Abstract
Pathogenicity of the enterobacterium Erwinia chrysanthemi (Dickeya dadantii), the causative agent of soft-rot disease in many plants, is a complex process involving several factors whose production is subject to temporal regulation during infection. PecS is a transcriptional regulator that controls production of various virulence factors. Here, we used microarray analysis to define the PecS regulon and demonstrated that PecS notably regulates a wide range of genes that could be linked to pathogenicity and to a group of genes concerned with evading host defenses. Among the targets are the genes encoding plant cell wall-degrading enzymes and secretion systems and the genes involved in flagellar biosynthesis, biosurfactant production, and the oxidative stress response, as well as genes encoding toxin-like factors such as NipE and hemolysin-coregulated proteins. In vitro experiments demonstrated that PecS interacts with the regulatory regions of five new targets: an oxidative stress response gene (ahpC), a biosurfactant synthesis gene (rhlA), and genes encoding exported proteins related to other plant-associated bacterial proteins (nipE, virK, and avrL). The pecS mutant provokes symptoms more rapidly and with more efficiency than the wild-type strain, indicating that PecS plays a critical role in the switch from the asymptomatic phase to the symptomatic phase. Based on this, we propose that the temporal regulation of the different groups of genes required for the asymptomatic phase and the symptomatic phase is, in part, the result of a gradual modulation of PecS activity triggered during infection in response to changes in environmental conditions emerging from the interaction between both partners.
The pathogenicity of Erwinia chrysanthemi is mainly associated with the production of a set of pectinases that, by cleaving the pectic component of plant cell walls, progressively dissolve the outer barrier of plant cells and enable the bacteria to multiply and disseminate within the leaf and petiole (13, 34). This process may lead to the complete disorganization of parenchymatous tissue and gives rise to soft-rot symptoms. Symptom progression depends on the aggressiveness of the bacterial strain, the susceptibility of the plant host, and environmental conditions, among which temperature and humidity are particularly critical (41). E. chrysanthemi displays a broad host range, and this might be explained by the fact that these bacteria, owing to the wide range of their enzymatic activities, are able to attack any parenchymatous tissue.
Analysis of the infection process of the E. chrysanthemi model 3937 strain (the genome sequence of which is available at https://asap.ahabs.wisc.edu/asap/logon.php) clearly shows that infection involves first a colonization phase, where the bacteria reside and multiply within the intercellular spaces without causing any symptoms (13, 34). This phase of colonization is followed by the symptomatic phase of the disease only when environmental conditions are favorable for massive bacterial multiplication and production of plant cell wall-degrading enzymes (41). E. chrysanthemi produces different types of pectinases: pectin methyl esterases, pectin acetyl esterases, exo- and endopectate lyases, exopolygalacturonases, and a rhamnogalacturonate lyase (25, 51). All of these enzymes have the potential to degrade different parts of pectin, including the linear and branched regions, but are not equally involved in symptom initiation and spreading (3, 5, 27). In addition, the contribution of the individual enzymes to symptom development is independent of the relative in vitro specific activity of the enzymes (59). Importantly, the synthesis of pectate lyases is accurately controlled by a set of regulators, including KdgR, PecT, CRP, H-NS, Fur, ExpR, PecS, GacA, and RsmA-RsmB, that respond to metabolic stimuli or environmental conditions (9, 10, 14, 19, 26, 35, 48, 49, 54). Whereas the three major repressors KdgR, PecS, and PecT act independently on pectate lyase synthesis, some hierarchy exists, for example, between H-NS and PecT (35), between ExpR and PecS (48), between GacA and RsmA-RsmB (64), and between GacA and PecT (26). The effect of several regulatory mutations on pathogenicity has been tested, and it appears that some mutants differ from the wild type in the length of the latency period. Interestingly, the pecS mutant provokes symptoms more rapidly and with more efficiency than the wild-type strain (50).
The PecS protein belongs to the MarR family of transcriptional factors whose DNA-binding capacity is attenuated by specific anionic lipophilic ligands (usually phenolic compounds) (57). The precise signal that is perceived by PecS is currently unknown. PecS was first identified as a regulator controlling the production of pectinases, cellulase, and indigoidine, a blue pigment involved in defense against reactive oxygen species (49). PecS was later shown to regulate the out genes, which encode the type II secretion system responsible for pectinase and cellulase secretion (44); the hrpN gene, which encodes a protein secreted via the type III secretion system (36); and the divergent fliE gene and fliFGHIJKLMNOPQR operon involved in the biogenesis of the flagellar hook-basal body complex (53). Interestingly, PecS appears to be a horizontally acquired regulator. It is not widely present in soft-rot Erwinia species. It is fascinating that it plays such an important role in regulation since many of the genes it controls are widespread in these bacterial species. In addition to the PecS targets described above, genetic and physiological analyses strongly suggest that PecS controls the production of other virulence factors. For example, PecS was shown to overproduce a noncharacterized biosurfactant, a compound thought to be used by pathogenic bacteria to adhere to the host surface (S. Reverchon, unpublished data). Moreover, it was previously shown that the ind pecS mutant displayed higher resistance to the oxidative stressor hydrogen peroxide than the ind mutant (50), indicating that PecS exerts a regulatory influence over other genes involved in resistance to oxidative stress. Attempts to identify these additional PecS targets by scanning of the E. chrysanthemi genome sequence with the PecS consensus DNA-binding site (C11G10A9N8W7T6C5G4T3A2)T1A0T1(T2A3C4G5A6N7N8N9C10G11) (where the subscripts indicate positions of the bases and parentheses indicate palindromic-like components the sequence) defined from a SELEX experiment (53) were not very successful. Only the divergent fliE gene and fliFGHIJKLMNOPQR operon were identified by probing the E. chrysanthemi genome with the consensus sequence (53). This was probably due to a relatively high degree of degeneracy in the consensus sequence. Moreover, one cannot rule out the existence of PecS target genes regulated by an indirect mechanism. In this study, we have used DNA microarrays to investigate the extent of PecS involvement in the control of gene expression in E. chrysanthemi. We have now established that PecS notably regulates a wide range of genes that could be linked to pathogenicity and to a group of genes concerned with evading host defenses.
MATERIALS AND METHODS
Bacterial strains and cell growth conditions.
Bacterial strains, plasmids, and oligonucleotides used in this work are described in Tables 1 and 2. The E. chrysanthemi wild-type strain is 3937 and the pecS::MuCmr mutant is A3953. Bacteria were grown in M63 minimal medium supplemented with 0.2% glucose as the unique carbon source. M63 was prepared as described by Miller (33). Aerobic cultures were grown at 30°C with shaking (150 rpm) in 500-ml flasks containing 50 ml of medium. The A600 was determined in a PRIM Secoman spectrophotometer.
TABLE 1.
Bacterial strains, plasmids, and phage used in this study
| Strain, plasmid, or phage | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | λ− φ80lacZΔM15 Δ(lacZYA-argF)U196 recA1 endA1 hsdR17 (rK− mK−) supE44 thi-1 gyrA relA1 | Stratagene |
| E. chrysanthemi strains | ||
| 3937 | Wild-type strain isolated from S. ionantha | |
| A3953 | pecS::MudIIPR13 Cmr | 49 |
| A4587 | nipE-uidA-Kmr | This work |
| A4921 | pecS::MudIIPR13 CmrnipE-uidA-Kmr | This work |
| A4961 | ahpC-uidA-Kmr | This work |
| A4987 | pecS::MudIIPR13 CmrahpC-uidA-Kmr | This work |
| A4952 | virK-uidA-Kmr | This work |
| A4978 | pecS::MudIIPR13 CmrvirK-uidA-Kmr | This work |
| A4946 | rhlA-uidA-Kmr | This work |
| A4979 | pecS::MudIIPR13 CmrrhlA-uidA-Kmr | This work |
| A4889 | ΔavrLM-uidA-Kmr | This work |
| A4920 | pecS::MudIIPR13 Cmr ΔavrLM-uidA-Kmr | This work |
| A5009 | fliC-uiDA-Kmr | V. Shevchik, personal communication |
| A3997 | prtE-uidA-Kmr | 26 |
| Ech176 | prtE::MudII1734 Kmr | F. Van Gijsegem, unpublished data |
| A4111 | vfmE::Cmr | S. Reverchon, unpublished data |
| A4496 | vfmE-uiDA-Kmr | S. Reverchon, unpublished data |
| Plasmids | ||
| pGEM-T | AprlacZ′ | Promega |
| pUIDK1 | pBR322 derivative harboring the uidA-Kmr cassette | 2 |
| pSC3154 | pGEM-T carrying the nipE gene | This work |
| pSR3323 | pGEM-T carrying the rhlA gene | This work |
| pSR3326 | pGEM-T carrying the virK gene | This work |
| pSR3329 | pGEM-T carrying the ahpC gene | This work |
| pSLF3 | pUC18 carrying the border regions flanking the avrLM gene cluster | This work |
| pSR3333 | pGEM-T carrying the nipE regulatory region | This work |
| pSR3325 | pGEM-T carrying the rhlA regulatory region | This work |
| pSR3328 | pGEM-T carrying the virK regulatory region | This work |
| pSR3332 | pGEM-T carrying the ahpC regulatory region | This work |
| pSR3334 | pGEM-T carrying the avrL regulatory region | This work |
| pSR1802 | pBluescript carrying the celZ regulatory region | 44 |
| Phage ΦEC2 | E. chrysanthemi generalized transducing phage | 47 |
TABLE 2.
Primers used in this study
| Primer function and gene or primer name | Primer directiona | Primer sequence (5′→3′)b | Efficiencyc | Restriction site added |
|---|---|---|---|---|
| Expression studies | ||||
| rpoA | F | AAACCGCGCCTGGTAGATA | 1.78 | |
| R | CCTTTCAGGTTGAGCAGGAT | |||
| rhlA | F | GCATATTTCCGATCCTGCAC | 1.93 | |
| R | CCCAGGAAATCGACAGGATA | |||
| virK | F | CGTCCAAGTCGCTGAAAAGC | 1.93 | |
| R | CCTGATAACGCATGAACTGC | |||
| nipE | F | CAACATTCCCCCTCTGGATT | 1.94 | |
| R | GTGCAACGTGTTGGATTTGT | |||
| avrL | F | GCTTGTCGCAGGCAGTTTAC | 1.94 | |
| R | CGGGAACTTGATCTGGTTGA | |||
| avrM | F | GATTATCGGCGGCTTATTCA | 1.96 | |
| R | CGGCCTCGACATCTTTTATG | |||
| ahpC | F | AGAAAAGCATCGAAGGCAAA | 1.92 | |
| R | CATGCCAGGCTTTGTGAGTA | |||
| fabI | F | CATCTTCCGCAACATCACAC | 1.94 | |
| R | ACGGTAAGCGCATTCTGGTA | |||
| indA | F | TACGGCCAGAAAGATTGAGG | 1.95 | |
| R | GCCAGAATCACCGGAATATC | |||
| fadR | F | GGATTCGCGGAAGAGTACAT | 1.80 | |
| R | GGTTTACCGTGCTGAATCGT | |||
| rsmA | F | GAGTTGGCGAAACCCTCAT | 1.96 | |
| R | GCTGAGACTTCTCTGCCTGAA | |||
| hns | F | GCGAAGCACTTAAGATTCTAAACA | 1.86 | |
| R | CTTTACCGGCAACTTTGCTT | |||
| rpmE2 | F | CGGGTATCCATCCCAACTAC | 1.94 | |
| R | CGCTACCTTCTTTGGCGTAT | |||
| ffh | F | TGGTGCGTGATTTCATCAAT | ||
| R | CTTACCAACGCTGGTGGTTT | 1.92 | ||
| Gene cloning | ||||
| rhlAdeb | GCTCTAGACTGTCTGAAGACACAGTCAGATTC | XbaI | ||
| rhlAstop | CCGTGATAGCGCTGGTTATTC | |||
| virKdeb | GCTCTAGACTGCTGAGTCCCGACAGCGG | XbaI | ||
| virKstop | CGATGGCTGCCGGAACCGGC | |||
| nipEdeb | CAGATGTTCGAGTTGGCG | |||
| nipEstop | CGCCAGTTACAGCCAGAC | |||
| ahpCdeb | GCTCTAGATGGGCCGCATCTGTGCCG | XbaI | ||
| ahpCstop | CGTCCAGCGTCGCCACTAAC | |||
| avr1B1 | CAACGATATCATCCGAATCATG | |||
| avr1B2 | CTTAAGTCCGTTTTCCGAGCATG | |||
| avr2B1 | CTTAAGTGCTGGATGGAACGGTATTCAC | |||
| avr2B2 | TGATCAGGCGAAATTCGGCACGCAGTTC | |||
| Regulatory region cloning | ||||
| RRrhlA5′ | GCTCTAGACTGTCTGAAGACACAGTCAGATTC | XbaI | ||
| RRrhlA3′ | GGAATTCAATAATGGTTTCATGTGCAGG | EcoRI | ||
| RRvirK5′ | GCTCTAGACTGCTGAGTCCCGACAGCGG | XbaI | ||
| RRvirK3′ | GGAATTCGACTTGGACGCCACGGCC | EcoRI | ||
| RRnipE5′ | GCTCTAGAGAATATTATCCCTCATGGGTG | XbaI | ||
| RRnipE3′ | GGAATTCCAGTTTGTCAAAATCATCG | EcoRI | ||
| RRavrL5′ | GCTCTAGACAACGATATCATCCGAATCATG | XbaI | ||
| RRavrL3′ | GGAATTCGCATAAGCGGCTCTCGCCGCC | EcoRI | ||
| RRaphC5′ | GCTCTAGATGGTGCCGCATCTGTTGCCG | XbaI | ||
| RRahpC3′ | GGAATCGTAAAGTCAGCCGGATAGAAG | EcoRI | ||
| mfhR5′ | CCGTTTTGGTGAATTGCGCAAAG | |||
| mfhR3′ | GCAGATCTTTTAGCCTTGCGCCCGCGCG | BglII | ||
| Primer extension | ||||
| rhlA-pExt1 | GACAATGCATTTTTCTATGG | |||
| virK-pExt1 | GCAGCACAGGGGTAATACG | |||
| nipE-pExt1 | GCCAGTACAGCTGTCTTCTT | |||
| avrL-pExt1 | GCGCCCCAGAGAAATATGC | |||
| ahpC-pExt2 | GTAAAGTCAGCCGGATAGAAG |
F, forward; R, reverse.
Restriction sites are underlined.
Primer PCR efficiency was estimated via a calibration dilution curve and slope calculation. Efficiency = 10−1/slope.
RNA extraction.
Despite the fact that PecS targets are maximally expressed in stationary phase, RNA extraction was performed from cultures of strains 3937 and A3953 grown to late exponential phase (A600 of 0.8) because the quality of RNA obtained during this period was better than that obtained in the stationary phase. Moreover, we previously demonstrated that pecS mutation similarly affects its target gene expression at the end of exponential phase and in the stationary phase. Indeed, although the expression of the target genes is more important during the stationary phase of growth, the deregulation ratios observed in a pecS mutant were approximately the same during the late exponential phase and in the stationary phase (49). Cells were harvested by centrifugation, and total RNA was extracted from the pellets using the frozen acid-phenol method described by Maes and Messens (30) and then treated with DNase I. Absence of genomic DNA contamination was checked by PCR with all primer pairs used for quantitative reverse transcription-PCR (qRT-PCR) (Table 2). Isolated RNA was quantified on the basis of its absorption at 260 nm using an ND 100 Nanodrop spectrophotometer, visualized on an agarose gel to check quality, and stored at −80°C until further use.
Microarray design and analysis.
The microarrays used in this study were custom designed and produced by NimbleGen Systems, Inc. (Madison, WI), based on the annotated sequence (version number 6) of E. chrysanthemi (available at https://asap.ahabs.wisc.edu/asap/logon.php), which comprised 4,753 coding sequences (CDS). The microarrays consisted of 24-mer oligonucleotides with 20 perfect match probes per CDS, duplicated in two blocks on the array. For microarray analyses, cDNA was synthesized, labeled, and hybridized by NimbleGen Systems, Inc. For each strain, two independent biological replicates were tested.
Data analysis.
Each microarray experiment was repeated independently twice (biological replicates). Expression data were normalized using quantile normalization (6) and summarized to one expression value per gene and per block for each of the four arrays using the robust multichip average algorithm (20). To detect differentially expressed genes, we used the LIMMA (linear model for microarray analysis) package (version 2.4.13) from the R Bioconductor project (16, 56). Data were fitted to a linear model with a strain effect and a biological replicate effect, and the within-array replicates were used to improve the precision of gene variance estimations and to increase statistical power (duplicate correlation function) (56). We applied the empirical Bayes method implemented in LIMMA to moderate standard errors across genes (55). P values were adjusted for multiple testing by controlling the false discovery rate (FDR), according to Benjamini and Hochberg's method (4). Using this model, 437 genes had an FDR-adjusted P value of <0.05 for the strain effect (pecS mutant versus wild-type strain). From this list of genes, we further decided to focus our attention on those that were estimated to differ by at least ±33% (i.e., a log2-fold change of ±0.41) between the pecS mutant and the wild-type strain.
Real-time RT-PCR.
After the array data were obtained, three additional RNA samples were extracted from each strain, and their qualities were measured in the same way as for RNA samples used for the microarray experiments. RT was performed using SuperScript II reverse transcriptase (Invitrogen) with 1 μg of total RNA and 50 ng of random hexamer primers, according to the manufacturer's protocol (first strand cDNA synthesis). One microliter of the RT reaction mixture was added as a template to the Qbiogen Sybr Green mix for PCR with gene-specific primers. Primers used in this work are listed in Table 2. The thermal cycling reactions were performed using a LightCycler from Roche according to the following conditions: an initial step at 95°C for 10 min, followed by 35 cycles at 95°C for 15 s, 55°C for 15 s, and 72°C for 20 s. Two housekeeping genes, rpoA and ffh, were used as standards to obtain normalized target gene expression ratios (42, 61). The statistical program used to analyze the data was the Relative Expression Software Tool (43). We observed that rpoA expression is relatively stable under all conditions tested. Furthermore, the ffh gene was selected as a reference gene for real-time RT-PCR for accurate normalization based on studies performed in the related plant pathogen Pectobacterium atrosepticum (58). Expression levels of the rpoA and ffh genes are similar in the wild-type strain and in the pecS mutant (−1.09- and 1.00-fold changes in microarray, respectively, and −1.03- and 1.08-fold changes in qRT-PCR, respectively). In addition, a total of 2.5 × 105 copies of GeneAmplimer pAW 109 RNA (Applied Biosystems) was added to the RT reaction mixture and used as a control for the retrotranscription efficiency (63). The specificity of the PCR primers was verified with a melting curve analysis.
Primer extension analysis.
Primer extensions were performed as previously described (37). The transcription start sites of ahpC, avrL, rhlA, nipE, and virK were determined with 10 μg of total RNA extracted from E. chrysanthemi strains 3937 and A3953. Primers used for specific detection of mRNA were 5′ end labeled (primers ahpC-pExt2, avrL-pExt1, nipE-pExt1, virK-pExt1, and rhlA-pExt1) (Table 2). The extension products were resolved on a 6% sequencing gel and visualized by autoradiography on Amersham MP film. The length of each transcript was identified using the corresponding sequencing profile as a reference.
Band shift assays.
The rhlA, ahpC, virK, nipE, and avrL promoter/operator regions and the mfhR nonspecific fragment derived from the coding region of a transcriptional regulator not controlled by PecS were generated by PCR with primers RRrhla5′-RRrhla3′, RRahpC5′-RRahpC3′, RRvirK5′-RRvirK3′, RRnipE5′-RRnipE3′, RRavrL5′-RRavrL3′, and mfhR5′-mfhR3′ (Table 2) and with genomic DNA from E. chrysanthemi as a template. The resulting PCR fragments were cloned into pGEMT using a TA cloning kit from Promega. DNA probes digested with appropriate restriction enzymes were labeled, as previously described (37), and purified after electrophoresis on agarose gel using a Qiagen gel extraction kit. Each labeled DNA probe (about 100,000 cpm) and the purified PecS regulator (15 to 800 nM) were incubated for 20 min at 30°C in 20 μl of 12 mM HEPES-NaOH (pH 7) containing 4 mM Tris-HCl (pH 7), 75 mM KCl, 1 mM dithiothreitol, 4 μg of acetylated bovine serum albumin, 1 μg of nonspecific synthetic DNA poly(dI-dC)·poly(dI-dC), and 5% glycerol (vol/vol). The reaction mixtures were then submitted to electrophoresis on a 4% nondenaturing polyacrylamide gel in TA buffer (6.75 mM Tris-HCl, 3.3 mM sodium acetate, pH 7.4). Bands were detected by autoradiography on Amersham MP film.
DNase I footprinting.
About 100,000 cpm of DNA probe, labeled at one end, was incubated for 20 min at 30°C with the PecS purified protein in the gel shift assay buffer. The reaction mixtures were adjusted to 10 mM MgCl2, 5 mM CaCl2, and 0.1% Nonidet P-40 before the addition of DNase I (2 × 10−3 units of RNase-free DNase I; Roche). Digestion was performed at 30°C for 45 s and stopped by the addition of 25 μl of 50 mM EDTA. The reaction volume was adjusted to 200 μl with TE buffer (10 mM Tris [pH 8.0]-1 mM EDTA) containing 0.2% sodium dodecyl sulfate. Reaction mixtures were incubated for 30 min at 42°C with 20 μg of proteinase K. After phenol-chloroform extraction, DNA fragments contained in the supernatant were ethanol precipitated and separated by electrophoresis on a 6% sequencing gel. The digestion profile was revealed by autoradiography.
Construction of E. chrysanthemi rhlA, ahpC, nipE, virK, and ΔavrLM mutations.
The uidA-Kmr cassette includes a promoterless uidA gene that encodes β-glucuronidase and the Tn903 aphAI gene responsible for kanamycin resistance (2). Insertion of a uidA-Kmr cassette into a gene in the correct orientation generates a transcriptional fusion. For the construction of the rhlA, ahpC, nipE, and virK fusions, the corresponding genes were amplified using the primer pairs rhlAdeb-rhlAstop, ahpCdeb-ahpCstop, nipEdeb-nipEstop, and virKdeb-virKstop, and the resulting PCR fragments were cloned into the pGEMT plasmid. The uidA-Kmr cassette was introduced into the unique Eco47III site in nipE, into the unique HpaI site in rhlA, into the unique AgeI site in virK, and into the unique StuI site in ahpC. For the construction of the ΔavrLM-uidA-Kmr fusion, the DNA fragments flanking the avrLM cluster were amplified using the avr1B1-avr1B2 and avr2B1-avr2B2 primer pairs and cloned in the pGEMT and pGEMTeasy vectors, respectively. These border fragments were then cloned in the EcoRI (avrB2) and SalI-SphI sites of pUC18, resulting in the pSLF3 plasmid carrying a 0.6-kb DNA fragment harboring an exact deletion of the avrLM coding sequences. The uidA-Kmr cassette was introduced between these border fragments after a SalI-SacI double digestion, creating a glucuronidase fusion under the control of the avrL promoter. Primers used are listed in Table 2.
Plasmids harboring rhlA-uidA, ahpC-uidA, virK-uidA, nipE-uidA, and ΔavrLM-uidA fusions were selected, and the resulting insertions were introduced into the E. chrysanthemi chromosome by marker exchange recombination (52).
Biosurfactant production and swarming motility assays.
To visualize the production of biosurfactant, bacterial strains were inoculated by stabbing 0.7% semisolid agar plates, and the halo size of surfactant production was examined after growth for 8 h at 30°C. For a swarming motility assay on the surface of a solid medium, bacterial strains were inoculated onto LB medium-1% agar plates and incubated at 30°C for 24 h.
H2O2 susceptibility assay.
E. chrysanthemi strains were grown in LB medium to stationary phase and diluted to an optical density at 600 nm of 0.1 in LB medium. Ten microliters of bacterial suspensions was then spotted on LB agar plates supplemented with various concentrations of H2O2 ranging from a final concentration of 0 to 2,000 μM (0, 50, 100, 250, 500, 750, 1,000, 2,000 μM). Following incubation at 30°C for 24 h, the lowest concentration resulting in no growth was deemed to be the MIC as defined by Andrews (1). Results are shown as the mean ± standard deviations of triplicate values obtained from three independent experiments.
Virulence assays on chicory leaves.
Prior to infection, chicory leaves were slightly wounded in their centers to define the inoculation sites. Thirty leaves were infected for each strain with 106 bacteria (10 μl of a suspension at 108 bacteria per ml). After incubation in a dew chamber for 24 h at 30°C, the length of rotted tissue was measured to estimate the disease severity.
Virulence assays on Saintpaulia ionantha.
Pathogenicity tests on one-month-old potted S. ionantha cv. Blue Rhapsody cuttings were performed according to Lebeau et al. (26). Bacteria cells grown in LB agar medium for 16 h at 30°C were suspended in a 100 mM KCl solution to a concentration of 107 or 108 CFU/ml. One hundred microliters of the resulting suspension was inoculated into one leaf per plant by needle-free syringe infiltration after the lower side of the leaf was subjected to wounding. Plants were incubated under tropical conditions (day/night temperature of 28°C/26°C; 16 h of light; relative humidity of ±100%). Eighteen plants were tested for each bacterial strain. Progression of the symptoms was scored daily for 6 days. At each day from day 2 (i.e., when the first maceration of whole leaves was observed) to day 6, the proportion of full symptomatic leaves was compared between wild-type and mutant groups using a Fisher's exact test. Statistical significance was defined as a P value of <0.05. The assays were carried out independently in triplicates.
Microarray data accession number.
Microarray data from this study were submitted to the European Bioinformatics Institute (http://www.ebi.ac.uk/arrayexpress) under accession number E-TABM-428.
RESULTS AND DISCUSSION
Characterization of the PecS regulon in E. chrysanthemi.
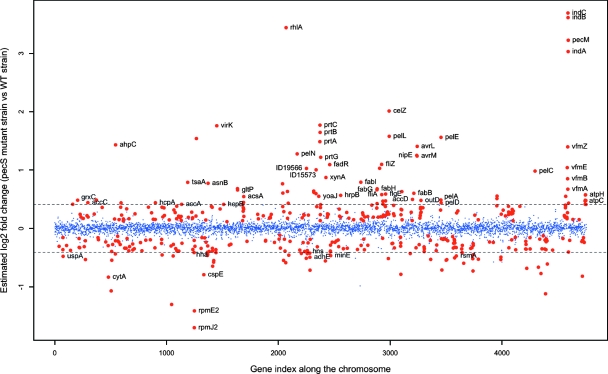
In order to elucidate the magnitude of the complete PecS regulon, we used CDS pangenomic microarrays to compare the E. chrysanthemi 3937 transcriptome with that of strain A3953, which has an insertion in pecS. The two strains were grown to late exponential phase in M63 minimal medium supplemented with 0.2% glucose, a carbon source allowing expression of most known PecS targets (cellulase, indigoidine, flagella, and pectate lyases). In addition, the two strains exhibited the same growth rate in M63 minimal medium supplemented with 0.2% glucose. For each strain, two independent cultures were prepared, and the total bacterial RNA was extracted and subsequently hybridized to separate arrays. Repeatability was excellent between biological replicates. Indeed, the correlation coefficient between signal intensities of any replicate was ∼0.98. Differentially expressed genes were identified using the LIMMA package from the R Bioconductor program, taking advantage of the within-array replicates. We detected a total of 437 genes that exhibited significant differential expression in the pecS mutant strain compared to the wild-type strain, with P values adjusted for multiple testing (FDR) of <0.05 (259 genes have an FDR of <0.01). The relative changes in gene expression ranged from a 12-fold upregulation of the ind genes, involved in indigoidine biosynthesis, to a 2.6-fold downregulation of the ykgMO (rpmE2-rpmJ2) operon, encoding paralogues of 50S ribosomal proteins L31 and L36 (Fig. 1). Among the PecS target genes, about 53% are upregulated, and the remaining 47% are downregulated. This is consistent with previous observations, which have shown that PecS could regulate gene expression either as a repressor (44, 45, 49, 53) or as an antirepressor (38).
FIG. 1.
Expression ratios of genes in pecS mutant versus wild-type strain along the chromosome. Blue points represent genes that were expressed similarly in pecS and wild-type (WT) strains. Red points represent genes that were differentially expressed in the pecS mutant versus the wild-type strain with an FDR-adjusted P value of <0.05. Genes with a greater than ±1.33-fold difference in the expression ratio fall outside of the field marked by dotted lines. The gene index is organized so that gene 1 corresponds to the origin of replication, OriC.
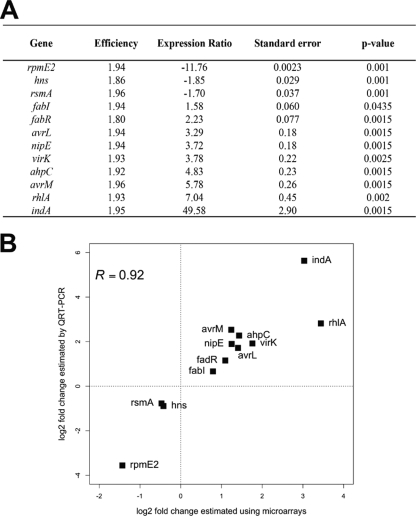
As only four genes have a relative change greater than ±1.33-fold and are not differentially expressed, based on P values (Fig. 1), we decided to define the PecS regulon as 134 genes with P values of <0.05 that were altered in expression more than 1.33-fold. On the basis of these criteria, most of the previously identified PecS target genes are included in the 134 genes retained. Indeed, 18% of these genes (25 out of 134) have already been shown to be regulated by PecS, such as pelD or outD (relative change ratios of 1.36 and 1.40, respectively). This constituted an internal validation of the assays (Fig. 1; see also Table S1 in the supplemental material). In order to validate several new PecS targets, 12 genes were selected, based on their putative function or the degree of PecS regulation, for subsequent analysis using qRT-PCR (Fig. 2A). Transcript levels were measured from new biological replicates and, thus, the real-time RT-PCR results are independent from the results obtained with microarray analysis. Ratios of the relative change in gene expression observed by microarrays ranged from −2.63 (rpmE2) to 8.18 (indA) whereas values determined by qRT-PCR ranged between −11.76 (rpmE2) and 49.58 (indA) (Fig. 2B). Overall, the qRT-PCR confirmed the microarray data since the correlation coefficient (R) calculated between relative changes obtained by microarray analysis and RT-PCR analysis is 0.92 (Fig. 2B).
FIG. 2.
Validation of microarray results by qRT-PCR. (A) Expression ratios of genes in pecS mutant versus the wild-type strain measured by qRT-PCR. Expression of each gene was normalized to the expression of the two housekeeping genes, rpoA and ffh. A positive expression ratio indicates upregulated genes in the pecS background, and a negative expression ratio indicates downregulated genes in the pecS background. Standard errors were calculated from the data from three independent biological replicates. (B) Comparison of gene expression measurements by microarray hybridization and real-time qRT PCR. The correlation coefficient (R) is given.
PecS regulates proteins homologous to secreted proteins from other plant-associated bacteria.
To get an insight into the PecS regulon, differentially expressed genes were clustered according to gene ontology. A total of 71% of all PecS-dependent genes retained in our analysis are associated with gene ontology terms (i.e., a biological process, molecular function, or cellular component). Cellular localization clustering of gene products shows that extracellular localization is overrepresented. Globally, 20% (27 genes) of the 134 retained PecS target genes encode either exported proteins or secretion systems that are known, or thought, to play an important role in the interaction between E. chrysanthemi and plant hosts.
Among the differentially expressed genes, 12 encoded secreted degradative enzymes (PrtA, PrtB, PrtC, PrtG, CelZ, XynA, PelA, PelC, PelD, PelE, PelL, and PelN) (Table 3), illustrating the overrepresentation of genes encoding proteins with plant cell wall catabolism functions (9%). Six of these genes (pelA, pelC, pelD, pelE, pelL, and celZ) have been previously identified as targets of PecS using a gene fusion approach (28, 49). This indicates that, in addition to the pel and celZ genes, pecS also controls expression of the genes encoding the four metalloproteases PrtA,-B, -C, and -G (threefold change) and the xylanase XynA (1.83-fold change). In addition to the genes encoding the degradative enzymes, we have identified those encoding proteins AvrL (GeneID 19143; 2.65-fold change) and AvrM (GeneID 15381; 2.36-fold change), which have been recently described as constituents of the E. chrysanthemi secretome (22). AvrL and AvrM are similar to the avirulence protein AvrXa from Xanthomonas campestris, which confers avirulence to most Arabidopsis thaliana accessions (39). Two proteins, HcpA (GeneID 17270; 1.38-fold change) and HcpB (GeneID 15893; 1.33-fold change), similar to the hemolysin-coregulated proteins (Hcps) ECA4275 and ECA2866 from Erwinia atroseptica, were upregulated in the pecS mutant. The Hcps ECA4275 and ECA2866 belong to the secretome of E. atroseptica and were specifically induced by plant host extract (31). Moreover, a bacterial strain overexpressing these Hcps was shown to have increased virulence (31). Five additional target genes are predicted by PSORT (15) to encode exported proteins and are related to proteins from other plant-associated bacteria: GeneID 19566 and GeneID 15573 (2.04-fold change for both genes), YoaJ (GeneID 14642; 1.46-fold change), VirK (GeneID 20481; 3.4-fold change), and NipE (GeneID 15387; 2.38-fold change). GeneID 19566 and GeneID 15573 displayed, respectively, 63% and 70% identity with the proteins of unknown function ECA2469 and ECA2267 from E. atroseptica, and they could be involved in the interaction between Erwinia species and plant hosts. Moreover, YoaJ, which is present in Erwinia carotovora, was annotated as a putative endoglucanase and was controlled by quorum sensing (40). VirK is a protein of unknown function from Agrobacterium tumefaciens. Interestingly, a VirK homologue was also found in Ralstonia solanacearum and is regulated by the master regulator HrpG, which also controls hrp genes as well as cellulase and pectinase genes (60). Finally, NipE was characterized as a necrosis-inducing protein in E. carotovora. This last protein is produced when E. carotovora is grown on solid medium at acid pH, which points to its involvement in the early steps of infection (32). NipE has been identified for the first time in Phytophthora, where it is named Nep1 (for necrosis and ethylene-inducing peptide 1). This protein seems to be widespread in both plant-necrotic fungi and plant-necrotic bacteria (17). Nep1-like proteins induce specific plant cell death and play dual roles in plant-pathogen interactions as toxin-like virulence factors and as triggers of plant innate immune responses (46). However, under the conditions tested, no special phenotype with regard to virulence could be attributed to the virK, nipE, or ΔavrLM mutations (see below).
TABLE 3.
A subset of genes whose expression is dependent on PecS in E. chrysanthemi that have been selected for their potential link with plant interaction
| Gene product category and gene name | GeneID no. | Gene product description | Fold changeb | FDR-adjusted P value |
|---|---|---|---|---|
| Secreted proteins and secretion systems | ||||
| avrL | 19143 | Avirulence protein L | 2.65 | 3.33E-06 |
| avrM | 15381 | Avirulence protein M | 2.36 | 8.21E-06 |
| celZa | 18772 | Endo-1,4-β-glucanase | 4.04 | 2.04E-07 |
| hcpA | 17270 | Haemolysin-coregulated protein | 1.36 | 1.82E-03 |
| hcpB | 15893 | Haemolysin-coregulated protein | 1.33 | 5.31E-03 |
| hrpBa | 19592 | HrpB, type III protein secretion system | 1.48 | 5.38E-05 |
| nipE | 15387 | Similar to necrosis-inducing protein | 2.38 | 2.95E-06 |
| outDa | 18230 | Type II secretory pathway component, OutD | 1.40 | 2.72E-03 |
| pelAa | 19643 | Pectate lyase PelA | 1.40 | 1.03E-03 |
| pelCa | 20837 | Pectate lyase PelC | 1.98 | 7.88E-06 |
| pelDa | 19648 | Pectate lyase PelD | 1.36 | 9.01E-03 |
| pelEa | 19646 | Pectate lyase PelE | 2.95 | 1.83E-06 |
| pelL | 18773 | Pectate lyase PelL | 2.99 | 1.15E-06 |
| pelN | 19391 | Pectate lyase PelN | 2.43 | 1.15E-06 |
| prtA | 20373 | Secreted metalloprotease A | 2.80 | 3.42E-06 |
| prtB | 47107 | Secreted metalloprotease B | 3.13 | 2.51E-07 |
| prtC | 20371 | Secreted metalloprotease C | 3.41 | 2.51E-07 |
| prtG | 17123 | Secreted metalloprotease G | 2.33 | 6.62E-06 |
| virK | 20481 | Similar to VirK of A. tumefaciens | 3.39 | 3.48E-07 |
| xynA | 19026 | Xylanase | 1.83 | 1.12E-03 |
| yoaJ | 14642 | Endoglucanase | 1.46 | 9.78E-03 |
| 19566 | Unknown exported protein | 2.04 | 1.37E-04 | |
| 15573 | Unknown exported protein | 2.01 | 8.70E-04 | |
| Oxidative stress response proteins | ||||
| acsA | 18857 | Achromobactin biosynthetic protein AcsA | 1.46 | 1.12E-03 |
| acsC | 18859 | Achromobactin biosynthetic protein AcsC | 1.36 | 1.03E-03 |
| acsF | 18866 | Diaminobutyrate-2-oxoglutarate transaminase | 1.33 | 4.13E-03 |
| ahpC | 18570 | Alkyl hydroperoxide reductase small subunit | 2.70 | 1.43E-05 |
| asnB | 46967 | Asparagine synthase B | 1.71 | 1.52E-02 |
| atpA | 14787 | Membrane-bound ATP synthase, F1 sector, alpha-subunit | 1.49 | 3.61E-04 |
| atpB | 14794 | F0F1-type ATP synthase, subunit a | 1.39 | 1.40E-03 |
| atpC | 14784 | F0F1-type ATP synthase, epsilon subunit | 1.39 | 7.71E-04 |
| atpD | 14785 | F0F1-type ATP synthase, beta subunit | 1.33 | 1.59E-04 |
| atpE | 14792 | F0F1-type ATP synthase, subunit c | 1.39 | 1.67E-04 |
| atpF | 14791 | F0F1-type ATP synthase, subunit b | 1.40 | 1.43E-04 |
| atpG | 14786 | F0F1-type ATP synthase, gamma subunit | 1.40 | 1.37E-04 |
| atpH | 14789 | F0F1-type ATP synthase, delta subunit | 1.49 | 2.97E-04 |
| atpI | 14795 | F0F1-type ATP synthase, subunit I | 1.33 | 8.40E-04 |
| grxC | 20419 | Lipid hydroperoxide reductase | 1.40 | 4.90E-04 |
| indAa | 16084 | Indigoidine biosynthesis | 8.18 | 3.26E-08 |
| indBa | 16083 | Indigoidine biosynthesis | 12.24 | 4.16E-09 |
| indCa | 16081 | Indigoidine synthase | 12.94 | 3.26E-08 |
| groL/mopA | 18669 | Chaperonin GroEL | 1.35 | 1.94E-02 |
| tsaA | 17551 | Peroxiredoxin | 1.73 | 4.28E-03 |
| yfeH | 15572 | Similar to putative cytochrome oxidase from E. coli/Shigella | 1.51 | 9.18E-04 |
| Motility proteins | ||||
| flgE | 17935 | Flagellar biosynthesis, hook protein | 1.51 | 1.27E-02 |
| flgF | 17936 | Flagellar biosynthesis, cell-proximal portion of basal-body rod | 1.39 | 3.72E-02 |
| fliA | 19722 | Alternative sigma factor F | 1.50 | 6.53E-03 |
| fliC | 19735 | Flagellin, filament structural protein | 1.38 | 9.13E-03 |
| fliD | 17963 | Flagellar biosynthesis; filament capping protein; enables filament assembly | 1.34 | 2.26E-03 |
| fliY | 17013 | Amino acid ABC transporter, periplasmic amino acid-binding protein | −1.45 | 6.03E-05 |
| fliZ | 19721 | Regulatory protein | 2.14 | 2.40E-03 |
| rhlA | 19839 | Acyltransferase | 10.87 | 3.43E-07 |
| 14536 | Methyl-accepting chemotaxis protein | 2.04 | 8.21E-06 | |
| 18765 | Methyl-accepting chemotaxis protein | −1.43 | 2.19E-03 | |
| Regulatory proteins | ||||
| fadR | 18979 | Negative regulator for fad regulon and positive activator of fabA | 2.13 | 6.29E-06 |
| hha | 19540 | Hemolysin expression modulating protein | −1.34 | 1.51E-03 |
| hns | 20646 | H-NS protein; DNA-binding protein HLP-II (HU, BH2, HD, NS); pleiotropic regulator | −1.34 | 5.83E-04 |
| rsmA | 19320 | Posttranscriptional RNA-binding regulatory protein RsmA | −1.39 | 8.65E-03 |
| vfmE | 16073 | DNA-binding transcriptional regulatory protein VfmE | 2.06 | 3.48E-07 |
Previously shown as regulated by PecS.
Positive values represent genes upregulated in the pecS mutant, whereas negative values represent genes downregulated in the pecS mutant compared to the wild-type strain.
PecS has been previously shown to be involved in the regulation of the out genes, encoding the type II secretion system responsible for pectinase and cellulase secretion (44). Our microarray experiments confirmed the regulation of the type II secretion system Out by PecS since outD was differentially expressed between the wild-type and pecS strains, with a 1.4-fold change. Moreover, PecS is also involved in the regulation of the hrp genes, encoding a type III secretion system, since a 1.48-fold change was detected for hrpB.
PecS and bacterial oxidative stress response.
Plants induced an oxidative stress in response to bacterial infection. PecS was shown to strongly repress the expression of the indA, indB, and indC genes involved in the biosynthesis of the blue pigment indigoidine, a major factor in the resistance of E. chrysanthemi to oxidative stress (50). In accordance with the previous data, we have shown here that the ind genes are among the most upregulated in the pecS mutant (ratio equal to at least 8). Interestingly, indigoidine is synthesized from glutamine as a precursor, and the gltP gene encoding a glutamate/aspartate symporter (GeneID 20737) is also upregulated 1.58-fold in the pecS mutant. In addition, four other genes possibly involved in resistance to oxidative stress were also upregulated in the pecS mutant. These are grxC (GeneID 20419), encoding a peroxiredoxin/glutaredoxin; yfeH (GeneID 15572), encoding a protein similar to a cytochrome oxidase; and tsaA (GeneID 17551) and ahpC (GeneID 18570), both encoding alkyl hydroxyperoxide reductases (Table 3). Previous genetic analysis suggested that, in addition to the ind genes, PecS exerts a regulatory influence over other genes involved in the resistance to oxidative stress (50). We therefore hypothesized that some of the above mentioned genes might encode these additional systems. To assess this hypothesis, we cloned ahpC, which is the most strongly regulated of these genes (2.7-fold change), and we constructed the corresponding mutant by reverse genetics. The MIC of H2O2 was evaluated for the wild-type strain and its derivatives, pecS, ahpC, and ahpC-pecS mutants. MICs of H2O2 for the wild-type and ahpC strains were 500 μM ± 0 and 750 μM ± 0 for pecS and ahpC-pecS. The pecS mutant is clearly more resistant to oxidative stress than the wild-type strain. However, inactivation of ahpC did not decrease the MICs, indicating that the other peroxidases tsaA and grxC, detected as deregulated in the pecS background by microarray analysis, might have a compensatory effect. Further investigations should clarify this assertion by analyzing the phenotype of the double mutants ahpC tsaA and ahpC grxC. In addition, all the proteins constituting the bacterial ATP synthase are slightly upregulated in the pecS mutant (1.4-fold change) (Table 3). In an indirect manner, the increase in oxygen reduction by the ATP synthase could contribute to resistance to oxidative stress. Moreover, asnB, encoding asparaginase, and groL, encoding chaperonin GroEL, are part of the PecS regulon. Increases in AsnB and the chaperonin GroEL have been previously observed in response to oxidative stress in Mycobacterium (12). Therefore, the increased expression of these targets in the pecS mutant strain may influence the oxidative stress response. Finally, the high-affinity iron uptake system, Acs (achromobactin), is also upregulated in the pecS mutant (Table 3). This system is required for bacterial iron nutrition, but it is also described as having an antioxidant activity in planta (7). Overall, these data suggest that, by controlling various key systems involved in resistance to oxidative stress, PecS might play an essential role in the detoxification of different oxidative stressor compounds produced by the plant host during infection.
PecS and motility genes.
In accordance with the increased motility observed in the pecS mutant (53), seven genes involved in the biogenesis of the flagellum have been identified among the upregulated genes. These include the fliAZY and flgEF operons (1.5-fold change), fliC (1.38-fold change), and fliD (1.34-fold change) (Table 3). FliA is a specific sigma factor for flagellum synthesis whereas FliZ is an activator involved in the stimulation of the class II genes in the regulatory cascade of flagellum synthesis (24). Moreover, in the human-pathogenic bacterium Salmonella enterica serovar Typhimurium, FliZ regulates the expression of genes responsible for pathogenicity and host invasion (21). The four other genes identified in this analysis encode structural components of the flagellum: fliC encodes the flagellin, FliD is involved in the polymerization of flagellin, and the two genes organized in the operon flgEF encode proteins of the basal body rod. Previously, PecS was shown to directly repress the expression of the fliE gene and divergent fliFGHIJKLMNOPQR operon (53). This microarray analysis confirmed that PecS effects are not restricted to these two operons but that PecS has a broader effect on the regulation of flagellum synthesis in E. chrysanthemi.
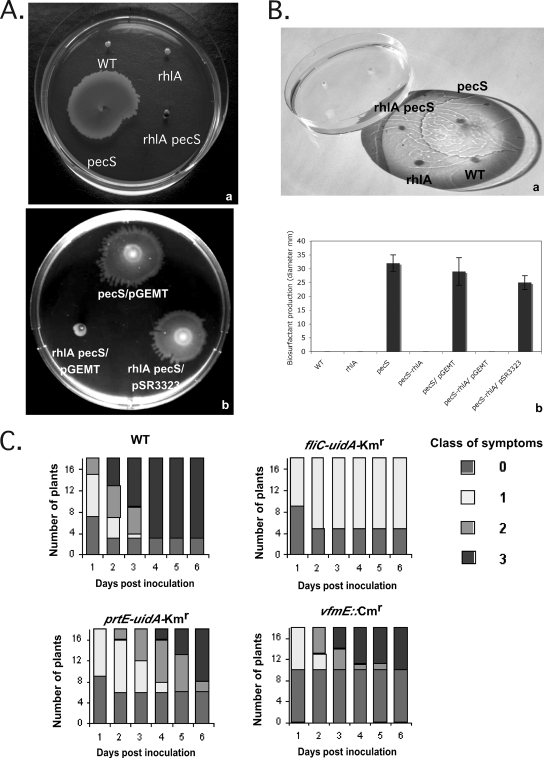
The regulatory overlaps between virulence genes and flagella have been observed in Salmonella (23) and in enteropathogenic Escherichia coli (18). This connection probably reflects a need for the pathogen to coordinate its physical motility with the expression of genes involved in niche invasion and adaptation. We showed that motility is also required for E. chrysanthemi virulence since the symptoms caused by the nonmotile fliC mutant remained confined to the leaf area infiltrated with the bacterial inoculum, and no systemic infections were observed with this mutant (Fig. 3C). Also upregulated by a factor of 11-fold in the pecS mutant was the gene (GeneID 19839) encoding an acyltransferase (Table 3). The corresponding protein displays 43% identity and 65% similarity with the RhlA protein from Pseudomonas aeruginosa, which is involved in the synthesis of surface-active β-d-(β-d-hydroxyalkanoyloxy) alkanoic acids (HAA), the precursors of rhamnolipid surfactants promoting swarming motility (11, 65). In order to investigate the potential role of rhlA in E. chrysanthemi, we constructed the corresponding mutant by reverse genetics in both the wild type and pecS. We further compared the biosurfactant production of the pecS mutant in semisolid medium and its swarming motility on the surface of a solid medium with that of the parental strain and the double rhlA pecS mutant. As shown in Fig. 3A and B, the pecS mutant produced biosurfactant promoting swarming motility, whereas the wild-type strain did not. In addition, introduction of an rhlA mutation in the pecS background abolished biosurfactant production and swarming motility. The swarming motility and the biosurfactant production of the double rhlA pecS mutant were restored nearly to the pecS strain level by introducing the plasmid pSR3323 containing the rhlA gene (Fig. 3A and B, panel b). These results demonstrated that rhlA is indeed essential for the production of extracellular surfactant and swarming, presumably by catalyzing the formation of surface-active HAA. Recently, RhlA-dependent surfactant biosynthesis was also reported in Serratia, and in this bacteria RhlA was also required for the multicellular swarming phenotype (62).
FIG. 3.
Phenotypic analysis. (A) Swarming motility of various bacterial strains (a). Bacterial strains were inoculated onto LB-1% agar plates and incubated at 30°C for 24 h. Swarming motility of the rhlA pecS double mutant complemented with plasmid pSR3323 expressing RhlA is shown (b). (B) Biosurfactant production of various bacterial strains. Bacterial strains were inoculated by stabbing 0.7% semisolid agar plates and incubated at 30°C for 8 h. A wet puddle appears on the surface of the agar medium for the pecS strain producing biosurfactant (a). The size of the halo of this secreted substance is a direct assay of the surfactant production (b). Data shown are the means ± standard deviations of at least three independent experiments. (C) Interactions between the fliC, prtE, and vfmE mutants and S. ionantha plants. Infection was performed on a single leaf per plant by infiltration of 107 bacteria in Saintpaulia. Symptom occurrence was scored daily for a week. Symptoms are classified in four stages: stage 0, no symptoms; stage 1, rotting confined to the infiltrated zone; stage 2, maceration of the leaf limb; stage 3, maceration of the whole leaf including the petiole. At least three independent experiments were performed, and for each, mutant scores were different from wild-type scores, with a P of < 0.05. Results of a typical experiment are presented. Results obtained with the prtE-uidA-Kmr and vfmE::Cmr mutants were confirmed with independent mutants (prtE::MudII1734 and vfmE-uidA-Kmr). For the fliC-uidA-Kmr mutant, two independent transductants were analyzed, and similar results were obtained. WT, wild type.
PecS and regulators.
The microarray analysis shows that several PecS-controlled genes encode regulatory proteins (Table 3). This suggests that for several PecS targets the regulation could be indirect. For example fadR is upregulated by a factor of 2 in the pecS mutant. This gene encodes a transcriptional regulator that represses genes involved in the fatty acid biosynthetic pathway. Accordingly, 8 of the 12 genes implicated in this pathway are members of the PecS regulon: accA, accC, and accD, which are involved in the initial steps of fatty acid biosynthesis; and fabB, fabI, fabH, fabD, and fabG, which are involved in fatty acid elongation. This suggests that PecS indirectly regulates fatty acid biosynthesis genes, and this could be mediated by FadR. Acceleration of fatty acid synthesis in the pecS mutant could offer a correction for the drain on the pathway due to the formation of HAA by RhlA and ensure that the supply of fatty acid for membrane phospholipid synthesis is not compromised.
Three genes that encode proteins with global regulatory roles were downregulated in the absence of PecS. The hns and hha genes (−1.33-fold change), whose products can form heteromeric complexes and regulate virulence genes in response to temperature in several enterobacteria (29), and the rsmA gene (−1.39-fold change), whose product is a posttranscriptional regulatory RNA-binding protein that controls virulence gene expression in Erwinia species (10), were downregulated. This suggests that some targets revealed by transcriptome analysis could be subject to indirect regulation by PecS via these global regulators. In addition, this shows that PecS can act at multiple levels within a regulatory hierarchy. For example H-NS regulates pectate lyases (35) as does PecS, which also interacts with promoters of pel genes. Moreover, RsmA destabilizes pel mRNA. The reduced expression of rsmA in the pecS background could contribute to the overproduction of pectate lyases observed in the pecS mutant. The VfmE regulator, which belongs to the AraC family, was derepressed by a factor of 2.04 in the pecS mutant. This regulator activates the expression of plant cell wall-degrading enzymes (Reverchon et al., unpublished results). This is the first report that PecS regulates other regulatory proteins and might be at the top of several hierarchies. The possible interplay of the H-NS, Hha, RsmA, and VfmE regulators with PecS is interesting since it has now become evident that regulation of bacterial virulence gene expression is a complex process (8).
PecS binds the rhlA, ahpC, nipE, virK, and avrL promoters.
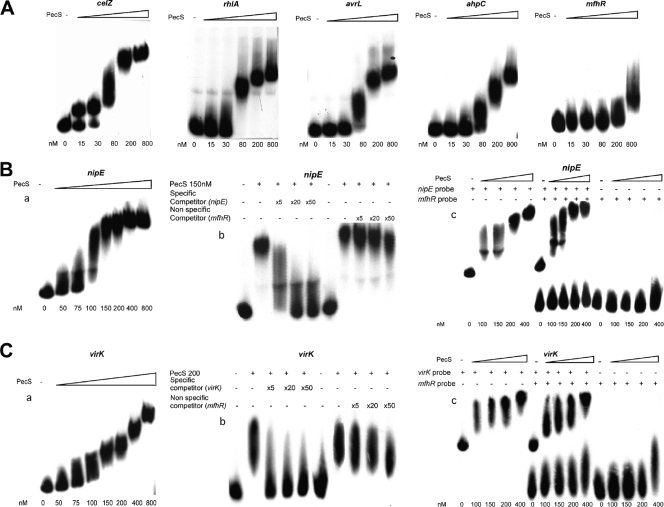
PecS is known to regulate pectate lyases and cellulase by directly binding the pel and celZ promoters (44); however, as PecS also controls the expression of regulatory genes (Table 3), PecS could have indirect effects on the transcription of other genes. To determine whether PecS directly interacts with some genes identified by microarray analysis, we examined the ability of purified PecS to bind promoters of a subset of PecS-regulated loci. These genes were selected based on an interest in their putative function and the degree of PecS-dependent regulation observed by microarray analysis. We selected five genes likely to be important for plant-bacteria interaction (rhlA, ahpC, nipE, virK, and avrL). In vitro interaction between the purified PecS protein and the regulatory regions of these five genes was investigated. The promoter of celZ was used as a positive control for PecS-binding, and the mfhR nonspecific fragment derived from the coding region of a transcriptional regulator not controlled by PecS (−1.03-fold change by microarray analysis) was used as a negative control. Using electrophoretic mobility shift assays (EMSAs), we found that PecS specifically interacts with the five targets, albeit with different affinities. rhlA and ahpC display the highest affinity, followed by avrL and, finally, nipE and virK (Fig. 4). To further demonstrate the specificity of binding of PecS to the nipE and virK regulatory regions, excess unlabeled mfhR fragment was incubated with PecS and either 32P-labeled nipE probe or 32P-labeled virK probe (Fig. 4B and C, panels b). Up to 50-fold molar excess of unlabeled mfhR fragment, acting here as a nonspecific competitor, did not block the binding of PecS to the nipE or virK probe, whereas binding was increasingly reduced in the control assay using the cold specific competitor DNA. Simultaneous titrations with PecS and both 32P-labeled nipE probe and 32P-labeled mfhR probe or both 32P-labeled virK probe and 32P-labeled mfhR probe were also performed (Fig. 4B and C, panels c). In these simultaneous titrations, nipE and virK probes were shifted whereas the mfhR probe was not. These findings, therefore, suggest that PecS is capable of regulating the expression of the rhlA, ahpC, nipE, virK, and avrL genes by direct binding to the promoter regions.
FIG. 4.
Band shift assay for PecS-DNA binding. (A) Labeled DNA probes, corresponding to promoter regions of rhlA, avrL, and ahpC, were incubated with increasing concentrations of PecS, as indicated at the bottom. The celZ regulatory region was used as a positive control (44), and the mfhR nonspecific fragment derived from the coding region of a transcriptional regulator not controlled by PecS was used as a negative control. (B) Specificity of PecS binding on nipE regulatory region. EMSA shows binding of PecS to the 400-bp nipE regulatory region (region −300 to +100 relative to the translation start site) with an increasing PecS concentration (0, 50, 75, 100, 150, 200, 400, and 800 nM) in the presence of 30 fmol of radiolabeled probe (a). In the reactions mixtures containing 30 fmol of radiolabeled probe and 150 nM PecS, competition assays were performed with the molar excesses of the unlabeled 400-bp nipE regulatory region specific competitor indicated above the lanes (×0, ×5, ×20, and ×50) and with the indicated molar excesses of unlabeled nonspecific competitor DNA derived from a 250-bp fragment of the mfhR coding region (b). Simultaneous titrations were performed with PecS and both 32P-labeled nipE probe and 32P-labeled mfhR probe (c). (C) Specificity of PecS binding on virK regulatory region. EMSA shows binding of PecS to the 440-bp virK regulatory region (region −370 to +70 relative to the translation start site) with an increasing PecS concentration (0, 50, 75, 100, 150, 200, 400, and 800 nM) in the presence of 30 fmol of radiolabeled probe (a). In the reaction mixtures containing 30 fmol of radiolabeled probe and 200 nM PecS, competition assays were performed with molar excesses of the unlabeled 440-bp virK regulatory region specific competitor indicated above the lanes (×0, ×5, ×20, and ×50) and with the indicated molar excesses of unlabeled nonspecific competitor DNA derived from a 250-bp fragment of the mfhR coding region (b). Simultaneous titrations were performed with PecS and both 32P-labeled virK probe and 32P-labeled mfhR probe (c).
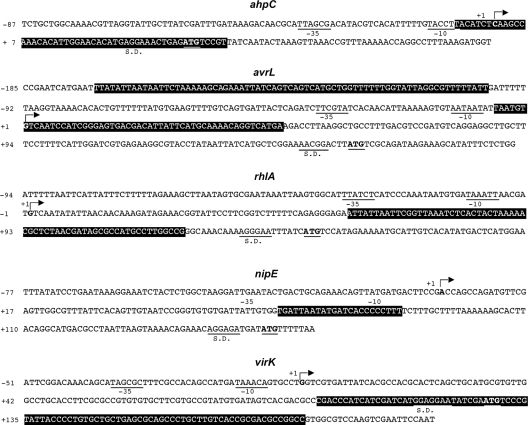
To confirm this assertion, the transcription start point of each gene was determined by primer extension. In each case, a unique transcription start point was identified which was preceded by −10 and −35 sequences that could represent σ70 RNA polymerase DNA binding sites (Fig. 5; see also Fig. S1A in the supplemental material). Finally, the regions of the DNA segment that interact with PecS were identified by DNase I footprinting analysis (Fig. 5; see also Fig. S1B in the supplemental material). A unique clear footprint was obtained with the rhlA, ahpC, nipE, and virK operators. Consistent with the EMSA results, the protection on rhlA and ahpC was observed at lower PecS concentrations, whereas the protection on nipE and virK was seen at the highest concentrations. For ahpC, the protected area overlaps the +1 transcriptional start point, suggesting that PecS inhibits transcription initiation at this promoter. In the case of the rhlA, nipE, and virK operators, the footprints are located downstream from the +1 transcriptional start sites, which suggests that PecS inhibits transcriptional elongation of these genes. Finally, two footprints were obtained at high PecS concentrations for avrL, one overlapping the +1 transcriptional start site and another one spanning the upstream promoter region. This suggests that PecS represses avrL gene expression by inhibition of transcript elongation and by competing with an unidentified regulator for occupation of the upstream portion of the regulatory region. Overall, these results indicate that PecS directly represses rhlA, ahpC, nipE, virK and avrL gene expression. In these different regions protected by PecS, no clear motif resembling the PecS consensus defined by SELEX (53) could be identified, suggesting that in vivo PecS recognizes a DNA structural conformation motif more than a specific primary sequence.
FIG. 5.
Sequence of the ahpC, avrL, rhlA, nipE, and virK regulatory regions. The transcription start sites are indicated by arrows and by a bold character, and the −10, −35 RNA polymerase-binding regions are underlined. The initiation codons are in bold letters and are underlined. The PecS binding sites are boxed in black. The Shine-Dalgarno sequence (S.D.) corresponding to the ribosome binding sites are underlined.
Pathogenicity of the rhlA, ahpC, nipE, virK, ΔavrLM, fliC, prtE, and vfmE mutants.
Several PecS-controlled genes have been shown to encode factors involved in E. chrysanthemi pathogenicity, such as the genes encoding several pectinases, the out genes encoding the type II secretion system required for secretion of these enzymes into the extracellular medium, and the genes involved in the biosynthesis of the antioxidant indigoidine. To examine whether other virulence factors could be identified among the new PecS target genes, pathogenicity tests were performed with two levels of inoculum (106 and 107 bacteria) in S. ionantha plants, the host from which our 3937 strain was isolated, and with one level of inoculum (106 bacteria) on chicory leaves. Under our experimental conditions, no differences were observed in the virulence of the rhlA, ahpC, nipE, virK, and ΔavrLM mutants compared with the symptoms caused by the wild-type parent, regardless of the inoculum size and the plant host tested (data not shown). The lack of phenotypes could be due to the presence of genes with overlapping functions in the genome. For example in the ahpC mutant, tsaA and grxC encoding peroxidases might have compensatory effects. Moreover, testing an inappropriate plant host could explain the absence of phenotypes. Indeed, in E. carotovora subsp. carotovora the Nip− mutant strain showed reduced virulence in a potato tuber assay but was unaffected in virulence in potato stem and on other tested host plants (lettuce, eggplant, cauliflower, broccoli, and celery) (32).
Mutants affected in three other PecS targets, namely, a fliC mutant impaired in flagellin production and, thus, nonmotile; prtE mutants defective in the type I secretion system responsible for the secretion of the four metalloproteases PrtA, PrtB, PrtC, and PrtG; and mutants defective in the vfmE regulatory gene, all showed altered virulence compared to the wild-type parent. The fliC mutant was able to cause maceration of the leaf area infiltrated with the bacterial inoculum, but the symptoms very rarely spread outside this area. Both prtE and vfmE mutants exhibited delayed symptoms, and, for the vfmE mutants, more plants remained symptomless (Fig. 3C). Thus, some new PecS target genes are shown to encode virulence determinants.
Conclusion.
In order to establish the extent of the PecS regulon and to understand the PecS function in the different stages of E. chrysanthemi pathogenesis, we have undertaken a transcriptomic analysis of the pecS mutant strain compared to the wild-type strain. The data presented here show that PecS exerts wide-ranging effects on gene expression in E. chrysanthemi, justifying its description as a global regulator. Functional clustering reveals that the majority of PecS targets encode proteins involved in motility, secretion systems, stress response, and secreted proteins. Thus, PecS regulates a wide range of genes that could be linked with adaptation to the interaction with the host plant, i.e., virulence factors and genes governing protective functions to evade host defenses.
Upon entering a host plant, E. chrysanthemi cells colonize the intercellular spaces of the cortical parenchyma and migrate within the cell walls without causing any severe injury to the cellular structures (13, 34). During this colonization phase there is no production of plant cell wall-degrading enzymes (26), but bacteria have to adapt to the apoplast environment, which is an acidic, low-nutrient medium with a low availability of iron.
After the colonization phase, the bacteria may reside latently in the plant intercellular spaces without provoking any symptoms or may start the disease process. Thus, disease caused by E. chrysanthemi is an intricate process with two successive phases, an asymptomatic phase and a symptomatic phase, that require the temporal expression of different groups of genes. We postulate that this temporal regulation is, in part, the result of a gradual modulation of PecS function triggered during the asymptomatic phase in response to changes in environmental conditions emerging from the interaction between the partners. To date, the mechanism of PecS inactivation remains unknown; it could be due to a modulation in PecS synthesis, in PecS degradation, or in PecS activity. As PecS belongs to the MarR family of transcriptional regulators whose DNA-binding capacity is attenuated by specific anionic lipophilic ligands (57), we suppose that PecS activity could be modulated by a lipophilic signal produced by the plant in response to bacterial invasion. In the early phase of colonization, this signal is probably present in low concentrations in the apoplast and results in a partial inactivation of PecS activity, leading to the derepression of the target genes weakly or moderately controlled by PecS such as fliC for motility; ahpC, tsaA, and grxC for the oxidative stress response; and acs for achromobactin siderophore biosynthesis to compete with the plant cells for iron assimilation. During the late asymptomatic phase, the plant perceives the bacterium as an intruder and probably induces a sustained production of the signal. This leads to a strong inactivation of PecS, which would, in turn, result in production of large amounts of the radical scavenger pigment indigoidine, the cellulase CelZ, and also the biosurfactant whose synthesis is directed by RhlA, all of which were strongly and directly repressed by PecS. This production of the coregulated virulence factors accompanied by the significant production of pectinases allows the development of soft-rot symptoms and an efficient propagation of the bacteria in the host tissues. This would result in production of nutriments for bacterial multiplication. Thus, PecS regulation is a key to providing both optimal nutrition and resistance to the hostile conditions encountered in macerating tissues. Under our laboratory conditions, which are very favorable to disease development, the loss of PecS regulation provokes the overexpression of achromobactin and of the antioxidant genes, giving a survival advantage to the pecS mutant in planta. The overproduction of plant cell wall-degrading enzymes, such as pectinases, proteases, xylanase, cellulase, and probably other toxin-like virulence factors, such as NipE and Hcps, confers a greater aggressivity to the pecS mutant. However, under natural conditions, the premature production of plant cell wall-degrading enzymes in the pecS mutant might be deleterious in the dynamic interplay between bacterial colonization and plant defense activation. Based on these observations, we propose a pivotal role of PecS in the switch from the asymptomatic to the symptomatic lifestyle.
The synthesis of virulence factors must be finely regulated to ensure their expression at appropriate stages of infection. This is illustrated by the very complex regulation of pectinase-encoding genes. Several regulators are involved, suggesting very strict control to prevent their expression at the onset of infection. This microarray investigation has pointed out that PecS controls two global regulatory proteins already known to be involved in pectinase regulation: H-NS and RsmA (10, 35). In addition, expression of vfmE encoding a new regulator of plant cell wall-degrading enzymes is also dependent on PecS. This suggests a much more complex regulatory network for the hierarchical induction of virulence factors in E. chrysanthemi. Future investigations should clarify how the action of PecS is integrated with the other regulators during the infection process.
Supplementary Material
Acknowledgments
We thank all the members of the Lyon and Paris Erwinia groups for their helpful discussions, Vladimir Shevchik for providing the E. chrysanthemi fliC mutant before publishing, Vincent Geoghegan and Valerie James for the English corrections, Loic Desquilbet for his advice in the statistical analysis of pathogenicity test data, and Géraldine Effantin for technical support. We acknowledge members of the International Erwinia Consortium for the exchange of unpublished data concerning the E. chrysanthemi 3937 genome and Thomas Lautier for initiating us with qRT-PCR experiments.
This work was supported by grants from the Centre National de la Recherche Scientifique, the Ministère de l'Education Nationale et de la Recherche, and the Programme Microbiologie (ACIM-2-17).
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andrews, J. 2001. Determination of minimum inhibitory concentrations J. Antimicrob. Chemother. 485-16. [DOI] [PubMed] [Google Scholar]
- 2.Bardonnet, N., and C. Blanco. 1992. ′uidA-antibiotic-resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol. Lett. 72243-248. [DOI] [PubMed] [Google Scholar]
- 3.Beaulieu, C., M. Boccara, and F. Vangijsegem. 1993. Pathogenic behavior of pectinase-defective Erwinia chrysanthemi mutants on different plants. Mol. Plant-Microbe Interact. 6197-202. [Google Scholar]
- 4.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B 57289-300. [Google Scholar]
- 5.Boccara, M., A. Diolez, M. Rouve, and A. Kotoujansky. 1988. The role of the individual pectate lyases of Erwinia chrysanthemi strain 3937 in pathogenicity on Saintpaulia plants. Physiol. Mol. Plant Pathol. 3395-104. [Google Scholar]
- 6.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19185-193. [DOI] [PubMed] [Google Scholar]
- 7.Boughammoura, A., T. Franza, A. Dellagi, C. Roux, B. Matzanke-Markstein, and D. Expert. 2007. Ferritins, bacterial virulence and plant defence. Biometals 20347-353. [DOI] [PubMed] [Google Scholar]
- 8.Brencic, A., and S. C. Winans. 2005. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 69155-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castang, S., S. Reverchon, P. Gouet, and W. Nasser. 2006. Direct evidence for the modulation of the activity of the Erwinia chrysanthemi quorum-sensing regulator ExpR by acylhomoserine lactone pheromone. J. Biol. Chem. 28129972-29987. [DOI] [PubMed] [Google Scholar]
- 10.Cui, Y., A. Chatterjee, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J. Bacteriol. 1775108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deziel, E., F. Lepine, S. Milot, and R. Villemur. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 1492005-2013. [DOI] [PubMed] [Google Scholar]
- 12.Dosanjh, N. S., M. Rawat, J. H. Chung, and Y. Av-Gay. 2005. Thiol specific oxidative stress response in Mycobacteria. FEMS Microbiol. Lett. 24987-94. [DOI] [PubMed] [Google Scholar]
- 13.Fagard, M., A. Dellagi, C. Roux, C. Perino, M. Rigault, V. Boucher, V. E. Shevchik, and D. Expert. 2007. Arabidopsis thaliana expresses multiple lines of defense to counterattack Erwinia chrysanthemi. Mol. Plant-Microbe Interact. 20794-805. [DOI] [PubMed] [Google Scholar]
- 14.Franza, T., I. Michaud-Soret, P. Piquerel, and D. Expert. 2002. Coupling of iron assimilation and pectinolysis in Erwinia chrysanthemi 3937. Mol. Plant-Microbe Interact. 151181-1191. [DOI] [PubMed] [Google Scholar]
- 15.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21617-623. [DOI] [PubMed] [Google Scholar]
- 16.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gijzen, M., and T. Nurnberger. 2006. Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 671800-1807. [DOI] [PubMed] [Google Scholar]
- 18.Grant, A. J., M. Farris, P. Alefounder, P. H. Williams, M. J. Woodward, and C. D. O'Connor. 2003. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48507-521. [DOI] [PubMed] [Google Scholar]
- 19.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50213-257. [DOI] [PubMed] [Google Scholar]
- 20.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4249-264. [DOI] [PubMed] [Google Scholar]
- 21.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 3081-90. [DOI] [PubMed] [Google Scholar]
- 22.Kazemi-Pour, N., G. Condemine, and N. Hugouvieux-Cotte-Pattat. 2004. The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics 43177-3186. [DOI] [PubMed] [Google Scholar]
- 23.Kelly, A., M. D. Goldberg, R. K. Carroll, V. Danino, J. C. Hinton, and C. J. Dorman. 2004. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 1502037-2053. [DOI] [PubMed] [Google Scholar]
- 24.Kutsukake, K., T. Ikebe, and S. Yamamoto. 1999. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet. Syst. 74287-292. [DOI] [PubMed] [Google Scholar]
- 25.Laatu, M., and G. Condemine. 2003. Rhamnogalacturonate lyase RhiE is secreted by the out system in Erwinia chrysanthemi. J. Bacteriol. 1851642-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebeau, A., S. Reverchon, S. Gaubert, Y. Kraepiel, E. Simond-Cote, W. Nasser, and F. Van Gijsegem. 2008. The GacA global regulator is required for the appropriate expression of Erwinia chrysanthemi 3937 pathogenicity genes during plant infection. Environ. Microbiol. 10545-559. [DOI] [PubMed] [Google Scholar]
- 27.Lojkowska, E., C. Dorel, P. Reignault, N. Hugouvieuxcottepattat, and J. Robertbaudouy. 1993. Use of Gus fusion to study the expression of Erwinia chrysanthemi pectinase genes during infection of potato tubers. Mol. Plant-Microbe Interact. 6488-494. [Google Scholar]
- 28.Lojkowska, E., C. Masclaux, M. Boccara, J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1995. Characterization of the pelL gene encoding a novel pectate lyase of Erwinia chrysanthemi 3937. Mol. Microbiol. 161183-1195. [DOI] [PubMed] [Google Scholar]
- 29.Madrid, C., J. M. Nieto, and A. Juarez. 2002. Role of the Hha/YmoA family of proteins in the thermoregulation of the expression of virulence factors. Int. J. Med. Microbiol. 291425-432. [DOI] [PubMed] [Google Scholar]
- 30.Maes, M., and E. Messens. 1992. Phenol as grinding material in RNA preparations. Nucleic Acids Res. 204374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattinen, L., R. Nissinen, T. Riipi, N. Kalkkinen, and M. Pirhonen. 2007. Host-extract induced changes in the secretome of the plant pathogenic bacterium Pectobacterium atrosepticum. Proteomics 73527-3537. [DOI] [PubMed] [Google Scholar]
- 32.Mattinen, L., M. Tshuikina, A. Mae, and M. Pirhonen. 2004. Identification and characterization of Nip, necrosis-inducing virulence protein of Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 171366-1375. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Murdoch, L., J. C. Corbel, D. Reis, Y. Bertheau, and B. Vian. 1999. Differential cell wall degradation by Erwinia chrysanthemi in petiole of Saintpaulia ionantha. Protoplasma 21054-74. [Google Scholar]
- 35.Nasser, W., and S. Reverchon. 2002. H-NS-dependent activation of pectate lyases synthesis in the phytopathogenic bacterium Erwinia chrysanthemi is mediated by the PecT repressor. Mol. Microbiol. 43733-748. [DOI] [PubMed] [Google Scholar]
- 36.Nasser, W., S. Reverchon, R. Vedel, and M. Boccara. 2005. PecS and PecT coregulate the synthesis of HrpN and pectate lyases, two virulence determinants in Erwinia chrysanthemi 3937. Mol. Plant-Microbe Interact. 181205-1214. [DOI] [PubMed] [Google Scholar]
- 37.Nasser, W., J. Robert-Baudouy, and S. Reverchon. 1997. Antagonistic effect of CRP and KdgR in the transcription control of the Erwinia chrysanthemi pectinolysis genes. Mol. Microbiol. 261071-1082. [DOI] [PubMed] [Google Scholar]
- 38.Nasser, W., V. E. Shevchik, and N. Hugouvieux-Cotte-Pattat. 1999. Analysis of three clustered polygalacturonase genes in Erwinia chrysanthemi 3937 revealed an anti-repressor function for the PecS regulator. Mol. Microbiol. 34641-650. [DOI] [PubMed] [Google Scholar]
- 39.Parker, J. E., C. E. Barber, M. J. Fan, and M. J. Daniels. 1993. Interaction of Xanthomonas campestris with Arabidopsis thaliana: characterization of a gene from X. c. pv. raphani that confers avirulence to most A. thaliana accessions. Mol. Plant-Microbe Interact. 6216-224. [DOI] [PubMed] [Google Scholar]
- 40.Pemberton, C. L., N. A. Whitehead, M. Sebaihia, K. S. Bell, L. J. Hyman, S. J. Harris, A. J. Matlin, N. D. Robson, P. R. Birch, J. P. Carr, I. K. Toth, and G. P. Salmond. 2005. Novel quorum-sensing-controlled genes in Erwinia carotovora subsp. carotovora: identification of a fungal elicitor homologue in a soft-rotting bacterium. Mol. Plant-Microbe Interact. 18343-353. [DOI] [PubMed] [Google Scholar]
- 41.Perombelon, M. C. M. 2002. Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol. 511-12. [Google Scholar]
- 42.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Praillet, T., W. Nasser, J. Robert-Baudouy, and S. Reverchon. 1996. Purification and functional characterization of PecS, a regulator of virulence-factor synthesis in Erwinia chrysanthemi. Mol. Microbiol. 20391-402. [DOI] [PubMed] [Google Scholar]
- 45.Praillet, T., S. Reverchon, and W. Nasser. 1997. Mutual control of the PecS/PecM couple, two proteins regulating virulence-factor synthesis in Erwinia chrysanthemi. Mol. Microbiol. 24803-814. [DOI] [PubMed] [Google Scholar]
- 46.Qutob, D., B. Kemmerling, F. Brunner, I. Kufner, S. Engelhardt, A. A. Gust, B. Luberacki, H. U. Seitz, D. Stahl, T. Rauhut, E. Glawischnig, G. Schween, B. Lacombe, N. Watanabe, E. Lam, R. Schlichting, D. Scheel, K. Nau, G. Dodt, D. Hubert, M. Gijzen, and T. Nurnberger. 2006. Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 183721-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Resibois, A., M. Colet, M. Faelen, E. Schoonejans, and A. Toussaint. 1984. Phi-Ec2, a new generalized transducing phage of Erwinia chrysanthemi. Virology 137102-112. [DOI] [PubMed] [Google Scholar]
- 48.Reverchon, S., M. L. Bouillant, G. Salmond, and W. Nasser. 1998. Integration of the quorum-sensing system in the regulatory networks controlling virulence factor synthesis in Erwinia chrysanthemi. Mol. Microbiol. 291407-1418. [DOI] [PubMed] [Google Scholar]
- 49.Reverchon, S., W. Nasser, and J. Robert-Baudouy. 1994. pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol. Microbiol. 111127-1139. [DOI] [PubMed] [Google Scholar]
- 50.Reverchon, S., C. Rouanet, D. Expert, and W. Nasser. 2002. Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J. Bacteriol. 184654-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robert-Baudouy, J., W. Nasser, G. Condemine, S. Reverchon, V. E. Shevchik, and N. Hugouvieux-Cotte-Pattat. 2000. Pectic enzymes of Erwinia chrysanthemi, regulation and role in pathogenesis, p. 221-268. In G. Stacey and N. T. Keen (ed.), Plant-microbe interactions, vol. 5. The American Phytopathological Society, St. Paul, MN. [Google Scholar]
- 52.Roeder, D. L., and A. Collmer. 1985. Marker-exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi. J. Bacteriol. 16451-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rouanet, C., S. Reverchon, D. A. Rodionov, and W. Nasser. 2004. Definition of a consensus DNA-binding site for PecS, a global regulator of virulence gene expression in Erwinia chrysanthemi and identification of new members of the PecS regulon. J. Biol. Chem. 27930158-30167. [DOI] [PubMed] [Google Scholar]
- 54.Sepulchre, J. A., S. Reverchon, and W. Nasser. 2007. Modeling the onset of virulence in a pectinolytic bacterium. J. Theor. Biol. 244239-257. [DOI] [PubMed] [Google Scholar]
- 55.Smyth, G. K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3Article3. [DOI] [PubMed] [Google Scholar]
- 56.Smyth, G. K., J. Michaud, and H. S. Scott. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 212067-2075. [DOI] [PubMed] [Google Scholar]
- 57.Sulavik, M. C., L. F. Gambino, and P. F. Miller. 1995. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol. Med. 1436-446. [PMC free article] [PubMed] [Google Scholar]
- 58.Takle, G. W., I. K. Toth, and M. B. Brurberg. 2007. Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant Biol. 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tardy, F., W. Nasser, J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1997. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J. Bacteriol. 1792503-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valls, M., S. Genin, and C. Boucher. 2006. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS Pathog. 2e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williamson, N. R., P. C. Fineran, W. Ogawa, L. R. Woodley, and G. P. Salmond. 2008. Integrated regulation involving quorum sensing, a two-component system, a GGDEF/EAL domain protein and a post-transcriptional regulator controls swarming and RhlA-dependent surfactant biosynthesis in Serratia. Environ. Microbiol. 101202-1217. [DOI] [PubMed] [Google Scholar]
- 63.Wisniewski, J. P., and P. M. Rogowsky. 2004. Vacuolar H+-translocating inorganic pyrophosphatase (Vpp1) marks partial aleurone cell fate in cereal endosperm development. Plant Mol. Biol. 56325-337. [DOI] [PubMed] [Google Scholar]
- 64.Yang, S., Q. Peng, Q. Zhang, X. Yi, C. J. Choi, R. M. Reedy, A. O. Charkowski, and C. H. Yang. 2008. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937). Mol. Plant-Microbe Interact. 21133-142. [DOI] [PubMed] [Google Scholar]
- 65.Zhu, K., and C. O. Rock. 2008. RhlA converts β-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the β-hydroxydecanoyl-β-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J. Bacteriol. 1903147-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.