Abstract
Anabaena sp. strain PCC 7120 is a filamentous cyanobacterium that fixes N2 in specialized cells called heterocysts, which differentiate from vegetative cells in a process that requires the nitrogen control transcription factor NtcA. 2-Oxoglutarate-stimulated binding of purified NtcA to wild-type and modified versions of the ntcA gene promoter from Anabaena sp. was analyzed by mobility shift and DNase I footprinting assays, and the role of NtcA-binding sites in the expression of the ntcA gene during heterocyst differentiation was studied in vivo by using an ntcA-gfp translational fusion and primer extension analysis. Mutation of neither of the two identified NtcA-binding sites eliminated localized expression of ntcA in proheterocysts, but mutation of both sites led to very low, nonlocalized expression.
Nitrogen fixation in many cyanobacteria, including Anabaena sp. strain PCC 7120, requires differentiation of a specific cell type, called a heterocyst, in which the machinery for nitrogen fixation is confined (6, 27). The differentiation of heterocysts begins shortly after nitrogen deprivation and, under our experimental conditions, requires about 24 h to complete. Heterocyst differentiation depends on the NtcA protein, a transcriptional regulator belonging to the cyclic AMP receptor protein (CAP [or CRP]) family (5, 8, 26). In the absence of combined nitrogen, NtcA activates the expression of genes required for heterocyst differentiation and nitrogen fixation (7). In order to activate gene expression, NtcA binds upstream from the transcriptional starts (7), and a consensus sequence for NtcA binding (GTAN8TAC) has been defined (7, 11). Binding of NtcA to DNA and NtcA-mediated transcriptional activation are influenced by the C-to-N balance of the cell, and 2-oxoglutarate has been identified as a positive effector (10, 21, 22, 24, 25). Heterocyst differentiation in Anabaena sp. strain PCC 7120 also requires HetR (2, 3). The hetR gene encodes a positive-acting factor that exhibits autoprotease (28) and DNA-binding (9) activities in vitro. Induction of expression of ntcA and hetR is mutually dependent in the context of heterocyst differentiation (13), and induction of both genes is mostly localized to cells developing into heterocysts (2, 16).
The ntcA gene is transcribed from a complex promoter that produces three transcriptional start points (TSPs) (13, 16, 18) (see the sequence of the promoter region in Fig. 3B). Promoters P1, P2, and P3 produce TSPs at positions −49, −136, and −180, respectively, with respect to the translational start of ntcA. P2 functions independently of the nitrogen source, whereas P1 functions in the presence of combined nitrogen, but its use transiently increases after 6 to 12 h of nitrogen deprivation in an NtcA- and HetR-dependent manner (13). Finally, P3 functions only in the absence of combined nitrogen and its use is transiently induced after 6 to 12 h of nitrogen deprivation in an NtcA- and HetR-dependent manner (13). The promoter of the ntcA gene contains two putative NtcA-binding sites separated by 40 nucleotides and centered at positions −143.5 (GTAN8AAC) and −103.5 (GTAN8TAC) with respect to the translational start of ntcA. (We will refer to them as “distal” and “proximal,” respectively, according to their position with respect to the ntcA open reading frame.) The distal site overlaps the −10 hexamer of the P2 promoter (13). In vitro binding of NtcA to the distal site, but not to the proximal site, has been demonstrated by DNase I protection footprinting assays (18). However, two retarded bands have been observed in electrophoretic mobility shift assays (EMSAs) with NtcA and a fragment from the ntcA promoter (10). In this study, we used a combination of in vivo and in vitro approaches in order to understand the operation of the complex Anabaena ntcA promoter region, which is functional in the two different cell types, vegetative cells and heterocysts, of a nitrogen-fixing filament.
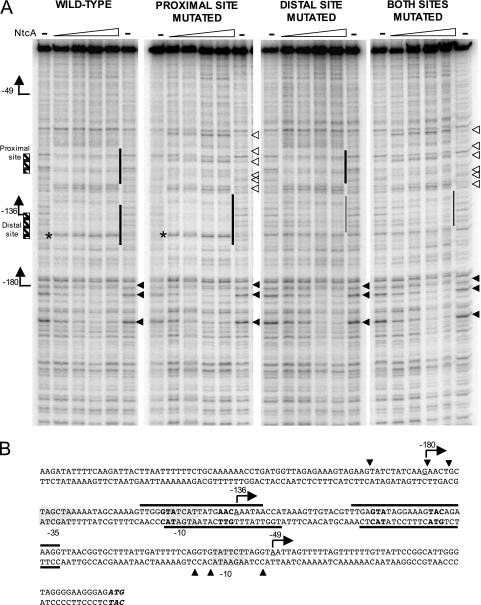
FIG. 3.
Sequences of the ntcA promoter region protected by NtcA in the presence of 2-oxoglutarate. Arrows labeled −49, −136, and −180 indicate the position and direction of TSPs corresponding to P1, P2, and P3, respectively. (A) DNase I protection footprint assays carried out with purified NtcA and wild-type and mutated versions of the ntcA promoter region. The fragments were amplified by PCR using oligonucleotides NA8 (32P labeled) and NA14 (unlabeled). Assay mixtures contained 2.5 fmol of labeled fragment and 0.068, 0.137, 0.274, or 0.412 μM of purified NtcA in the presence of 0.6 mM 2-oxoglutarate. Windows of protection due to NtcA binding (vertical lines), positions protected in all four fragments (solid triangles), and positions altered in the presence of NtcA in fragments containing a mutated proximal site (open triangles) are indicated. Thick vertical lines indicate regions protected in the presence of every NtcA concentration tested. Thin vertical lines indicate that mutation of the distal site did not completely eliminate protection of the sequence corresponding to that site. The positions of NtcA-binding sites (striped boxes) are also indicated on the left. Asterisks denote a position hypersensitive to DNase I digestion as referred to in the text. (B) Sequence of the ntcA promoter, from position −253 to the translational start of the ntcA gene (ATG). Triplets corresponding to the NtcA-binding sites are indicated in boldface. The positions around the NtcA-binding sites protected in both strands of the wild-type fragment are indicated with horizontal lines above or below the sequence. Putative −10 and −35 determinants are indicated with gray boxes. Solid triangles point to positions protected in all four fragments. The positions of TSPs for P1, P2, and P3 are underlined.
Methods.
Anabaena sp. (also known as Nostoc sp.) strain PCC 7120 and its derivatives (see Table S1 in the supplemental material) were grown photoautotrophically at 30°C in BG110C medium (BG11 medium [19] without NaNO3 and supplemented with 10 mM NaHCO3) supplemented with 6 mM NH4Cl plus 12 mM N-Tris (hydroxymethyl) methyl-2-aminoethanesulfonic acid (TES)-NaOH buffer (pH 7.5) and bubbled with a mixture of CO2 and air (1% [vol/vol]). Media were supplemented with 2 μg ml−1 of streptomycin and 2 μg ml−1 of spectinomycin in the case of strains bearing the PntcA-gfp fusions. For determination of green fluorescent protein (GFP) fluorescence and for RNA isolation, cells growing exponentially (about 3 to 5 μg chlorophyll a ml−1) in BG110C medium supplemented with NH4Cl were harvested by filtration at room temperature and either used directly or washed with BG110C (nitrogen-free) medium, resuspended in BG110C medium, and further incubated under culture conditions for the number of hours indicated in each experiment. Total DNA (4) and RNA (13) from Anabaena sp. strain PCC 7120 and its derivatives were isolated as previously described. Standard molecular biology methods were used (1). The plasmids used in this study are listed in Table S1 in the supplemental material. Overexpression of NtcA in Escherichia coli cells bearing plasmid pCSAM61 (13) and purification of NtcA using HiTrap heparin HP columns (Pharmacia Biotech) were carried out as described previously (22). Images of radioactive gels were obtained and analyzed by using a Cyclone storage phosphor system and Optiquant image analysis software (Packard). The accumulation of GFP reporter was analyzed by laser confocal microscopy as previously described (16) by collection of emissions in a window of 500 to 570 nm. All images were collected using the same settings, so that the intensities can be compared. (see also the supplemental material.)
Binding of NtcA to wild-type and mutated versions of the ntcA promoter.
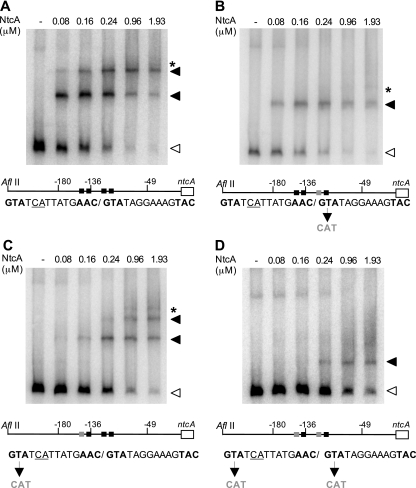
As a first step to understand the role of the two putative NtcA-binding sites present in the promoter region of the ntcA gene, we prepared modified versions of this region bearing mutations in one or both sites, and carried out EMSA with purified NtcA protein following procedures previously described (11). Three mutated versions of the ntcA upstream region (positions −603 to +18 with respect to the translational start of ntcA) were generated, with changes in the sequence of the proximal (GTAN8TAC→CATN8TAC), distal (GTAN8AAC→CATN8AAC), or both NtcA-binding sites. As previously observed (10), binding of NtcA to a wild-type fragment produced two retarded bands that likely correspond to binding of NtcA to one (lower retarded band) or both (upper retarded band) binding sites (Fig. 1A). Mutation of the proximal site resulted, as expected, in the absence of the upper retarded band that likely corresponds to binding of NtcA to two sites (Fig. 1B). However, mutation of the distal site did not abolish binding of NtcA to this site, since the upper retarded band was still observed, although at higher NtcA concentrations than in the case of the wild-type fragment (compare Fig. 1A and C). Consistently, NtcA was still able to bind to the fragment containing mutations in both NtcA-binding sites (Fig. 1D). The presence of faint bands appearing when a high concentration of NtcA was used (Fig. 1A, B, and C) suggests low-affinity binding of NtcA to additional positions. In fact, imperfect NtcA-binding sites are present in the DNA fragment used, just upstream from positions −180 (GTAN8TAT) and −49 (GTAN8TAA) (Fig. 3B). However, because mutation of the distal site did not abolish NtcA binding to that site (see above), the retarded band shown in Fig. 1D (fragment with both sites mutated) could result from binding of NtcA to the mutated distal site.
FIG. 1.
Binding of purified NtcA to wild-type and mutated versions of the ntcA promoter region. EMSA was carried out with a wild-type fragment (A) or with fragments bearing mutations in the proximal (B), distal (C), or both (D) NtcA-binding sites. A scheme of the fragment used in each case, including the positions of TSPs −180, −136, and −49, together with the sequences of wild-type and mutated NtcA-binding sites present in each fragment, is shown in panels A through D. Mutations introduced in each fragment are shown in gray under the wild-type sequence. The positions of free DNA fragments (open triangles), retarded bands (black triangles), and faint bands appearing when a high concentration of NtcA was used (asterisks), as referred to in the text, are indicated. Assay mixtures contained 0.1 fmol of labeled DNA fragment.
In the case of fragments containing a mutated proximal site (Fig. 1B), the retarded band presumably corresponding to binding of NtcA to only one site was already observed at the lowest NtcA concentration used, suggesting that NtcA exhibits high affinity for the distal site. In contrast, binding of NtcA to the proximal site required larger amounts of NtcA (lower retarded band in Fig. 1C), indicating lower affinity of NtcA for this site. Thus, the lower retarded band observed with the wild-type fragment (Fig. 1A) should correspond mostly to binding of NtcA to the distal site. Binding of NtcA to all four fragments could be competed by addition of an unlabeled fragment corresponding to the glnA promoter, which contains a high-affinity NtcA-binding site, confirming that the observed shifts were in fact due to specific NtcA binding (not shown).
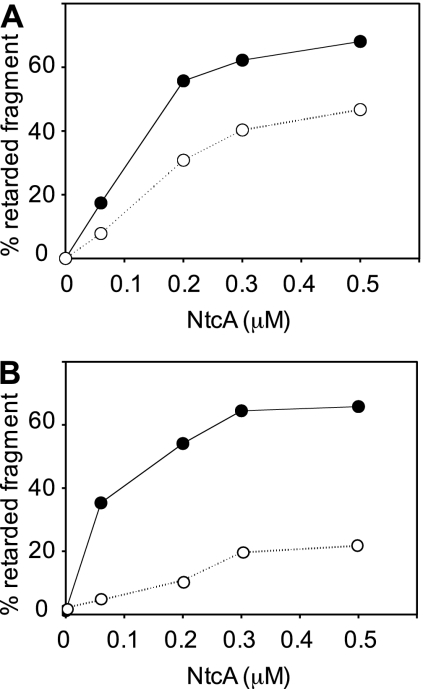
Parallel mobility shift assays were carried out in the absence or presence of 0.6 mM 2-oxoglutarate in order to test the responsiveness of NtcA binding to this effector (24) (see Fig. S1 in the supplemental material). The amount of fragment in retarded complexes corresponding to binding to only one NtcA-binding site was quantified in assays carried out with fragments mutated in the proximal (see Fig. S1B in the supplemental material) or distal (see Fig. S1C in the supplemental material) NtcA-binding site. The results shown in Fig. 2 indicate that binding to both sites was positively influenced by 2-oxoglutarate. Although NtcA alone showed higher affinity for the distal than for the proximal site, affinity appeared to be slightly higher for the proximal than for the distal site in the presence of 2-oxoglutarate. It is worth noting that the distal site (GTAN8AAC) does not exactly match the consensus for NtcA-binding sites (GTAN8TAC). However, the presence of nucleotides C and A in the second and third positions of the 8-nucleotide spacing between triplets (underlined in Fig. 1) most likely contributes to NtcA binding to this site (7, 23).
FIG. 2.
Effect of 2-oxoglutarate on binding of NtcA to DNA fragments containing the ntcA promoter region bearing mutations in the proximal (A) or distal (B) NtcA-binding sites. The percentage of fragment retarded (one band in panel A and two bands in panel B) in parallel assays carried out in the absence (open circles) or presence (closed circles) of 2-oxoglutarate (see Fig. S1 in the supplemental material) is represented with respect to the NtcA concentration in the assays.
The responsiveness to the metabolic signal effector makes the proximal site a good candidate for an NtcA activator site of P1 in response to the cellular condition of nitrogen deprivation, which in cyanobacteria results in high levels of 2-oxoglutarate (14). The high affinity of NtcA for the distal site in the absence of 2-oxoglutarate suggests that this site would be the one preferably occupied under cellular conditions in which levels of NtcA are low. However, because binding of NtcA to the distal site would most likely be incompatible with binding of RNA polymerase to, and transcription from, P2, and because no repression of transcription from P2 is observed in cells overexpressing NtcA (15), binding of NtcA to this site in vivo could be relatively low with respect to RNA polymerase binding to P2.
To determine the DNA sequences interacting with NtcA in wild-type and mutant versions of the ntcA promoter, binding of NtcA in the presence of 2-oxoglutarate was also analyzed by DNase I protection footprinting assays carried out as described previously (22) (Fig. 3 and see Fig. S2 in the supplemental material). The DNA fragments used were PCR amplified with oligonucleotides NA8 and NA14 and plasmid pCSEL18 (wild-type promoter), pCSEL35 (proximal site mutated), pCSEL44 (distal site mutated), or pCSEL47 (both sites mutated) as a template. One of the two oligonucleotides used for the PCRs was end labeled with [γ-32P]dATP and polynucleotide kinase. Oligonucleotide NA8 was labeled in the experiment shown in Fig. 3A, whereas oligonucleotide NA14 was labeled in the experiment shown in Fig. S2 in the supplemental material. Two regions (far-left panel in Fig. 3A), each corresponding to one NtcA-binding site, were protected in the wild-type fragment in the presence of every NtcA concentration tested. The area protected by NtcA around the distal site included the −10 determinant for the P2 promoter, as well as the position corresponding to TSP2 (−136) (Fig. 3B). Mutation of the proximal site eliminated protection of the corresponding sequences (second and far right panels in Fig. 3A). Mutation of the distal site did not completely eliminate protection of sequences corresponding to that site (third and far right panels in Fig. 3A), thus indicating that NtcA could still bind to those positions, although a larger amount of NtcA was required in order to see a window of protection. This observation is consistent with the fact that retarded bands that might be due to NtcA binding to the distal site were observed in EMSA with fragments bearing mutations in such a site (Fig. 1).
Binding of NtcA in vitro to either site did not seem to rely on binding to the other site, although cooperative effects cannot be excluded in vivo. Interestingly, binding of NtcA modified the pattern of DNase I digestion in the area corresponding to the proximal binding site, even when that site was mutated (either alone or in combination with a mutation of the distal site) (Fig. 3A and see Fig. S2 in the supplemental material). This modification could be due to conformational changes induced by binding of NtcA to other positions, including the distal site. Binding of NtcA to the distal (Fig. 3A) or the proximal (see Fig. S2 in the supplemental material) site produced a band hypersensitive to DNase I digestion, corresponding to the T of the GTA triplet (Fig. 3A and see Fig. S2 in the supplemental material). Such hypersensitivity, also observed in the case of NtcA binding to the glnA and rbcL promoters (17), indicates that NtcA binding, similar to CRP binding (20), might produce conformational changes in the DNA backbone. Additional subtle changes that would correspond to partial protection of sequences located upstream from the distal NtcA-binding site (around TSP3) (Fig. 3A) and downstream from the proximal NtcA binding site (upstream of TSP1) (see Fig. S2 in the supplemental material) were observed with all four DNA fragments used in footprinting experiments. Sequences protected by NtcA binding in both strands are summarized in Fig. 3B.
In vivo operation of mutated versions of the ntcA promoter.
We have previously analyzed the expression of an ntcA-gfp translational fusion from the wild type and shortened versions of the ntcA promoter (16). Our previous results indicate that (i) there is a certain level of operation of P1 in the absence of NtcA in all cells of the filament (13); (ii) in the absence of P3, the fragment covering positions −143 to −182 exerts a negative effect on expression of P1 (16); and (iii) P1 alone appears capable of driving localized induction of ntcA in differentiating cells in the absence of the fragment from positions −143 to −182, but in its presence, localized expression of P1 in proheterocysts requires P3 (strain CSEL7) (16). We have now prepared constructs in which the ntcA-gfp translational fusion is expressed from modified versions of the ntcA promoter (PntcA-gfp). In order to preserve any autoregulatory effect of NtcA, all of the PntcA-gfp constructs were integrated, through homologous recombination, in the nucA-nuiA region of the α megaplasmid of Anabaena sp. strain PCC 7120, so that expression of the ntcA gene in the chromosome remained unaltered. Constructs bearing translational fusions of the mutated versions of the ntcA promoter with the gfp gene (GFP-mut2) were prepared as described by Olmedo-Verd et al. (16; see the supplemental material for details). Strains CSEL10, CSEL11, and CSEL12 contained the gfp gene fused to mutated versions of the ntcA promoter, including mutations in the proximal, distal, or both NtcA-binding sites, respectively. Strain CSEL6 bearing the wild-type version of the ntcA promoter translationally fused to gfp (16) was used as a control.
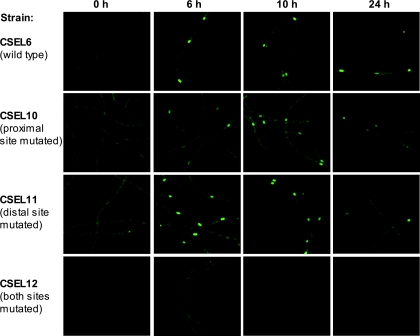
Ammonium-grown filaments were subjected to nitrogen deprivation, and expression of GFP was analyzed by means of fluorescence imaging and primer extension. GFP fluorescence in strains carrying different versions of the PntcA-gfp fusion is shown in Fig. 4. Mutations eliminating NtcA binding to the proximal site (strain CSEL10) or reducing NtcA binding to the distal site (strain CSEL11) did not abolish localized increase of GFP fluorescence. However, mutation of both NtcA-binding sites resulted in low, nonlocalized expression of the fusion (strain CSEL12).
FIG. 4.
GFP fluorescence of strains carrying the PntcA-gfp fusions integrated in the nucA-nuiA genomic region of the Anabaena α megaplasmid. GFP fluorescence micrographs of ammonium-grown filaments (panels labeled 0 h) or ammonium-grown filaments incubated for 6, 10 or 24 h in a medium lacking combined nitrogen are shown for the indicated CSEL strains. The NtcA-binding site or sites mutated in each strain are indicated.
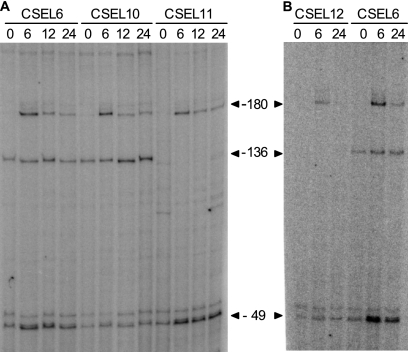
In order to determine which promoters were active in each strain, primer extension assays of ntcA-gfp transcripts were carried out as described previously (12) using oligonucleotide GFP4 (5′CAAGAATTGGGACAACTCC3′; complementary to positions +46 to +28 with respect to the translational start of the gfp gene). Results from one representative experiment are shown in Fig. 5. When the products of primer extension were compared to those obtained in the control strain bearing a wild-type PntcA-gfp fusion (strain CSEL6), induction of P1 could not be observed in strain CSEL10, indicating that NtcA binding to the proximal site is required for P1 induction. In contrast, expression from P1 was unaltered (or slightly increased) in strain CSEL11, suggesting that binding of NtcA to the proximal site is sufficient for P1 activation. This is consistent with previous results showing high P1 expression from a fragment bearing the proximal NtcA binding site and only half of the distal site (strain CSEL8 [16]). The location of the proximal binding site with respect to TSP1 (centered at position −54.5) does not, however, correspond to that observed in class II (around −42) or class I (near −93, −83, −72, or −62) bacterial activated promoters. As expected, because mutation of the distal NtcA-binding site affects the −10 determinant of P2, no transcript corresponding to TSP2 was observed in strain CSEL11. Utilization of P3 was unaltered in strains CSEL10 and CSEL11. Finally, mutation of both NtcA-binding sites (strain CSEL12) led to a basal level of expression from P1 (as observed in strain CSEL10), no expression from P2 (as observed in strain CSEL11), and relatively low expression from P3. The basis for the observed effect of mutations of both NtcA-binding sites on the utilization of P3 expression is currently unknown. It is worth mentioning that all PntcA-gfp constructs analyzed are integrated in the α megaplasmid, so that expression of the ntcA gene and levels of the NtcA protein remain unaltered with respect to those of the wild-type strain.
FIG. 5.
Primer extension analysis of the expression of the PntcA-gfp fusion in strains CSEL6, CSEL10, CSEL11, and CSEL12. Primer extension was carried out with primer GFP4 and RNA isolated from filaments grown with ammonium (lanes labeled 0) or from filaments grown with ammonium, washed, and incubated for 6, 12, or 24 h in the absence of combined nitrogen. The positions of TSPs −49, −136, and −180, which were generated from promoters P1, P2, and P3, respectively, are indicated. The results shown in panels A and B correspond to two different step-down experiments.
Concluding remarks.
Taking into account present and previous results (13, 16), a model for the operation of the ntcA complex promoter region can be suggested as follows. Basal operation of the ntcA promoter (i.e., in the presence of combined nitrogen) would include expression from P2 and basal expression from P1 and would not require NtcA or HetR (13). When the cells are subjected to nitrogen deficiency, which results in an increase in the level of 2-oxoglutarate, binding of low, preexisting amounts of NtcA protein to the proximal NtcA-binding site would lead to increased transcription from P1. This may result in increased transcription in all cells of the filament. Then the presence of HetR and/or other HetR-dependent elements in specific cells of the filament that are becoming heterocysts would promote transient expression from P3, which is required for transient localized induction of P1 in the complete promoter fragment (strain CSEL7 [16]). Transient induction of P1 and P3 exhibits similar timing and dependence on NtcA and HetR (Fig. 5; see also reference 13) and might thus be somewhat coupled. Expression from P1 and P3 likely accounts for the approximately fivefold transient increase in the amount of ntcA transcripts previously shown to take place in whole filaments of the wild-type strain, but not in the hetR mutant, during nitrogen step down (13). Accordingly, we have observed a similar fivefold increase in the amount of gfp transcripts in strain CSEL6 by means of Northern hybridization (not shown). Occupancy of the proximal NtcA-binding site appears to play a key role in determining the promoter(s) to be used and the total rate of transcription of ntcA under different nitrogen regimens and in the two cell types of the nitrogen-fixing filament.
Supplementary Material
Acknowledgments
We thank José Enrique Frías and Ignacio Luque for critically reading the manuscript and Alicia Orea for help with confocal microscopy.
This work was supported by grants BFU2004-00872 and BFU2007-60457 from the Ministerio de Educación y Ciencia, Spain.
Footnotes
Published ahead of print on 19 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2007. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, NY.
- 2.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 977-84. [DOI] [PubMed] [Google Scholar]
- 3.Buikema, W. J., and R. Haselkorn. 1991. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 5321-330. [DOI] [PubMed] [Google Scholar]
- 4.Cai, Y., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 1723138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frías, J. E., E. Flores, and A. Herrero. 1994. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 14823-832. [DOI] [PubMed] [Google Scholar]
- 6.Golden, J. W., and H. S. Yoon. 2003. Heterocyst development in Anabaena. Curr. Opin. Microbiol. 6557-563. [DOI] [PubMed] [Google Scholar]
- 7.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrero, A., A. M. Muro-Pastor, A. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28469-487. [DOI] [PubMed] [Google Scholar]
- 9.Huang, X., Y. Dong, and J. Zhao. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. USA 1014848-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurent, S., H. Chen, S. Bédu, F. Ziarelli, L. Peng, and C.-C. Zhang. 2005. Nonmetabolizable analogue of 2-oxoglutarate elicits heterocyst differentiation under repressive conditions in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. USA 1029907-9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luque, I., E. Flores, and A. Herrero. 1994. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 132862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 1999. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J. Bacteriol. 1816664-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 441377-1385. [DOI] [PubMed] [Google Scholar]
- 14.Muro-Pastor, M. I., J. C. Reyes, and F. J. Florencio. 2001. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 27638320-38328. [DOI] [PubMed] [Google Scholar]
- 15.Olmedo-Verd, E., E. Flores, A. Herrero, and A. M. Muro-Pastor. 2005. HetR-dependent and -independent expression of heterocyst-related genes in an Anabaena strain overproducing the NtcA transcription factor. J. Bacteriol. 1871985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olmedo-Verd, E., A. M. Muro-Pastor, E. Flores, and A. Herrero. 2006. Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 1886694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramasubramanian, T. S., T.-F. Wei, and J. W. Golden. 1994. Two Anabaena sp. strain PCC 7120 DNA-binding factors interact with vegetative cell- and heterocyst-specific genes. J. Bacteriol. 1761214-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramasubramanian, T. S., T.-F. Wei, A. K. Oldham, and J. W. Golden. 1996. Transcription of the Anabaena sp. strain PCC 7120 ntcA gene: multiple transcripts and NtcA binding. J. Bacteriol. 178922-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain stories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1111-61. [Google Scholar]
- 20.Schultz, S. C., G. C. Shields, and T. A. Steitz. 1991. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 2531001-1007. [DOI] [PubMed] [Google Scholar]
- 21.Tanigawa, R., M. Shirokane, S.-I. Maeda, T. Omata, K. Tanaka, and H. Takahashi. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA 994251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valladares, A., E. Flores, and A. Herrero. 2008. Transcription activation by NtcA and 2-oxoglutarate of three genes involved in heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1906126-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vázquez-Bermúdez, M. F., E. Flores, and A. Herrero. 2002. Analysis of binding sites for the nitrogen-control transcription factor NtcA in the promoters of Synechococcus nitrogen-regulated genes. Biochim. Biophys. Acta 157895-98. [DOI] [PubMed] [Google Scholar]
- 24.Vázquez-Bermúdez, M. F., A. Herrero, and E. Flores. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 51271-74. [DOI] [PubMed] [Google Scholar]
- 25.Vázquez-Bermúdez, M. F., A. Herrero, and E. Flores. 2003. Carbon supply and 2-oxoglutarate effects on expression of nitrate reductase and nitrogen-regulated genes in Synechococcus sp. strain PCC 7942. FEMS Microbiol. Lett. 221155-159. [DOI] [PubMed] [Google Scholar]
- 26.Wei, T.-F., T. S. Ramasubramanian, and J. W. Golden. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 1764473-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu, X., J. Elhai, and C. P. Wolk. 2008. Transcriptional and developmental responses by Anabaena to deprivation of fixed nitrogen, p. 383-422. In A. Herrero and E. Flores (ed.), The cyanobacteria. Molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom.
- 28.Zhou, R., X. Wei, N. Jiang, H. Li, Y. Dong, K.-L. Hsi, and J. Zhao. 1998. Evidence that HetR protein is an unusual serine-type protease. Proc. Natl. Acad. Sci. USA 954959-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.