Summary
Inhibitor of growth 1 (ING1) is implicated in oncogenesis, DNA damage repair and apoptosis. Mutations within the ING1 gene and altered expression levels of ING1 are found in multiple human cancers. Here, we show that both DNA repair and apoptotic activities of ING1 require the interaction of the C-terminal plant homeodomain (PHD) finger with trimethylated at Lys 4 histone H3 (H3K4me3). The ING1 PHD finger recognizes methylated H3K4 but not other histone modifications as revealed by the peptide microarrays. The molecular mechanism of the histone recognition is elucidated based on a 2.1 Å resolution crystal structure of the PHD-H3K4me3 complex. The K4me3 occupies a deep hydrophobic pocket formed by the conserved Y212 and W235 residues that make cation–π contacts with the trimethylammonium group. Both aromatic residues are essential in the H3K4me3 recognition, as substitution of these residues with Ala disrupts the interaction. Unlike the wild type ING1, the W235A mutant, overexpressed in the stable clones of melanoma cells or in HT1080 cells, was unable to stimulate DNA repair after UV irradiation or promote DNA-damage induced apoptosis, indicating that H3K4me3 binding is necessary for these biological functions of ING1. Furthermore, N216S, V218I and G221V mutations, found in human malignances, impair the ability of ING1 to associate with H3K4me3 or to induce nucleotide repair and cell death, linking the tumorigenic activity of ING1 with epigenetic regulation. Together, our findings reveal the critical role of the H3K4me3 interaction in mediating cellular responses to genotoxic stresses and offer new insight into the molecular mechanism underlying the tumor suppressive activity of ING1.
Keywords: PHD finger, ING1, histone, structure, cancer
Introduction
Inhibitor of growth 1 (ING1) is a member of the recently discovered family of tumor suppressors implicated in oncogenesis, control of DNA damage repair, cellular senescence and apoptosis.1–5 ING1 was identified in a screen designed to isolate genes with a decreased expression level in cancer cells.1 Various functions of ING1 are linked to the p53-dependent signaling pathways. Overexpressed ING1 activates p53 target genes including the inhibitor of cell cycle progression p21WAF1 and proapoptotic factor Bax, and promotes p53-induced apoptosis.2,4,6–8 Recent studies have implicated ING1 in the control of cellular responses to DNA damage and other genotoxic stresses.9–11 As the DNA damage occurs, ING1 translocates to the nucleolus, where it either triggers cell cycle arrest followed by DNA damage repair or induces apoptosis if the damage can not be repaired. ING1 is also critical in chromatin remodeling and associates with and influences the activity of Sin3a/HDAC1/2 histone deacetylase complexes.5,12,13
The human ING1 gene encodes several protein isoforms including p27ING1, p33ING1 and p47ING1, with the p33ING1 isoform (referred to as ING1 in this study) being the most widely expressed. ING1 is a small 279-residue modular protein, conserved from yeast to humans. It contains an amino-terminal sin3-associated protein 30 (SAP30) interacting domain (SAID), a nuclear localization signal (NLS), and a carboxy-terminal plant homeodomain (PHD) finger. The SAID domain is shown to associate with a SAP30 subunit of chromatin-modifying Sin3a/HDAC1/2 complexes.5 The NLS motif is necessary for the nuclear localization of ING1. Since the discovery of ING1, four homologous proteins (ING2–ING5) have been identified in mammals, plants and yeast. While all ING members contain the PHD finger, their unique amino-terminal regions bind distinct effectors, engaging these proteins in diverse biological processes and linking them to different forms of cancer. For example, ING1 and ING2 are found as stable components of histone deacetylase complexes, whereas ING3, ING4 and ING5 associate with histone acetyltransferase complexes.5,14 Together, these findings define the ING family of tumor suppressors as critical regulators of chromatin acetylation that bridge epigenetic regulation with cancer development.
The ING1 PHD finger contains 55 residues and belongs to a large family of zinc binding RING finger domains. The PHD module was first identified in a plant HAT3.1 protein in 1993 (ref. 15), and has since been found in several hundred eukaryotic proteins (SMART). The PHD fingers share a Cys4-HisCys3 consensus sequence that coordinates two zinc ions in a cross-braced arrangement. Atomic resolution structures of the ligand-free16–19 and ligand-bound PHD fingers20–23 show topological similarities to the RING24 and FYVE25,26 domains. Although the PHD module was identified a decade ago, its biological role became clear only recently. It has been found that the PHD finger of transcriptional cofactor p300 localizes to nucleosomes and has a function related to chromatin.27 The ACF1 PHD fingers are reported to associate with the central domain of core histones.28 The AIRE PHD domains appear to bind the ATTGGTTA DNA sequence motif.29 A subset of PHD modules including those of ING2 and BPTF have been shown to recognize methylated histone H3K4me3 (ref. 20,21,30,31), whereas the BHC80 PHD finger binds unmodified H3 (ref. 23). Thus, it remains unclear if the PHD family has a common function, or whether it is possible to predict a binding partner based on the primary sequence of this domain.
A wealth of experimental data points to the major function of ING1 as a type II tumor suppressor. The expression level of ING1 varies in many human malignancies, and is particularly reduced in brain, breast and gastrointestinal tumors and lymphoblastic leukemia.32,33 Mutations within the ING1 gene lead to the genomic instability, rapid cancer progression and a poorer survival rate for cancer patients. The missense and silent mutations are found in 19.6 % malignant melanoma, 12.9% esophageal squamous cell cancer (SCC) and 13% head and neck SCC.33,34 Particularly, Cys215 and Asn216 are replaced by Ser, Val218 by Ile and Gly221 by Val in human malignancies.34–36 Each of these residues is found in the ING1 PHD finger, raising a possibility that the cancer mutations disrupt the normal biological function of this module. Additionally, a loss of nuclear localization and an increase of cytoplasmic localization of ING1 has been observed in melanoma, ductal breast and papillary thyroid carcinomas and lymphoblastic leukaemia.37–39 To understand the molecular basis of histone recognition by the ING1 PHD finger and to further our knowledge of tumorigenic mechanisms, we determined a 2.1 Å resolution crystal structure of the PHD finger of human ING1 in complex with a H3K4me3 peptide and elucidated the role of cancer specific mutations in the histone binding and function of ING1.
Results and Discussion
The ING1 PHD finger recognizes H3K4me3
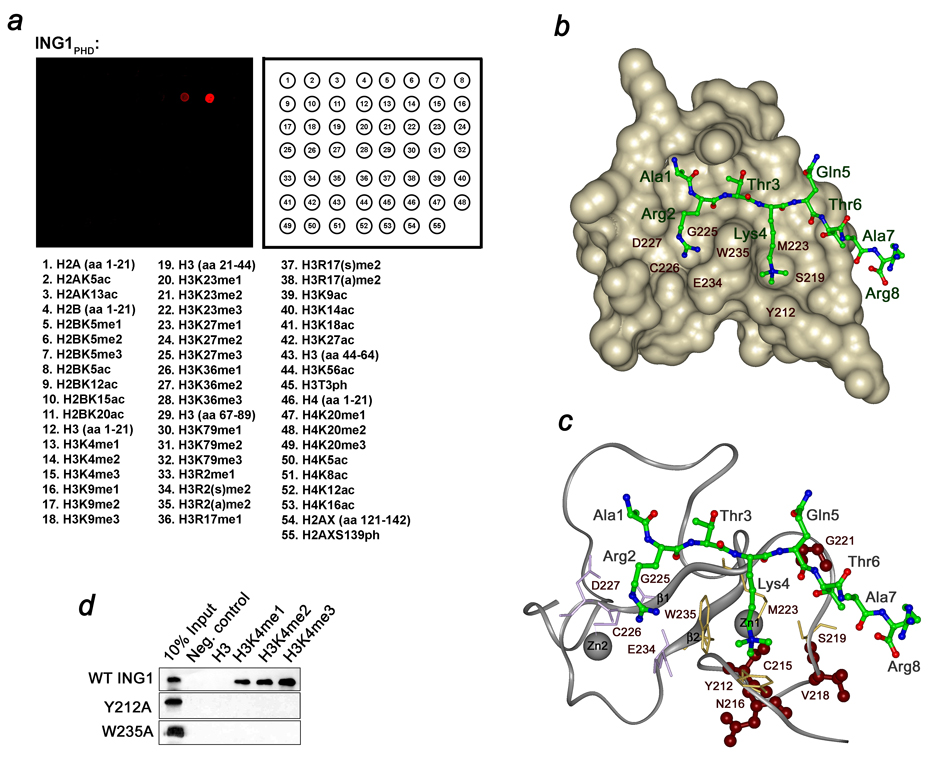
The ING1 tumor suppressor contains a carboxy-terminal plant homeodomain (PHD) module of unknown structure and function. To determine the binding partner of the PHD finger, it was tested by in vitro peptide microarrays (Fig. 1a). The biotinylated peptides corresponding to methylated, acetylated, phosphorylated and unmodified histones H2A, H2B, H3 and H4 were incubated with GST-fusion ING1 PHD. Subsequently the peptide-bound protein was detected using anti-GST antibody and fluorescein-conjugated secondary antibody. Of fifty five peptides screened, the H3K4me3 peptide was distinctly recognized by ING1 PHD. Additionally a weaker interaction was observed with H3K4me2 but not with other modified or unmodified histone peptides, pointing to a high specificity of the ING1 PHD finger.
Figure 1. The ING1 PHD finger recognizes H3K4me3.
(a) Peptide microarrays containing the indicated histone peptides were probed with GST-ING1 PHD. Red spots indicate binding. me, methylation; ac, acetylation; ph, phosphorylation; s, symmetric; a, asymmetric. (b) Structure of the ING1 PHD finger in complex with the histone H3K4me3 peptide. The PHD finger is shown as solid surface. The histone peptide is depicted as ball-and-stick model with C, O and N atoms colored green, red and blue, respectively. (c) The PHD finger is shown as a ribbon with residues mutated in human cancers colored brown. (d) Interactions of the GST-fusion wild type and mutant ING1 PHD fingers with biotinylated histone peptides examined by western blot experiments.
To estimate the strength of association with the histone tails, binding affinities of the ING1 PHD finger toward mono-, di- and tri-methylated H3K4 peptides were measured by tryptophan fluorescence spectroscopy and NMR. The KD values were found to be 419 µM, 17.3 µM and 3.3 µM for H3K4me1, H3K4me2 and H3K4me3, respectively (Table 1). Comparable affinities, in the range of 1–3 µM, are exhibited by PHD fingers of ING2 and BPTF for H3K4me3.20,21 The robust binding of the ING1 PHD finger suggests that H3K4me3 represents its biological target.
Table 1.
Binding affinities of the wild type and mutant ING1 PHD fingers for methylated H3K4. Determined by atryptophan fluorescence and bNMR.
| ING1 PHD finger | Peptide | Kd (µM) |
|---|---|---|
| wt ING1 | H3K4me3 | 3.3 ± 1.6a |
| wt ING1 | H3K4me2 | 17.3 ± 3.9a |
| wt ING1 | H3K4me1 | 419 ± 71b |
| N216S | H3K4me3 | 36 ± 4a |
| V218I | H3K4me3 | 112 ± 11a |
| G221V | H3K4me3 | 114 ± 21b |
| Y212A | H3K4me3 | 243 ± 16b |
| W235A | H3K4me3 | >9,000b |
Structure of the ING1 PHD-H3K4me3 complex
To establish the molecular basis of the histone recognition, we determined the crystal structure of the ING1 PHD finger in complex with an H3K4me3 peptide containing residues 1–12 of the histone H3 (Table 2). The structure of ING1 PHD comprises long loops stabilized by two zinc-binding clusters and a small antiparallel double-stranded β sheet (Fig. 1b,c). The H3K4me3 peptide adopts an extended conformation and pairs with the existing β sheet, forming the third antiparallel β strand. The peptide lies in a large binding site involving one third of the PHD finger residues. The fully extended side chain of trimethylated Lys 4 occupies the deep pocket formed by two aromatic (Y212 and W235) and two aliphatic (S219 and M223) residues of the PHD finger. The Y212 and W235 residues make cation–π contacts with the trimethylammonium group of Lys 4. The peptide’s Arg 2 is bound in the adjacent pocket and is surrounded by G225, C226, D227 and E234.
Table 2.
Data collection and refinement statistics of the H3K4me3-bound ING1 PHD finger.
| Data Collection | ||||
|---|---|---|---|---|
| Native | ||||
| Space group | P3221, a=b=49.06, c=52.62Å, a=β=90°, γ=120°, one molecule per A.U. | |||
| Resolution (Å) | 2.1 | |||
| Wavelength (Å) | 1.257 | |||
| Redundancy 1 | 9.35 (9.78) | |||
| Completeness (%) | 99.9 (100) 1 | |||
| Rmerge 2 | 0.071 (0.253) | |||
| I/σ(I) | 16.3 (5.4) | |||
| Refinement Statistics (|F|>0) | ||||
| Resolution (Å) | 24.53-2.10 | |||
| Rworking % | 24.01 | |||
| Rfree3% | 26.35 | |||
| Number of protein atoms | 479 | |||
| Number of non-protein atoms | 41 water molecules and 2 zinc ions | |||
| R.m.s.d. from ideal bond length (Å) | 0.006 | |||
| R.m.s.d. from ideal bond angle (o) | 1.091 | |||
| Average B-value for all atoms (Å 2) | 48.257 | |||
| Ramachandran statistics | Most favored 35 residues | Additionally allowed 3 residues | Generously allowed 1 residues | Disallowed 41 residue |
Numbers in parenthesis represent values for the highest resolution bin.
Rmerge = Σ| Iobs − Iavg | / ΣIavg.
Rfree was calculated with 10% of reflections.
Residue Glu234 is clearly in a conformation which Phi/Psi values fall into the disallowed region of the Ramachandran plot.
To examine whether the aromatic Y212 and W235 residues are essential, they were substituted with alanine, and the corresponding mutant proteins were tested by Western blot analysis. As shown in Figure 1d, the Y212A and W235A mutants were unable to bind methylated H3K4 peptides, indicating that these residues are required for the K4me3 recognition. A similar aromatic cage around Lys 4 is formed by the BPTF, ING2 and RAG2 PHD fingers20,21,40 and CHD1 double chromodomains41. This implies that the general mechanism of H3K4me3 recognition is conserved among such structurally unrelated modules as PHD and chromodomain. Two other primary characteristics of the H3K4me3 binding is formation of the β sheet-type hydrogen bonds between the peptide and the β1 strand of PHD, and extensive complementary surface interactions.
H3K4me3 binding of the ING1 PHD finger is necessary for both DNA repair and apoptosis
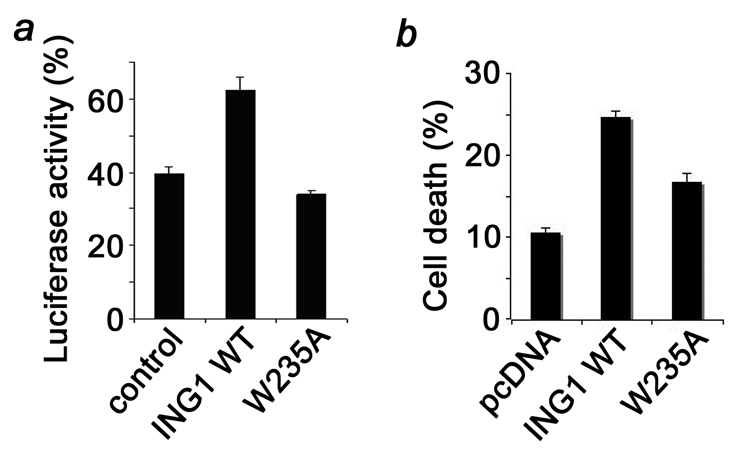
ING1 mediates the cellular response to genotoxic stresses and is critical for maintaining the genomic stability of the cell. Depending on the severity of the DNA damage, ING1 either triggers cell cycle arrest followed by DNA damage repair or induces apoptosis if the damage can not be repaired. We tested the importance of H3K4me3 recognition in regulating these functions of ING1 using two approaches in which DNA damage was stimulated at different levels. We first measured the DNA repair rate by the host-cell-reactivation assay (Fig. 2a). A UV-damaged plasmid containing the renilla luciferase reporter gene (pRL-CMV) was cotransfected with either vector, full length wild type ING1 or ING1 W235A mutant into human melanoma cell line, MMRU. The activity of the reporter gene was used as an indicator of the extent of repair. Our data showed that MMRU cells overexpressing wild type ING1 had a significantly higher repair rate of UV-damaged plasmid (62.4%) compared with the vector control (39.6%; P < 0.01, t test). The W235A mutant, which is defective in H3K4me3 binding, had the repair rate similar to vector’s and substantially lower than wild type ING1’s (33.9%; P < 0.01, t test). Thus, recognition of the H3K4me3 epigenetic mark by ING1 is required to promote the repair of UV-damaged DNA.
Figure 2. The function of ING1 requires its H3K4me3 binding activity.
(a) Assessment of the repair rate of UV-damaged DNA by luciferase reporter assay in MMRU cells cotransfected with undamaged or damaged pRL-CMV luciferase plasmid and pcDNA3-vector (control), wild-type ING1 expression vectors and ING1W235A mutant. Forty hours after transfection, luciferase activity was measured with a luminometer. Columns, mean from triplicates; bars, SD. The experiment was repeated twice with similar results. (b) Stimulation of apoptosis by overexpressed full-length wild type ING1 and W235A mutant. Cell death was measured in HT1080 cells transfected with 1 µg of the indicated expression vectors. The cells were first treated with Doxorubicin and then trypsinized and stained with trypan blue. The error bars represent the mean ± s.e.m. from three independent experiments.
To examine whether the ability of ING1 to induce cell death depends on the interaction of its PHD finger with methylated H3, we measured the apoptosis level in HT1080 cells, which were transfected with full length wild type or W235A mutant ING1 and treated with a chemotherapy drug Doxorubicin (Fig. 2b). Overexpression of ING tumor suppressors followed by the Doxorubicin treatment is known to promote DNA damage-induced apoptosis30,42. As shown in Figure 2b, the cell death level was increased upon overexpression of wild type ING1 (24.6%; P < 0.01, t test) but not the W235A mutant (16.7%; P = 0.02, t test), suggesting that intact PHD is necessary for robust ING1-dependent induction of DNA damage-triggered apoptosis.
Human cancer mutations of ING1 are found in the histone binding region
The ING1 residues, mutated in human cancers including C215, N216, V218 and G221, are colored brown in Figure 1c. All four residues are located in or near the H3K4me3 binding site, and contribute to the interaction. The C215 residue is involved in coordination of one of the zinc ions, therefore its replacement with any other residue would disrupt the three dimensional structure of the PHD finger and abolish the interaction with H3K4me3. The N216 residue stabilizes a turn at the amino-terminus by forming hydrogen bonds to L214 and Q217. It is also located underneath Y212 and may help position the aromatic ring of Y212 orthogonal to the protein surface for the maximum cation - π contact. The V218 residue is next to S219, which forms a wall of the binding groove for trimethylated Lys 4. G221 is involved in restraining Thr 6 of the histone peptide. The amide group of Thr 6 donates a hydrogen bond to the carbonyl group of G221, while the amide group of G221 is hydrogen bonded to the side chain hydroxyl group of Thr 6.
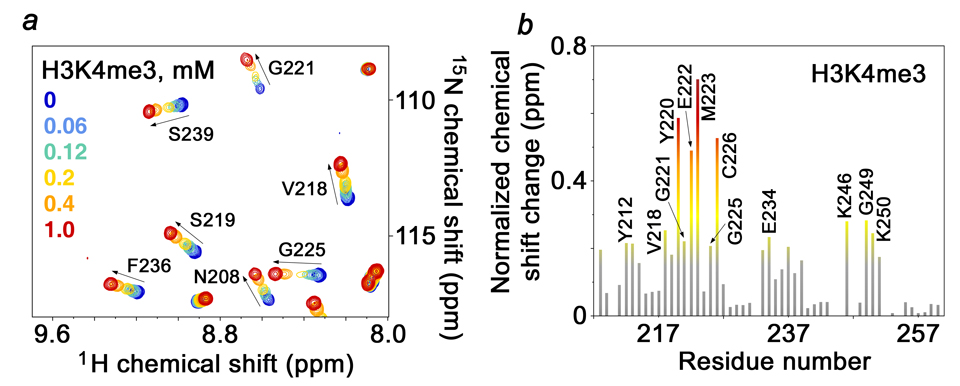
Cancer specific mutations decrease H3K4me3 binding
Initially, the role of cancer relevant residues in the interaction of the wild type ING1 PHD finger with H3K4me3 was suggested by NMR titration experiments (Fig. 3). Substantial chemical shift changes were observed in 1H,15N HSQC spectra of the 15N-labeled PHD finger when the H3K4me3 peptide was gradually added. The largest resonance perturbations were seen in the Y212, C213, V218, Y220, G221, M223, G225, C226, E234, K246, G249 and K250 residues, which comprise the binding sites for trimethylated Lys 4 and Arg 2 and/or are involved in complimentary surface interactions and restraining the peptide backbone (Fig. 3b). Notably, the cancer specific V218 and G221 residues were significantly perturbed upon addition of the histone peptide suggesting that these residues are essential for the H3K4me3 interaction.
Figure 3. The cancer specific residues of ING1 are involved in the interaction with H3K4me3.
(a) Six superimposed 1H,15N heteronuclear single quantum coherence (HSQC) spectra of PHD (0.2 mM), collected during titration of a H3K4me3 peptide, are color-coded according to the ligand concentration (inset). (b) The histogram displays normalized 1H,15N chemical shift changes observed in the corresponding (a) spectra of the PHD finger. The normalized52 chemical shift change was calculated using equation [(ΔδH)2 + (ΔδN/5)2]0.5, where δ is the chemical shift in parts per million (ppm). Colored bars indicate a significant change being greater than average plus one half standard deviation.
To address whether the cancer mutants of ING1 are able to recognize H3K4me3, N216, V218 and G221 were substituted for the residues found in human malignancies. The N216S, V218I and G221V mutant PHD fingers were generated and their interactions with H3K4me3 were examined by NMR and tryptophan fluorescence. A similar pattern of resonance perturbations induced by H3K4me3 was seen in 1H,15N HSQC spectra of the wild type ING1 and cancer specific N216S, V218I and G221V mutants (data not shown), indicating that the histone binding site remains largely unchanged, however all three mutants showed a significant decrease in their binding affinities (Table 1). Replacement of G221 with Val and V218 with Ile had the most disruptive effect on the interaction (Kd of 114 µM and 112 µM, respectively). This reveals the importance of two hydrogen bonding contacts involving G221 and the surface complementarity at the PHD-H3K4me3 interface in the strong anchoring to the histone tail. Substitution of N216 with Ser reduced the binding affinity by 12-fold, pointing to the essential role of this residue in stabilization of the Lys 4 groove (Table 1). As expected, mutation of the Y212 or W235 residues, involved in caging trimethylated Lys 4, disrupted the H3K4me3 association, attesting to a major contribution of these aromatic residues to the binding energetics.
The cancer specific mutations disrupt the DNA repair and apoptotic functions of ING1
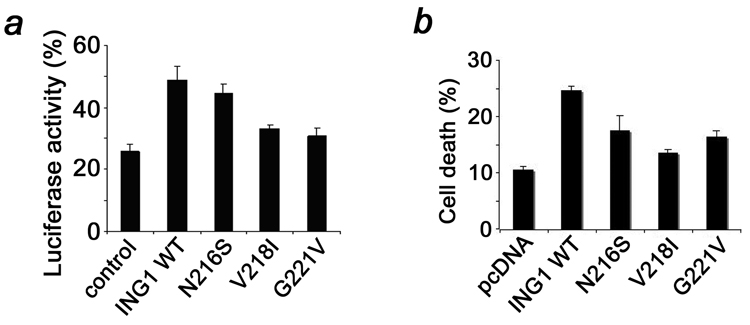
To determine the significance of cancer mutations in the biological functions of ING1, full length N216S, V218I and G221V mutant proteins were produced and their abilities to stimulate DNA repair and cell death were measured by luciferase reporter and apoptotic assays (Fig. 5). All three mutants and particularly V218I and G221V showed a substantial decrease in the DNA repair efficiency and in augmentation of DNA-damage induced apoptosis, suggesting that the cancer specific mutations alter these biological activities of ING1. Together, our data demonstrate the significance of the H3K4me3 binding for the proper functioning of ING1 and suggest that the disruption of this interaction may lead to cancer.
Figure 5. The cancer specific mutations disrupt the DNA repair and apoptotic functions of ING1.
(a) Evaluation of the DNA repair efficiency of ING1wt (48.9%; P = 0.01, t test), N216S (44.7%; P = 0.2, t test), V218I (33.1%; P < 0.01, t test), and G221V (30.8%; P < 0.01, t test) by luciferase reporter assay in MMRU cells. (b) Stimulation of apoptosis by overexpressed full-length N216S (17.5%; P = 0.06, ttest), V218I (13.5%; P= 0.03, t test) and G221V (16.4%; P < 0.01, t test) mutants of ING1 in HT1080 cells.
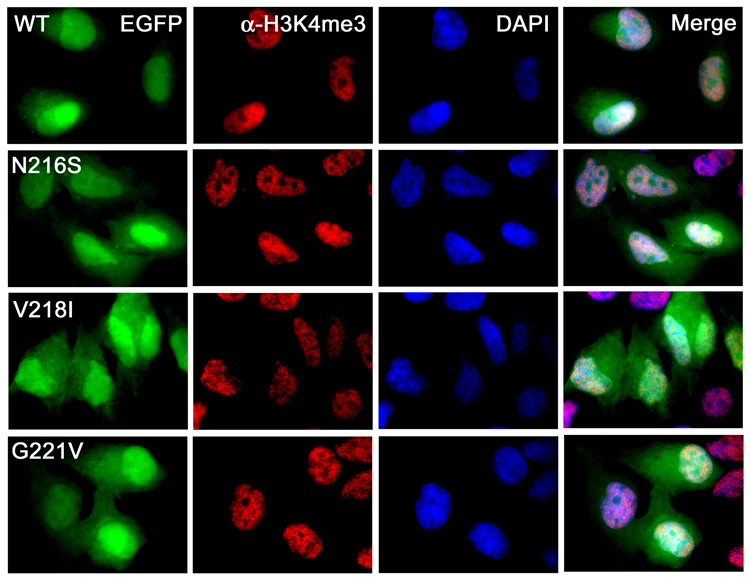
Intracellular localization of ING1 in tumor cells is often shifted from nuclear to cytoplasmic.34,37,38 To test whether the cancer specific mutations have an effect on localization of ING1 PHD in vivo, we transfected HeLa cells with Enhanced Green Fluorescent Protein (EGFP)-fusions of either wild type ING1 PHD, or the N216S, V218I and G221V mutants. The overexpressed proteins were then examined by fluorescence microscopy. As shown in Figure 3, the wild type EGFP-ING1 PHD finger localized predominantly to the nucleus, accumulating at the chromatin regions enriched in H3K4me3, with only a weak diffuse staining observed in the cytoplasm. However, the mutant EGFP-ING1 PHD fingers were dispersed in the cytosol more than the wild type protein, indicating that the H3K4me3 interaction stabilizes ING1 at the methylated chromatin and that the cancer specific mutations may contribute to the elevated cytoplasmic level of ING1 in cancer cells.
Conclusions
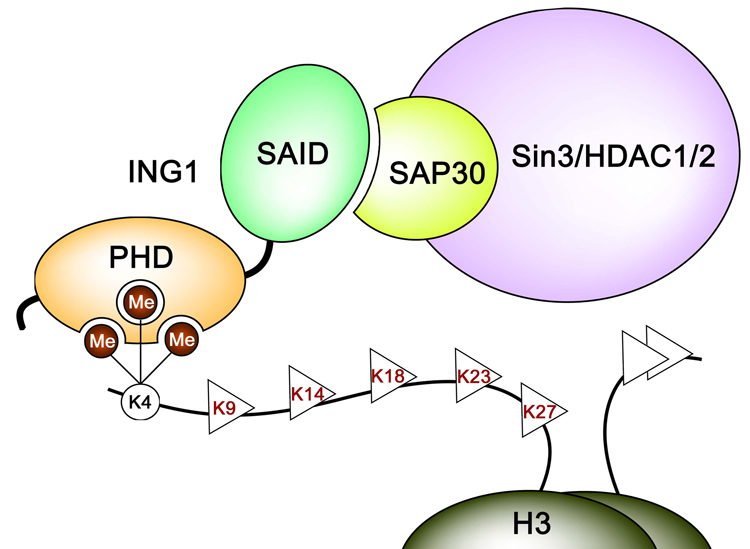
ING1 mediates cellular stress responses such as DNA repair and apoptosis to prevent tumorigenesis. Here we show that both functions require the specific binding of the PHD finger to histone H3K4me3 and that the disruption of this interaction may cause cancer. Recent reports suggest that ING1 is centrally involved in chromatin remodeling. While the PHD finger of ING1 binds methylated histone tails, the amino-terminal SAID domain associates with Sin3a/HDAC1/2 histone deacetylase complexes.5,12,13,20,30 Thus ING1 may bridge or stabilize the Sin3a/HDAC1/2 complexes to the nucleosome for subsequent deacetylation of acetylated histone residues, linking DNA repair, apoptosis and tumorigenic functions with chromatin regulation and gene transcription (Fig. 6).
Figure 6.
A model of the ING1 functioning. Binding of the C-terminal PHD finger and the N-terminal SAID domain of ING1 to H3K4me3 and a SAP30 subunit of the Sin3a/HDAC1/2, respectively, tethers the histone modifying complex to the nucleosome for subsequent deacetylation of acetylated lysine residues of histone H3.
Methods
Mutagenesis
The N216S, V218I, G221V, Y212A and W235A mutants of the ING1 PHD finger (residues 200–279) were generated using QuikChange (Stratagene). The sequences were confirmed by DNA sequencing.
Expression and Purification of Proteins
The wild type and mutant ING1 PHD proteins (residues 200–279 and 202–261) were expressed in E. coli BL21(DE3) pLysS (Stratagene) grown in zinc-enriched LB media or 15NH4Cl-supplemented (Isotec) M9-minimal media. Bacteria were harvested by centrifugation after IPTG induction and lysed using a French press. The GST-fusion proteins were purified on a glutathione Sepharose 4B column (Amersham), cleaved with PreScission protease (Amersham) and concentrated in Millipore concentrators (Millipore). The proteins were further purified by FPLC and concentrated into Tris or d11-Tris (150 mM NaCl, 10 mM d10-dithiothreitol in 7% 2H2O/H2O, pH 6.5). The N216S, V218I, G221V, Y212A ING1 mutant proteins appear to be structured based on the dispersion of amide resonances in 1H,15N HSQC spectra. Substitution of the W235 residue in the similar sequence of the ING2 PHD finger does not perturb the fold.20
Peptide microarray
Peptide microarray experiments were performed as described previously.43 Briefly, biotinylated histone peptides were printed in six replicates onto a streptavidin-coated slide (ArrayIt) using a VersArray Compact Microarrayer (Bio-Rad). After a short blocking incubation with biotin (Sigma), the slides were incubated with GST-fusion ING1 PHD in peptide binding buffer (50 mM Tris-HCl, pH 7.5, 150mM NaCl, 0.1% Nonidet P-40, 20% fetal bovine serum) overnight at 4 °C with gentle agitation. After washing with the same buffer, slides were probed first with anti-GST antibody and then fluorescein-conjugated secondary antibody and visualized with a GenePix 4000 scanner (Molecular Devices).
Fluorescence spectroscopy
Tryptophan fluorescence spectra were recorded at 25°C on Fluoromax3 spectrofluorometer. The samples of 1–10 µM wild type and mutant PHD fingers containing progressively increased concentration (up to 5 mM) of histone tail peptides were excited at 295 nm. Emission spectra were recorded between 305 and 405 nm with a 0.5 nm step size and a 1 s integration time and averaged over 3 scans. The KDs were determined by a nonlinear least-squares analysis using the equation: ΔI=(ΔImax*L)/(KD+L), where L is concentration of the histone peptide, ΔI is observed change of signal intensity, and ΔImax is the difference in signal intensity of the free and bound states of the protein. The KD value was averaged over two experiments for the H3K4me2 binding and over three separate experiments for other interactions.
X-ray crystallography
The PHD finger of human ING1 (1.0 mM) was combined with H3K4me3 peptide (residues 1–12) in a 1:1.5 molar ratio prior to crystallization. Crystals of the complex were grown using the hanging drop vapor diffusion method at 4°C by mixing 1.5 µl of the protein-peptide solution with 1 µl of a well solution containing 0.01 M NiCl2·6H2O, 0.1 M Tris (pH 7.0) and 20% polyethylene glycol monomethyl ether 2K (PEGMME 2K). Crystals grew in a trigonal space group P3221 with unit cell parameters of a=b=49.06 Å, and c=52.62 Å, α=β=90°, γ=120°, one molecule per asymmetric unit. Crystals were soaked for 15 minutes in the well solution supplemented with 25% PEGMME 2K before being flash cooled in liquid nitrogen. A complete native data set diffracting to 2.1 Å was collected at 100K on a "NOIR-1" MBC system detector at beamline 4.2.2 at the Advanced Light Source in Berkeley, CA. Data were processed with DTREK44 and statistics are shown in Table 2. The structure was determined by rigid body refinement in CNS45 using the crystal structure of the ING2 PHD/H3K4me3 complex (PDB 2G6Q) as a model. Residues 209–259 of the PHD domain and 1–8 of the peptide were built into the density using program O46 without any ambiguity. Refinement was performed using program CNS45.
NMR spectroscopy
NMR experiments were performed at 298 K on Varian INOVA 600 and 500 MHz spectrometers using unlabeled or uniformly 15N-labeled ING1 PHD finger. The spectra were processed with NMRPipe47 and analyzed using PIPP48, nmrDraw, and in-house software programs. Sequential assignments were made using 3D 15N-edited NOESY-HSQC49,50, HSQC-NOESY-HSQC and TOCSY-HSQC51. The histone peptide binding was characterized by monitoring chemical shift changes in 1H,15N HSQC spectra of 0.2 mM wild type or mutant proteins as the H3K4me3 peptide was added stepwise. The dissociation constants (Kds) were determined by a nonlinear least-squares analysis using Xmgr program and the equation: Δδ=Δδmax((([L]+[P]+KD)−sqrt(sqr([L]+[P]+KD)−(4*[P]*[L])))/(2*[P])), where [L] is concentration of the peptide, [P] is concentration of PHD, Δδ is observed chemical shift change, and Δδmax is the difference in chemical shifts of the free and the ligand-bound protein. The synthetic histone peptide NH2-ARTK(me3)QTARKSTG-COOH was synthesized at the Biophysics Core Facility of the University of Colorado Health Sciences Center.
Host-Cell-Reactivation Assay
The pRL-CMV plasmid, which contains a gene encoding for renilla luciferase, was irradiated with UVC at 800 J/m2 using an UV cross-linker (UltraLum, Claremont, CA). The irradiated plasmid or non-irradiated control plasmid was then co-transfected with wild-type ING1or ING1 mutants N216S, V218I, G221V, or W235A. Forty hours after transfection, cells were lysed by passive lysis buffer, and reporter enzyme level was analyzed with a luciferase assay kit (Promega, Madison, WI). All reactions were performed in triplicates. The percentage luciferase activity was calculated as the fraction of irradiated plasmid over non-irradiated control plasmid.
Cell-Culture and Western analyses were carried out as described.42
In vivo localization of the wild type and mutant EGFP ING1 PHD finger
EGFP fusion constructs of the wild type ING1 PHD finger and Y212A, N216S, V218I and G221V mutants were amplified by Polymerase Chain Reaction (PCR) using corresponding pGEX-6P plasmids, cleaved with BglII and EcoRI restriction enzymes and cloned into pEGFP-C1 vector (BD Clontech). The HeLa cells were grown in Dulbecco Modified Essential Medium (DMEM) (Mediatech) supplied with 10% Fetal Bovine Serum (FBS) (Invitrogen). The cells were transfected with pEGFP PHD, pEGFP Y212A PHD, pEGFP N216S PHD, pEGFP V218I PHD or pEGFP G221V PHD plasmids using Effectene reagent (Qiagen) and 24 hours later were replated onto 25 mm glass coverslips. 72 hours after transfection the cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in Phosphate Buffered Saline (PBS), stained with DAPI (Molecular Probes) and with rabbit polyclonal anti-H3K4me3 primary Abs (Abcam) followed by donkey anti-rabbit Alexa555 conjugated secondary Abs (Molecular Probes). Coverslips with stained cells were mounted on glass slides with Mowiol 488 (EMD Calbiochem). Microscopic imaging of fixed cells was performed on Olympus IX-81 inverted microscope equipped with 100W mercury lamp, 100x oil immersion objective lens, dual filter wheels, and standard DAPI, GFP and TexasRed filter sets (Chroma). Fluorescence acquisition was performed with SlideBook v.4.2 software (Intelligent Imaging Innovation).
Figure 4.
The PHD-H3K4me3 binding stabilizes ING1 at the methylated chromatin. Intracellular localization of the wild type (WT) and mutant EGFP-ING1 PHD fingers (green) in HeLa cells. The cells were stained with anti-H3K4me3 antibodies (red) and DAPI (blue). The ratio of the nuclear vs. cytoplasmic EGFP [2.28 (wild type), 1.92 (N216S), 1.75 (V218I) and 1.84 (G221V)] was obtained by averaging the fluorescence intensity in ten or more cells.
Acknowledgements
We thank D. Bentley and F. Davrazou for discussions and J. Nix at the ALS for helping with X-ray data collection. This research was supported by grants from the NIH, GM070358 and GM073913 (V.V.V), GM079641 (O.G.) and CA113472 and GM071424 (T.G.K.), Canadian Institutes of Health Research (G.L.) and the University of Colorado Cancer Center (T.G.K.). P.V.P is a recipient of the NIH Ruth L. Kirschstein NRSA Predoctoral Fellowship, K.S.C. is an American Cancer Society Postdoctoral Fellow and T.G.K. is a NARSAD Young Investigator.
Abbreviations
- ING1
Inhibitor of growth 1
- PHD
plant homeodomain
- H3K4me3
histone H3 trimethylated at Lys 4
- SAID
sin3-associated protein 30 (SAP30) interacting domain
- EGFP
Enhanced Green Fluorescent Protein
Footnotes
ACCESSION CODES:
The coordinates have been deposited in the Protein Data Bank under accession number 2QIC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet. 1996;14:415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- 2.Garkavtsev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol Cell Biol. 1997;17:2014–2019. doi: 10.1128/mcb.17.4.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helbing CC, Veillette C, Riabowol K, Johnston RN, Garkavtsev I. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res. 1997;57:1255–1258. [PubMed] [Google Scholar]
- 4.Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature. 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 5.Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1) Mol Cell Biol. 2002;22:835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeremski M, Hill JE, Kwek SS, Grigorian IA, Gurova KV, Garkavtsev IV, Diatchenko L, Koonin EV, Gudkov AV. Structure and regulation of the mouse ing1 gene. Three alternative transcripts encode two phd finger proteins that have opposite effects on p53 function. Journal of Biological Chemistry. 1999;274:32172–32181. doi: 10.1074/jbc.274.45.32172. [DOI] [PubMed] [Google Scholar]
- 7.Leung KM, Po LS, Tsang FC, Siu WY, Lau A, Ho HT, Poon RY. The candidate tumor suppressor ING1b can stabilize p53 by disrupting the regulation of p53 by MDM2. Cancer Res. 2002;62:4890–4893. [PubMed] [Google Scholar]
- 8.He GH, Helbing CC, Wagner MJ, Sensen CW, Riabowol K. Phylogenetic analysis of the ING family of PHD finger proteins. Mol Biol Evol. 2005;22:104–116. doi: 10.1093/molbev/msh256. [DOI] [PubMed] [Google Scholar]
- 9.Cheung KJ, Jr, Bush JA, Jia W, Li G. Expression of the novel tumour suppressor p33(ING1)is independent of p53. Br J Cancer. 2000;83:1468–1472. doi: 10.1054/bjoc.2000.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung KJ, Jr, Mitchell D, Lin P, Li G. The tumor suppressor candidate p33(ING1) mediates repair of UV-damaged DNA. Cancer Res. 2001;61:4974–4977. [PubMed] [Google Scholar]
- 11.Scott M, Boisvert FM, Vieyra D, Johnston RN, Bazett-Jones DP, Riabowol K. UV induces nucleolar translocation of ING1 through two distinct nucleolar targeting sequences. Nucleic Acids Res. 2001;29:2052–2058. doi: 10.1093/nar/29.10.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skowyra D, Zeremski M, Neznanov N, Li M, Choi Y, Uesugi M, Hauser CA, Gu W, Gudkov AV, Qin J. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J Biol Chem. 2001;276:8734–8739. doi: 10.1074/jbc.M007664200. [DOI] [PubMed] [Google Scholar]
- 13.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Schindler U, Beckmann H, Cashmore AR, Schindler TF. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant Journal. 1993;4:137–150. doi: 10.1046/j.1365-313x.1993.04010137.x. [DOI] [PubMed] [Google Scholar]
- 16.Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. Journal of Molecular Biology. 2000;304:723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- 17.Capili AD, Schultz DC, Rauscher IF, Borden KL. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. Embo J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwan AHY, Gell DA, Verger A, Grossley M, Matthews JM, Mackay JP. Engineering a protein scaffold from a PHD finger. Structure. 2003;11:803–813. doi: 10.1016/s0969-2126(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 19.Bottomley MJ, Stier G, Pennacchini D, Legube G, Simon B, Akhtar A, Sattler M, Musco G. NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1): Insights into apeced disease. J Biol Chem. 2005 doi: 10.1074/jbc.M413959200. in press. [DOI] [PubMed] [Google Scholar]
- 20.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barlow PN, Luisi B, Milner A, Elliott M, Everett R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. Journal of Molecular Biology. 1994;237:201–211. doi: 10.1006/jmbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- 25.Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- 26.Kutateladze TG, Overduin M. Structural mechanism of endosome docking by the FYVE domain. Science. 2001;291:1793–1796. doi: 10.1126/science.291.5509.1793. [DOI] [PubMed] [Google Scholar]
- 27.Ragvin A, et al. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J. Mol. Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 28.Eberharter A, Vetter I, Ferreira R, Becker PB. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. Embo J. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purohit S, Kumar PG, Laloraya M, She JX. Mapping DNA-binding domains of the autoimmune regulator protein. Biochem Biophys Res Commun. 2005;327:939–944. doi: 10.1016/j.bbrc.2004.12.093. in press. [DOI] [PubMed] [Google Scholar]
- 30.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 32.Toyama T, Iwase H, Watson P, Muzik H, Saettler E, Magliocco A, DiFrancesco L, Forsyth P, Garkavtsev I, Kobayashi S, Riabowol K. Suppression of ING1 expression in sporadic breast cancer. Oncogene. 1999;18:5187–5193. doi: 10.1038/sj.onc.1202905. [DOI] [PubMed] [Google Scholar]
- 33.Campos EI, Chin MY, Kuo WH, Li G. Biological functions of the ING family tumor suppressors. Cell Mol Life Sci. 2004;61:2597–2613. doi: 10.1007/s00018-004-4199-4. [DOI] [PubMed] [Google Scholar]
- 34.Campos EI, Martinka M, Mitchell DL, Dai DL, Li G. Mutations of the ING1 tumor suppressor gene detected in human melanoma abrogate nucleotide excision repair. Int J Oncol. 2004;25:73–80. [PubMed] [Google Scholar]
- 35.Gunduz M, Ouchida M, Fukushima K, Hanafusa H, Etani T, Nishioka S, Nishizaki K, Shimizu K. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Research. 2000;60:3143–3146. [PubMed] [Google Scholar]
- 36.Chen L, Matsubara N, Yoshino T, Nagasaka T, Hoshizima N, Shirakawa Y, Naomoto Y, Isozaki H, Riabowol K, Tanaka N. Genetic alterations of candidate tumor suppressor ING1 in human esophageal squamous cell cancer. Cancer Research. 2001;61:4345–4349. [PubMed] [Google Scholar]
- 37.Nouman GS, Anderson JJ, Wood KM, Lunec J, Hall AG, Reid MM, Angus B. Loss of nuclear expression of the p33(ING1b) inhibitor of growth protein in childhood acute lymphoblastic leukaemia. J Clin Pathol. 2002;55:596–601. doi: 10.1136/jcp.55.8.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nouman GS, Anderson JJ, Mathers ME, Leonard N, Crosier S, Lunec J, Angus B. Nuclear to cytoplasmic compartment shift of the p33ING1b tumour suppressor protein is associated with malignancy in melanocytic lesions. Histopathology. 2002;40:360–366. doi: 10.1046/j.1365-2559.2002.01369.x. [DOI] [PubMed] [Google Scholar]
- 39.Nouman GS, Angus B, Lunec J, Crosier S, Lodge A, Anderson JJ. Comparative assessment expression of the inhibitor of growth 1 gene (ING1) in normal and neoplastic tissues. Hybrid Hybridomics. 2002;21:1–10. doi: 10.1089/15368590252917584. [DOI] [PubMed] [Google Scholar]
- 40.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 42.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 43.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 45.Brunger AT, et al. Crystallography & NMR system: A new software suite for macronolacular structure determination. Acta Crystallogr. D Biol. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 47.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 48.Garret DS, Powers R, Gronenborn AM, Clore GM. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 49.Zuiderweg ER, Fesik SW. Heteronuclear three-dimensional NMR spectroscopy of the inflammatory protein C5a. Biochemistry. 1989;28:2387–2391. doi: 10.1021/bi00432a008. [DOI] [PubMed] [Google Scholar]
- 50.Marion D, Kay LE, Sparks SW, Torchia DA, Bax A. Three-dimensional heteronuclear NMR of nitrogen-15 labeled proteins. J. Am. Chem. Soc. 1989;111:1515–1517. [Google Scholar]
- 51.Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, Clore GM. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemistry. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 52.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]