Abstract
In 3T3-L1 cells, HuR is constitutively expressed and prior to induction of differentiation localized predominantly to the nucleus. Within minutes of induction of differentiation, nuclear HuR binds to its target ligand mRNAs, and the complexes appear to move to the cytosol. One ligand mRNA is the CCAAT/enhancer-binding protein β (C/EBPβ) message. To examine the function and importance of the HuR-C/EBPβ interaction, retroviral expression constructs were created in which the HuR binding site was altered by deletion (βdel) or deletion and substitution (βd/s). Expression of these constructs in murine embryonic fibroblasts resulted in significant adipose conversion relative to those cells expressing wild type C/EBPβ. C/EBPβ protein content was increased markedly in both βdel and βd/s, which correlated with the acquisition of the adipocyte phenotype. Analysis of the βd/s cell line demonstrated a robust expression of C/EBPα coincident with peroxisome proliferator-activated receptor γ expression. Total C/EBPβ mRNA accumulation indicated no difference between cells harboring either the wild type C/EBPβ cDNA or βd/s construct. However, cytosolic C/EBPβ mRNA in the cells expressing the βd/s construct was maintained at levels between 2- and 7-fold greater than in the cells expressing the wild type construct. Alteration in mRNA half-life was not responsible for the increased accumulation. Mechanistically, these data suggest that HuR binding results in nuclear retention of the C/EBPβ mRNA and is consistent with HuR control, at least in part, of mRNA processing.
Adipocyte differentiation is a complex process regulated in large part by the temporally controlled expression and activation of numerous transcription factors (1). Among these proteins, the CCAAT/enhancer-binding protein β (C/EBPβ)3 and peroxisome proliferator-activated receptor (PPAR) families of transcriptional activators have been identified as critical to initiation of the differentiation process as well as to maintenance of the adipocyte phenotype (1). Functional roles for these factors have been established at least in part through use of the 3T3-L1 preadipocyte model system (1). When 3T3-L1 preadipocytes are induced to differentiate, the cells reenter the cell cycle and undergo mitotic clonal expansion followed by growth arrest and expression of the adipocyte phenotype (1). During this process, C/EBPβ is expressed coincidentally with induction of differentiation and is essential not only for mitotic clonal expansion but also for the transcriptional activation of PPARγ and C/EBPα genes (2–6). The indispensable nature of appropriate C/EBPβ expression was demonstrated in studies with C/EBPβ-/- murine embryonic fibroblasts (MEFs), which when treated with the differentiation inducers could neither reenter the cell cycle and undergo mitotic clonal expansion nor express the adipocyte phenotype (4). Similar results were observed in 3T3-L1 cells expressing a dominant-negative C/EBP (5).
Messenger RNA export from the nucleus, mRNA turnover, and translation initiation are important control points in the post-transcriptional regulation of gene expression. At least in part, control of these processes is exerted through recognition of cis elements in the mRNA by specific binding proteins. One of these proteins is HuR, a 36-kDa protein that belongs to the Hu/ELAV (embryonic lethal abnormal vision) family of RNA-binding proteins (7). HuR is ubiquitously expressed, localized predominantly to the nucleus, and has been demonstrated to shuttle between the nucleus and cytoplasm. The shuttling activity suggests but has not yet proved that HuR functions by binding to nascent mRNAs in the nucleus and protecting them from degradation by actively participating in their nucleocytoplasmic transport (8–23). Once in the cytosol, there is compelling evidence to suggest that HuR functions to control the stability and translational efficiency of its ligand mRNAs (8–26). Recent data have also supported a role for HuR in the regulation of polyadenylation by competitively inhibiting the binding of the cleavage and polyadenylation specificity factor, thereby attenuating polyadenylation and nuclear export (27). It is not clear that any one mRNA is subjected to all four HuR-mediated regulations, i.e. 1) control of polyadenylation, 2) translocation to the cytosol, 3) stability, and 4) translational initiation/efficiency. It is important to realize that HuR is a regulatory protein involved in the post-transcriptional processing of certain mRNAs and that the particular function(s) may depend on the particular message.
In 3T3-L1 preadipocytes, HuR is constitutively expressed and localized predominantly to the nucleus (28). Within 30 min of exposure to the differentiation stimulus, the HuR content in the cytosol increases, consistent with HuR regulating the availability of relevant mRNAs for translation. We have demonstrated previously (28) that one of the relevant mRNAs forming a nuclear complex with HuR upon induction of the differentiation program and translocating to the cytosol as a messenger ribonucleoprotein (mRNP) complex with HuR is the C/EBPβ message.
In this study, we begin to address the functional significance of the interactions between HuR and the C/EBPβ 3′-untranslated region (UTR). Using non-adipogenic MEF-3T3 cells, we examine the effects of conditional ectopic expression of C/EBPβ and mutants unable to bind HuR on the metabolism of the C/EBPβ message.
EXPERIMENTAL PROCEDURES
Materials—Dulbecco's modified Eagle's medium (DMEM), Opti-MEM I reduced serum medium, RNase inhibitor, Lipofectamine 2000, and reagents for molecular biology were purchased from Invitrogen. The BD Retro-X Universal packaging system, the RevTet-Off™ system, the MEF/3T3 Tet-Off cell line, and the vesicular stomatitis virus G protein (VSV-G) and enhanced green fluorescent protein expression vectors were obtained from BD Biosciences/Clontech. Bovine calf serum and fetal calf serum were purchased from Hyclone Laboratories (Logan, UT). The 3T3-L1 cells used in this work were obtained from Howard Green (Harvard University, Boston, MA). The BCA protein assay kit, the NE-PER™ cell fractionation kit, and HALT™ protease inhibitor mixture were from Pierce. The QuikChange-XL site-directed mutagenesis kit and XL-10 Gold Ultracompetent cells were purchased from Stratagene (Cedar Creek, TX). The plasmid midi kit was obtained from Qiagen (Valencia, CA). The MAXIScript T7 kit was from Ambion (Austin, TX). All other chemicals were of reagent grade and purchased from Sigma. The pBluescript-C/EBPβ plasmid DNA was obtained from Steve Farmer (Boston University, Boston, MA). The 3A2 monoclonal antibody directed against HuR, as well as the VSV-G, C/EBPβ, and PPARγ antibodies, was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The β-tubulin antibody was provided by Ted Bertrand (Brody School of Medicine, Greenville, NC).
Isolation of Nuclear and Cytosolic Fractions—The NE-PER™ cell fractionation kit was used to isolate nuclear and cytosolic fractions from the cells following the manufacturer's instructions with minor modifications as described previously (28). For isolation of RNA, 4 wells of a 6-well cluster plate were used as a source of material for the separation. In addition to the standard protease inhibitors, RNase inhibitor was added to the lysate. Incubation time for the initial extract was increased from 10 to 15 min. The isolated fractions were stored at -80 °C until used.
Western Blot Analysis—Western blot analysis was performed as described previously (28).
Preparation of C/EBPβ Retroviral Expression Constructs— The full-length (1.5 kb) C/EBPβ cDNA construct (βwt) in pBluescript was the starting material. The mutant C/EBPβ cDNA construct with point mutations flanking the HuR binding site (βpm) was created by induction of mutations at bases 1288 and 1389 with mutation primers 5′-GTAATCACCTTAAAGATCTTCCTGCGGGGTTGTTGA-3′ and 5′-GTCTTATTATTTTTTTTGTATTATATAAAAAAGATCTATTTCTATGAGAAAAGAGGCGTATG-3′, respectively (primer for the sense strand shown). This resulted in the creation of BglII restriction sites at positions 1288 and 1389. Removal of the 101-base BglII fragment followed by ligation resulted in the generation of the mutant C/EBPβ cDNA construct with the HuR binding site deleted (βdel). Replacement of the BglII fragment with a 113-base BglII fragment derived from the GLUT1 3′-UTR (bases 1758–1871) resulted in the creation of the mutant C/EBPβ cDNA construct with the HuR binding site deleted and substituted (βd/s). Each of the constructs (βwt, βpm, βdel, and βd/s) was excised from pBluescript and ligated into the EcoRI/BamHI sites of the donor vector pDNR-1r. The constructs were then excised from the HindIII/SalI site of pDNR-1r and ligated into the HindIII/SalI site of pRevTRE. Confirmation of mutations and appropriate orientation was obtained by sequencing at the University of Tennessee, Knoxville, Molecular Biology Resource Facility.
Riboprobe Preparation—To prepare RNA probes for gel shift assays, regions of the various β constructs were isolated by PCR as double-stranded templates under the control of the T7 promoter. The T7 polymerase binding site was included by addition of the sequence 5′-GGATCCTAATACGACTCACTATAGGGAG-3′ to the 5′ end of a forward primer. The forward primers for the constructs were as follows: βwt, 5′-GCGGGGTTGTTGATGTTT-3′, which was used for both βwt and βpm; βdel, 5′-TTTCGGGACTTGATGCAA-3′; and βd/s, 5′-AGCAGTGAAGTCCAGGAG-3′. The reverse primer (β-Reverse, 5′-CTTTAATGCTCGAAACGG-3′) was utilized with all four constructs.
RNA Gel Shift Analysis—Radiolabeled riboprobes were prepared using the MAXIScript T7 kit following the manufacturer's instructions. Probes were generated using 100 μCi of 800 Ci/mmol [α-32P]UTP/reaction. The probe was gel-purified and extracted using the manufacturer's extraction buffer. Gel shift assays were performed using 100,000 cpm of probe/assay. Approximately 2 μg of protein was incubated with the probe in the presence of binding buffer (10 mm Hepes, pH 7.6, 3 mm MgCl2, 40 mm KCl, 5% glycerol, and 1 mm dithiothreitol) for 1 h, heparin was then added to a final concentration of 1 mg/ml, and the incubation was continued for an additional 15 min at room temperature. An equal volume of 2× native gel sample buffer (200 mm Tris-HCl, 20% glycerol, and 0.005% bromphenol blue, pH 8.6) was added to the reaction mixture, and the sample was added to a native 4% acrylamide gel in 1× Tris borate/EDTA buffer, pH 8.0, that had been prerun for 20 min at 250 V. Gels were dried under vacuum and exposed to x-ray film. When supershifts were performed, antibody (2 μl) was added to samples after the 1-h binding incubation, and the binding reaction was continued for an additional 30 min at room temperature.
Cell Culture and Transfections—GP2-293 packaging cells were maintained in growth medium consisting of DMEM with 10% fetal bovine serum (FBS) on collagen I-coated plates. Upon reaching 90% confluency, the medium was changed to Opti-MEM I reduced serum medium with 10% FBS (no antibiotics). Co-transfections of the particular β construct along with an expression construct for VSV-G were performed utilizing Lipofectamine 2000 according to the manufacturer's recommendations. All transfections were accompanied by an enhanced green fluorescent protein transfection efficiency control. At 6 h post-transfection, the medium was changed to fresh DMEM with 10% FBS. At 48 h after the medium change, the virus particle-containing supernatants were harvested, filtered through a 0.45-μm filter, and either used immediately or stored at -80 °C. A second harvest was carried out at 96 h. Production of virus particles was confirmed by Western blot analysis for VSV-G in the culture supernatants.
Transductions—The multipotential MEFs (MEF/3T3 Tet-Off cell line) express the tetracycline-controlled transactivator and were cultured in growth medium consisting of DMEM containing 10% calf serum and 100 ng/ml doxycycline on 6-well plates. Cells were plated and transduced at 15–20% confluency by addition of 2 ml of virus particle-containing medium. Plates were centrifuged at room temperature for 90 min at 2000 rpm, in effect pelleting the virus particles onto the cell membrane. Following an overnight incubation, the medium was changed to DMEM with 10% FBS and 10 ng/ml doxycycline. For cells that would follow the differentiation protocol, doxycycline was removed at 50% confluency. Cells were allowed to reach confluency, and then the medium was changed to DMEM supplemented with 10% FBS, 10 μg/ml insulin, 1 μm dexamethasone, 0.5 mm 3-isobutyl-1-methylxanthine, and 10 μm troglitizone, with the control set of transductants receiving the 10 ng/ml doxycycline to repress transduced gene expression. At 48 h following the initial insulin/dexamethasone/3-isobutyl-1-methylxanthine treatment, the medium was replaced with DMEM with 10% FBS, 10 μg/ml insulin, 10 μm troglitizone, and 10 ng/ml doxycycline where applicable. 48 h later, the medium was replaced with DMEM with 10% FBS, 2.5 μg/ml insulin, 10 μm troglitizone, and 10 ng/ml doxycycline where applicable. Notably, because of the high transduction efficiency (always in excess of 90%, similar to reports by Neal and Clipstone (29)), no drug selection was required. In every experiment, control transductions were performed in duplicate using a construct in which expression of enhanced green fluorescent protein was quantified by flow cytometry.
Real-time PCR—Real-time PCR analysis was performed essentially as we have described previously (30). Briefly, total RNA (0.5 μg, integrity demonstrated by ethidium-stained agarose gels) was subjected to reverse transcription with random primers and reverse transcriptase from the iScript™ cDNA synthesis kit (Bio-Rad). Quantitative real-time PCR was performed with specific primers designed for each gene with the Beacon Designer tool (Bio-Rad). Amplification and detection were done with the iCycler IQ real-time PCR detection system with IQ SYBR Green Supermix (Bio-Rad). Standard curves were prepared for each target gene, and PCR efficiency was determined to be in excess of 90% for all primer sets. Threshold temperatures were selected automatically, and all amplifications were followed by melt-curve analysis, i.e. plot of the negative first derivative of the fluorescence versus temperature with the software assigning the melt temperature. Single-melt temperatures were recorded in all cases. All primers were subjected to Blast search to ensure specificity and fold analysis to eliminate any primers with potential to form secondary structure. To calculate relative C/EBPβ mRNA, the threshold cycle (CT) determined for the cells transduced with the empty vector (EV) (endogenous C/EBPβ) was subtracted from the average CT for βwt and βd/s (ΔCT), thus correcting for the minor levels of endogenous expression. A standard curve was generated for each real-time PCR determination using a dilution series (50 ng, 33.3 ng, 11.1 ng, 3.7 ng, 367 pg, 120 pg) of total RNA from 3T3-L1 adipocytes (day 2). A plot of log starting quantity (ng) on the x axis and the CT on the y axis was utilized to determine the arbitrary C/EBPβ mRNA levels of the unknown samples. Real-time PCR analyses with 90% or higher efficiency were utilized for quantification. For multiple independent runs after normalization to β-laminin expression, results were corrected for endogenous C/EBPβ expression using the CT values obtained from the cells harboring the EV.
C/EBPβ mRNA Half-life Determination—The MEF-3T3 cells were transduced with the series of constructs and induced to differentiate as described above. At 24 h after induction of differentiation, doxycycline was added at a final concentration of 0.2 μm to the cultures to stop transcription. Total RNA was then isolated with respect to time, and analysis of C/EBPβ mRNA content was carried out using real-time PCR as described above. The data were plotted as log RNA content versus time. The equation y = ae-bx was fitted to the data, and half-lives were calculated.
Polyadenylation of the C/EBPβ mRNA—Polyadenylation of the C/EBPβ mRNA was determined using the primer/adapter reverse transcription-PCR method as described by Huarte et al. (31). The primer/adapter utilized for reverse transcription was the following: 5′-(dT)12GCGGCCGGCGCCTCGAGCG-3′. For the PCR, the forward primer was 5′-AAACGTGGCTGAGCGCGTGT-3′, and the reverse primer was the primer/adapter described above.
RESULTS
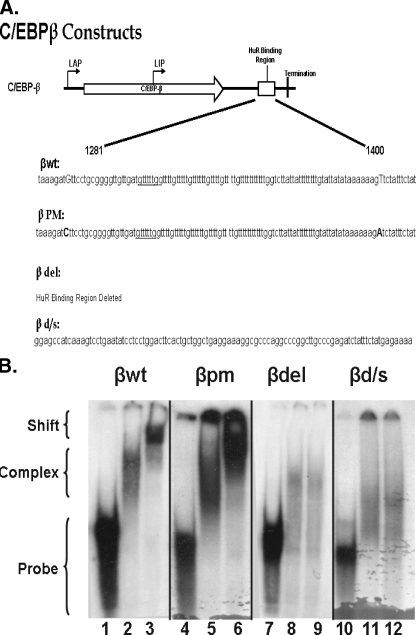
Ectopic Expression of Both Wild Type and Mutant C/EBPβ mRNAs in the Multipotential Precursor MEF-3T3 Cells—Our previous work (28) demonstrated that upon exposure of the cells to the differentiation inducers there is a rapid formation of a nuclear HuR-C/EBPβ complex followed by a translocation of the complex to the cytosol. Our recent detailed analysis demonstrated the presence of a single binding site for HuR in the C/EBPβ mRNA. That site is in the AU-rich element (ARE) in the 3′-UTR of the message (32). Therefore, to address the function of HuR in the translocation and expression of C/EBPβ, we created constructs that expressed wild type C/EBPβ as well as mutants that could not bind HuR (Fig. 1A): 1) full-length wild type C/EBPβ cDNA (βwt), 2) C/EBPβ cDNA with point mutations at each end of the ARE (βpm), 3) deletion of the ARE (βdel), and 4) deletion of the ARE and substitution with a sequence that did not contain a HuR binding site (βd/s).
FIGURE 1.
RNA gel shift analysis of HuR binding to wild type and mutant C/EBPβ 3′-UTR AREs. A, a schematic of C/EBPβ mRNA indicating the approximate translation initiation sites for liver activating protein 1 and 2 (LAP) and liver inhibitory protein (LIP) forms of C/EBPβ, the HuR binding domain, and termination of translation is shown. The graphic below describes the sequence alterations in the mutants. In βwt, the uppercase bases indicate the sequence altered to form βpm, in which the uppercase bases indicate the mutations used to create BglII restriction sites in βwt. βdel is a religation of the construct after removal of the BglII fragment. βd/s consists of removal of the BglII fragment and insertion of a 101-base fragment that does not bind HuR. B, cytosolic extracts and a radiolabeled probe corresponding to the ARE (βwt and βpm), βd/s, or the region flanking (βdel) the C/EBPβ 3′-UTR were used to perform RNA gel shift and super shift analysis. Lanes 1, 4, 7, and 10, probe alone; lanes 2, 5, 8, and 11, probe plus 10 μg of adipocyte cytosolic extract; lanes 3, 6, 9, and 12, supershift of complex formed as in lanes 2, 5, 8, and 11 using the 3A2 anti-HuR monoclonal antibody. The gel shifts shown were performed at the same time, and separation was achieved on two separate gels, βwt andβdel on one andβpm andβd/s on the second. The arrangement was for logical presentation. These data are representative of two individual gel shifts with each probe as the alterations were made, using two individual preparations of cytosolic extracts.
Riboprobes (∼150 bases in length) were prepared containing the ARE for the βwt and βpm constructs; the probe for the βd/s construct contained the substituted region (Fig. 1A), whereas the probe for the βdel construct contained 75 bases flanking either side of the deleted ARE. Gel shift assays were performed with adipocyte total cell lysates as a source of HuR protein to determine the ability of the probes to bind HuR. In Fig. 1B, lanes 1, 4, 7, and 10 display the various probes in the absence of added protein; lanes 2, 5, 8, and 11 show the probes + protein; and lanes 3, 6, 9, and 12 display the interaction of the probes with protein and HuR antibody. As demonstrated in lanes 1–3, the βwt ARE forms a complex with HuR and is recognized by the HuR antibody, as we have demonstrated previously (28, 32). Lanes 4–6 display the βpm riboprobe in which single base changes at each terminus of the ARE were made, and, as shown in the figure, binding was not altered. However, when the ARE was deleted (βdel; lanes 7–9) or deleted and substituted with a similar sized fragment (βd/s; lanes 10–12), HuR binding was lost. We do note that proteins other than HuR must bind to both βdel and βd/s as judged by the loss of the probe band and appearance of complexes of higher mobility. There is minimal homology between the βdel and βd/s probes; the sequence 60gccctgagtaatcacctg77 within βd/s exhibits 13 identical matches (boldface) within an 18-base region between positions 13 and 30 of the βdel probe. We cannot address whether this homology is significant enough to be responsible for the complexes observed on the gel or if the complexes are the result of nonspecific binding. Our data do, however, support that these complexes do not involve HuR.
The constructs were then subcloned into the pRevTRE vector, a retroviral tetracycline/doxycycline-regulated expression system, RevTet-Off™, and packaged by co-transfection with pVSV-G (encoding the vesicular stomatitis virus glycoprotein) into the GP2–293 pantropic packing cell line using Lipofectamine® 2000. The virus particles produced were then used to transduce MEF-3T3 cells (see “Experimental Procedures”), which express HuR at normal levels but very low amounts of endogenous C/EBPβ. This approach is similar to that of Farmer and colleagues (33, 34), in which conditional ectopic expression of C/EBPβ in the multipotential NIH 3T3 cells in the presence of the differentiation induction mixture induced PPARγ and stimulated adipogenesis. Preliminary experiments confirmed that, similar to our previous studies with 3T3-L1 cells (28), the C/EBPβ mRNA was a ligand for HuR in the transduced MEF-3T3 cells.4
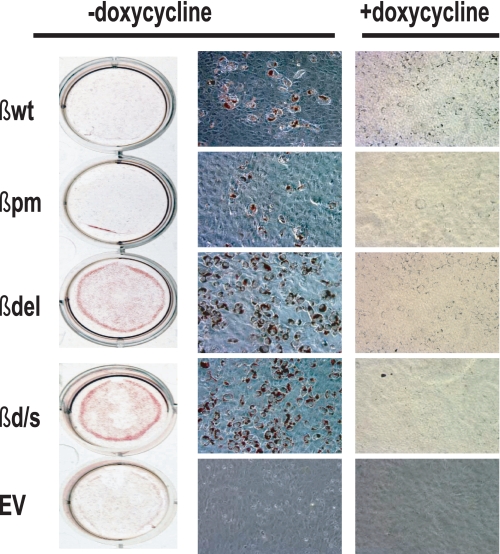
We then examined the effect of expression of the C/EBPβ constructs on the ability of the MEF-3T3 cells to differentiate. As shown in Fig. 2, based on oil red O staining at 8 days after induction of differentiation, expression of all four constructs led to the accumulation of triacylglycerol to some degree, with the greatest amount present in the βdel and βd/s. This was unexpected as these two constructs, as demonstrated in Fig. 1B, do not bind HuR. The absence of triacylglycerol deposits in the cells maintained in doxycycline and those transduced with the EV supports the premise that expression of the various β constructs is responsible for the altered phenotype.
FIGURE 2.
Expression of the adipocyte phenotype in MEF-3T3 cells transduced with expression constructs for wild type C/EBPβ and mutants that no longer bind HuR. MEF-3T3 cells were transduced with pRevTRE expression vector containing either the C/EBPβ cDNA or constructs altered at the HuR binding site (Fig. 1A). Expression of the constructs was up-regulated by removal of doxycycline from the culture medium, and the cells were induced to differentiate as described under “Experimental Procedures.” Cultures maintained in doxycycline resulted in suppression of construct expression and served as the negative controls. At day 8 post-induction of differentiation, the accumulated triacylglycerol in the cells was stained with oil red O. The data presented are representative of an experiment performed independently at least four times.
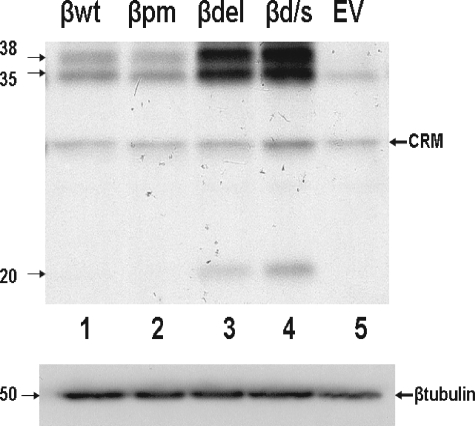
The expression level of C/EBPβ protein in the five transduced cell lines on day 8 after induction of differentiation is shown in Fig. 3. 25 μg of total cellular protein was analyzed by Western blotting, and full-length C/EBPβ (liver activating protein, 35- and 38-kDa species) was observed to be expressed significantly (greater than 3-fold) above endogenous (cells carrying the EV) levels in all four cell lines transduced with a β construct. The cells carrying the βd/s and βdel constructs expressed both the 35- and 38-kDa forms of C/EBPβ at ∼8-fold above EV and were the only cell lines in which the dominant-negative isoform liver inhibitory protein (20 kDa) was expressed at detectable levels. The mRNAs generated by the βd/s and βdel constructs did not bind HuR (Fig. 1B) yet expressed C/EBPβ protein at levels significantly above those found endogenously or in cells containing the βwt or βpm constructs. Thus, HuR binding to the ARE in the C/EBPβ message is not necessary for either movement of the message to the cytosol or expression of C/EBPβ protein. The enhanced expression driven by the βd/s and βdel constructs is consistent with HuR mediating an attenuation of expression when bound to the C/EBPβ ARE.
FIGURE 3.
C/EBPβ protein accumulates to a greater degree in cells expressing a C/EBPβ mRNA that does not bind HuR. Upper panel, total cell lysates were prepared from cells transduced with the indicated construct, and 25 μg of total protein was subjected to Western blot analysis for C/EBPβ. Molecular masses are indicated in kDa. CRM indicates cross-reacting material observed in this analysis. Lower panel, the blot was stripped and reprobed with β-tubulin, which served as a loading control. The experiment was performed twice with identical results.
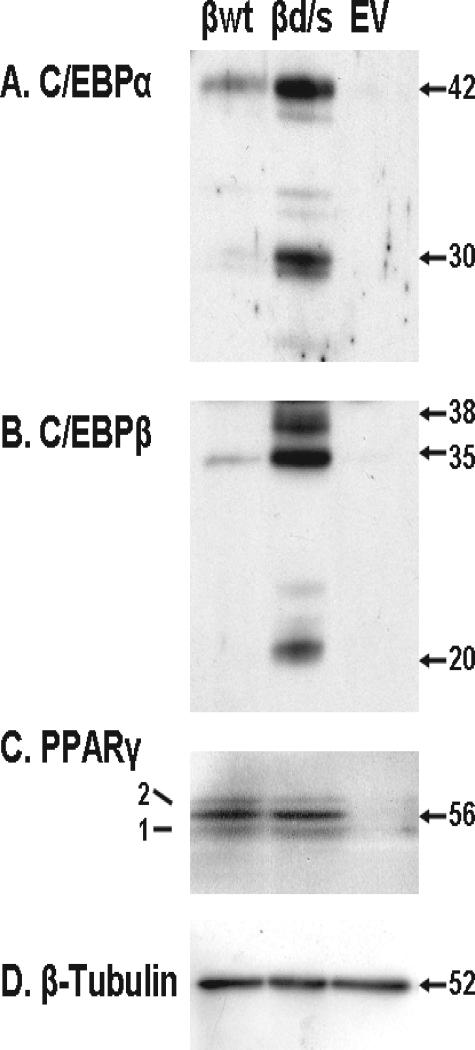
Further experimentation was carried out with only three of the constructs, βwt, βd/s, and EV. As the βd/s cell line accumulated more triacylglycerol, appearing to undergo a more thorough differentiation, and expressed more C/EBPβ protein, we next examined other markers of differentiation. The Western blot analysis for C/EBPα and PPARγ displayed in Fig. 4 indicates that increased expression of C/EBPβ in the βd/s cells resulted in increased expression of C/EBPα relative to either the βwt or EV cell lines. PPARγ levels were found to be identical in both βwt and βd/s, suggesting that even the low levels of C/EBPβ found in the βwt cells was sufficient to drive maximal expression of PPARγ.
FIGURE 4.
C/EBPα is expressed to a greater degree in the βd/s cells. At 8 days post-induction of differentiation, cell extracts were prepared, and 25 μg of protein of the three cell lines indicated was subjected to Western blot analysis for C/EBPα, C/EBPβ, and PPARγ. Molecular masses are indicated in kDa. B–D are from the same plot probed sequentially for C/EBPβ, PPARγ, and β-tubulin. A is from a separate Western blot. The experiment was performed twice with identical results.
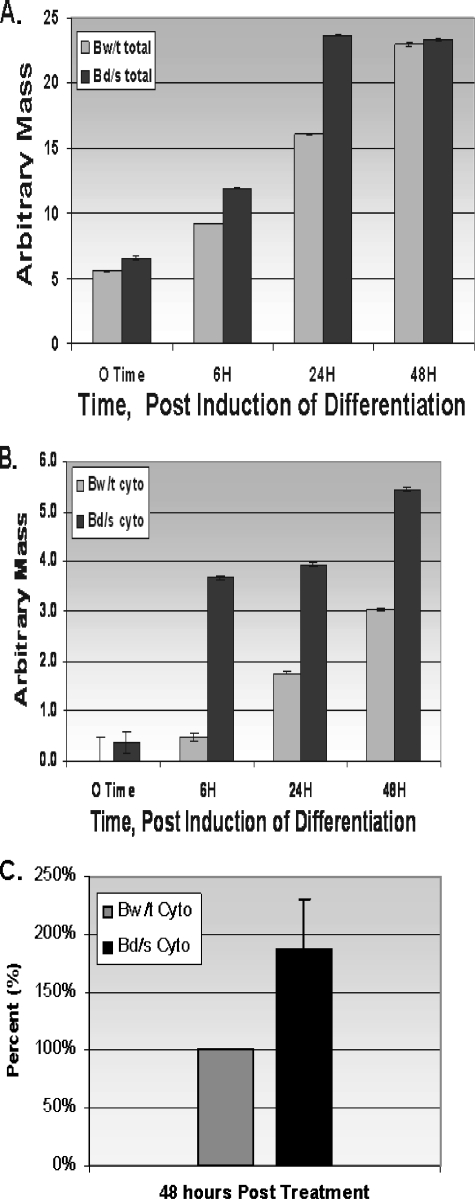
Accumulation of C/EBPβ mRNA in βwt and βd/s Cell Lines— We examined the accumulation of total cellular C/EBPβ mRNA after removal of doxycycline and exposure of the cells to the differentiation mixture. The results shown in Fig. 5A indicate that total RNA in both βwt and βd/s cells accumulated in a similar manner. The loss of the HuR-binding ARE in βd/s had no effect on the accumulation of total cellular message. This might be expected as the constructs were driven by the same tetracycline/doxycycline-regulated promoter.
FIGURE 5.
C/EBPβ mRNA from βwt and βd/s cell lines. A, total RNA was isolated with respect to time after removal of doxycycline from the culture medium and exposure of the cells to the differentiation protocol. Accumulation of total C/EBPβ mRNA was determined by real-time PCR analysis as described under “Experimental Procedures.” The experiment was performed three times with identical results. B, cytosolic accumulation of C/EBPβ mRNA in βwt (Bw/t) and βd/s (Bd/s) cell lines is shown. The cytosolic fraction (cyto) of the cells was isolated using the Pierce NE-PER™ kit as we have described previously (29). Cytosolic RNA was isolated using the TRIZol™ reagent. Accumulation of C/EBPβ mRNA was determined by real-time PCR analysis as described under “Experimental Procedures.” The experiment was performed three times with similar results. Data are plotted as the mean ± S.D. C, the levels of βwt and βd/s mRNAs in the cytosol at the 48-h time point (five independent determinations) are shown. Normalization was to β-laminin mRNA, the expression of which does not change over the time course of the experiment, followed by correction for endogenous expression as discussed under “Experimental Procedures.” βwt content was defined as 100%, and βd/s is expressed relative to that value.
We next examined the appearance and accumulation of C/EBPβ mRNA in the cytosolic compartment. Using the Pierce NE-PER™ kit, we isolated cytosolic and nuclear compartments prior to and after removal of doxycycline and induction of differentiation, as we have described previously (28). The real-time PCR data shown in Fig. 5B indicate that at all time points βd/s mRNA accumulated in the cytosol to a greater degree than the βwt message. Thus, the loss of the ability to bind HuR at the canonical ARE (present in βwt, but absent in βd/s) did not hinder the movement of the C/EBPβ mRNA into the cytosol. The previous data (Fig. 5A) indicated that total cellular C/EBPβ mRNA accumulated to a similar degree in both βwt and βd/s. These data (Fig. 5B) would suggest that a greater percentage of the total βd/s mRNA is in the cytosol, available for translation and driving the accumulation of C/EBPβ protein shown in Figs. 3 and 4. To confirm this hypothesis, we selected the 48-h time point and performed five independent isolations and subsequent determinations of the cytosolic versus nuclear distribution of βwt and βd/s. Those data are displayed in Fig. 5C and demonstrate that there is approximately twice the βd/s mRNA in the cytosol relative to the βwt mRNA. We note that in separate experiments a minimum 2-fold differential has been demonstrated to be maintained at least through day 8 of the differentiation program. Thus, in the absence of HuR binding, more C/EBPβ mRNA accumulates in the cytosol.
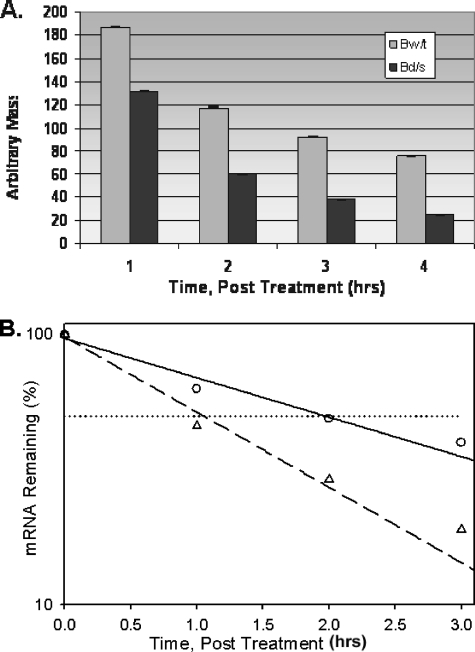
C/EBPβ mRNA Half-life—The accumulation of the βd/s mRNA in the cytosol could be a result of stabilization of the message upon removal of the ARE. To examine for this possibility, half-life determinations were made 24 h after induction of differentiation (Fig. 6). The βwt and βd/s mRNAs exhibited half-lives of 120 and 60 min, respectively, calculated using exponential decay regression. With the consideration that the βd/s mRNA has a more rapid half-life, the increased cytosolic accumulation is all the more significant.
FIGURE 6.
C/EBPβ mRNA half-life of βwt and βd/s cell lines. At 24 h after induction of differentiation, doxycycline was added to the culture medium, and total RNA was isolated with respect to time. Real-time PCR analysis was utilized to quantify the C/EBP βwt (Bw/t) and βd/s (Bd/s) mRNAs remaining after addition of the doxycycline. The half-lives of the C/EBPβ mRNAs were determined graphically using a plot of log[mRNA] versus time as described under “Experimental Procedures.” A, results of the real-time analysis. Results are plotted as the mean ± S.D. of two independent experiments analyzed in triplicate. B, half-life plot of real-time results. βwt (○) and βd/s (▵) mRNAs remaining after addition of doxycycline with respect to time are shown. The equation y = ae-bx was fitted to the data, and half-lives were calculated. The experiment was performed twice with similar results.
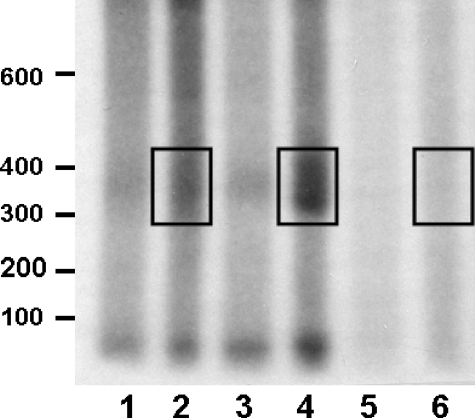
C/EBPβ mRNA Polyadenylation—The data presented to this point are consistent with involvement of HuR in nuclear processing of the C/EBPβ message. It would appear that once this mRNP complex forms, its translocation to the cytosol is attenuated. To investigate the influence of HuR on nuclear processing of the C/EBPβ mRNA, we examined nuclear polyadenylation of the C/EBPβ transcripts from the βwt and βd/s cell lines. To accumulate the data presented in Fig. 7, RNA was isolated from the nuclear fraction and subjected to reverse transcription using an oligo(dT) primer/adapter followed by PCR using a forward primer located 298 nucleotides upstream of the site of poly(A) addition of the C/EBPβ mRNA in conjunction with the 32P-labeled oligo(dT) primer/adapter. Reverse transcription-PCR with these primers of the mRNA from the βd/s cells (Fig. 7, lane 4) produced a smear of products ranging in size from ∼300 to almost 400 nucleotides. The minimal size predicted was 329 nucleotides (298 bp of C/EBPβ plus 31 nucleotides of the primer/adapter). Whereas there is evidence of polyadenlylation with βwt (Fig. 7, lane 2), densitometric analysis indicated that it is ∼35% that found in the βd/s. Notably, there is no evidence of polyadenylation occurring when mRNA was isolated from cells containing the empty vector (Fig. 7, lane 6). The data suggest that in the absence of HuR binding, the C/EBPβ mRNA is more extensively polyadenylated, leading to translocation to the cytosol. However, in βwt, which binds HuR, polyadenylation appears to occur to a lesser extent, with approximately one-third of the RNA (relative to βd/s) reaching the cytosol. Similar results were obtained using an RNase H-based approach for determination of poly(A) tail size for total cellular RNA.5
FIGURE 7.
Identification of a differential nuclear polyadenylation of C/EBPβ mRNA from βwt and βd/s cell lines. Poly(A) tail lengths on βwt and βd/s were determined by reverse transcription-PCR poly(A) analysis of a preparation of total nuclear RNA from βwt and βd/s cell lines. Reverse transcription was primed with an oligo(dT) primer/adapter as described, and poly(A) tail lengths on the βwt and βd/s mRNAs were determined by PCR using a 5′-32P-labeled primer/adapter for the reverse primer and a gene-specific primer for the forward primer. The products of the PCRs were visualized by autoradiography after electrophoresis on 6% polyacrylamide/urea gels. Size markers are in nucleotides. The regions corresponding to polyadenylation of the C/EBPβ mRNA are boxed for comparison. Lanes 2, 4, and 6 contain the full PCRs, whereas lanes 1, 3, and 5 contain 20% of the reactions. The autoradiogram presented is representative of an experiment performed 3 times with similar results.
DISCUSSION
As preadipocytes differentiate, controlled expression of C/EBPβ is essential to acquisition of the adipose phenotype. Transcriptional activation of the C/EBPβ gene in 3T3-L1 cells occurs within minutes of exposure to the differentiation inducers and is controlled, at least in part, by the cAMP-response element-binding protein (35, 36). In the differentiation program of 3T3-L1 cells, C/EBPβ first controls the entry of the cells into mitotic clonal expansion, and then the expression of C/EBPα and PPARγ (2–6, 33, 34). The timing of expression during these processes is critical because C/EBPβ is promitotic and C/EBPα is antimitotic, and thus C/EBPβ expression must attenuate as C/EBPα expression initiates. This study describes a critical early post-transcriptional regulation initiated in the nucleus involving formation of a HuR-C/EBPβ mRNA complex. Formation of this mRNP appears to control the rate of C/EBPβ mRNA translocation to the cytosol but is not essential for the translocation process itself. In our previous work, we identified the HuR binding site in the C/EBPβ 3′-UTR and demonstrated that it is the only site within the entire message (32). As evidenced in the data presented in Fig. 5, deletion of this HuR binding site did not disrupt nuclear to cytoplasmic translocation of the message. Indeed, in the absence of HuR binding, ∼2–7-fold (dependent on the time point) more C/EBPβ mRNA was localized to the cytosol. Conceivably, accumulation of βd/s in the cytosol could be a consequence of deletion of the ARE instability element, resulting in a stabilized message. However, as displayed in Fig. 6, the βd/s mRNA actually has a shorter half-life than the βwt message, making its accumulation more difficult. Overall, our data are consistent with C/EBPβ mRNA translocation to the cytosol occurring more readily when HuR is not bound, consistent with HuR functioning as a “brake” or attenuator of movement of the complex to the cytosol. When HuR binds to the C/EBPβ message in the nucleus, movement to the cytosol is not prohibited, simply diminished. The C/EBPβ mRNA reaches the cytosol in lower quantities and is translated into protein. The differentiation program is initiated and maintained. When the binding site is altered such that HuR cannot bind, more C/EBPβ mRNA per unit time is found in the cytosol. The presence of more message drives the overexpression of C/EBPβ protein, resulting in overexpression of C/EBPα protein and a more robust differentiation program.
Until recently, potential nuclear functions of the Hu proteins have not been addressed. However, several recent reports have supported roles for HuR as well as the neuronal specific Hu proteins in the nuclear processing of mRNAs with respect to the regulation of splicing as well as polyadenylation (27, 37, 38). The C/EBPβ message is derived from an intronless gene, thus, regulation at the level of splicing is not an option. However, the HuR binding site is in close proximity to the polyadenylation signal and is flanked by uridylate-rich regions. The sequence flanking the HuR binding site in the C/EBPβ mRNA is similar to that previously described for the non-neuronal alternative 3′-terminal exon 4 of the calcitonin/calcitonin gene-related peptide message. On that message, HuR was demonstrated to inhibit both cleavage and polyadenylation by competing for binding to the RNA with subunits of the cleavage and polyadenylation specificity factor. Our data are consistent with HuR serving this same function when bound to the C/EBPβ mRNA. This potential mechanism is currently under investigation.
Whereas these results specifically define a nuclear function for HuR in the metabolism of the C/EBPβ message, our previous data describing cytosolic HuR-C/EBPβ mRNP particles suggest that HuR functions in both compartments. HuR may be involved in other aspects of regulating the expression of C/EBPβ. The apparent disparity between the levels of increased βd/s mRNA and C/EBPβ protein levels has led us to focus on the involvement of HuR in the control of translational efficiency, and those studies are currently in progress.
Our current data describe the involvement of HuR in the metabolism of the C/EBPβ message and indicate that formation of the mRNP particle in the nucleus controls the quantity of message entering the cytosol. On the same time frame, HuR binds to other ligand mRNAs that may also play critical roles in the differentiation process. For example, β-actin can be found in both nuclear and cytosolic complexes with HuR 30 min after induction of differentiation.5 The function of that interaction remains unknown. Although it was not the goal of this study, the data presented demonstrate very effectively that the interaction between HuR and C/EBPβ mRNA is not necessary for initiation of the differentiation program. Thus, our previously described attenuation of the differentiation process by small interfering RNA-mediated suppression of HuR was not a consequence of the loss of interaction with C/EBPβ mRNA. Other ligand mRNAs that are dependent on interaction with HuR to perhaps stabilize the message or increase its translational efficiency to produce a functional protein must be involved. In attempting to identify these critical mRNAs, we are focusing on the immediate early gene mRNAs, including c-fos, Krox20, fosB, and c-jun, all of which have apparent HuR binding sites. At least c-fos and c-jun have been demonstrated to be ligands for HuR in other systems (16). Moreover, we have previously documented activation of the transient expression of these immediate early genes within minutes of exposure to the differentiation mixture (39), making them potentially ideal targets for HuR binding.
Finally, it is interesting that genetic approaches have identified mouse strains with mutations in the HuR binding motif of inflammatory mRNAs that correlate with the development of autoimmunity (40). On the basis of these observations, we suggest that, at least in some cases, alterations in the ability of HuR to bind the C/EBPβ mRNA could lead to overexpression of C/EBPβ protein and result in the onset of adipogenesis and potentially the disease state of obesity. This we believe merits investigation.
Acknowledgments
We gratefully acknowledge the expert laboratory support of Melinda Carver, Becky Keener, Ashley Ferguson, and Mitch Harris. We also thank Dr. Joseph M. Chalovich for aid in determination of the mRNA half-lives and Drs. Kira Gantt and Tania Kastelic for thoughtful criticism on reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK55169. This work was also supported by American Diabetes Association Grant 7-03-RA-76 and Brody Brothers Foundation Grant MT7753. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: C/EBP, CCAAT/enhancer-binding protein; MEF, murine embryonic fibroblast; UTP, uridine 5′-triphosphate; VSV-G, vesicular stomatitis virus G protein; ARE, AU-rich element; EV, empty vector; PPAR, peroxisome proliferator-activated receptor; UTR, untranslated region; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; mRNP, messenger ribonucleoprotein.
M. Carver and P. H. Pekala, unpublished data.
V. A. Karschner and P. H. Pekala, unpublished data.
References
- 1.Otto, T. C., and Lane, M. D. (2005) Mol. Biol. 40 229-242 [DOI] [PubMed] [Google Scholar]
- 2.Zuo, Y., Qiang, L., and Farmer, S. R. (2006) J. Biol. Chem. 281 7960-7967 [DOI] [PubMed] [Google Scholar]
- 3.Hamm, J. K., Park, B. H., and Farmer, S. R. (2001) J. Biol. Chem. 276 18464-18471 [DOI] [PubMed] [Google Scholar]
- 4.Tang, Q.-Q., Otto, T. C., and Lane, M. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 850-855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, J. W., Tang, Q.-Q., Vinson, C., and Lane, M. D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 43-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, J. W., Klemm, D. J., Vinson, C., and Lane, M. D. (2004) J. Biol. Chem. 279 4471-4478 [DOI] [PubMed] [Google Scholar]
- 7.Robinow, S., Campos, A. R., Yao, K. M., and White, K. (1988) Science 242, 1570-1572 [DOI] [PubMed] [Google Scholar]
- 8.Fan, X. C., and Steitz, J. A. (1998) EMBO J. 17 3448-3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myer, V. E., Fan, X. C., and Steitz, J. A. (1997) EMBO J. 16 2130-2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy, N. S., Chung, S., Furneaux, H., and Levy, A. P. (1998) J. Biol. Chem. 273 6417-6420 [DOI] [PubMed] [Google Scholar]
- 11.Ma, W. J., Cheng, S., Campbell, C., Wright, A., and Furneaux, H. (1996) J. Biol. Chem. 271 8144-8151 [DOI] [PubMed] [Google Scholar]
- 12.Peng, S. S., Chen, C. Y., Xu, N., and Shyu, A. B. (1998) EMBO J. 17 3461-3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King, P. H., Fuller, J. J., Nabors, L. B., and Detloff P. J. (2000) Gene (Amst.) 242 125-131 [DOI] [PubMed] [Google Scholar]
- 14.Ford, L. P., Watson, J., Keene, J. D., and Wilusz, J. (2001) EMBO J. 20 1134-1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallouzi, I. E., Brennan, C. M., Stenberg, M. G., Swanson, M. S., Eversole, A., Maizels, N., and Steitz, J. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 3073-3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keene, J. D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon, D. A., Tolley, N. D., King, P. H., Nabors, L. B., McIntyre, T. M., Zimmerman, G. A., and Prescott, S. M. (2001) J. Clin. Investig. 108 1657-1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldberg-Cohen, I., Furneaux, H., and Levy, A. P. (2002) J. Biol. Chem. 277 13635-13640 [DOI] [PubMed] [Google Scholar]
- 19.Loflin, P., and Lever, J.E. (2001) FEBS Lett. 509 267-271 [DOI] [PubMed] [Google Scholar]
- 20.Dean, J. L., Wait, R., Mahtani, K. R., Sully, G., Clark, A. R., and Saklatvala, J. (2001) Mol. Cell. Biol. 21 721-730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirigiti, P., Bai, Y., Yang, Y. F., Li, X., Li, B., Brewer, G., and Machida, C. A. (2001) Mol. Pharmacol. 60 1308-1324 [DOI] [PubMed] [Google Scholar]
- 22.Brennan, C. M., Gallouzi, I. E., and Steitz, J. A. (2000) J. Cell Biol. 151 1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallouzi, I. E., and Steitz, J. A. (2001) Science 294 1895-1901 [DOI] [PubMed] [Google Scholar]
- 24.Jain, R. G., Andrews, L. G., McGowan, K. M., Pekala, P. H., and Keene, J. D. (1997) Mol. Cell. Biol. 7 954-962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gantt, K. R., Jain, R. G., Dudek, R. W., and Pekala, P. H. (2004) Biochem. Biophys. Res. Commun. 313 619-622 [DOI] [PubMed] [Google Scholar]
- 26.Antic, D., Lu, N., and Keene, J. D. (1999) Genes Dev. 13 449-461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu, H., Zhou, H.-L., Hasman, R. A., and Lou, H. (2007) J. Biol. Chem. 282 2203-2210 [DOI] [PubMed] [Google Scholar]
- 28.Gantt, K., Cherry, J., Tenney, R., Karschner, V., and Pekala, P. H. (2005) J. Biol. Chem. 280 24765-24774 [DOI] [PubMed] [Google Scholar]
- 29.Neal, J. W., and Clipstone, N. A. (2003) J. Biol. Chem. 278 17246-17254 [DOI] [PubMed] [Google Scholar]
- 30.Gantt, K. R., Cherry, J., Richardson, M., Karschner, V., Atasoy, U., and Pekala, P. H. (2006) J. Cell. Biochem. 99 565-574 [DOI] [PubMed] [Google Scholar]
- 31.Huarte, J., Stutz, A., O'Connell, M. L., Gubler, P., Belin, D., Darrow, A. L., Strickland, S., and Vassalli, J.-D. (1992) Cell 69 1021-1030 [DOI] [PubMed] [Google Scholar]
- 32.Jones, H., Carver, M., and Pekala, P. H. (2007) Biochem. Biophys. Res. Commun. 355 217-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, Z., Xie, Y., Morrison, R. F., Bucher, N., and Farmer, S. R. (1998) J. Clin. Investig. 101 22-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison, R., and Farmer, S. R. (1999) J. Biol. Chem. 274 17088-17097 [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez, G. A., and Montminy, M. R. (1986) Cell 59 675-680 [DOI] [PubMed] [Google Scholar]
- 36.Klemm, D. J., Roesler, W. J., Boras, T., Colton, L. A., Felder, K., and Reusch, J. E. (1998) J. Biol. Chem. 273 917-923 [DOI] [PubMed] [Google Scholar]
- 37.Zhu, H., Hasman, R. A., Barron, V. A., Luo, G., and Lou, H. (2006) Mol. Biol. Cell 17 5105-5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, H., Hinman, M. N., Hasman, R. A., Mehta, P., and Lou, H. (2008) Mol. Cell. Biol. 28 1240-1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens, J. M., Butts, M., Stone, R., Pekala, P. H., and Bernlohr, D. A. (1993) Mol. Cell. Biochem. 123 63-71 [DOI] [PubMed] [Google Scholar]
- 40.DiMarco, S., Hel, Z., Lachance, C., Furneaux, H., and Radzioch, D. (2001) Nucleic Acids Res. 29 863-871 [DOI] [PMC free article] [PubMed] [Google Scholar]