreases with MAFP inhibitor bound without the lipid interface present, which implies that it is the binding substrate in the active site. This interaction may help to orient the other residues in this region to interact with the head groups of a membrane surface. Region 466-470 is near Trp-464 that could penetrate into the hydrophobic core of the membrane, as proposed in previous studies (38). Previous work has shown that mutations to residues Arg-467 and Lys-273 decreases binding to negatively charged phospholipids (24).
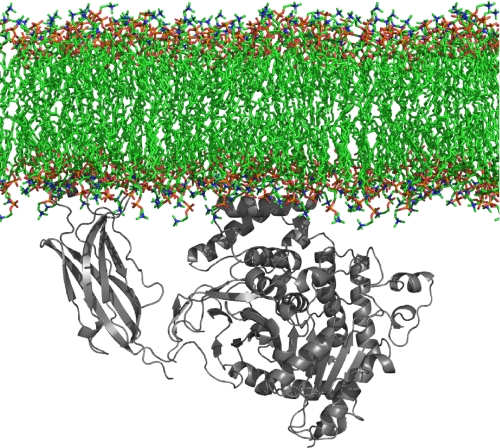
From these results, we have generated a model of the initial step in Ca2+-mediated membrane binding before opening of the lid shown in Fig. 7. This model corresponds to our deuterium exchange data, where region 35-39 and 96-98 along with Trp-464 penetrate the membrane surface, and electrostatic contacts between Lys-273, Arg-274, and Arg-467 help bind the zwitterionic head groups on the membrane surface. Interactions between the catalytic domain alone are not sufficient to bind membrane phospholipid, with Ca2+ binding causing the enzyme to translocate to the membrane surface through the C2 domain, and this binding is increased through electrostatic and hydrophobic interactions between the membrane surface and the catalytic domain. Large regions of the catalytic domain do not penetrate the membrane surface, based upon the observation that there are no decreases in deuterium exchange in areas of the i-face of the enzyme upon lipid surface binding. Upon this binding, the lid region is opened through hydrophobic contacts between the enzyme and a single phospholipid diffusing into the active site, substrate binds in the open form of the enzyme in the active site, and free arachidonic acid is liberated.
FIGURE 7.
Hypothetical model of initial membrane binding before lid opening. Ca2+ binding at the C2 domain causes translocation of the enzyme to the surface and penetration of the hydrophobic core of the membrane. There seems to be no penetration of the catalytic domain, and its interactions with the surface are mediated through electrostatic contacts from regions 268-279 and 466-470 to PC head groups. The model was generated using PyMOL.
We plan to study the interactions of various other lipid activators of this enzyme using these same techniques and to further characterize the structure of the open lid form of the enzyme. This work suggests interesting possibilities such as being able to isolate the open conformation of the enzyme by using inhibitor-enzyme complexes. Further study using deuterium exchange along with crystallography and molecular modeling techniques offers the potential to further define the interaction of the enzyme with membrane surfaces.
This work represents the first DXMS study of an intracellular lipase binding a lipid surface. This study shows lipid binding effects of the entire enzyme instead of only the C2 domain as has been previously demonstrated with other techniques. It also uses inhibitor binding to probe the structure of the open conformation of the enzyme.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM20501 (to E. A. D.) and CA099835, CA118595, GM037684, AI0220221, and AI022160 (to V. L. W.). This work was also supported by Discovery Grant UC10591 from the University of California Industry.-University Cooperative Research Program (to V. L. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains three supplemental figures.
Footnotes
The abbreviations used are: GIVA, group IVA; PLA2, phospholipase A2; DXMS, deuterium exchange mass spectrometry; PC, phosphatidylcholine; PAPC, 1-palmitoyl-2-arachidonoyl-sn-phosphatidylcholine; PDB, protein data bank.
References
- 1.Schaloske, R. H., and Dennis, E. A. (2006) Biochim. Biophys. Acta 1761 1246-1259 [DOI] [PubMed] [Google Scholar]
- 2.Six, D. A., and Dennis, E. A. (2000) Biochim. Biophys. Acta 1488 1-19 [DOI] [PubMed] [Google Scholar]
- 3.Clark, J. D., Lin, L. L., Kriz, R. W., Ramesha, C. S., Sultzman, L. A., Lin, A. Y., Milona, N., and Knopf, J. L. (1991) Cell 65 1043-1051 [DOI] [PubMed] [Google Scholar]
- 4.Kramer, R. M., Roberts, E. F., Manetta, J., and Putnam, J. E. (1991) J. Biol. Chem. 266 5268-5272 [PubMed] [Google Scholar]
- 5.Funk, C. D. (2001) Science 294 1871-1875 [DOI] [PubMed] [Google Scholar]
- 6.Bonventre, J. V., Huang, Z., Taheri, M. R., O'Leary, E., Li, E., Moskowitz, M. A., and Sapirstein, A. (1997) Nature 390 622-625 [DOI] [PubMed] [Google Scholar]
- 7.Nagase, T., Uozumi, N., Ishii, S., Kume, K., Izumi, T., Ouchi, Y., and Shimizu, T. (2000) Nat. Immunol. 1 42-46 [DOI] [PubMed] [Google Scholar]
- 8.Uozumi, N., Kume, K., Nagase, T., Nakatani, N., Ishii, S., Tashiro, F., Komagata, Y., Maki, K., Ikuta, K., Ouchi, Y., Miyazaki, J., and Shimizu, T. (1997) Nature 390 618-622 [DOI] [PubMed] [Google Scholar]
- 9.Nalefski, E. A., Sultzman, L. A., Martin, D. M., Kriz, R. W., Towler, P. S., Knopf, J. L., and Clark, J. D. (1994) J. Biol. Chem. 269 18239-18249 [PubMed] [Google Scholar]
- 10.Dessen, A., Tang, J., Schmidt, H., Stahl, M., Clark, J. D., Seehra, J., and Somers, W. S. (1999) Cell 97 349-360 [DOI] [PubMed] [Google Scholar]
- 11.Perisic, O., Fong, S., Lynch, D. E., Bycroft, M., and Williams, R. L. (1998) J. Biol. Chem. 273 1596-1604 [DOI] [PubMed] [Google Scholar]
- 12.Stahelin, R. V., Subramanian, P., Vora, M., Cho, W., and Chalfant, C. E. (2007) J. Biol. Chem. 282 20467-20474 [DOI] [PubMed] [Google Scholar]
- 13.Subramanian, P., Vora, M., Gentile, L. B., Stahelin, R. V., and Chalfant, C. E. (2007) J. Lipid Res. 48 2701-2708 [DOI] [PubMed] [Google Scholar]
- 14.Mosior, M., Six, D. A., and Dennis, E. A. (1998) J. Biol. Chem. 273 2184-2191 [DOI] [PubMed] [Google Scholar]
- 15.Six, D. A., and Dennis, E. A. (2003) J. Biol. Chem. 278 23842-23850 [DOI] [PubMed] [Google Scholar]
- 16.Cho, W. (2001) J. Biol. Chem. 276 32407-32410 [DOI] [PubMed] [Google Scholar]
- 17.Perisic, O., Paterson, H. F., Mosedale, G., Lara-Gonzalez, S., and Williams, R. L. (1999) J. Biol. Chem. 274 14979-14987 [DOI] [PubMed] [Google Scholar]
- 18.Ball, A., Nielsen, R., Gelb, M. H., and Robinson, B. H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 6637-6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bittova, L., Sumandea, M., and Cho, W. (1999) J. Biol. Chem. 274 9665-9672 [DOI] [PubMed] [Google Scholar]
- 20.Malkova, S., Long, F., Stahelin, R. V., Pingali, S. V., Murray, D., Cho, W., and Schlossman, M. L. (2005) Biophys. J. 89 1861-1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malmberg, N. J., Van Buskirk, D. R., and Falke, J. J. (2003) Biochemistry 42 13227-13240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nalefski, E. A., McDonagh, T., Somers, W., Seehra, J., Falke, J. J., and Clark, J. D. (1998) J. Biol. Chem. 273 1365-1372 [DOI] [PubMed] [Google Scholar]
- 23.Stahelin, R. V., Rafter, J. D., Das, S., and Cho, W. (2003) J. Biol. Chem. 278 12452-12460 [DOI] [PubMed] [Google Scholar]
- 24.Das, S., and Cho, W. (2002) J. Biol. Chem. 277 23838-23846 [DOI] [PubMed] [Google Scholar]
- 25.Das, S., Rafter, J. D., Kim, K. P., Gygi, S. P., and Cho, W. (2003) J. Biol. Chem. 278 41431-41442 [DOI] [PubMed] [Google Scholar]
- 26.Roussel, A., Miled, N., Berti-Dupuis, L., Riviere, M., Spinelli, S., Berna, P., Gruber, V., Verger, R., and Cambillau, C. (2002) J. Biol. Chem. 277 2266-2274 [DOI] [PubMed] [Google Scholar]
- 27.Schrag, J. D., Li, Y., Cygler, M., Lang, D., Burgdorf, T., Hecht, H. J., Schmid, R., Schomburg, D., Rydel, T. J., Oliver, J. D., Strickland, L. C., Dunaway, C. M., Larson, S. B., Day, J., and McPherson, A. (1997) Structure (Camb.) 5 187-202 [DOI] [PubMed] [Google Scholar]
- 28.Schrag, J. D., Li, Y. G., Wu, S., and Cygler, M. (1991) Nature 351 761-764 [DOI] [PubMed] [Google Scholar]
- 29.Burke, J. E., Karbarz, M. J., Deems, R. A., Li, S., Woods, V. L., Jr., and Dennis, E. A. (2008) Biochemistry 47 6451-6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamuro, Y., Anand, G. S., Kim, J. S., Juliano, C., Stranz, D. D., Taylor, S. S., and Woods, V. L., Jr. (2004) J. Mol. Biol. 340 1185-1196 [DOI] [PubMed] [Google Scholar]
- 31.Mandell, J. G., Falick, A. M., and Komives, E. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 14705-14710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brudler, R., Gessner, C. R., Li, S., Tyndall, S., Getzoff, E. D., and Woods, V. L., Jr. (2006) J. Mol. Biol. 363 148-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoofnagle, A. N., Resing, K. A., Goldsmith, E. J., and Ahn, N. G. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 956-961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wales, T. E., and Engen, J. R. (2006) Mass Spectrom. Rev. 25 158-170 [DOI] [PubMed] [Google Scholar]
- 35.Hsu, Y. H., Burke, J. E., Stephens, D. L., Deems, R. A., Li, S., Asmus, K. M., Woods, V. L., Jr., and Dennis, E. A. (2008) J. Biol. Chem. 283 9820-9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucas, K. K., and Dennis, E. A. (2005) Prostaglandins Other Lipid Mediat. 77 235-248 [DOI] [PubMed] [Google Scholar]
- 37.Hixon, M. S., Ball, A., and Gelb, M. H. (1998) Biochemistry 37 8516-8526 [DOI] [PubMed] [Google Scholar]
- 38.Evans, J. H., and Leslie, C. C. (2004) J. Biol. Chem. 279 6005-6016 [DOI] [PubMed] [Google Scholar]
- 39.Huang, Z., Liu, S., Street, I., Laliberte, F., Abdullah, K., Desmarais, S., Wang, Z., Kennedy, B., Payette, P., Riendeau, D., Weech, P., and Gresser, M. (1994) Mediat. Inflamm. 3 307-308 [Google Scholar]
- 40.Leslie, C. C. (1991) J. Biol. Chem. 266 11366-11371 [PubMed] [Google Scholar]
- 41.Reynolds, L., Hughes, L., Louis, A. I., Kramer, R. A., and Dennis, E. A. (1993) Biochim. Biophys. Acta 1167 272-280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.