Abstract
Samples from infected root canals of 43 teeth with chronic apical periodontitis were analyzed for the presence and relative levels of 83 oral bacterial species and/or phylotypes using a reverse-capture checkerboard hybridization assay. Associations between the most frequently detected taxa were also recorded. The most prevalent taxa were Olsenella uli (74%), Eikenella corrodens (63%), Porphyromonas endodontalis (56%), Peptostreptococcus anaerobius (54%), and Bacteroidetes oral clone X083 (51%). When prevalence was considered only for bacteria present at levels >105, Bacteroidetes clone X083 was the most frequently isolated bacterium (37%), followed by Parvimonas micra (28%), E. corrodens (23%), and Tannerella forsythia (19%). The number of target taxa per canal was directly proportional to the size of the apical periodontitis lesion, with lesions >10 mm in diameter harboring a mean number of approximately 20 taxa. Several positive associations for the most prevalent taxa were disclosed for the first time and may have important ecological and pathogenic implications. In addition to strengthening the association of several cultivable named species with chronic apical periodontitis, the present findings using a large-scale analysis allowed the inclusion of some newly named species and as-yet-uncultivated phylotypes in the set of candidate pathogens associated with this disease.
Chronic apical periodontitis is arguably one of the most common forms of biofilm-induced diseases that affect humans (9). The disease develops after dental pulp necrosis and infection as a result of caries, trauma, or iatrogenic clinical procedures. The environmental conditions in the necrotic root canal are conducive to the establishment of a microbiota conspicuously dominated by anaerobic bacteria. Bacterial profiles of the endodontic microbiota vary from individual to individual (37); i.e., each individual harbors a unique microbiota in terms of species richness and abundance. This indicates that apical periodontitis has a heterogeneous etiology, where no single species can be considered to be the main endodontic pathogen, and multiple bacterial combinations can play a role in disease causation.
Early studies of the microbiota associated with apical periodontitis were conducted using broad-range culture methods. Those studies were followed by a generation of studies employing molecular detection methods such as species-specific PCR and the original checkerboard DNA-DNA hybridization assay to target cultivable bacteria previously isolated from infected canals or from other oral diseased sites. These methods allowed the inclusion of some culture-difficult species in the set of candidate endodontic pathogens. The adoption of 16S rRNA gene clone library analysis allowed an even more comprehensive broad-range investigation of bacterial communities in endodontic infections. By this technique, not only cultivable species but also as-yet-uncultivated and uncharacterized bacteria can be identified. Studies using the 16S rRNA gene clone library analysis have revealed that 40 to 55% of the bacterial taxa found in primary endodontic infections have not been cultivated and validly named (20, 30). However, technical difficulties and high cost can make it difficult to analyze a large number of samples by this method. Cataloging bacterial species in the oral cavity by clone libraries provides 16S rRNA gene sequence data that can be used to design oligonucleotide probes or primers to target both cultivable and as-yet-uncultivated bacteria. Primers are used in PCR assays, which are, however, restricted by the need to perform several individual reactions to survey several samples for the presence of several species and phylotypes. Probes can be used in molecular biology techniques suitable for large-scale clinical studies, including the reverse-capture checkerboard hybridization assay.
The present study was undertaken to evaluate the presence and relative levels of 83 bacterial taxa in necrotic root canals of teeth with chronic apical periodontitis by using a reverse-capture checkerboard hybridization assay. Target taxa for investigation included cultivable species previously linked to endodontic infections as well as newly characterized species and as-yet-uncultivated phylotypes that have been recently detected in clone libraries from periodontal (19, 22) and endodontic (20, 30) infections. Some of them were never previously found in infected root canals, and others have never been tested against a large number of samples. Associations between the most frequently detected taxa were also calculated.
MATERIALS AND METHODS
Subjects and sample collection.
Ethical approval for the study was granted by the Ethics Committee of the Estácio de Sá University, Rio de Janeiro, Brazil. Root canal samples were taken from 43 patients presenting to the endodontic clinic at Estácio de Sá University for evaluation and treatment of apical periodontitis. Only single-rooted teeth from adult patients older than 22 years of age, all of them having necrotic pulps and radiographic evidence of apical periodontitis lesion, were included in this study. The size of each lesion was calculated by taking the average of the lesions' largest dimension and the extent in the direction perpendicular to the largest dimension. All cases were asymptomatic at the time of treatment, seven of which had associated sinus tract. Selected teeth showed an absence of periodontal pockets deeper than 4 mm.
Samples were taken from the necrotic root canals under strict aseptic conditions and after a two-step disinfection protocol of the operative field with 2.5% NaOCl as previously described (37). Endodontic files with the handle cut off and paper points used for sampling of the canals were transferred into cryotubes containing TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.6]) and immediately frozen at −20°C. Furthermore, samples were brought to room temperature, and DNA was extracted using the QIAamp DNA Mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. DNA from a panel of several oral bacterial species was also prepared to serve as controls (39).
Oligonucleotide probes.
Six taxon-specific probes and two universal probes were described in previous studies (3, 5). The other probes used in this study were designed as follows: 16S rRNA gene sequences of each of the target bacteria were retrieved from the GenBank database and aligned with the sequences of their nearest neighbors in the phylogenetic tree. Potential probes with a melting temperature of approximately 51°C to 52°C were designed from variable areas. A BLAST-based algorithm (1) was then used to verify their uniqueness. Probes were synthesized with multiple thymidines at the 5′ end and were tested against purified DNA from the panel of oral species. No cross-reactions were observed for the probes used in this study, except for a TM7 group-specific probe (5′-CCC GTC AAT TCC TTT ATG TTT TA-3′), which was excluded from the panel. Probe sequences are depicted in Appendix S1 in the supplemental material.
Checkerboard hybridization assay.
The PCR-based reverse-capture checkerboard assay was carried out as described previously by Paster et al. (21), with some modifications. Initially, whole-genomic DNA extracts from clinical samples were used as templates in a 16S rRNA gene-based PCR protocol consisting of two steps. First, a practically full-length 16S rRNA gene fragment was amplified from 5 μl of the DNA extracts using universal primers 8f and 1492r. Next, 1 μl of the resulting PCR product from each sample was used to run two sets of partial 16S rRNA gene amplification, one using primers digoxigenin-8f and 519r and the other using primers digoxigenin-515f and 1492r. Primers 8f and 1492r were described previously by Paster et al. (21), while primers 519r and 515f were modified from the sequences reported previously by Hutter et al. (16) to accommodate a broader range of oral species and/or phylotypes. Therefore, two different fragments, which together encompass the nearly full-length 16S rRNA gene, were obtained for each sample. This two-step heminested approach was used to achieve a better performance of PCR, particularly for samples with low numbers of bacteria (25). Since three different checkerboard runs had to be performed for each sample (only 30 probes fit on each checkerboard), the first PCR products were used as templates for three subsequent sets of labeled heminested amplification with the two primer pairs. Use of the same PCR product for the subsequent labeled reactions reduced the variation in the samples (8).
All PCR amplifications were performed in a 50-μl reaction mixture containing a 1 μM concentration of each primer, 5 μl of 10× PCR buffer, 3 mM MgCl2, 2 U of Tth DNA polymerase, and 0.2 mM of each deoxyribonucleoside triphosphate (all reagents were from Biotools, Madrid, Spain). Negative controls consisting of sterile ultrapure water instead of sample were included with each batch of samples analyzed.
The temperature profile for the first PCR using primers 8f and 1492r was 95°C for 1 min; 26 cycles at 94°C for 45 s, 50°C for 45 s, and 72°C for 1.5 min; and 72°C for 15 min. Cycling conditions for the second round of amplification using primer pair digoxigenin-8f/519r or digoxigenin-515f/1492r included 95°C for 5 min and 28 cycles at 94°C for 30 s, 55°C for 1 min, 72°C for 1.5 min, and 72°C for 20 min. Amplicons were separated by electrophoresis in agarose gels and viewed under UV transillumination.
Labeled PCR products obtained with primer pairs 8f/519r and 515f/1492r were mixed using equal proportions of each one (45 μl) and used in the checkerboard assay to determine the presence and levels of 83 bacterial taxa. 16S rRNA gene probes were synthesized with multiple thymidines at the 5′ end. Two lanes in each membrane contained standards at 105 and 106 cells, which were treated the same way as the clinical samples. Probes were randomly distributed along three different membranes. Each membrane shared the two universal probes, which served as controls. A total of 1,350 hybridizations can be performed simultaneously using a single membrane. Overall, 3,569 hybridizations were carried out in this study, excluding the universal probes.
The checkerboard assay was performed using the Minislot-30 and Miniblotter-45 system (Immunetics, Cambridge, MA) (3, 8, 21, 25). First, 100 pmol of probe in Tris-EDTA buffer (10 mM Tris HCl, 1 mM EDTA [pH 8.0]) was introduced into the horizontal wells of the Minislot apparatus and cross-linked to the Hybond N+ nylon membrane (Amersham Pharmacia Biotech, Buckinghamshire, England) by UV irradiation using a Stratalinker 1800 apparatus (Stratagene, La Jolla, CA) on the auto-cross-link position. The polythymidine tails are preferentially cross-linked to the nylon, leaving the specific probe available for hybridization (21). The membrane was then prehybridized at 55°C for 1 h. Subsequently, 90 μl of the labeled PCR products with 50 μl of 55°C preheated hybridization solution was denatured at 95°C for 5 min and loaded onto the membrane using the Miniblotter apparatus. Hybridization was carried out at 54°C for 2 h.
After hybridization, the membrane was washed and blocked in a buffer with casein. The membrane was sequentially incubated in antidigoxigenin antibody conjugated with alkaline phosphatase (Roche Molecular Biochemicals, Mannheim, Germany) and ultrasensitive chemiluminescent substrate CDP Star (Roche Molecular Biochemicals). Finally, a square of X-ray film was exposed to the membrane in a cassette for 20 min in order to detect the hybrids.
Data analysis.
The prevalence of the target species and/or phylotypes was recorded as the percentage of cases examined. The obtained chemiluminescent signals were also evaluated using ImageJ (http://rsb.info.nih.gov/ij/) and converted to counts by comparison with standards at known concentrations run on each membrane. Because of the recognized difficulties in inferring absolute counts for PCR-amplified samples and because estimates had to be made for counting as-yet-uncultivated phylotypes, counts were transformed into semiquantitative data and categorized as follows: a level below detection (or absence), a level of <105 bacteria, a level of 105 bacteria, a level of >105 to <106 bacteria, a level of 106 bacteria; and a level of >106 bacteria. A heavy infection was considered to be levels of >105.
Relative risk (RR) with a 95% confidence interval was used to examine pairs of the most prevalent taxa for associations. RR calculations were based on the detection of the target taxa in each subject, irrespective of their proportional recovery in samples. Moreover, data on the presence and absence of taxa found in at least five samples were used for the calculation of Euclidean distances, percent disagreement, or Pearson coefficient and then subjected to cluster analysis by Ward's method or the unweighted-pair group method using arithmetic averages to determine bacterial associations in complexes.
RESULTS
All sample extracts were positive for PCR amplification using the broad-range 16S rRNA gene primers, indicating that bacteria were present in all examined samples and that significant inhibitors of the PCR were not present. Negative controls yielded no amplicons.
The results of the reverse-capture checkerboard analysis revealed that 72 of the 83 oligonucleotide probes tested were reactive with one or more clinical samples. All the 43 samples were positive for at least two taxon-specific probes. The number of taxa per infected canal ranged from 2 to 45 (mean, 14.3; median, 12). Root canals of teeth with lesions <5 mm in diameter (n = 14) harbored a mean number of 11.7 taxa. Canals of teeth with lesions that were 5 to <10 mm in diameter (n = 15) showed a mean number of 16 taxa, while the canals associated with lesions larger than 10 mm (n = 14) presented a mean number of 19.9 taxa. Three canals harbored more than 40 taxa, and they were all associated with lesions ≥10 mm in diameter. Canals of teeth with sinus tracts exhibited a mean number of 16.7 taxa.
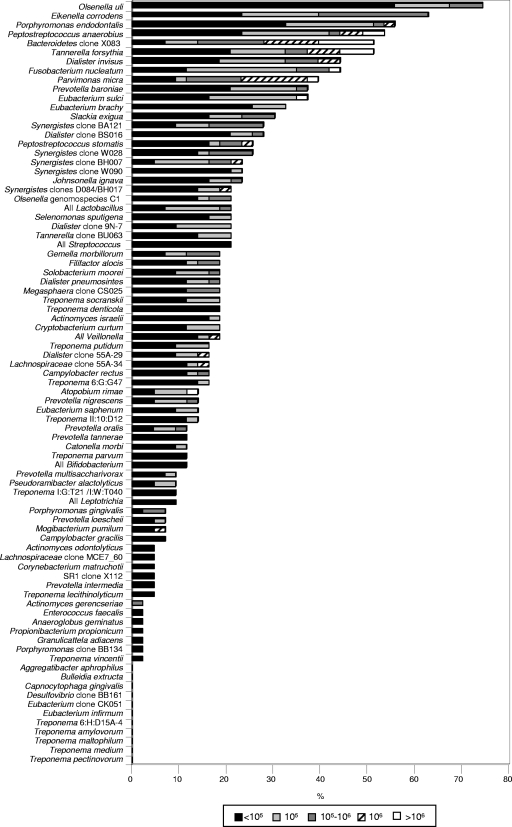
Taxa detected more frequently included Olsenella uli (32/43 cases [74%]), Eikenella corrodens (27/43 [63%]), Porphyromonas endodontalis (24/43 [56%]), Peptostreptococcus anaerobius (23/43 [54%]); Bacteroidetes oral clone X083 (22/43 [51%]), Tannerella forsythia (22/43 [51%]), Dialister invisus (19/43 [44%]), and Fusobacterium nucleatum (19/43 [44%]). Prevalence values for the 83 taxa are depicted in Fig. 1.
FIG. 1.
Stacked bar chart of frequency of detection and levels of bacterial species and phylotypes in root canal samples of teeth with chronic apical periodontitis from 43 individuals. The total length of each bar stack indicates the percentage of positive samples, i.e., the prevalence of bacterial species and phylotypes. Different shades within each bar indicate the percentages of samples containing different levels of the species.
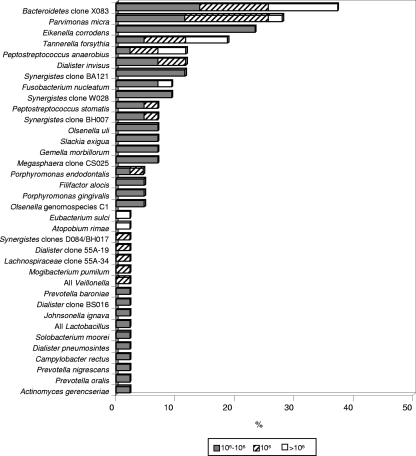
The mean number of bacterial taxa present at high counts (>105) in infected canals ranged from 0 to 10 (mean, 2.6; median, 2). Thirty-five cases showed at least one target taxon at levels above 105. Of the 36 taxa detected at levels >105, the most frequently found ones were Bacteroidetes oral clone X083 (16/43 [37%]), Parvimonas micra (12/43 [28%]), E. corrodens (10/43 [23%]), T. forsythia (8/43 [19%]), P. anaerobius (5/43 [12%]), D. invisus (5/43 [12%]), and Synergistes oral clone BA121 (5/43 [12%]). Figure 2 depicts the prevalences of bacterial taxa encountered at levels >105.
FIG. 2.
Stacked bar chart of frequency of detection of bacterial species and phylotypes in root canal samples at levels above 105. Different shades within each bar indicate the percentages of samples containing different levels of the species.
Several pairs of bacterial taxa were positively associated (RR > 1). Strong positive associations occurred between O. uli and Eubacterium sulci; Bacteroidetes clone X083 and P. micra, Prevotella baroniae, or P. anaerobius; and T. forsythia and E. sulci or P. micra. P. micra and P. baroniae were positively related to all the other taxa except F. nucleatum. T. forsythia was the only species that showed positive associations with all the other most prevalent taxa. Other positive associations are shown in Table 1.
TABLE 1.
Bacterial associations in infected root canals of teeth with chronic apical periodontitis as determined by RR calculation
| Bacterium | RR (95% confidence interval)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. corrodens | P. endodontalis | P. anaerobius | Bacteroidetes clone X083 | T. forsythia | D. invisus | F. nucleatum | P. micra | P. baroniae | E. sulci | |
| O. uli | 1.0 (0.7-1.8) | 1.0 (0.6-2.1) | 1.2 (0.7-2.7) | 1.5 (0.8-3.8) | 2.2 (0.9-6.3) | 1.3 (0.6-3.3) | 1.8 (0.8-5.4) | 1.6 (0.7-4.8) | 1.5 (0.6-4.5) | 5.2 (1.2-30.0) |
| E. corrodens | 1.0 (0.6-1.8) | 2.8 (1.3-6.8) | 1.0 (0.6-2.0) | 1.6 (0.8-3.3) | 1.0 (0.5-2.1) | 1.0 (0.5-2.1) | 1.4 (0.7-3.4) | 1.3 (0.6-3.2) | 1.8 (0.8-4.7) | |
| P. endodontalis | 0.7 (0.4-1.3) | 1.4 (0.8-2.6) | 1.4 (0.8-2.6) | 1.4 (0.7-2.8) | 1.1 (0.6-2.2) | 1.9 (0.9-4.5) | 2.4 (1.0-6.3) | 1.0 (0.5-2.3) | ||
| P. anaerobius | 3.0 (1.5-6.1) | 1.5 (0.8-2.8) | 1.0 (0.5-1.9) | 1.0 (0.5-1.9) | 2.1 (0.9-4.9) | 1.4 (0.7-3.3) | 1.4 (0.7-3.3) | |||
| Bacteroidetes clone X083 | 1.7 (0.9-3.1) | 1.6 (0.8-3.3) | 0.7 (0.4-1.4) | 4.4 (1.7-13.0) | 4.1 (1.6-12.3) | 1.0 (0.4-2.1) | ||||
| T. forsythia | 1.3 (0.7-2.6) | 1.6 (0.8-3.3) | 3.1 (1.3-7.9) | 2.1 (0.9-5.0) | 4.1 (1.6-12.3) | |||||
| D. invisus | 0.9 (0.5-1.8) | 1.1 (0.5-2.3) | 1.3 (0.6-2.7) | 1.0 (0.4-2.1) | ||||||
| F. nucleatum | 0.5 (0.2-1.2) | 0.6 (0.2-1.3) | 2.8 (1.2-6.4) | |||||||
| P. micra | 2.5 (1.2-5.4) | 1.5 (0.7-3.2) | ||||||||
| P. baroniae | 1.3 (0.6-2.7) | |||||||||
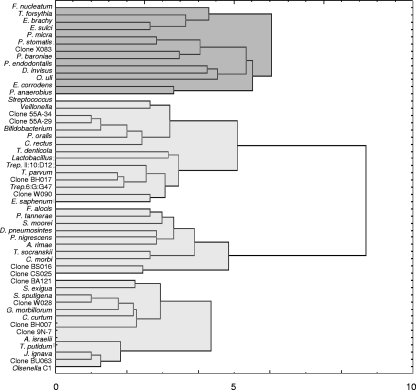
Cluster analysis using different distance measures and linkage techniques revealed four clusters, with only minor differences among the different methods. The Ward's method with Euclidean distances was selected to display the results (Fig. 3). A major cluster composed almost exclusively of the most prevalent taxa stood out against the other three. Several of the positive associations observed in RR calculations were confirmed in this broader analysis.
FIG. 3.
Dendrogram of a cluster analysis of 51 bacterial taxa present in primary root canal infections of teeth with chronic apical periodontitis using the Euclidean distance calculation and Ward's method. The bacterial cluster on top (dark gray) is composed almost exclusively of the most prevalent taxa.
DISCUSSION
Although the original checkerboard approach has already been used in endodontic microbiology research (4, 11, 32, 38, 39, 42), this is the first study to use the reverse-capture checkerboard assay to investigate the presence and relative levels of a large panel of target taxa in infected root canals of teeth with chronic apical periodontitis. This approach has important advantages over the original checkerboard method. One advantage relates to the PCR amplification of samples previously to hybridization, which increases the method's sensitivity compared to that of the original assay, even though the recent introduction of multiple-displacement amplification may have overcome the problem of the low sensitivity of the original checkerboard (4). In fact, the most significant advantages are related to the use of oligonucleotide probes instead of whole genomic probes. Oligonucleotide probes display higher specificity, since all the probe sequences in a panel can be designed to have similar melting temperature values. This allows a stringent hybridization temperature to be used for all probes in an individual membrane (21). Moreover, mismatches are not tolerated due to the considerable reduction of bond strength between short probes and the target (17). Oligonucleotide probes still have the advantage that they can be designed to detect both cultivable and as-yet-uncultivated bacteria, while in the original checkerboard method with whole genomic probes, only cultivable species are targeted.
In the present study, several cultivable species were among the most frequently detected taxa, including O. uli, E. corrodens, P. endodontalis, and P. anaerobius. O. uli was present in about three-fourths of the samples, indicating that this species is a very common member of the microbiota associated with chronic apical periodontitis. O. uli has only recently been recognized as a member of the endodontic microbial consortium of teeth with apical periodontitis (6, 20, 34). E. corrodens has been detected in endodontic infections mainly by molecular methods (28), and the frequency at which this species was found in this study is probably the highest ever reported for E. corrodens in endodontic infections. Even when presence at high levels was regarded, E. corrodens was one of the most prevalent taxa. P. endodontalis has been found in endodontic infections by culture (14), but its association with apical periodontitis has been strengthened by findings from molecular studies, whereby the highest prevalence values have been reported (10, 12, 35). In this study, P. endodontalis was found in about one-half of the infected canals. In spite of being among the most prevalent taxa, both O. uli and P. endodontalis were usually found at low levels and consequently not as the dominant taxa in the mixed consortium. Although the low levels of infection may apparently put into question a pathogenic role for these taxa in chronic apical periodontitis, at least an ecological role in the mixed consortium cannot be disregarded.
Other cultivable species frequently encountered in this study included P. anaerobius, T. forsythia, F. nucleatum, and P. micra. P. anaerobius has often been reported to occur in infected root canals (20, 43). In the present study, this species was found in 23 cases, 5 of which showed it as being one of the dominant taxa. The important periodontal pathogen T. forsythia was previously detected in endodontic infections only by molecular biology techniques (7, 33, 38), and the high prevalence observed in the present study, in many cases at high counts, confirms the species association with apical periodontitis. F. nucleatum and P. micra have long been recognized as putative endodontic pathogens by studies using culture and molecular methods (10, 13, 38, 43).
Some newly named species, such as P. baroniae, D. invisus, and Peptostreptococcus stomatis, were also found at somewhat high frequencies. Interestingly, Prevotella baroniae was the most prevalent of the seven Prevotella species targeted in this study. Prevotella baroniae is likely a synonym with Prevotella sp. clones PUS9.180 and E9_42-E4 and has been recently linked to acute apical abscesses (18, 30). D. invisus was originally found in infected root canals of teeth with chronic apical periodontitis (20). Findings for this species are in line with findings from several other recent studies that demonstrated the presence of D. invisus in association with both asymptomatic and symptomatic endodontic infections (24, 27, 29, 34). P. stomatis (or Peptostreptococcus sp. clone CK035) is a newly named species that had been previously detected in infected canals only by studies using clone library analysis (29-31). This is the first study evaluating the occurrence of this species in a large number of root canal samples. P. stomatis was detected in about one-fourth of the cases and as one of the most dominant taxa in the community in some of them.
Treponema species are examples of culture-difficult bacteria that have been identified in endodontic infections only by molecular methods (2, 26, 38). All 10 cultivable and 4 as-yet-uncharacterized oral treponemes were targeted in this study. Of the nine treponemes detected, T. denticola and T. socranskii were the most prevalent species, which is in agreement with data from previous studies (2, 26). The other most frequently found Treponema species and/or phylotypes included Treponema sp. strain 6:G:G47 (16%), T. putidum (16%), Treponema sp. strain II:10:D12 (14%), and T. parvum (12%). No Treponema species was found at levels >105.
Because clone libraries from endodontic infections reveal that approximately one-half of the detected taxa still remain to be cultivated and characterized, a comprehensive analysis of the microbiota involved with different forms of apical periodontitis should include these bacteria (20, 30). An initial and essential step toward the recognition of as-yet-uncultivated bacteria as candidate endodontic pathogens is evaluating their prevalence in a large number of diseased cases to look for association. Ten of the 22 as-yet-uncultivated phylotypes targeted in this study were detected in at least 20% of the cases: Bacteroidetes clone X083, five Synergistes and two Dialister phylotypes, Olsenella sp. genomospecies C1, and Tannerella sp. clone BU063. Bacteroidetes clone X083 occurred in one-half of the canals and was the most prevalent taxon at levels >105. These findings suggest that this phylotype can be a candidate endodontic pathogen that was previously overshadowed by inherent limitations of culture methods. Synergistes bacteria have been only recently disclosed in endodontic infections by molecular biology techniques (20, 34), with clone BA121 being the most frequently detected phylotype (34). This was corroborated by the present findings. Other four as-yet-uncultivated Synergistes phylotypes were also often detected, indicating that these bacteria are also common members of endodontic infections and that their occurrence had been underrated by culture studies. Overall, these association results indicate that several as-yet-uncultivated phylotypes should be included in the set of candidate endodontic pathogens and that efforts should be directed toward the development of culture media and strategies to cultivate and study some important features of these bacteria, including pathogenicity and susceptibility to antimicrobial agents.
Endodontic bacteria fall into eight bacterial phyla, namely, Firmicutes, Bacteroidetes, Spirochaetes, Fusobacteria, Actinobacteria, Proteobacteria, Synergistes, and TM7 (20, 29, 30, 34). The panel of taxa targeted in this study included representatives of all these phyla except TM7. In fact, a TM7 group-specific probe had to be removed from the panel because of cross-reactivity to several other species (data not shown). Clone X112 from the Sulfur River 1 (SR1) phylum was originally detected in subgingival samples from patients with marginal periodontitis (22) and was included in our panel. This clone was detected in two infected canals. This is the first report of a member of phylum SR1 in infected canals. Although this finding expands the diversity of endodontic bacteria to include another phylum, the fact that clone X112 was detected at low prevalences and even so as very weak signals indicates that it is not an important member of endodontic infections.
Culture studies have demonstrated that primary endodontic infections are characterized by a mixed consortium dominated by anaerobic bacteria and composed of a mean number of 2.6 to 5.4 taxa per canal (23, 36, 43, 44). Nonetheless, broad-range molecular analyses of the root canal microbiota of teeth with chronic apical periodontitis have revealed higher figures: 7 taxa in denaturing gradient gel electrophoresis analyses (37), 11 taxa in terminal restriction fragment length polymorphism analyses (30), 10 to 12 taxa in clone library analyses (29, 30), and 20 taxa in combined culturing and clone library analyses (20). Therefore, data from culture studies tend to underestimate the number of bacterial taxa in infected canals, and this can be a result of difficulties or even impossibilities in cultivating a significant proportion of the endodontic microbiota. This was confirmed in the present study, where a mean number of about 14 taxa were present per canal. This value is almost the same as that reported previously by Brito et al. (4), when using the original checkerboard to detect 77 cultivable species in nonamplified samples from asymptomatic primary infections. Considering that only target species can be detected by the checkerboard technology, this figure might well be larger to include nontarget taxa and taxa at levels below the detection limits of the assay. The mean number of taxa per canal was clearly in direct proportion to the lesion size: small lesions (<5 mm) harbored 11.7 taxa, lesions from 5 to <10 mm harbored 16 taxa, and lesions >10 mm harbored about 20 species. These differences in species richness help explain the long-held concept that the endodontic treatment of teeth with large lesions has a lower success rate than treatment of teeth with small or no lesions (41).
The necrotic root canal affords bacteria space and a moist, warm, nutritious, and anaerobic environment, which is by and large protected from host defenses. Even so, only a restricted assortment of oral bacteria is found in an individual infected canal. This indicates that selective pressures that favor the establishment of some species and inhibit others occur in the root canal system (43). One important ecological factor that helps to determine the composition of the root canal microbiota includes bacterial interactions. Positive bacterial interactions enhance the survival capacity of the interacting bacteria and enable different species to coexist in habitats where neither could exist alone. Positive interactions can also result in enhanced pathogenicity due to additive or synergistic effects. In the present study, several positive associations were depicted for some taxa for the first time, particularly the newly named species and as-yet-uncultivated phylotypes. The most prevalent taxon, O. uli, was positively related to several other taxa and showed no negative associations. Some strong positive associations were observed for both Bacteroidetes clone X083 and T. forsythia, with the latter being positively associated with all the other taxa tested.
This study also for the first time examined broader bacterial associations using cluster analysis, and four major complexes were disclosed. One of these complexes involved 13 taxa, all of which were very frequently detected. Cluster analyses of conventional checkerboard identifications have been used in periodontal microbiology research to evaluate bacterial associations in subgingival (40) and supragingival (15) plaque. However, direct comparisons between the endodontic complexes observed in the present study and the periodontal complexes cannot be performed because of the differences in the habitats and the inclusion of several new and as-yet-uncultivated taxa in the checkerboard panel used in this study. The factors dictating ecological interrelationships and the resulting pathogenic implications of the various combinations observed in the present study remain to be determined. Also, future research should address the association of specific bacterial pairs or complexes with symptoms and other clinical conditions in endodontics.
The knowledge of the bacterial diversity involved with apical periodontitis has been substantially refined and redefined after about 1 decade of application of molecular biology methods to endodontic microbiology research. In addition to strengthening the association of several cultivable named species with chronic apical periodontitis, the present findings using a large-scale analysis allowed the inclusion of some newly named species and as-yet-uncultivated phylotypes in the set of candidate pathogens associated with this disease.
Supplementary Material
Acknowledgments
This study was supported by grants 470417/2004-8 (I.N.R.), 300693/2005-2 (I.N.R.), and 304552/2006-2 (J.F.S.) from the Conselho Nacional de Desenvolvimento Científico e Tecnológico, a Brazilian governmental institution.
We are grateful to Bruce Paster for providing a detailed protocol of the checkerboard method and for his valuable advice and to Marlei Gomes da Silva for technical assistance.
Footnotes
Published ahead of print on 3 September 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumgartner, J. C., S. U. Khemaleelakul, and T. Xia. 2003. Identification of spirochetes (treponemes) in endodontic infections. J. Endod. 29794-797. [DOI] [PubMed] [Google Scholar]
- 3.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 401001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brito, L. C., F. R. Teles, R. P. Teles, E. C. Franca, A. P. Ribeiro-Sobrinho, A. D. Haffajee, and S. S. Socransky. 2007. Use of multiple-displacement amplification and checkerboard DNA-DNA hybridization to examine the microbiota of endodontic infections. J. Clin. Microbiol. 453039-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byun, R., M. A. Nadkarni, K. L. Chhour, F. E. Martin, N. A. Jacques, and N. Hunter. 2004. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J. Clin. Microbiol. 423128-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavez de Paz, L. E., A. Molander, and G. Dahlen. 2004. Gram-positive rods prevailing in teeth with apical periodontitis undergoing root canal treatment. Int. Endod. J. 37579-587. [DOI] [PubMed] [Google Scholar]
- 7.Conrads, G., S. E. Gharbia, K. Gulabivala, F. Lampert, and H. N. Shah. 1997. The use of a 16s rDNA directed PCR for the detection of endodontopathogenic bacteria. J. Endod. 23433-438. [DOI] [PubMed] [Google Scholar]
- 8.Corby, P. M., J. Lyons-Weiler, W. A. Bretz, T. C. Hart, J. A. Aas, T. Boumenna, J. Goss, A. L. Corby, H. M. Junior, R. J. Weyant, and B. J. Paster. 2005. Microbial risk indicators of early childhood caries. J. Clin. Microbiol. 435753-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figdor, D. 2002. Apical periodontitis: a very prevalent problem. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 94651-652. [DOI] [PubMed] [Google Scholar]
- 10.Fouad, A. F., J. Barry, M. Caimano, M. Clawson, Q. Zhu, R. Carver, K. Hazlett, and J. D. Radolf. 2002. PCR-based identification of bacteria associated with endodontic infections. J. Clin. Microbiol. 403223-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatti, J. J., J. M. Dobeck, C. Smith, R. R. White, S. S. Socransky, and Z. Skobe. 2000. Bacteria of asymptomatic periradicular endodontic lesions identified by DNA-DNA hybridization. Endod. Dent. Traumatol. 16197-204. [DOI] [PubMed] [Google Scholar]
- 12.Gomes, B. P., R. C. Jacinto, E. T. Pinheiro, E. L. Sousa, A. A. Zaia, C. C. Ferraz, and F. J. Souza-Filho. 2005. Porphyromonas gingivalis, Porphyromonas endodontalis, Prevotella intermedia and Prevotella nigrescens in endodontic lesions detected by culture and by PCR. Oral Microbiol. Immunol. 20211-215. [DOI] [PubMed] [Google Scholar]
- 13.Gomes, B. P., E. T. Pinheiro, C. R. Gade-Neto, E. L. Sousa, C. C. Ferraz, A. A. Zaia, F. B. Teixeira, and F. J. Souza-Filho. 2004. Microbiological examination of infected dental root canals. Oral Microbiol. Immunol. 1971-76. [DOI] [PubMed] [Google Scholar]
- 14.Haapasalo, M., H. Ranta, K. Ranta, and H. Shah. 1986. Black-pigmented Bacteroides spp. in human apical periodontitis. Infect. Immun. 53149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haffajee, A. D., S. S. Socransky, M. R. Patel, and X. Song. 2008. Microbial complexes in supragingival plaque. Oral Microbiol. Immunol. 23196-205. [DOI] [PubMed] [Google Scholar]
- 16.Hutter, G., U. Schlagenhauf, G. Valenza, M. Horn, S. Burgemeister, H. Claus, and U. Vogel. 2003. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 14967-75. [DOI] [PubMed] [Google Scholar]
- 17.Juretschko, S., A. M. Buccat, and T. R. Fritsche. 2004. Applications of fluorescence in situ hybridization in diagnostic microbiology, p. 3-18. In D. H. Persing, F. C. Tenover, J. Versalovic, Y.-W. Tang, E. R. Unger, D. Relman, and T. J. White (ed.), Molecular microbiology. Diagnostic principles and practice. ASM Press, Washington, DC.
- 18.Khemaleelakul, S., J. C. Baumgartner, and S. Pruksakorn. 2002. Identification of bacteria in acute endodontic infections and their antimicrobial susceptibility. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 94746-755. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, P. S., A. L. Griffen, M. L. Moeschberger, and E. J. Leys. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 433944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munson, M. A., T. Pitt-Ford, B. Chong, A. Weightman, and W. G. Wade. 2002. Molecular and cultural analysis of the microflora associated with endodontic infections. J. Dent. Res. 81761-766. [DOI] [PubMed] [Google Scholar]
- 21.Paster, B. J., I. M. Bartoszyk, and F. E. Dewhirst. 1998. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 20223-231. [Google Scholar]
- 22.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 1833770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters, L. B., P. R. Wesselink, and A. J. van Winkelhoff. 2002. Combinations of bacterial species in endodontic infections. Int. Endod. J. 35698-702. [DOI] [PubMed] [Google Scholar]
- 24.Rôças, I. N., J. C. Baumgartner, T. Xia, and J. F. Siqueira, Jr. 2006. Prevalence of selected bacterial named species and uncultivated phylotypes in endodontic abscesses from two geographic locations. J. Endod. 321135-1138. [DOI] [PubMed] [Google Scholar]
- 25.Rôças, I. N., M. Hulsmann, and J. F. Siqueira, Jr. 2008. Microorganisms in root canal-treated teeth from a German population. J. Endod. 34926-931. [DOI] [PubMed] [Google Scholar]
- 26.Rôças, I. N., J. F. Siqueira, Jr., A. F. Andrade, and M. Uzeda. 2003. Oral treponemes in primary root canal infections as detected by nested PCR. Int. Endod. J. 3620-26. [DOI] [PubMed] [Google Scholar]
- 27.Rôças, I. N., and J. F. Siqueira, Jr. 2006. Characterization of Dialister species in infected root canals. J. Endod. 321057-1061. [DOI] [PubMed] [Google Scholar]
- 28.Rôças, I. N., and J. F. Siqueira, Jr. 2006. Culture-independent detection of Eikenella corrodens and Veillonella parvula in primary endodontic infections. J. Endod. 32509-512. [DOI] [PubMed] [Google Scholar]
- 29.Saito, D., R. de Toledo Leonardo, J. L. M. Rodrigues, S. M. Tsai, J. F. Hofling, and R. B. Gonçalves. 2006. Identification of bacteria in endodontic infections by sequence analysis of 16S rDNA clone libraries. J. Med. Microbiol. 55101-107. [DOI] [PubMed] [Google Scholar]
- 30.Sakamoto, M., I. N. Rôças, J. F. Siqueira, Jr., and Y. Benno. 2006. Molecular analysis of bacteria in asymptomatic and symptomatic endodontic infections. Oral Microbiol. Immunol. 21112-122. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto, M., J. F. Siqueira, Jr., I. N. Rôças, and Y. Benno. 2007. Bacterial reduction and persistence after endodontic treatment procedures. Oral Microbiol. Immunol. 2219-23. [DOI] [PubMed] [Google Scholar]
- 32.Sassone, L., R. Fidel, L. Figueiredo, S. Fidel, M. Faveri, and M. Feres. 2007. Evaluation of the microbiota of primary endodontic infections using checkerboard DNA-DNA hybridization. Oral Microbiol. Immunol. 22390-397. [DOI] [PubMed] [Google Scholar]
- 33.Siqueira, J. F., Jr., and I. N. Rôças. 2003. Bacteroides forsythus in primary endodontic infections as detected by nested PCR. J. Endod. 29390-393. [DOI] [PubMed] [Google Scholar]
- 34.Siqueira, J. F., Jr., and I. N. Rôças. 2005. Uncultivated phylotypes and newly named species associated with primary and persistent endodontic infections. J. Clin. Microbiol. 433314-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siqueira, J. F., Jr., I. N. Rôças, J. C. Oliveira, and K. R. Santos. 2001. Molecular detection of black-pigmented bacteria in infections of endodontic origin. J. Endod. 27563-566. [DOI] [PubMed] [Google Scholar]
- 36.Siqueira, J. F., Jr., I. N. Rôças, S. S. M. Paiva, K. M. Magalhães, and T. Guimarães-Pinto. 2007. Cultivable bacteria in infected root canals as identified by 16S rRNA gene sequencing. Oral Microbiol. Immunol. 22266-271. [DOI] [PubMed] [Google Scholar]
- 37.Siqueira, J. F., Jr., I. N. Rôças, and A. S. Rosado. 2004. Investigation of bacterial communities associated with asymptomatic and symptomatic endodontic infections by denaturing gradient gel electrophoresis fingerprinting approach. Oral Microbiol. Immunol. 19363-370. [DOI] [PubMed] [Google Scholar]
- 38.Siqueira, J. F., Jr., I. N. Rôças, R. Souto, M. de Uzeda, and A. P. Colombo. 2000. Checkerboard DNA-DNA hybridization analysis of endodontic infections. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 89744-748. [DOI] [PubMed] [Google Scholar]
- 39.Siqueira, J. F., Jr., I. N. Rôças, R. Souto, M. Uzeda, and A. P. Colombo. 2001. Microbiological evaluation of acute periradicular abscesses by DNA-DNA hybridization. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 92451-457. [DOI] [PubMed] [Google Scholar]
- 40.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25134-144. [DOI] [PubMed] [Google Scholar]
- 41.Strindberg, L. Z. 1956. The dependence of the results of pulp therapy on certain factors. Acta Odontol. Scand. 14(Suppl. 21)1-175. [Google Scholar]
- 42.Sunde, P. T., L. Tronstad, E. R. Eribe, P. O. Lind, and I. Olsen. 2000. Assessment of periradicular microbiota by DNA-DNA hybridization. Endod. Dent. Traumatol. 16191-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sundqvist, G. 1992. Associations between microbial species in dental root canal infections. Oral Microbiol. Immunol. 7257-262. [DOI] [PubMed] [Google Scholar]
- 44.Wittgow, W. C., Jr., and C. B. Sabiston, Jr. 1975. Microorganisms from pulpal chambers of intact teeth with necrotic pulps. J. Endod. 1168-171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.