Abstract
Signal transducer and activator of transcription 3 (STAT3) is a latent transcription factor mainly activated by the interleukin-6 cytokine family. Previous studies have shown that activated STAT3 recruits p300, a coactivator whose intrinsic histone acetyltransferase activity is essential for transcription. Here we investigated the function of the STAT3 NH2-terminal domain and how its interaction with p300 regulates STAT3 signal transduction. In STAT3-/- mouse embryonic fibroblasts, a stably expressed NH2 terminus-deficient STAT3 mutant (STAT3-ΔN) was unable to efficiently induce either STAT3-mediated reporter activity or endogenous mRNA expression. Chromatin immunoprecipitation assays were performed to determine whether the NH2-terminal domain regulates p300 recruitment or stabilizes enhanceosome assembly. Despite equivalent levels of STAT3 binding, cells expressing the STAT3-ΔN mutant were unable to recruit p300 and RNA polymerase II to the native socs3 promoter as efficiently as those expressing STAT3-full length. We previously reported that the STAT3 NH2-terminal domain is acetylated by p300 at Lys-49 and Lys-87. By introducing K49R/K87R mutations, here we found that the acetylation status of the STAT3 NH2-terminal domain regulates its interaction with p300. In addition, the STAT3 NH2-terminal binding site maps to the p300 bromodomain, a region spanning from amino acids 995 to 1255. Finally a p300 mutant lacking the bromodomain (p300-ΔB) exhibited a weaker binding to STAT3, and the enhanceosome formation on the socs3 promoter was inhibited when p300-ΔB was overexpressed. Taken together, our data suggest that the STAT3 NH2-terminal domain plays an important role in the interleukin-6 signaling pathway by interacting with the p300 bromodomain, thereby stabilizing enhanceosome assembly.
STATs2 are cytoplasmic transcription factors that can be activated by the IL-6 cytokine family, a group of homologous peptides that include IL-6, oncostatin M (OSM), IL-11, and leukemia-inhibitory factor (1, 2). The classic signaling pathway initiated by the IL-6 cytokine family is via ligand binding to cognate high affinity α chain receptors, e.g. IL-6Rα or OSMRα (3), that then complex with the ubiquitously expressed transmembrane protein gp130 β-subunit. The ligand·Rα·gp130 complex activates the cytoplasmic Janus and Tyk tyrosine kinases that phosphorylate the cytoplasmic domain of gp130, thereby providing docking sites for STAT1 and STAT3 isoforms (1, 2, 4, 5). The recruited STATs are then phosphorylated by the same Janus/Tyk kinases at a specific tyrosine on the COOH-terminal transactivation domain (TAD), inducing their subsequent dimerization, nuclear translocation, and specific binding to IL-6 response elements (6). After the nuclear translocation of hetero- or homodimeric STATs, the magnitude of IL-6 signaling is further regulated by the recruitment of coactivators with histone acetyltransferase activity.
The crucial role of histone acetyltransferases in inducing chromatin remodeling and transcription activation has long been recognized (7). Several proteins with intrinsic histone acetyltransferase activity have been identified, including GCN5 (8), p300/CREB-binding protein (CBP) homologs (9), p300/CBP-associated factor (10), and TAFII250 (11). Histone acetyltransferases activate transcription by one or more of the following ways. 1) They are able to relax core nucleosome structure by acetylating the NH2-terminal histone tails (12). 2) They can directly acetylate transcription factors and alter their transcription activities (13–17). 3) They function as scaffold proteins to recruit other coactivators to the transcriptional apparatus (18, 19). 4) They serve as bridging factors to physically connect sequence-specific transcription factors with multiple components in the basal transcription machinery (20, 21). p300 and its homolog CBP are potent transcriptional coactivators that are actively involved in all of the four processes mentioned above. They have been shown to interact with several transcription factors, such as MyoD (17), p53 (22), and E2F1 (23), and regulate their activity by reversible acetylation.
There is strong evidence demonstrating that the histone acetyltransferase activity of p300 is required for STAT3 target gene activation (24–26). Our previous studies have shown that overexpression of the p300 inhibitor adenovirus 12S E1A significantly inhibits the IL-6-induced activation of human angiotensinogen (hAGT), a vasoactive peptide and acute phase protein controlled by STAT3 (24). The ectopic expression of p300 enhances the induction of hAGT reporter gene stimulated by IL-6 (24). Conversely expression of a p300 mutant deficient in histone acetyltransferase activity functions as a dominant-negative inhibitor and strongly inhibits STAT3-dependent transcription (24). Moreover IL-6-inducible p300-STAT3 association causes an increase in histone H4 acetylation on the hAGT promoter (24), indicating that p300 recruitment augments STAT3 transactivation by acetylating histone tails, thereby relaxing chromatin structure.
p300 interacts with STAT3 within both its COOH-terminal TAD and NH2-terminal domain (25, 26), and this phenomenon has also been confirmed for STAT1 and STAT2 (27, 28). The STAT family shares a highly conserved modular structure that includes an NH2-terminal domain, a coiled coil domain, a DNA-binding domain, a linker domain, an SH2 domain, and a COOH-terminal TAD (29). The coiled coil domain is actively involved in protein-protein interaction (30), and the SH2 domain mediates the STAT3 dimerization via intermolecular Tyr(P)-SH2 interactions (31). The COOH-terminal TAD contains a conserved single tyrosine residue that is phosphorylated in STAT activation and facilitates transcriptional activation (32). The function of the NH2-terminal domain in STAT3, however, is poorly understood. In this study we investigated the STAT3 NH2-terminal function by stably expressing an NH2 terminus-deleted mutant (STAT3-ΔN) in STAT3-/- MEFs. Both OSM-inducible γ-FBG reporter gene and endogenous mRNA expression including socs3, c-fos, and p21 were significantly reduced in response to STAT3-ΔN expression. Because the STAT3 NH2-terminal domain is involved in p300 binding, the defective activity observed in STAT3-ΔN is probably caused by the reduced cooperation between STAT3 and p300. This hypothesis was then tested in native chromatin by chromatin immunoprecipitation (ChIP) assays that revealed a reduction in OSM-inducible p300 recruitment to the socs3 promoter in MEFs stably expressing STAT3-ΔN. At the same time, there was a decrease in RNA pol II binding to the socs3 promoter, indicating the STAT3 NH2-terminal domain not only stabilizes coactivator association but also facilitates the assembly of transcription preinitiation complex.
Recent studies identified STAT3 not only as a binding partner of p300 but also as a substrate for its acetylation where p300 modifies STAT3 at multiple sites. A single acetylation on Lys residue Lys-685 localized in the COOH-terminal TAD is required for STAT3 dimerization and the subsequent DNA binding activity (26). Our laboratory independently identified two other Lys residues, Lys-49 and Lys-87, in the STAT3 NH2-terminal domain that are also inducibly acetylated by p300 in response to IL-6 and OSM (25). Although these NH2-terminal acetylations have no effect on STAT3 DNA binding activity, they are essential for STAT3-dependent transcription because K49R/K87R substitutions significantly inhibit STAT3 target gene expression (25). We also noticed that the K49R/K87R mutations decrease the association between p300 and STAT3, indicating that the inducible NH2-terminal acetylations may augment STAT3-p300 interaction. In this study, we further investigated the interaction between the STAT3 NH2-terminal domain and p300 and found that the acetyl-Lys mimic substitutions (K49Q/K87Q) increase the STAT3 NH2-terminal binding to p300, confirming the hypothesis that the NH2-terminal acetylations stabilize the STAT3-p300 interaction. We also discovered that the STAT3 NH2-terminal binding site maps to the p300 bromodomain. The deletion of the bromodomain in p300 molecule decreased its ability to cooperate with STAT3. In addition, the bromodomain-deficient p300 mutant (p300-ΔB) exhibited weaker chromatin binding. Taken together, we propose a model in which the IL-6- or OSM-inducible acetylations of STAT3 on Lys residues 49 and 87 trigger the recognition of the NH2-terminal domain by the p300 bromodomain, resulting in a strengthened recruitment of p300 to the promoter of the STAT3 target gene, thereby facilitating subsequent enhanceosome assembly.
EXPERIMENTAL PROCEDURES
Materials—Polyclonal antibodies of p300 (N-15), STAT3 (c-20), and RNA pol II (N-20) were purchased from Santa Cruz Biotechnology, Inc. Antibodies against V5 and FLAG were obtained, respectively, from Invitrogen and Sigma. Recombinant mouse OSM was provided by R&D Systems.
Cell Cultures—The human HepG2 and HEK 293 cell lines (ATCC, Manassas, VA) were cultured as described before (25, 33). STAT3+/- and STAT3-/- MEFs were generous gifts from Dr. Stephanie Watowich at the University of Texas M. D. Anderson Cancer Center. MEFs were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin and maintained at 37 °C in a humidified atmosphere of 5% CO2. Cells were serum-starved for at least 16 h before treatment with OSM (20 ng/ml) or IL-6 (10 ng/ml).
Plasmid Construction—The γ-FBG-LUC reporter and pEFF6-V5-STAT3 expression vectors including full length (FL), STAT3-ΔN (aa 133–770), and V5-STAT3-130 (aa 1–130) were constructed as described previously (33). FLAG-tagged WT-124, KR-124, and KQ-124 were produced by PCR using pEFF6-V5-STAT3-WT, STAT3-K48R/K87R, and STAT3-K48Q/K87Q (25) as templates, respectively. The primers used for this purpose were: sense primer 5′-CATCGATGGATCCATGGACTACAAAGACGATGACGATAAGGCCCAATGGAATCAGCTACAG-3′ and antisense primer 5′-CGTACCTCTAGACTACTGGGCCGCAGTGGCTGCAGTCTG-3′. The PCR products were digested with BamHI and XbaI endonucleases, gel-purified, and cloned into pECFP-Nuc (Clontech) restricted with the same endonucleases.
The expression vector used for PXFS-FLAG-p300 constructs was described previously (33). The full-length p300 cDNA was amplified from pCMVβ p300 (34) using the following primers: sense 5′-CAGTCTAGACGTAAGCTTGCCGAGAATGTGGTGGAACCGG-3′ and antisense 5′-CTCGTAGATATCCTAGTGTATGTCTAGTGTACTC-3′. The PCR product was restricted with HindIII and EcoRV and cloned into the PXFS-FLAG vector. The four NH2 terminus-deleted truncations (p300-ΔN1, p300-ΔN2, p300-ΔN3, and p300-ΔN4) were produced from p300-FL by PCR using the same antisense primer, 5′-CATGAACTACTCTAACAGTGACC-3′, and different sense primers as follows: sense 5′-GGTTCTGGAGCAAAGCTTGCTGATCCAGAGAAGCGCAAGCTCATCC-3′ for p300-ΔN1, sense 5′-CCATCCACTACTAAGCTTCGGAAACAGTGGCACGAAGATATTAC-3′ for p300-ΔN2, sense 5′-GTCAAAGAAAAAGCTTTTCAAACCCAGAAGAACTACGACAG-3′ for p300-ΔN3, and sense 5′-GCGGAAGAAAGATGAAGCTTATCTGTGTCCTTCACCATGAGATC-3′ for p300-ΔN4. All the PCR fragments were restricted by HindIII and XbaI and cloned into the PXFS-FLAG plasmid restricted with the same endonucleases. The two COOH terminus-deleted truncations (p300-ΔC1 and p300-ΔC2) were amplified from p300-FL using the same sense primer as for p300-FL, 5′-ACGTCTAGACGTAAGCTTGCCGAGAATGTGGTGGAACCGG-3′, and antisense primer 5′-GATATCCTAGATCTCATGGTGAAGGACACAG-3′ for p300-ΔC1 or antisense primer 5′-GATATCCTACGGAGATGACTGGGTAGCT-3′ for p300-ΔC2. The PCR products were digested with HindIII and EcoRV and ligated into the PXFS-Straw-FLAG plasmid restricted with the same endonucleases. p300-ΔB mutant was generated by two steps. First the fragment containing the sequences encoding aa 1–994 was produced by using the primer set sense 5′-CAGTCTAGACGTAAGCTTGCCGAGAATGTGGTGGAACCGG-3′ and antisense 5-CTGAAATAAGCTTCGGCTGAGTATCTGCTGG-3′ from p300-FL, producing a PCR product overhang with a HindIII site on both the 5′- and 3′-ends. This PCR product was then restricted with HindIII and fused into pXFS-FLAG-ΔN4 upstream of the sequences encoding aa 1255 of p300, finally producing a plasmid encoding p300 mutant with the internal region spanning from aa 995 to aa 1255 deleted (p300-ΔB).
Transient Transfection and Luciferase Activity Assay—Transient transfection using Lipofectamine PLUS reagent (Invitrogen) was performed in triplicate plates according to the manufacturer's instructions. For the reporter assay, cells were plated into 6-well plates and cotransfected with the γ-FBG-LUC reporter gene, the indicated expression vectors, and the transfection efficiency control plasmid pSV2PAP. 24 h later, cells were stimulated with OSM or IL-6. Both luciferase and alkaline phosphatase activities were measured 48 h after transfection. For the co-immunoprecipitation assay, the indicated expression plasmids were cotransfected into 10-cm2 dishes using the same protocol, and cells were harvested for protein extraction 24 h after transfection.
Preparation of Subcellular Extracts—Sucrose cushion-purified nuclear extract (NE) was prepared as described before (25). In brief, cells were harvested in phosphate-buffered saline and centrifuged. Pellets were resuspended in a double cell volume of solution A and centrifuged to obtain the supernatants as cytoplasmic fraction. The nuclear pellets were purified in solution B and lysed in solution C. After centrifugation at 12,000 × g at 4 °C for 20 min, the supernatants were saved as NE. The protein concentrations were measured by Coomassie dye binding (protein reagent, Bio-Rad).
Western Immunoblot—Proteins were fractionated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA). Membranes were blocked in 5% milk for 0.5–1 h and incubated with indicated primary Ab at 4 °C overnight. Membranes were washed in Tris-buffered saline, 0.1% Tween 20 and incubated with secondary Ab at 20 °C for 1 h. Signals were detected by the enhanced chemiluminescence assay (ECL; Amersham Biosciences) or visualized by the Odyssey Infrared Imaging System. β-Actin was used as a loading control.
Co-immunoprecipitation Assay—1–2 mg of protein was precleared with 40 μl of protein A-Sepharose beads (Sigma) for 1 h at 4 °C. Immunoprecipitation was performed in the presence of 5 μg of the indicated primary Ab at 4 °C overnight. Immune complexes were captured by adding 50 μl of protein A-Sepharose beads (50% slurry) and rotated at 4 °C for 2 h. After the supernatant was discarded, protein A-Sepharose beads were washed with cold phosphate-buffered saline four to five times, and immunoprecipitates were fractionated by SDS-PAGE.
Two-step ChIP Assay—Two-step ChIP was performed as described previously (35). In brief, 4–6 × 106 MEFs cells/100-mm dish were washed twice with phosphate-buffered saline after stimulation. Protein-protein cross-linking was first performed with disuccinimidyl glutarate (Pierce) followed by protein-DNA cross-linking with formaldehyde. After cells were washed and collected in 1 ml of phosphate-buffered saline, pellets were lysed by SDS lysis buffer and sonicated four times for 15 s each at setting 4 with a 10-s break on ice until DNA fragment lengths were between 200 and 1000 bp. Equal amounts of DNA were immunoprecipitated overnight at 4 °C with 4 μg of the indicated Abs in ChIP dilution buffer. Immunoprecipitates were collected with 40 μl of protein A magnetic beads (Dynal Inc., Brown Deer, WI) and washed sequentially with ChIP dilution buffer, high salt buffer, LiCl wash buffer, and finally in 1× Tris-EDTA buffer. DNA was eluted in 250 μl of elution buffer for 15 min at room temperature. Samples were decross-linked in decross-linking mixture at 65 °C for 2 h. DNA was phenol/chloroform-extracted, precipitated by 100% ethanol, and used for real time PCR.
Quantitative Real Time PCR (Q-RT-PCR)—Total cellular RNA was extracted by Tri Reagent (Sigma). 2 μg of RNA was used for reverse transcription using the SuperScript III First-Strand Synthesis System from Invitrogen. 2 μl of cDNA products was amplified in a 20-μl reaction system containing 10 μl of iQ SYBR Green Supermix (Bio-Rad) and 400 nm primer mixture. The primers were designed by PrimerExpress v2.0 and are shown in Table 1. All the reactions were processed in a MyiQ Single Color Real-Time PCR thermocycler using a two-step plus melting curve program, and the results were analyzed by the iQ5 program (Bio-Rad). The -fold change of mRNA expression was calculated by normalizing the absolute amount to internal control glyceraldehyde-3-phosphate dehydrogenase and comparing with untreated cells.
TABLE 1.
Primer sets used for Q-RT-PCR
| Target genes | Sense | Antisense |
|---|---|---|
| socs3 | 5′-CCGCGGGCACCTTTC-3′ | 5′-TTGACGCTCAACGTGAAGAAGT-3′ |
| c-fos | 5′-CCTGCCCCTTCTCAACGA-3′ | 5′-TCCACGTTGCTGATGCTCTT-3′ |
| p21 | 5′-TTCCGCACAGGAGCAAAGT-3′ | 5′-CGGCGCAACTGCTCACT-3′ |
For quantitative real time genomic PCR, the primer set used for the ChIP assay was first tested by performing quantitative PCR on serial dilutions of mouse genomic DNA (from 40 ng to 25 μg) isolated from MEFs. A standard curve was generated by plotting the logarithm of the genomic DNA concentration versus the threshold cycle (Ct value). This standard curve confirmed that ChIP-quantitative PCR results were within the linear range of amplification (not shown). The -fold change of DNA in each immunoprecipitate was determined by the comparative Ct method. First the Ct value of each sample was normalized to the corresponding input DNA reference, producing the ΔCt value (ΔCt = Ctsample - Ctinput). Next the signals in OSM-treated samples were compared with those of untreated samples by calculating the ΔΔCt value (ΔΔCt = ΔCttreated - ΔCtuntreated). Here ΔCttreated was the Ct value of OSM-treated samples normalized to input DNA, and ΔCtuntreated represented the Ct value of OSM-untreated samples normalized to input DNA. Finally we calculated the -fold change relative to that amount in unstimulated cells as 2-ΔΔCt. The primer set used for quantitative real time genomic PCR to amply the STAT3 binding sites on the socs3 promoter included sense primer 5′-CGCGCACAGCCTTTCAGT-3′ and antisense primer 5′-CCCCCGATTCCTGGAACT-3′.
RESULTS
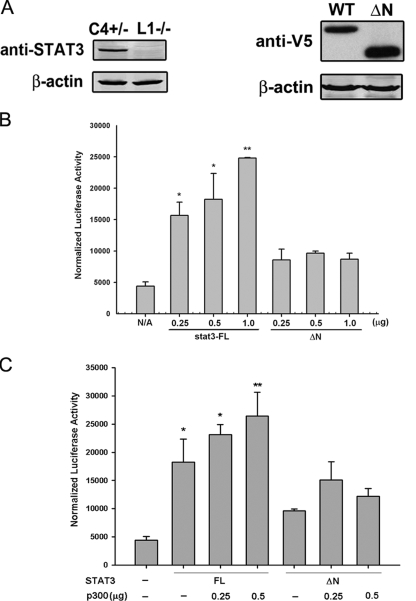
The STAT3 NH2-terminal Domain Is Required for OSM-inducible Transcription—Based on our findings that the STAT3 NH2-terminal domain may play an important role in STAT3 function, we generated a V5 epitope-tagged STAT3 mutant (V5-STAT3-ΔN) in which the entire NH2-terminal region (aa 1–133) was deleted and tested its transcriptional activity in luciferase reporter assays. For this purpose, STAT3-/- MEFs were used to avoid the interference from endogenous STAT3. First the loss of STAT3 in STAT3-/- MEFs was confirmed by Western immunoblot (Fig. 1A, left panel). STAT3-/- MEFs were then transiently transfected with V5-STAT3-FL or V5-STAT3-ΔN, and ectopically expressed STAT3 was measured by Western immunoblot with anti-V5 Ab. As seen in Fig. 1A, right panel, the expression of V5-STAT3-ΔN was not affected by the removal of the NH2-terminal domain.
FIGURE 1.
The STAT3 NH2-terminal domain is required for OSM-inducible transcription. A, the left panel is a Western immunoblot showing the endogenous STAT3 level in STAT3+/- MEFs (C4+/-) and the loss of STAT3 expression in STAT3-/- MEFs (L1-/-). In the right panel, the expression of transiently transfected V5-STAT3-FL and V5-STAT3-ΔNin STAT3-/- MEFs was measured by Western immunoblot with anti-V5 Ab. β-Actin was used as a loading control. B, STAT3-/- MEFs were transfected with the γ-FBG-LUC reporter gene together with different doses of V5-STAT3-FL or V5-STAT3-ΔN expression vectors and treated with OSM for 24 h prior to the reporter gene assay. Shown is mean and standard deviation (S.D.) normalized luciferase reporter activity. C, different doses of pCMVβ p300 were cotransfected with V5-STAT3-FL or V5-STAT3-ΔN together with γ-FBG-LUC reporter followed by OSM stimulation for 24 h. Data shown are means ± S.D. from three independent transfections. The data were analyzed by Student's t test. The luciferase reporter activities in V5-STAT3-FL-transfected MEFs were compared with those in V5-STAT3-ΔN-transfected MEFs using a Student's t test. *, p value <0.05; **, p value <0.01. N/A, not applicable.
Different doses of V5-STAT3-FL or V5-STAT3-ΔN were then cotransfected with the reporter gene γ-FBG-LUC, which contains the three native STAT3 binding sites from the γ-FBG gene. Because MEFs express low levels of IL-6Rα and OSM has been shown to induce STAT3-p300 cooperation in STAT3-/- MEFs (36), OSM was used in these experiments to activate STAT3 signal transduction. Reporter activity was measured in the presence or absence of OSM. As little as 0.25 μg of V5-STAT3-FL was able to increase γ-FBG-LUC activity by about 3-fold, and a further increase to 5-fold was observed when the amount of V5-STAT3-FL expression vector was increased (Fig. 1B). In contrast to the behavior of V5-STAT3-FL, V5-STAT3-ΔN was able to induce only a 1.5-fold increase in reporter activity, and a dose-dependent response was not observed (Fig. 1B). Our findings that the STAT3 NH2-terminal domain is sufficient to associate with p300 raised the possibility that this domain regulates STAT3 signaling by mediating p300 recruitment. Therefore, we next tested whether the STAT3 NH2-terminal domain was required for the functional cooperation between STAT3 and p300. To this end, different doses of p300 expression vectors were cotransfected with V5-STAT3-FL or the V5-STAT3-ΔN mutant into STAT3-/- MEFs. p300 further enhanced γ-FBG-LUC activity induced by V5-STAT3-FL; however, in the presence of V5-STAT3-ΔN, p300 was unable to rescue the lower levels of reporter activity (Fig. 1C).
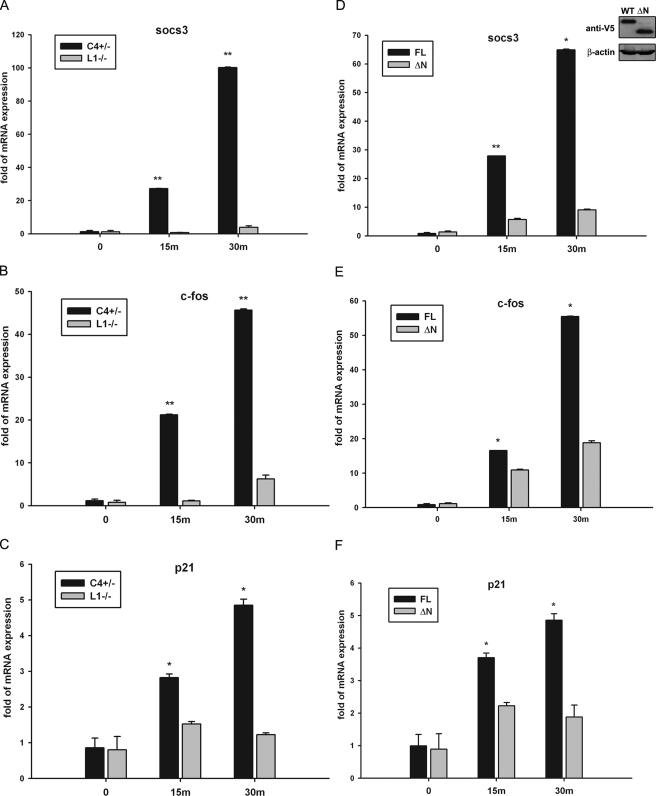
The STAT3 NH2-terminal Domain Is Required for the OSM-inducible mRNA Expression—To further explore the role of the STAT3 NH2 terminus, we analyzed the expression of endogenous STAT3 target genes in MEFs by Q-RT-PCR. Socs3, c-fos, and p21, well established STAT3-dependent genes (37–40), were all significantly and rapidly induced by OSM in STAT3+/- MEFs with a 100-fold induction of socs3 expression, for example, observed within 30 min of stimulation. This highly inducible up-regulation was dramatically decreased in STAT3-/- MEFs where less than a 5-fold induction of socs3 was observed, indicating that STAT3 was a crucial transactivator for the induction of these genes (Fig. 2, A–C). To investigate how the NH2-terminal deletion affects STAT3 target gene expression, we stimulated a population of STAT3-/- MEFs stably expressing V5-tagged STAT3-FL or STAT3-ΔN mutant with OSM. In cells expressing STAT3-FL, all three STAT3 target genes were strongly activated by OSM. Conversely cells expressing V5-STAT3-ΔN, despite its expression being equivalent to V5-STAT3-FL (Fig. 2D, inset), were defective in gene expression (Fig. 2D–F). Collectively these results indicate the necessity of the NH2-terminal domain for transcription activation by STAT3 in response to OSM signaling.
FIGURE 2.
The STAT3 NH2-terminal domain is required for OSM-inducible mRNA expression. A–C, STAT3+/- (C4+/-) and STAT3-/- (L1-/-) MEFs were treated with OSM (20 ng/ml) for 15 or 30 min. Whole cellular mRNA was isolated, and the expression of socs3, c-fos, and p21 was measured by Q-RT-PCR. The -fold change in OSM-treated cells over OSM-unstimulated control was obtained after correction for the amount of internal control, glyceraldehyde-3-phosphate dehydrogenase. Error bars are S.D. The mRNA induction in STAT3+/- MEFs was compared with that in STAT3-/- MEFS. D–F, STAT3-/- MEFs were transfected with V5-STAT3-FL or V5-STAT3-ΔN expression vectors. Positively transfected cells were selected by puromycin at 48 h and pooled. The inset in D is a Western immunoblot showing the stably expressed V5-STAT3-FL and V5-STAT3-ΔN after antibiotic selection. The V5-STAT3-FL- or V5-STAT3-ΔN-complemented STAT3-/- MEFs were stimulated by OSM (20 ng/ml) for 15 or 30 min (m). Shown is the result of Q-RT-PCR assays plotting the change of mRNA abundance in OSM-treated cells normalized to glyceraldehyde-3-phosphate dehydrogenase expressed as a -fold change relative to unstimulated controls. The mRNA induction in V5-STAT3-FL-complemented STAT3-/- MEFs was compared with that in V5-STAT3-ΔN-complemented STAT3-/- MEFs. The data were analyzed by Student's t test. *, p value <0.05; **, p value <0.01.
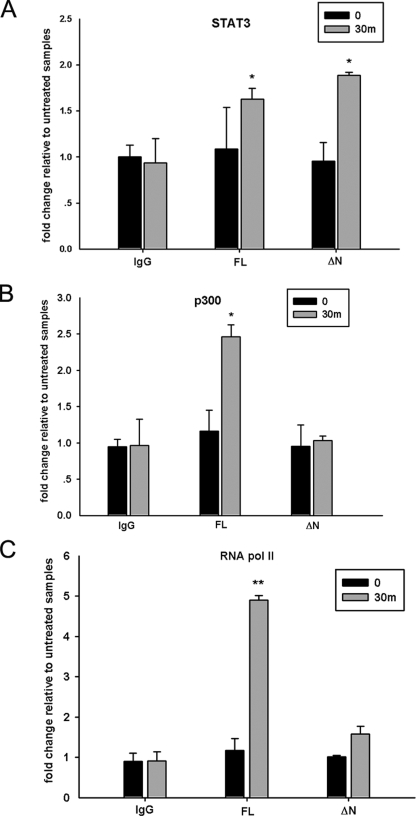
The NH2-terminal Deletion Affects Enhanceosome Assembly on the socs3 Promoter—To clarify whether the NH2 terminus of STAT3 regulates its transcription activity by mediating p300 recruitment, two-step ChIP assays were performed to analyze p300 binding to the socs3 promoter. Two STAT3 consensus binding regions have been identified on the socs3 promoter upstream of the transcription initiation site (37, 38). A primer pair amplifying both binding sites was designed and optimized by quantitative real time genomic PCR to show a linear dynamic range from 40 ng to 25 μg of DNA (“Experimental Procedures”). Next we examined the OSM-inducible STAT3 binding to the socs3 promoter in STAT3-/- MEFs complemented with V5-STAT3-FL or V5-STAT3-ΔN mutant in ChIP experiments by using anti-V5 as the primary Ab. This experiment revealed that both V5-STAT3-FL and V5-STAT3-ΔN inducibly associate with the socs3 promoter within 30 min after OSM stimulation (Fig. 3A). This finding was consistent with previous studies showing that NH2-terminal deletion did not affect the nuclear translocation or DNA binding activity of STAT3 (25, 41). The ChIP assays were extended further to analyze the effect of OSM on inducible p300 and RNA pol II binding to the socs3 promoter. As a result, we observed a 2.5-fold increase of p300 binding in response to OSM when STAT3-/- MEFs were complemented with V5-STAT3-FL, but no significant p300 recruitment was detected when V5-STAT3-ΔN mutant was stably expressed (Fig. 3B). In addition, the inducible loading of RNA pol II on the socs3 gene was also inhibited in MEFs stably expressing STAT3-ΔN (Fig. 3C). Our explanation for these results is that the reduced p300 binding caused by the NH2-terminal deletion affects the association of RNA pol II because of its direct interaction with p300 (42, 43).
FIGURE 3.
The STAT3 NH2-terminal domain regulates enhanceosome assembly on the socs3 promoter. STAT3-/- MEFs stably expressing V5-STAT3-FL or V5-STAT3-ΔN were treated with OSM (20 ng/ml) for 30 min (m), and two-step ChIP assay was performed by using Abs specifically recognizing V5 tag (A), p300 (B), or RNA pol II (C). The sequence of the socs3 promoter in the immunoprecipitates was amplified by quantitative real time genomic PCR using specific primers. Shown is the -fold change of signals in OSM-treated samples compared with those of untreated samples. Error bars are S.D. A, both V5-STAT3-FL and V5-STAT3-ΔN are induced by OSM to bind to the socs3 promoter. B, the OSM-inducible p300 recruitment to the socs3 promoter is inhibited in cells stably expressing STAT3-ΔN. C, the OSM-inducible RNA pol II association to the socs3 promoter is reduced in STAT3-ΔN-complemented MEFs. The results are expressed as means ± S.D. from triplicates. The data were analyzed by Student's t test. *, p value <0.05; **, p value <0.01.
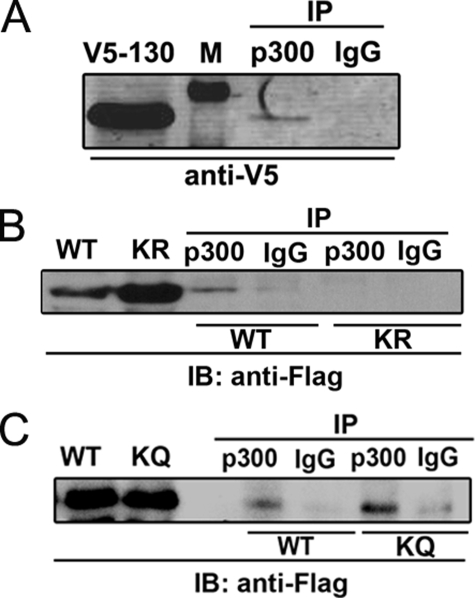
Association between the STAT3 NH2-terminal Domain and p300 Is Regulated by NH2-terminal Acetylation—To study the interaction between the STAT3 NH2-terminal domain and p300, we first sought to determine whether the STAT3 NH2 terminus is sufficient for p300 interaction. For this purpose, cells were cotransfected with pCMVβ p300 and a plasmid expressing the V5-tagged NH2-terminal domain of STAT3 (aa 1–130). Nuclear protein was recovered and immunoprecipitated by anti-p300 Ab followed by Western immunoblot using anti-V5 Ab. As seen in Fig. 4A, V5-STAT3 (aa 1–130) was specifically captured by p300 Ab but not by IgG. To determine the effect of STAT3 Lys-49/Lys-87 acetylations on p300 interaction, we generated FLAG-tagged acetylation-deficient (K49R/K87R) and pseudoacetylated (K49Q/K87Q) NH2-terminal mutants (aa 1–124) and examined their association with p300. In contrast to the FLAG-STAT3-WT (aa 1–124), the acetylation-deficient FLAG-STAT3 (aa 1–124)-K49R/K87R mutant was barely detectable in p300 immunoprecipitates (Fig. 4B). Instead the pseudoacetylated FLAG-STAT3 (aa 1–124)-K49Q/K87Q mutant exhibited stronger p300 binding despite the fact that its expression level was comparable to FLAG-STAT3-WT (aa 1–124) (Fig. 4C). These results indicate that the STAT3 NH2-terminal domain is sufficient for p300 interaction and that the Lys-49/Lys-87 acetylations increase this association.
FIGURE 4.
Interaction between the STAT3 NH2-terminal domain and p300. A, HepG2 cells were cotransfected with pEF6-V5-STAT3 (aa 1–130) with pCMVβ p300. Cells were treated with IL-6 (10 ng/ml) for 30 min before NE was prepared. 2 mg of NE was immunoprecipitated by anti-p300 Ab. p300-bound V5-STAT3 (aa 1–130) was detected by anti-V5 Ab in Western immunoblot. Lane 1 is lysate showing the expression of V5-STAT3 (aa 1–130). Lane 2 is protein standard (indicated by “M” in A). B, HepG2 cells were cotransfected with either pECFP-FLAG-STAT3-WT (aa 1–124) or pECFP-FLAG-STAT3-K49R/K87R mutant (aa 1–124) together with pCMVβ p300. NE was immunoprecipitated with anti-p300 Ab followed by Western immunoblot (IB) with anti-FLAG Ab. Lane 1 and lane 2 show FLAG-STAT3-WT (aa 1–124) and FLAG-STAT3-K49R/K87R (aa 1–124) expression in nuclear lysate. Lane 3 and lane 4 are immunoprecipitates (IP) of FLAG-STAT3-WT (aa 1–124) with anti-p300 Ab (lane 3) or IgG (lane 4). Lane 5 and lane 6 are immunoprecipitates of FLAG-STAT3-K49R/K87R mutant (aa 1–124) with anti-p300 Ab (lane 5) or IgG (lane 6). C, HepG2 cells were cotransfected with either pECFP-FLAG-STAT3-WT (aa 1–124) or pECFP-FLAG-STAT3-K49Q/K87Q mutant (aa 1–124) (KQ) together with pCMVβ p300. Immunoprecipitation was performed as described in Fig. 1B. Lane 1 and lane 2 show FLAG-STAT3-WT (aa 1–124) and FLAG-STAT3-K49Q/K87Q (aa 1–124) expression in nuclear lysate. Lane 3 and lane 4 are immunoprecipitates of FLAG-STAT3-WT (aa 1–124) with anti-p300 Ab (lane 3) or IgG (lane 4). Lane 5 and lane 6 are immunoprecipitates of FLAG-STAT3-K49Q/K87Q mutant (aa 1–124) with anti-p300 Ab (lane 5) or IgG (lane 6).
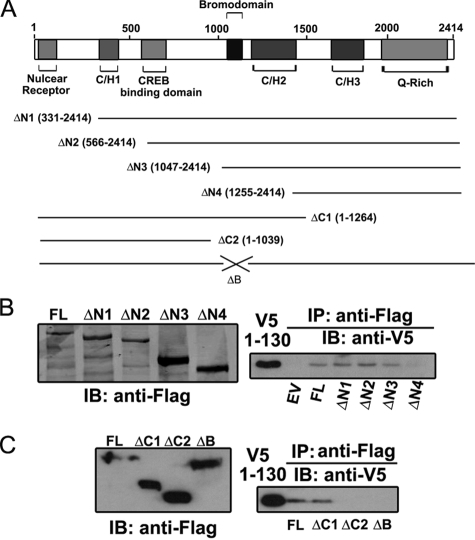
The Binding Site of STAT3 NH2-terminal Domain Maps to the p300 Bromodomain—To determine the domain of p300 responsible for the STAT3 NH2-terminal binding, we generated a panel of FLAG epitope-tagged p300 deletions. The related functional domains included in each truncation are shown in Fig. 5A. Each of the four NH2 terminus-deleted mutants (p300-ΔN1, -ΔN2, -ΔN3, and -ΔN4) or two COOH terminus-deleted mutants (p300-ΔC1 and -ΔC2) was individually coexpressed with V5-STAT3 (aa 1–130), and p300-bound NH2-terminal domain was examined by nondenaturing co-immunoprecipitation assays. STAT3 NH2-terminal binding was lost when the region between aa 1047 (p300-ΔN3) and aa 1255 (p300-ΔN4) was deleted (Fig. 5B, right panel). Importantly this region, from aa 1047 to aa 1255, contains the p300 bromodomain, spanning from aa 1053 to aa 1156. Consistent with this finding, the lack of bromodomain in ΔC2 (aa 1–1043) abrogated its interaction with the STAT3 NH2-terminal domain; however, the bromodomain-containing mutant ΔC1 (aa 1–1264) still complexed with the STAT3 NH2 terminus (Fig. 5C, right panel). To further confirm that the p300 bromodomain mediates its interaction with the STAT3 NH2-terminal region, an expression vector encoding p300 with an internal deletion of the bromodomain (p300-ΔB) was generated and tested. As we expected, no interaction was detected between p300-ΔB and the STAT3 NH2-terminal domain (Fig. 5C, right panel). Taken together, these data suggest that the p300 bromodomain binds the STAT3 NH2-terminal domain and that the IL-6- or OSM-inducible NH2-terminal acetylations further stabilize this interaction.
FIGURE 5.
The STAT3 NH2-terminal domain is associated with the p300 bromodomain. A, a schematic diagram showing different functional domains of p300 and all p300 mutants used in B and C. B and C, PXFS-FLAG-p300-FL, p300 NH2-terminal deletions (p300-ΔN1, -ΔN2, -ΔN3, and -ΔN4), COOH-terminal deletions (p300-ΔC1 and -ΔC2), or p300 mutant without the bromodomain (p300-ΔB) was individually cotransfected with pEF6-V5-STAT3 (aa 1–130) into HEK 293 cells. Immunoprecipitation (IP) was performed by anti-FLAG Ab, and the p300-associated STAT3 NH2 terminus was detected by anti-V5 Ab. The left panels are Western immunoblots (IB) showing the expression of each mutant in the cell lysate. The right panels show p300-associated STAT3 NH2 terminus. EV, the empty vector (PXFS-FLAG) was included as a negative control.
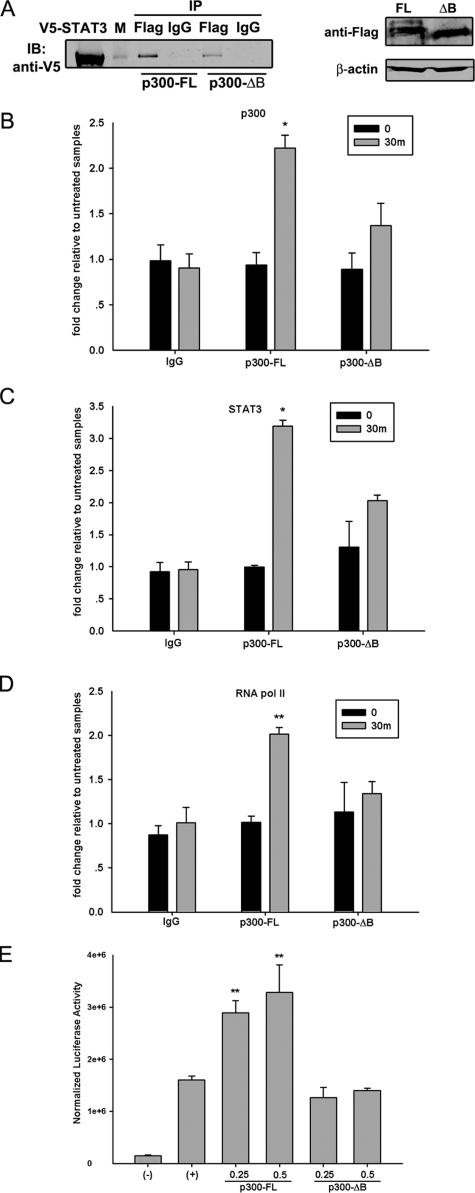
The p300 Bromodomain Facilitates STAT3-dependent Transcription—Because the p300 bromodomain was responsible for STAT3 NH2-terminal binding (Fig. 5), we proposed that the p300-STAT3 interaction would be affected when the p300 bromodomain was deleted. To test this, 293 cells were transfected with expression vectors encoding V5-STAT3 together with FLAG-tagged p300-FL or p300-ΔB mutant. The interaction between these two proteins was then measured by nondenaturing co-immunoprecipitation with anti-FLAG Ab followed by Western immunoblot with anti-V5 Ab. As we expected, the p300-ΔB mutant showed decreased interaction with STAT3 compared with that of p300-FL (Fig. 6A, left panel) despite the similar expression level of these two proteins (Fig. 6A, right panel). The remaining association observed between p300-ΔB and STAT3 indicates that multiple domains of p300 are involved in STAT3-p300 interaction. An earlier study using in vitro transcription reactions found that p300 forms a stable complex with chromatin templates that is mediated, at least in part, by the bromodomain (44). To confirm this finding in cellulo, ChIP experiments were performed to capture the binding of p300 to the endogenous socs3 promoter after p300-FL or p300-ΔB mutant was transiently transfected into STAT3+/- MEFs. As seen in Fig. 6B, the p300-ΔB mutant exhibited a weaker association with the socs3 promoter than did p300-FL. At the same time, reductions in STAT3 and RNA pol II binding were observed in p300-ΔB-transfected cells (Fig. 6, C and D). To further test the functional role of bromodomain, different doses of p300-FL or p300-ΔB mutant were cotransfected with γ-FBG-LUC reporter gene into HepG2 cells followed by IL-6 treatment for 24 h. As a result, the p300-FL expression further enhanced the IL-6-inducible reporter activity in a dose-dependent manner. The p300-ΔB mutant, however, failed to function as a transcriptional coactivator in this assay (Fig. 6E). Taken together, our results suggested that the bromodomain plays a critical role in p300 function because it mediates p300 interaction with STAT3 and facilitates enhanceosome formation.
FIGURE 6.
The p300 bromodomain facilitates STAT3-dependent transcriptional activation. A, HEK 293 cells were cotransfected with PXFS-FLAG-p300-FL or p300-ΔB mutant together with pEF6-V5-STAT3-FL. 48 h after transfection, 1 mg of whole cell extract was collected and immunoprecipitated (IP) by anti-FLAG Ab followed by Western immunoblot (IB) with anti-V5 Ab. The right panel is a Western immunoblot showing that p300-FL and p300-ΔB mutant had comparable expression levels. B, C, and D, STAT3+/- MEFs were transiently transfected with PXFS-FLAG-p300-FL or p300-ΔB mutant. 24 h after transfection, cells were treated with OSM (20 ng/ml) for 30 min (m) or left untreated. Two-step ChIP assays were performed by using antibodies specifically recognizing p300 (B), STAT3 (C), and RNA pol II (D). The signals in OSM-treated cells were compared with those in non-treated MEFs. E, HepG2 cells were transfected with different doses of PXFS-FLAG-p300-FL or p300-ΔB mutant together with γ-FBG-LUC reporter gene. 24 h later, cells were treated with IL-6 (10 ng/ml) for another 24 h, and then luciferase activity was measured. Data shown are means ± S.D. from three independent transfections. The data were analyzed by Student's t test. *, p value <0.05; **, p value <0.01.
DISCUSSION
STAT3 is the major transcription factor activated by the IL-6 family of cytokines. Although the STAT3 signaling from the cell membrane to the nucleus is well understood, the molecular events regulating gene transcription need more elucidation. Like many other transcription factors, the activated STAT3 recruits the p300 coactivator after nuclear translocation (24). The p300-induced acetylation on histone tails is coupled with chromatin remodeling, thereby enhancing target gene expression (24–26). The interaction between p300 and STAT3 is regulated via both the STAT3 NH2-terminal domain and the COOH-terminal TAD (26). In this study, we report that the NH2-terminal domain plays a critical role in STAT3-mediated signal transduction by regulating enhanceosome assembly at the promoter loci. This conclusion is supported by the following. 1) The NH2-terminal deletion inhibited OSM-induced reporter activity and abolished the cooperation between STAT3 and p300 (Fig. 1). 2) The expression of endogenous STAT3 target genes, including socs3, c-fos, and p21, was significantly reduced in STAT3-ΔN-complemented STAT3-/- MEFs (Fig. 2). 3) Significantly reduced loading of p300 and RNA pol II to the socs3 promoter was observed when STAT3-ΔN mutant was stably expressed in STAT3-/- MEFs (Fig. 3). These results suggest that the NH2-terminal deletion disrupts enhanceosome formation on the socs3 promoter, resulting in an inhibition in mRNA transcription.
The NH2-terminal domain of STATs comprises approximately the first 130 residues that assemble into an all-helical hooklike structure (45). Although it is highly conserved in the STAT family, the NH2-terminal domain is implicated in diverse functions, including STAT dimerization or tetramerization (46, 47), phosphorylation/dephosphorylation (48–50), and interaction of STAT with other transcription factors or coactivators (33, 51). One of the identified functions of the NH2-terminal region is regulating the cooperative binding of STAT dimer·dimer complexes on two tandemly arranged binding sites. This function is conserved in STAT1 (52), STAT4 (53), and STAT5 (54) and recently has also been confirmed in STAT3 when it activates the transcription of α2-macroglobulin gene (47). A tetrameric STAT3 complex is formed on the α2-macroglobulin enhancer sequence that is required for the maximum transcriptional activation (47). Two point mutations in the NH2-terminal region (W37A and Q66A) disrupt the tetramer binding (47). The murine socs3 promoter also contains two putative STAT3 binding sites that are highly conserved in the human SOCS3 promoter (37, 55). Both sites are required for the complete activation of the human SOCS3 promoter stimulated by leukemia-inhibitory factor (55). As for the murine socs3 promoter, the proximal site localized from nucleotides -72 to -64 is essential for leukemia-inhibitory factor-induced transactivation (37). The distal site from nucleotides -95 to -87, however, has not been directly tested. Although two STAT3 motifs on the murine socs3 promoter are tandemly linked, STAT3 binding is only detected on the proximal site, and no obvious dimer-dimer interaction is observed (41). This excludes the possibility that the defective activity of STAT3-ΔN mutant in activating the socs3 expression (Fig. 2) is caused by the disruption of STAT3 tetramerization.
Here we describe a new level of STAT3 NH2-terminal function achieved through its interaction with the p300 bromodomain. p300 is known to function as a bridging factor connecting sequence-specific transcription factors with the basal transcription machinery (56). The direct association between p300 and RNA pol II in mammary cells has long been known (42, 43). In this interaction, p300 specifically interacts with the unphosphorylated form of RNA pol II that is able to form the transcription preinitiation complex (42, 57). Our study showed that the STAT3 NH2-terminal deletion significantly decreases p300 recruitment to the socs3 promoter (Fig. 3B), resulting in an inhibition in RNA pol II binding (Fig. 3C). These results reveal the function of the NH2-terminal domain to integrate the enhancer binding proteins and facilitate the assembly of the transcription preinitiation complex.
The recent finding that STAT3 is also a direct target of p300 for acetylation draws attention to the study of p300-STAT3 interaction. STAT3 is acetylated by p300 at multiple lysine residues. The reversible acetylation on Lys-685 within TAD is essential for STAT3 dimerization and DNA binding ability (26). Our earlier studies identified another two acetylation sites (Lys residues 49 and 87) localized in the NH2-terminal domain of STAT3; acetylations of these sites are also indispensable for STAT3-mediated transactivation (25). Interestingly these NH2-terminal acetylations, however, have no effect on the inducible STAT3 binding to DNA (25).
In this study, we demonstrated that the STAT3 NH2-terminal domain is sufficient for p300 interaction (Fig. 4A) and showed that the NH2-terminal acetylations increase the p300-NH2-terminus interaction (Fig. 4, B and C). Here we also report the discovery that the p300 bromodomain is the site for NH2-terminal association (Fig. 5). The bromodomain represents a highly conserved protein module that is commonly found in many chromatin-associated proteins and nearly all histone acetyltransferases (58). The function of bromodomain as an acetyl-Lys-binding domain was discovered soon after its three-dimensional structure was elucidated. The solution structure of the bromodomain from p300/CBP-associated factor revealed an unusual left-handed four-helix bundle (59), a unique structural fold that was conserved in other chromatin-associated proteins, such as human TAFII250 (60) and the Saccharomyces cerevisiae Gcn5p (61). Although acetyl-Lys is the direct recognition site of the bromodomain, residues flanking both sides of acetyl-Lys are also important for the interaction and contribute to the ligand selectivity of bromodomains (62). The acetylation-enhanced interaction (Fig. 4, B and C) indicates that the acetylations on Lys residues 49 and 87 provide a binding motif for the bromodomain of p300, leading to a stable complex formation and efficient histone acetyltransferase recruitment to the promoter. There is also a possibility that the NH2-terminal acetylations induce conformational changes in residues flanking the acetyl-Lys that positively affect p300-STAT3 interaction. Interestingly the acetylation-dependent interaction is also observed between MyoD and the bromodomain of p300/CBP (17). p300/CBP selectively recognizes the acetylated form of MyoD in cells, and mutations of acetylation sites in MyoD decrease its ability to cooperate with p300/CBP (17). This suggests that acetylation is probably a common strategy that p300/CBP utilize to facilitate their interaction with non-histone proteins.
The bromodomain is also reported to mediate p300 binding to chromatin. p300 forms a stable complex with in vitro transcription template, and this association is regulated at least partially by the bromodomain (44). Our ChIP result in Fig. 6B confirmed this finding in cellulo by showing that the bromodomain deletion decreased p300 occupancy on the endogenous socs3 promoter. We also noticed that the bromodomain-deficient mutant (p300-ΔB) was still bound to STAT3 (Fig. 6A), although the interaction was significantly weaker than that of p300-FL and STAT3. These data indicate that more than one domain of p300 is involved in mediating p300-STAT3 interaction. It has been shown that TAD of STAT3 also associates with p300, although the binding site on p300 has not been identified yet (24, 26). Similarly the interaction between STAT1 and p300/CBP is also mediated by two contact regions. The STAT1 NH2-terminal region is recognized by the CREB-binding domain of p300/CBP, and the binding site of TAD maps to the p300/CBP domain that recruits adenovirus E1A protein (28).
In summary, we have provided evidence that the STAT3 NH2-terminal domain interacts with the p300 bromodomain in an acetylation-dependent manner. IL-6- or OSM-induced acetylations on Lys-49 and Lys-87 trigger the recognition of substrate by the p300 bromodomain, resulting in a strengthened association between p300 and STAT3. This interaction further stabilizes the recruitment of other transcription components like RNA pol II to efficiently initiate the transcription of STAT3 target genes.
Acknowledgments
We thank Dr. Stephanie Watowich from University of Texas M. D. Anderson Cancer Center and Dr. David Levy from New York University for generously providing STAT3+/- and STAT3-/- MEFs. Core laboratory support was provided by National Institutes of Health Grant P30 ES06676 (to J. Ward, University of Texas Medical Branch).
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL70925 (to A. R. B.) and N01 HV28184 (to A. Kurosky, University of Texas Medical Branch). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: STAT, signal transducer and activator of transcription; IL, interleukin; OSM, oncostatin M; Rα, receptor α chain; pol, polymerase; FBG, fibrinogen; TAD, transactivation domain; ChIP, chromatin immunoprecipitation; NE, nuclear extract; Q-RT-PCR, quantitative real time PCR; FL, full length; WT, wild type; MEF, mouse embryonic fibroblast; CREB, cAMP-response element-binding protein; CBP, CREB-binding protein; hAGT, human angiotensinogen; SH2, Src homology 2; HEK, human embryonic kidney; aa, amino acid(s); LUC, luciferase; Ab, antibody.
References
- 1.Heinrich, P. C., Behrmann, I., Muller-Newen, G., Schaper, F., and Graeve, L. (1998) Biochem. J. 334 297-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray, P. J. (2007) J. Immunol. 178 2623-2629 [DOI] [PubMed] [Google Scholar]
- 3.Hou, T., Tieu, B. C., Ray, S., Recinos, A., Cui, R., Tilton, R. G., and Brasier, A. R. (2008) Curr. Cardiol. Rev. 4 179-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ernst, M., and Jenkins, B. J. (2004) Trends Genet. 20 23-32 [DOI] [PubMed] [Google Scholar]
- 5.Heinrich, P. C., Behrmann, I., Haan, S., Hermanns, H. M., Muller-Newen, G., and Schaper, F. (2003) Biochem. J. 374 1-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darnell, J. E., Jr., Kerr, I. M., and Stark, G. R. (1994) Science 264 1415-1421 [DOI] [PubMed] [Google Scholar]
- 7.Bayle, J. H., and Crabtree, G. R. (1997) Chem. Biol. 4 885-888 [DOI] [PubMed] [Google Scholar]
- 8.Brownell, J. E., Zhou, J., Ranalli, T., Kobayashi, R., Edmondson, D. G., Roth, S. Y., and Allis, C. D. (1996) Cell 84 843-851 [DOI] [PubMed] [Google Scholar]
- 9.Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H., and Nakatani, Y. (1996) Cell 87 953-959 [DOI] [PubMed] [Google Scholar]
- 10.Yang, X. J., Ogryzko, V. V., Nishikawa, J., Howard, B. H., and Nakatani, Y. (1996) Nature 382 319-324 [DOI] [PubMed] [Google Scholar]
- 11.Mizzen, C. A., Yang, X. J., Kokubo, T., Brownell, J. E., Bannister, A. J., Owen-Hughes, T., Workman, J., Wang, L., Berger, S. L., Kouzarides, T., Nakatani, Y., and Allis, C. D. (1996) Cell 87 1261-1270 [DOI] [PubMed] [Google Scholar]
- 12.Workman, J. L., and Kingston, R. E. (1998) Annu. Rev. Biochem. 67 545-579 [DOI] [PubMed] [Google Scholar]
- 13.Yang, X., Gold, M. O., Tang, D. N., Lewis, D. E., Aguilar-Cordova, E., Rice, A. P., and Herrmann, C. H. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 12331-12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrera, J. E., Sakaguchi, K., Bergel, M., Trieschmann, L., Nakatani, Y., and Bustin, M. (1999) Mol. Cell. Biol. 19 3466-3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munshi, N., Merika, M., Yie, J., Senger, K., Chen, G., and Thanos, D. (1998) Mol. Cell 2 457-467 [DOI] [PubMed] [Google Scholar]
- 16.Soutoglou, E., Katrakili, N., and Talianidis, I. (2000) Mol. Cell 5 745-751 [DOI] [PubMed] [Google Scholar]
- 17.Polesskaya, A., Naguibneva, I., Duquet, A., Bengal, E., Robin, P., and Harel-Bellan, A. (2001) Mol. Cell. Biol. 21 5312-5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leo, C., and Chen, J. D. (2000) Gene (Amst.) 245 1-11 [DOI] [PubMed] [Google Scholar]
- 19.Xu, L., Glass, C. K., and Rosenfeld, M. G. (1999) Curr. Opin. Genet. Dev. 9 140-147 [DOI] [PubMed] [Google Scholar]
- 20.Hampsey, M., and Reinberg, D. (1999) Curr. Opin. Genet. Dev. 9 132-139 [DOI] [PubMed] [Google Scholar]
- 21.Shikama, N., Chan, H. M., Krstic-Demonacos, M., Smith, L., Lee, C. W., Cairns, W., and La Thangue, N. B. (2000) Mol. Cell. Biol. 20 8933-8943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu, W., and Roeder, R. G. (1997) Cell 90 595-606 [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Balbas, M. A., Bauer, U. M., Nielsen, S. J., Brehm, A., and Kouzarides, T. (2000) EMBO J. 19 662-671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray, S., Sherman, C. T., Lu, M., and Brasier, A. R. (2002) Mol. Endocrinol. 16 824-836 [DOI] [PubMed] [Google Scholar]
- 25.Ray, S., Boldogh, I., and Brasier, A. R. (2005) Gastroenterology 129 1616-1632 [DOI] [PubMed] [Google Scholar]
- 26.Yuan, Z. L., Guan, Y. J., Chatterjee, D., and Chin, Y. E. (2005) Science 307 269-273 [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharya, S., Eckner, R., Grossman, S., Oldread, E., Arany, Z., D'Andrea, A., and Livingston, D. M. (1996) Nature 383 344-347 [DOI] [PubMed] [Google Scholar]
- 28.Zhang, J. J., Vinkemeier, U., Gu, W., Chakravarti, D., Horvath, C. M., and Darnell, J. E., Jr. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 15092-15096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy, D. E., and Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3 651-662 [DOI] [PubMed] [Google Scholar]
- 30.Kammerer, R. A. (1997) Matrix Biol. 15 555-565, 567-568 [DOI] [PubMed] [Google Scholar]
- 31.Shuai, K., Horvath, C. M., Huang, L. H., Qureshi, S. A., Cowburn, D., and Darnell, J. E., Jr. (1994) Cell 76 821-828 [DOI] [PubMed] [Google Scholar]
- 32.Lutticken, C., Wegenka, U. M., Yuan, J., Buschmann, J., Schindler, C., Ziemiecki, A., Harpur, A. G., Wilks, A. F., Yasukawa, K., Taga, T., Kishimoto, T., Barbieri, G., Pellegrini, S., Sendtner, M., Heinrich, P., and Horn, H. (1994) Science 263 89-92 [DOI] [PubMed] [Google Scholar]
- 33.Hou, T., Ray, S., and Brasier, A. R. (2007) J. Biol. Chem. 282 37091-37102 [DOI] [PubMed] [Google Scholar]
- 34.Eckner, R., Ewen, M. E., Newsome, D., Gerdes, M., DeCaprio, J. A., Lawrence, J. B., and Livingston, D. M. (1994) Genes Dev. 8 869-884 [DOI] [PubMed] [Google Scholar]
- 35.Nowak, D. E., Tian, B., and Brasier, A. R. (2005) BioTechniques 39 715-725 [DOI] [PubMed] [Google Scholar]
- 36.Sun, W., Snyder, M., Levy, D. E., and Zhang, J. J. (2006) FEBS Lett. 580 5880-5884 [DOI] [PubMed] [Google Scholar]
- 37.Auernhammer, C. J., Bousquet, C., and Melmed, S. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 6964-6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul, C., Seiliez, I., Thissen, J. P., and Le Cam, A. (2000) Eur. J. Biochem. 267 5849-5857 [DOI] [PubMed] [Google Scholar]
- 39.Yang, E., Lerner, L., Besser, D., and Darnell, J. E., Jr. (2003) J. Biol. Chem. 278 15794-15799 [DOI] [PubMed] [Google Scholar]
- 40.Bellido, T., O'Brien, C. A., Roberson, P. K., and Manolagas, S. C. (1998) J. Biol. Chem. 273 21137-21144 [DOI] [PubMed] [Google Scholar]
- 41.Zhang, L., Badgwell, D. B., Bevers, J. J., III, Schlessinger, K., Murray, P. J., Levy, D. E., and Watowich, S. S. (2006) Mol. Cell. Biochem. 288 179-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho, H., Orphanides, G., Sun, X., Yang, X. J., Ogryzko, V., Lees, E., Nakatani, Y., and Reinberg, D. (1998) Mol. Cell. Biol. 18 5355-5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neish, A. S., Anderson, S. F., Schlegel, B. P., Wei, W., and Parvin, J. D. (1998) Nucleic Acids Res. 26 847-853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manning, E. T., Ikehara, T., Ito, T., Kadonaga, J. T., and Kraus, W. L. (2001) Mol. Cell. Biol. 21 3876-3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinkemeier, U., Moarefi, I., Darnell, J. E., Jr., and Kuriyan, J. (1998) Science 279 1048-1052 [DOI] [PubMed] [Google Scholar]
- 46.Chen, X., Bhandari, R., Vinkemeier, U., Van Den Akker, F., Darnell, J. E., Jr., and Kuriyan, J. (2003) Protein Sci. 12 361-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, X., and Darnell, J. E., Jr. (2001) J. Biol. Chem. 276 33576-33581 [DOI] [PubMed] [Google Scholar]
- 48.Chang, H. C., Zhang, S., Oldham, I., Naeger, L., Hoey, T., and Kaplan, M. H. (2003) J. Biol. Chem. 278 32471-32477 [DOI] [PubMed] [Google Scholar]
- 49.Mao, X., Ren, Z., Parker, G. N., Sondermann, H., Pastorello, M. A., Wang, W., McMurray, J. S., Demeler, B., Darnell, J. E., Jr., and Chen, X. (2005) Mol. Cell 17 761-771 [DOI] [PubMed] [Google Scholar]
- 50.Meyer, T., Hendry, L., Begitt, A., John, S., and Vinkemeier, U. (2004) J. Biol. Chem. 279 18998-19007 [DOI] [PubMed] [Google Scholar]
- 51.Zhang, X., Wrzeszczynska, M. H., Horvath, C. M., and Darnell, J. E., Jr. (1999) Mol. Cell. Biol. 19 7138-7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vinkemeier, U., Cohen, S. L., Moarefi, I., Chait, B. T., Kuriyan, J., and Darnell, J. E., Jr. (1996) EMBO J. 15 5616-5626 [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, X., Sun, Y. L., and Hoey, T. (1996) Science 273 794-797 [DOI] [PubMed] [Google Scholar]
- 54.John, S., Vinkemeier, U., Soldaini, E., Darnell, J. E., Jr., and Leonard, W. J. (1999) Mol. Cell. Biol. 19 1910-1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He, B., You, L., Uematsu, K., Matsangou, M., Xu, Z., He, M., McCormick, F., and Jablons, D. M. (2003) Biochem. Biophys. Res. Commun. 301 386-391 [DOI] [PubMed] [Google Scholar]
- 56.Chan, H. M., and La Thangue, N. B. (2001) J. Cell Sci. 114 2363-2373 [DOI] [PubMed] [Google Scholar]
- 57.Lu, H., Flores, O., Weinmann, R., and Reinberg, D. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 10004-10008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng, L., and Zhou, M. M. (2002) FEBS Lett. 513 124-128 [DOI] [PubMed] [Google Scholar]
- 59.Dhalluin, C., Carlson, J. E., Zeng, L., He, C., Aggarwal, A. K., and Zhou, M. M. (1999) Nature 399 491-496 [DOI] [PubMed] [Google Scholar]
- 60.Jacobson, R. H., Ladurner, A. G., King, D. S., and Tjian, R. (2000) Science 288 1422-1425 [DOI] [PubMed] [Google Scholar]
- 61.Owen, D. J., Ornaghi, P., Yang, J. C., Lowe, N., Evans, P. R., Ballario, P., Neuhaus, D., Filetici, P., and Travers, A. A. (2000) EMBO J. 19 6141-6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mujtaba, S., He, Y., Zeng, L., Farooq, A., Carlson, J. E., Ott, M., Verdin, E., and Zhou, M. M. (2002) Mol. Cell 9 575-586 [DOI] [PubMed] [Google Scholar]