Abstract
Patched (Ptc) is a 12-cross membrane protein that binds the secreted Hedgehog protein. Its regulation of the Hedgehog signaling pathway is critical to normal development and to a number of human diseases. This report analyzes features of sequence similarity and divergence in the Ptc protein family and identifies two subtypes distinguished by novel conserved domains. We used these results to propose a rational basis for classification. We show that one of the conserved sequence regions in the C-terminal domain of Ptch1 is responsible, at least in part, for rapid turnover. This sequence is absent in the stable Ptch2 protein.
The Hedgehog (Hh) signaling pathway participates in patterning organs of both vertebrates and insects, and misregulation of the pathway has been implicated in several human diseases. Genetic and biochemical studies have identified many components that contribute to the production of active secreted Hh protein and to the receipt of and response to Hh by receiving cells. However, key features of Hh signal transduction remain poorly understood. In particular, the mechanism by which Hh engages its receptor and activates signal transduction are unclear. Current models posit that Patched (Ptc), a transmembrane protein, engages Hh on the surface of Hh-responsive cells, but how it does so and how it regulates the downstream Hh signal transduction pathway is not understood. In humans, Ptch1 has been identified as a tumor suppressor (1, 2).
Although the Ptc tertiary structure has not been determined, secondary structure predictions suggest that Ptc proteins have 12 transmembrane-spanning domains, two large extracellular loops, and a C-terminal domain (CTD).3 Structure/function studies have ascribed distinct functions to some of these domains. First, indirect evidence that Ptc binds Hh has been reported for both vertebrate Ptch1 (3-5) and invertebrate Ptc (6-9). Based upon the phenotypes of loop 2 deletions, Hh binding has been attributed to the second extracellular loop (10). Second, five of the Ptc transmembrane segments appear to constitute a region that is conserved in the Neimann-Pick-1, hydroxymethylglutaryl-CoA reductase, and sterol regulatory element-binding protein cleavage-activating proteins (11-13) and that has been called a “sterol-sensing domain” (SSD). The NPC-1 family of proteins transports lipophilic molecules across membranes. Genetic studies suggest that Ptc negatively regulates Hh signal transduction unless bound by Hh.
Sequence analysis suggests that Ptc is structurally similar to the RND family of channels and transporters (14), and evidence has been reported for Hh-sensitive repression of signal transduction being effected by a transporter function of Ptc either for sterols (15, 16) or for vitamin D3 (17). Transporter activity would be consistent with the lack-of-function phenotype of Ptc protein mutants with altered residues that are conserved in and are required for activity of bacterial RND transporters (9) or of NPC-1 (18, 19). One RND family transporter, the proton-driven Escherichia coli AcrB protein, is a homotrimer with a central pore (20), and recent evidence suggests that Ptc is also a trimer (8).
A number of distinct functions are also encoded in the Ptc CTD. Activation of the Hh pathway correlates with relocalization of Ptc from the plasma membrane to endosomes (21-24), movement that is dependent upon the CTD. The ptc13 mutant has a missense change in the CTD (E1172K) and a partial loss-of-function phenotype; its endosomal localization is decreased (19). Deletion of the CTD also affects localization and results in plasma membrane accumulation (8, 24). The CTD can form a homotrimer, and it contains a PPAY sequence, a putative target site for the Nedd4 ubiquitin ligase (8). CTD deletions are more stable than the wild-type protein (8, 9). Several mutations in the CTD of human Ptc1 have been identified in human cancers (25).
Drosophila has a single ptc gene, but most vertebrates have two, customarily named Patched1 (Ptch1) and Patched2 (Ptch2). In general, these gene designations reflect the relative order of discovery, not function or sequence subtype. This study describes a bioinformatics analysis with the goal of establishing a basis for categorizing the members of the Ptc protein family. Because the CTD sequences are significantly less well conserved than other Ptc domains (8), their variability was used to define two subtypes. One (Group I) includes the invertebrate Ptc proteins and the Ptch1 proteins of human, mouse, dog, frog, chick, zebrafish, monkey, and rat as well as the Ptch2 protein of zebrafish. Group I CTDs are characterized by having two conserved domains, a PPXY sequence, and a size between 205 and 279 residues. The CTDs of Group II proteins have abbreviated versions of the two conserved domains, lack a PPXY sequence, and are shorter, containing between 75 and 109 residues; they likely represent a novel mammalian divergence. We also show that the PPXY sequence in the C-terminal cytoplasmic tail of Ptch1 is responsible, at least in part, for rapid turnover. The Ptch2 protein is relatively stable; it lacks a PPXY sequence.
EXPERIMENTAL PROCEDURES
Gene Predictions—The amino acid sequences of Ptc orthologs were collected from the GenBank™ Data Bank. To expand the set of Ptc CTD sequences, genomic sequences were also accessed using the UCSC Genome Browser (26) and Fly-Base. Using these sources and tools, we gathered the genomic sequences of putative Ptc orthologs based upon their homology to ptc genes in human, mouse, and Drosophila melanogaster.
Homologous genomic regions, plus 3 kb of additional sequence on either end, were analyzed with three gene prediction programs: Genescan (27), Augustus (28), and GeneID (29). Results from the programs varied slightly, and choices between different gene predictions were made as follows. The predictions were aligned with the sequences of “evolutionary relatives”: D. melanogaster Ptc for insects and mouse and human Ptc1 and Ptc2 for vertebrates. Alignments were created using ClustalW, tCOFFEE (30), or MUSCLE Version 3.7 (31). Results were displayed in Jalview (32). Because introns were more frequently mistaken for exons than the reverse and because we are concerned primarily with the CTD, predictions were chosen to minimize false inserts within the tail region.
mRNA Extraction and cDNA Formation—Seven Drosophila species were analyzed: D. ananassae, D. mojavensis, D. persimilis, D. pseudoobscura, D. virilis, D. simulans, and D. yakuba. RNA was extracted from 15 third-instar larvae and fractionated by selection with oligo(dT), and first-strand cDNA was synthesized using the RETROscript® kit (Ambion). Mid-neurula stage Xenopus tropicalis embryos (stages 14-18) were from R. Harland (University of California, Berkeley, CA); zebrafish embryos (18-somite stage) were from S. Amacher (University of California, Berkeley). mRNA was purified with the RNeasy minikit (Qiagen Inc.). cDNAs were synthesized using ∼2.0 μg of total mRNA and the FirstChoice® RLM-RACE kit (Ambion). Canis familiaris and Rhesus monkey (Macaca mulatta) cDNAs were purchased from BioChain Institute.
Primer Design for Reverse Transcription-PCR—Primers were designed based upon predicted CTD sequences and were chosen to span ∼300 bp of intron. PCR-amplified cDNA was purified after separation by agarose gel electrophoresis. Primer sequences were as follows: D. ananassae, GTCAAGTTCGAGGGCTTT and ACTCGTAAAGTTGTAGCTGCG (Rev1); D. simulans, CCTGCCCAACTATCCATC and CTCTTGCTGCTGCGCAC; D. mojavensis and D. virilis, CAAGACACTGATTGGACACATAC and Rev1; D. pseudoobscura, CGACCTGAAGATACCCAAGA and Rev1; D. yakuba, CTCAGATGCCCTTTTACCTTC and Rev1; C. familiaris, TGCTGGGAGTGCTGATG and GTTGGAGCTACCGCCC; M. mulatta, GTTTTGCTTCCCGTGCTTT and GTTGGAGCTGCTTCCCC; Danio rerio, TGCTATTCCTGTGGTCATCCT (first RLM-RACE forward primer) and TGCCTCCTCCTATGAACCAC and CTATTTACCTAGCATCTGGTTAGCTTG (PCR primers); and X. tropicalis TTTGCTGTCCTGGCAATTTT (first RLM-RACE forward primer) and ATGATTCCCAGCAGTGTTCC and TTATTTTGAGGCATTTCCCTGT (PCR primers).
Amplifications were performed using the High Fidelity PCR Master (Roche Applied Science). For sequencing, PCR-amplified ptc fragments were cloned into the pCRII-topo vector (Invitrogen). Six individual colonies of each species were sequenced.
Expression Constructs—Mouse Ptch1 and Ptch2 were cloned into pAcGFP-C3 (Clontech), generating fusion proteins with an N-terminal green fluorescent protein: Ptch1C2, Ptch1-(1-1162)/Ptch2-(1113-1171); Ptch2C1, Ptch2-(1-1112)/Ptch1-(1163-1377); Ptch1ΔC, Ptch1-(1-1162); and Ptch14A, PPPY (positions 1299-1302) to AAAA.
Western Analysis—NIH 3T3 cells were cultured in high-glucose Dulbecco's modified Eagle's medium with 10% (v/v) bovine calf serum. Cells (40,000 cells/cm2) were seeded in 6-well plates, incubated for 18 h (to 90-95% confluence), and transfected with Lipofectamine 2000 (Invitrogen). After 48 h, cycloheximide (100 μm; Sigma) was added. After the indicated periods, cells were rinsed with phosphate-buffered saline; detached with 500 μl of 0.25% trypsin/EDTA; and after the addition of 500 μl of medium, collected by centrifugation at 7000 rpm for 5 min. Cells were rinsed with 500 μl of cold phosphate-buffered saline and lysed with 100 μl of cold lysis buffer (0.5% Nonidet P-40, 50 mm NaCl, 50 mm Tris (pH 7.5) and Complete protease inhibitor mixture (Roche Applied Science)) by pipetting ∼30 times. After 30 min of agitation on a rocker at 4 °C, extracts were centrifuged at maximum speed for 30 min, and 20 μl of sample was mixed with 5 μl of 5× Laemmli sample buffer and incubated for 15 min at 4 °C. Twenty μl of each sample was loaded onto a 6% SDS-polyacrylamide gel (2% polyacrylamide stacking gel) that had been pre-run for 20 min at 70 V.
Western blots were prepared on polyvinylidene difluoride membranes by semidry electrotransfer, blocked with 5% dry milk in TBST (0.08% Tween 20, 150 mm NaCl, and 20 mm Tris) for 30 min at room temperature, and incubated overnight at 4 °C with rabbit anti-green fluorescent protein antibody (1:1000 in 5% dry milk/TBST). After washing three times with 5% dry milk/TBST for 1 h, membranes were incubated for 1.5 h with horseradish peroxidase-conjugated goat anti-rabbit IgG (diluted 1:1000 in 5% dry milk/TBST). Membranes were washed twice with 5% dry milk/TBST for 30 min and rinsed with TBST for 10 min. Detection was with the ECL Plus™ Western blotting detection system (Amersham Biosciences). Blots were quantified with NIH Image J. The intensity of each construct was compared with the tubulin loading control, and the ratio at 0 h was set at 100%.
RESULTS
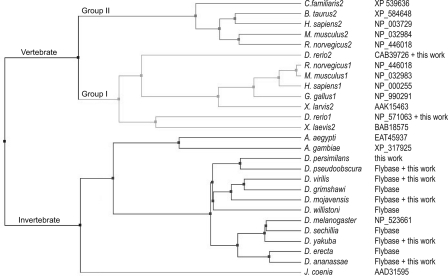
Conservation of Ptc Domains—Conservation of Ptc protein sequences during vertebrate and invertebrate evolution has been significant. To investigate how different regions of Ptc have either changed or been conserved, we assembled a group of invertebrate and vertebrate Ptc sequences for comparison. Thirteen sequences of vertebrate Ptc proteins were obtained from the NCBI Database (see Fig. 2). In addition, we prepared and partially sequenced Ptch cDNAs from C. familiaris, M. mulatta, and X. tropicalis; the total number of CTD sequences in our data set was 16. Thirteen sequences of invertebrate Ptc proteins were obtained from the NCBI and FlyBase Databases. We verified the Ptc CTD sequences for six of the Drosophila species (see Fig. 2).
FIGURE 2.
Phylogenetic tree of Ptc orthologs. The phylogenetic tree for the data set of vertebrate and invertebrate proteins was calculated using MUSCLE Version 3.7 (31) and Jalview Version 12.0.0 with default settings for alignment matrix and other parameters.
To estimate the general similarities of Ptc proteins, we first compared the sequences of D. melanogaster Ptc, which was the first Ptc protein identified, and human and mouse Ptch1 and Ptch2. Pairwise ClustalW comparisons of these proteins revealed the near identity of human and mouse Ptch1 proteins and of human and mouse Ptch2 proteins and the high degree of similarity (>50%) of these five proteins (Table 1). These alignments also revealed that the general topology of these proteins is similar, a result that is consistent with the similar distribution of predicted membrane-spanning domains in each protein (data not shown).
TABLE 1.
Similarity analysis of Ptc proteins
The calculated values of the percent similarity between full-length human (Homo sapiens), mouse (Mus musculus), and D. melanogaster Ptc proteins are given below.
| H. sapiens 1 | H. sapiens 2 | M. musculus 1 | M. musculus 2 | |
|---|---|---|---|---|
| H. sapiens 2 | 68 | |||
| M. musculus 1 | 97 | 61 | ||
| M. musculus 2 | 60 | 94 | 60 | |
| D. melanogaster | 51 | 51 | 51 | 51 |
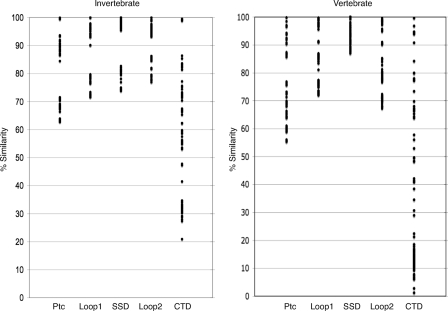
We extended this analysis to the entire data set and also focused on four domains: the predicted extracellular loops 1 and 2, the SSD, and the CTD. Pairwise comparisons of the full-length proteins and of four domains were made for both the invertebrate and vertebrate sequence data sets. These analyses revealed sequence similarities of >50% for the full-length proteins and >65% in both extracellular loops and the SSD. Variation in the CTDs was significantly higher (Fig. 1).
FIGURE 1.
Sequence variance in vertebrate and invertebrate Ptc proteins. The sequence similarity value of pairwise ClustalW alignments is plotted for the full-length proteins and for loop 1, the SSD, loop 2, and the CTD. Each circle represents a percent similarity value calculated for one of the pairwise comparisons among invertebrate (left) and vertebrate (right) proteins.
Vertebrate Ptch Proteins Have Either of Two CTD SubtypesFurther analysis of the vertebrate CTDs with ClustalW revealed two highly conserved subtypes that share >50% homology with each other (Table 2). The first group (Group I) includes dog, human, monkey, mouse, rat, and Xenopus laevis Ptch1 and X. tropicalis Ptc. Group II includes cow, dog, human, mouse, and rat Ptch2. The CTD sequences of X. laevis Ptch2 and zebrafish Ptch1 and Ptch2 have low homology to either group. Although we cannot evaluate why these sequences are so highly divergent (whether the differences are real or an artifact of isolation and cloning), as described below, the conservation they do have suggests that they should be included among the Group I orthologs.
TABLE 2.
Pairwise comparisons of vertebrate Ptch sequences
The percent values for sequence similarity between the CTDs in each Ptch protein in the data set are given below; values ≥50% are indicated in boldface.
| C. familiaris 1 | D. rerio 1 | D. rerio 2 | Gallus gallus 1 | H. sapiens 1 | M. mulatta | M. musculus 1 | Rattus norvegicus 1 | X. laevis 1 | X. laevis 2 | X. tropicalis | Bos taurus 2 | C. familiaris 2 | H. sapiens 2 | M. musculus 2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D. rerio 1 | 11 | ||||||||||||||
| D. rerio 2 | 17 | 31 | |||||||||||||
| G. gallus 1 | 75 | 11 | 18 | ||||||||||||
| H. sapiens 1 | 90 | 11 | 16 | 77 | |||||||||||
| M. mulatta | 91 | 11 | 16 | 78 | 97 | ||||||||||
| M. musculus 1 | 90 | 11 | 17 | 79 | 94 | 95 | |||||||||
| R. norvegicus 1 | 90 | 11 | 17 | 79 | 94 | 95 | 100 | ||||||||
| X. laevis 1 | 63 | 10 | 16 | 68 | 63 | 65 | 65 | 65 | |||||||
| X. laevis 2 | 36 | 16 | 12 | 37 | 37 | 38 | 38 | 38 | 37 | ||||||
| X. tropicalis | 49 | 2 | 3 | 53 | 48 | 50 | 50 | 50 | 67 | 29 | |||||
| B. taurus 2 | 13 | 17 | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 8 | ||||
| C. familiaris 2 | 11 | 12 | 9 | 11 | 12 | 12 | 12 | 12 | 10 | 11 | 8 | 76 | |||
| H. sapiens 2 | 13 | 19 | 15 | 13 | 13 | 13 | 14 | 14 | 14 | 13 | 8 | 85 | 73 | ||
| M. musculus 2 | 13 | 22 | 17 | 12 | 12 | 13 | 13 | 13 | 11 | 12 | 7 | 66 | 56 | 67 | |
| R. norvegicus 2 | 13 | 22 | 17 | 12 | 13 | 13 | 13 | 13 | 12 | 12 | 7 | 68 | 58 | 70 | 95 |
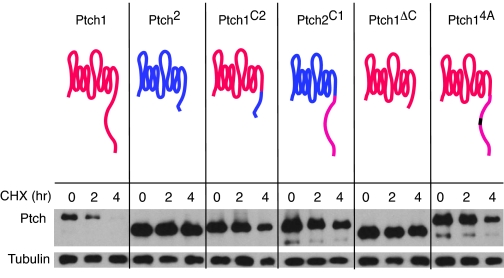
Two other features individuate these two groups (Table 3). First, Group I CTDs are significantly longer than Group II CTDs. With the exception of the two fish CTDs, all Group I CTDs exceed 205 residues. In contrast, none of the Group II CTDs is longer than 109 residues. Second, only the Group I CTDs include the PPXY sequence. In the D. melanogaster CTD, the PPXY residues have been implicated in the regulated turnover of the Ptc protein (8), and mutations in the PPXY sequence increase the protein half-life. CTD deletion mutants of both the D. melanogaster and mouse Ptc1 proteins also have reduced rates of degradation (Fig. 4) (8). The PPXY motif is not present in any of the Group II CTDs.
TABLE 3.
Characteristics of Group I and II CTDs
Listed are the number of residues in each CTD and the residue location of the PPXY sequence that is present in 9 of the 11 Group I proteins.
| Total length | PPXY | |
|---|---|---|
| Group I | ||
| C. familiaris 1 | 281 | 142 |
| D. rerio 1 | 60 | |
| D. rerio 2 | 58 | |
| G. gallus 1 | 272 | 139 |
| H. sapiens 1 | 276 | 142 |
| M. mulatta | 279 | 142 |
| M. musculus | 277 | 142 |
| R. norvegicus 1 | 277 | 142 |
| X. laevis 1 | 250 | 126 |
| X. laevis 2 | 262 | 176 |
| X. tropicalis | 205 | 72 |
| Group II | ||
| B. taurus 2 | 100 | |
| C. familiaris 2 | 109 | |
| H. sapiens 2 | 96 | |
| M. musculus 2 | 75 | |
| R. norvegicus 2 | 75 |
FIGURE 4.
The CTD regulates turnover of Ptch proteins. The stability of Ptch proteins in transfected NIH 3T3 cells was assessed by Western analysis of cells treated with cycloheximide (CHX) for the indicated times. Wild-type mouse Ptch1 and Ptch2, Ptch1 substituted with a Ptch2 CTD (Ptch1C2), Ptch2 substituted with a Ptch1 CTD (Ptch2C1), Ptch1 deleted of its CTD (Ptch1ΔC), and Ptch1 with the PPPY sequence in its CTD changed to AAAA (Ptch14A) are compared. Red, Ptch1 residues; blue, Ptch2 residues; black, mutated PPPY in CTD.
Conserved Sequences in the Ptc CTD—Phylogenetic relationships predicted by the relative degree of sequence similarity between Ptc proteins are consistent with a model in which Group I and II proteins represent distinct subtypes (Fig. 2). We generated phylogenetic trees by the method of “neighbor joining” based on BLOSUM62 for all of the Ptc proteins in our data set as well as for each of the four domains. Similar results were obtained for all five trees: the invertebrate Group I and II sequences were placed on separate branches in each calculation.
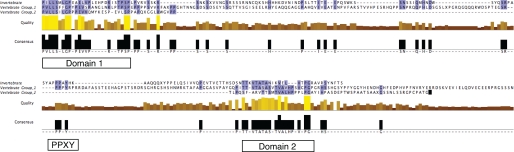
Sequence alignment performed by ClustalW, MUSCLE, and tCOFFEE identified two islands of significant conservation in the vertebrate Group I and II CTDs as well as the invertebrate CTDs. For purposes of illustration (Fig. 3), we used ClustalW to align vertebrate Group I and II CTDs and extracted a CTD “consensus” sequence for each type. We also generated an invertebrate CTD consensus sequence by aligning the D. melanogaster, Aedes aegypti, and Anopheles gambiae sequences. Fig. 3 presents an alignment of this “invertebrate CTD” consensus sequence with the consensus sequences of the vertebrate Group I and II CTDs. Two features of these alignments are noteworthy. First, the region of 30-40 residues at the immediate N terminus of the CTD is revealed as a domain of conservation in all three consensus sequences. We designated this region “Domain I.” Its conservation in the three sequences is >67% within the N-terminal 15 residues, and whereas the Group II homology does not extend further, the region of >50% conservation extends to 27 residues in the invertebrate Group I domains. Domain I of the zebrafish CTDs is the longer variety, consistent with a Group I designation. Second, “Domain II,” a more C-terminal region of conservation, extends for ∼21 residues and is also present in all three CTD types.
FIGURE 3.
Alignment of vertebrate Group I and II and invertebrate CTD sequences. Group I, Group II, and invertebrate CTD sequences were aligned using ClustalW and are displayed in Jalview, a multiple alignment editor that identifies conserved residues (blue, top rows); assesses “quality” of conservation (middle rows) by analyzing the proportion of identical conservative changes and non-conservation (scaled high (yellow), intermediate (brown), and low (black)); and defines an overall “consensus” (bottom rows). Islands of high sequence conservation (Domains I and II) and the PPPY sequence are indicated by boxes.
Role of the CTD in Regulating Turnover of Ptc1 and Ptc2The CTD of the D. melanogaster Ptc protein regulates both turnover and internalization (8). The wild-type protein is unstable, with a half-life of <2 h. Its CTD contains a potential target sequence (PPAY) for the Nedd4 ubiquitin ligase, and mutant protein that lacks either the C-terminal half of the CTD or the PPAY sequence is significantly more stable (8). Because our comparative sequence analysis (above) revealed that Group I Ptc proteins have a potential neddylation sequence (PPPY) but Group II proteins do not, we investigated whether the PPPY sequence in vertebrate Ptc proteins has a role in regulating turnover.
As shown in Fig. 4, Ptch1 was unstable (t½ = 1.7 h), whereas Ptch2 was stable (t½ > 18 h). Deleting the CTD of Ptch1 stabilized the protein (t½ > 17 h); swapping the CTDs of Ptch1 and Ptch2 stabilized Ptch1 (t½ = 5.8 h) and destabilized Ptch2 (t½ = 6.2 h). These properties indicate that the CTDs of Group I and II proteins help to determine lifetime. Mutating the PPPY sequence in the Ptch1 CTD to AAAA stabilized Ptch1 (t½ > 9 h). We conclude that the CTD of the vertebrate Ptc proteins plays a major role in regulating stability and that the PPPY sequence is an essential component of the mechanism that controls Ptc1 turnover.
DISCUSSION
The Ptc protein family is characterized by shared topology and high sequence conservation. All members are predicted to have 12 membrane-spanning domains, an SSD, two large extracellular loops, and a C-terminal cytoplasmic tail. Several lines of evidence suggest that two subtypes of Ptc protein compose this family. This evidence includes predicted phylogenetic relationships among the family members as well as separate comparisons of sequence similarities within extracellular loops 1 and 2, the SSD, and the CTD. Most Ptc proteins, including the invertebrate orthologs, are of the Group I type. Although the known family members represent products from the small and selected set of genomes that were available to us, it bears noting that Group II orthologs have been identified only in mammals. It follows that the Group II subtype may be unique to mammals and may represent a divergence in that lineage. Whereas it is clear that Group I proteins function as tumor suppressors and as key negative regulators of Hedgehog signal transduction, the function of the Group II proteins has not been elucidated. Studies of mouse Ptch2 mutants suggest that this Group II protein modulates tumorigenesis associated with Ptch1 haploinsufficiency (33), but how Ptch2 affects Ptch2 activity or downstream functions is not known.
Although sequence variance in the CTD is significantly greater than in other domains, the islands of conservation in it may represent regions with conserved function. We designated these regions Domain I, Domain II, and PPXY, and available genetic evidence suggests that all three are functionally important. Mutations in the D. melanogaster PPXY sequence increase the protein half-life (8), suggesting that this sequence motif functions to limit the lifetime of Ptc protein. In this work, we have shown that the PPPY sequence in mouse Ptch1 has a similar function. It is interesting to speculate that Group II proteins may not be regulated by the same mechanism and that the absence of the PPXY motif might indicate a different role for this protein. Domain I may also be critical to Ptch function. Among 12 CTD mutations that are listed in the PTCH Mutation Database of human mutations (25), three (T1195I, P1196S, and S1197F) alter conserved residues in Domain I. These three mutations were identified in patients with basal cell carcinoma, suggesting that these mutant proteins may not respond to Hh or its absence in a normal way. Presumably, the pathway is abnormally active in these mutant backgrounds, and given the negative role of Ptc, these mutations might decrease Ptc activity either by reducing the half-life of the protein or by affecting its localization. Although Domain II mutations in human Ptc have not been reported, studies of the D. melanogaster CTD with mutations in Domain II have revealed that Domain II is essential for self-association of the CTD (8). It remains for future work to investigate whether the Domain II sequences in vertebrate Group I and II proteins contribute a similar function.
Acknowledgments
We thank members of the Kornberg laboratory and Patricia Babbitt for technical help and comments on the manuscript, Michiel Bagnat for technical advice, and Bruce Macher, Laura Burrus, Ravinder Sehgal, and Diana Chu (San Francisco State University) for guidance and support.
This work was supported, in whole or in part, by National Institutes of Health Grant GM077407 (to T. B. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CTD, C-terminal domain; SSD, sterol-sensing domain; RLM-RACE, RNA ligase-mediated rapid amplification of cDNA ends.
References
- 1.Hahn, H., Wicking, C., Zaphiropoulous, P. G., Gailani, M. R., Shanley, S., Chidambaram, A., Vorechovsky, I., Holmberg, E., Unden, A. B., Gillies, S., Negus, K., Smyth, I., Pressman, C., Leffell, D. J., Gerrard, B., Goldstein, A. M., Dean, M., Toftgard, R., Chenevix-Trench, G., Wainwright, B., and Bale, A. E. (1996) Cell 85 841-851 [DOI] [PubMed] [Google Scholar]
- 2.Johnson, R. L., Rothman, A. L., Xie, J., Goodrich, L. V., Bare, J. W., Bonifas, J. M., Quinn, A. G., Myers, R. M., Cox, D. R., Epstein, E. H., Jr., and Scott, M. P. (1996) Science 272 1668-1671 [DOI] [PubMed] [Google Scholar]
- 3.Carpenter, D., Stone, D. M., Brush, J., Ryan, A., Armanini, M., Frantz, G., Rosenthal, A., and de Sauvage, F. J. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 13630-13634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marigo, V., Davey, R. A., Zuo, Y., Cunningham, J. M., and Tabin, C. J. (1996) Nature 384 176-179 [DOI] [PubMed] [Google Scholar]
- 5.Stone, D. M., Hynes, M., Armanini, M., Swanson, T. A., Gu, Q., Johnson, R. L., Scott, M. P., Pennica, D., Goddard, A., Phillips, H., Noll, M., Hooper, J. E., de Sauvage, F., and Rosenthal, A. (1996) Nature 384 129-134 [DOI] [PubMed] [Google Scholar]
- 6.Fuse, N., Maiti, T., Wang, B., Porter, J. A., Hall, T. M., Leahy, D. J., and Beachy, P. A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 10992-10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingham, P. W., Taylor, A. M., and Nakano, Y. (1991) Nature 353 184-187 [DOI] [PubMed] [Google Scholar]
- 8.Lu, X., Liu, S., and Kornberg, T. B. (2006) Genes Dev. 20 2539-2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taipale, J., Cooper, M. K., Maiti, T., and Beachy, P. A. (2002) Nature 418 892-897 [DOI] [PubMed] [Google Scholar]
- 10.Briscoe, J., Chen, Y., Jessell, T. M., and Struhl, G. (2001) Mol. Cell 7 1279-1291 [DOI] [PubMed] [Google Scholar]
- 11.Carstea, E. D., Morris, J. A., Coleman, K. G., Loftus, S. K., Zhang, D., Cummings, C., Gu, J., Rosenfeld, M. A., Pavan, W. J., Krizman, D. B., Nagle, J., Polymeropoulos, M. H., Sturley, S. L., Ioannou, Y. A., Higgins, M. E., Comly, M., Cooney, A., Brown, A., Kaneski, C. R., Blanchette-Mackie, E. J., Dwyer, N. K., Neufeld, E. B., Chang, T. Y., Liscum, L., Strauss, J. F., III, Ohno, K., Zeigler, M., Carmi, R., Sokol, J., Markie, D., O'Neill, R. R., van Diggelen, O. P., Elleder, M., Patterson, M. C., Brady, R. O., Vanier, M. T., Pentchev, P. G., and Tagle, D. A. (1997) Science 277 228-231 [DOI] [PubMed] [Google Scholar]
- 12.Hua, X., Nohturfft, A., Goldstein, J. L., and Brown, M. S. (1996) Cell 87 415-426 [DOI] [PubMed] [Google Scholar]
- 13.Osborne, T. F., and Rosenfeld, J. M. (1998) Curr. Opin. Lipidol. 9 137-140 [DOI] [PubMed] [Google Scholar]
- 14.Tseng, T. T., Gratwick, K. S., Kollman, J., Park, D., Nies, D. H., Goffeau, A., and Saier, M. H., Jr. (1999) J. Mol. Microbiol. Biotechnol. 1 107-125 [PubMed] [Google Scholar]
- 15.Corcoran, R. B., and Scott, M. P. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8408-8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dwyer, J. R., Sever, N., Carlson, M., Nelson, S. F., Beachy, P. A., and Parhami, F. (2007) J. Biol. Chem. 282 8959-8968 [DOI] [PubMed] [Google Scholar]
- 17.Bijlsma, M. F., Spek, C. A., Zivkovic, D., van de Water, S., Rezaee, F., and Peppelenbosch, M. P. (2006) PLoS Biol. 4 e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, V., Carrillo, G., Torroja, C., and Guerrero, I. (2001) Curr. Biol. 11 601-607 [DOI] [PubMed] [Google Scholar]
- 19.Strutt, H., Thomas, C., Nakano, Y., Stark, D., Neave, B., Taylor, A. M., and Ingham, P. W. (2001) Curr. Biol. 11 608-613 [DOI] [PubMed] [Google Scholar]
- 20.Murakami, S., Nakashima, R., Yamashita, E., and Yamaguchi, A. (2002) Nature 419 587-593 [DOI] [PubMed] [Google Scholar]
- 21.Capdevila, J., Pariente, F., Sampedro, J., Alonso, J. L., and Guerrero, I. (1994) Development (Camb.) 120 987-998 [DOI] [PubMed] [Google Scholar]
- 22.Denef, N., Neubuser, D., Perez, L., and Cohen, S. M. (2000) Cell 102 521-531 [DOI] [PubMed] [Google Scholar]
- 23.Incardona, J. P., Gruenberg, J., and Roelink, H. (2002) Curr. Biol. 12 983-995 [DOI] [PubMed] [Google Scholar]
- 24.Zhu, A. J., Zheng, L., Suyama, K., and Scott, M. P. (2003) Genes Dev. 17 1240-1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindstrom, E., Shimokawa, T., Toftgard, R., and Zaphiropoulos, P. G. (2006) Hum. Mutat. 27 215-219 [DOI] [PubMed] [Google Scholar]
- 26.Kent, W. J., Baertsch, R., Hinrichs, A., Miller, W., and Haussler, D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11484-11489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burge, C., and Karlin, S. (1997) J. Mol. Biol. 268 78-94 [DOI] [PubMed] [Google Scholar]
- 28.Stanke, M., Schoffmann, O., Morgenstern, B., and Waack, S. (2006) BMC Bioinformatics 7 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parra, G., Blanco, E., and Guigo, R. (2000) Genome Res. 10 511-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Notredame, C., Higgins, D. G., and Heringa, J. (2000) J. Mol. Biol. 302 205-217 [DOI] [PubMed] [Google Scholar]
- 31.Edgar, R. C. (2004) Nucleic Acids Res. 32 1792-1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clamp, M., Cuff, J., Searle, S. M., and Barton, G. J. (2004) Bioinformatics (Oxf.) 20 426-427 [DOI] [PubMed] [Google Scholar]
- 33.Lee, Y., Miller, H. L., Russell, H. R., Boyd, K., Curran, T., and McKinnon, P. J. (2006) Cancer Res. 66 6964-6971 [DOI] [PubMed] [Google Scholar]