Abstract
Although it is well established that mammary tumorigenesis converts transforming growth factor-β (TGF-β) from a tumor suppressor to a tumor promoter, the molecular, cellular and microenvironmental mechanisms underlying the dichotomous nature of TGF-β in mammary epithelial cells (MECs) remains to be determined definitively. Aberrant upregulation of the inducible cyclooxygenase, Cox-2, occurs frequently in breast cancers and is associated with increasing disease severity and the acquisition of metastasis; however, the impact of Cox-2 expression on normal and malignant MEC response to TGF-β remains unknown. We show here that TGF-β induced Cox-2 expression in normal MECs during their acquisition of an epithelial–mesenchymal transition (EMT) phenotype. Moreover, stable Cox-2 expression in normal MECs stimulated their invasion, EMT and anchorage-independent growth and inhibited their activation of Smad2/3 by TGF-β. Conversely, antagonizing TGF-β signaling in malignant, metastatic MECs significantly reduced their expression of Cox-2 as well as enhanced their activation of Smad2/3 by TGF-β. Along these lines, elevated Cox-2 expression elicited prostaglandin E2 (PGE2) production and the autocrine activation of EP receptors, which antagonized Smad2/3 signaling in normal and malignant MECs. Importantly, rendering normal and malignant MECs Cox-2 deficient inhibited their production of PGE2 and acquisition of an EMT morphology as well as potentiated their nuclear accumulation of Smad2/3 and transcription of plasminogen activator inhibitor-1 and p15 messenger RNA. Collectively, our findings establish Cox-2 as a novel antagonist of Smad2/3 signaling in normal and malignant MECs; they also suggest that chemotherapeutic targeting of Cox-2 may offer new inroads in restoring the tumor-suppressing activities of TGF-β in malignant, metastatic breast cancers.
Introduction
Among metazoan organisms, transforming growth factor-β (TGF-β) functions as an important regulator of cell growth and development (1). TGF-β signaling begins when ligand dimers bind to Ser/Thr protein receptor complexes composed of the transforming growth factor-β type I (TβR-I) and type II (TβR-II) receptors and in some circumstances to the accessory TGF-β type III receptor. Following its phosphorylation and activation by TβR-II, active TβR-I phosphorylates and stimulates the latent transcription factors, Smads 2 and 3, which subsequently bind and translocate to the nucleus with the co-Smad, Smad4 (1–3). The association of nuclear Smad2/3/4 complexes with additional transcriptional activators or repressors serves in regulating gene expression by TGF-β in a cell- and promoter-specific fashion. TGF-β also regulates cell behavior by activating Smad2/3-independent signaling systems in a cell- and context-specific manner. Included in this growing list of TGF-β-targeted effectors are the mitogen-activated protein kinases [MAPKs; e.g. extracellular signal-regulated kinase (ERK) 1/2, c-jun N-terminal kinase and p38 MAPK], phosphoinositide 3-kinase (PI3K)/AKT, the small guanosine triphosphate-binding proteins (e.g. Ras, RhoA, Rac1 and Cdc42) and nuclear factor kappa B (NF-κB), which collectively increase the complexity whereby TGF-β governs the actions of normal and malignant cells (1). Moreover, cross talk between Smad-dependent and -independent signaling inputs impact Smad2/3 function in multiple cellular compartments as well as contribute to the conversion of TGF-β from a tumor suppressor to a tumor promoter, particularly in cancers of the breast (1–3).
The conversion of mammary epithelial cells (MECs) from immotile, polarized phenotypes to highly motile, apolar morphologies is known as epithelial–mesenchymal transition (EMT), which represents a major determinant underlying how normal and malignant MECs sense and respond to TGF-β. Indeed, we recently showed that altered αvβ3 integrin expression (4–6) and aberrant coupling of TGF-β to NF-κB activation (7) both figure prominently in the oncogenic conversion of TGF-β during mammary tumorigenesis. Moreover, we also found that the induction of EMT by TGF-β facilitates its stimulation of NF-κB and proinflammatory gene expression in normal and malignant MECs (7). Along these lines, aberrant TGF-β activity and inflammation within mammary tumor microenvironments promotes their progression through the activation of tumor-associated fibroblasts and through the recruitment of innate and adaptive immune cells (1,2). Thus, chemotherapeutic targeting of the proinflammatory activities of TGF-β may prove useful in ameliorating the clinical course and outcome of metastatic breast cancer patients.
Inappropriate expression of the inducible cyclooxygenase, Cox-2, during mammary tumorigenesis is associated with the development of breast cancer inflammation, invasion, metastasis and angiogenesis and with the activation of tumor stroma and infiltrating macrophages (8–10). Indeed, whereas elevated Cox-2 expression promotes breast cancer cell metastasis to the lungs and bone (11,12), Cox-2 antagonism or deficiency suppresses the development and progression of mammary tumorigenesis (8–10,13). Cox-2 functions within the arachidonic acid pathway where it converts arachidonate to prostaglandin E2 (PGE2), a principle product and promoter of the tumorigenic activities of Cox-2 (14,15). Autocrine and paracrine PGE2 signaling stimulates the E-series of prostaglandin receptors (e.g. EPs 1–4), whose coupling to G proteins activates the 3′5′-cyclic adenosine monophosphate/protein kinase A, the PI3K/AKT and the ERK1/2 pathways as well as regulates the glycogen synthase kinase (GSK)-3β pathway (14). Given the striking parallels between oncogenic TGF-β signaling and Cox-2 in promoting mammary tumorigenesis, we hypothesized Cox-2 as a novel antagonist of MEC response to TGF-β. The aim of this study was to test this hypothesis and to determine how Cox-2 impacts the TGF-β signaling system in normal and malignant MECs.
Materials and methods
Materials
Recombinant human TGF-β1 was obtained from R&D Systems (Minneapolis, MN), whereas PGE2 and the chemical antagonists targeting TβR-I (inhibitor II), Cox-2 (NS-398) and GSK-3β (inhibitor XIII) were purchased from Calbiochem (San Diego, CA). The EP1/EP2 receptor antagonist AH6809 was obtained from Cayman Chemical (Ann Arbor, MI). The cDNA construct encoding kinase-dead GSK-3β (dnGSK-3β; pcDNA3-GSK-3β(K85A)-HA) was purchased from Addgene (Cambridge, MA), whereas lentiviral vectors (pLKO.1-puro) encoding for control (i.e. non-silencing short hairpin RNA (shRNA), catalog #RHS4346) or murine Cox-2 shRNA (catalog #RMM3981-97056325) were purchased from Open Biosystems (Huntsville, AL). All additional supplies or reagents were routinely available.
Cell culture and transgene expression
Normal murine NMuMG MECs and metastatic murine 4T1 breast cancer cells were obtained from American Type Culture Collection (Manassas, VA) and cultured as described previously (7). Engineering NMuMG cells to constitutively express Cox-2 commenced by polymerase chain reaction (PCR) amplifying the full-length murine Cox-2 cDNA using oligonucleotides containing BamHI (N-terminus) and XhoI (C-terminus) restriction sites. The resulting PCR product was ligated into corresponding sites located in mammalian expression vector, pcDNA3.1/Hyrog(+) (Invitrogen, Carlsbad, CA). The resulting Cox-2 insert was sequenced in its entirety on an Applied Biosystems 3730 DNA sequencing machine. Afterward, NMuMG cells were transfected overnight (20 μg/10 cm plate) with either pcDNA3.1/Hygro or pcDNA3.1-COX-2/Hygro and subsequently were subjected to hygromycin selection (300 μg/ml) for 2 weeks. Hygromycin-resistant NMuMG clones were isolated and screened for Cox-2 expression by immunoblotting with anti-Cox-2 antibodies as described below.
The creation of NMuMG and 4T1 cells lacking Cox-2 was accomplished by their overnight infection with control (i.e. non-silencing shRNA) or Cox-2 shRNA lentiviral (pLKO.1-puro) supernatants produced by 293T cells that were transiently transfected with lentiviral packaging vectors (i.e. pMD2.G, pRRE and pRSV) according to standard protocols (16). Cells expressing non-silencing and Cox-2 shRNAs were isolated by puromycin selection (5 μg/ml) for 14 days. Afterward, puromycin-resistant NMuMG and 4T1 clones were isolated and the extent of shRNA-mediated Cox-2 deficiency was monitored by immunoblotting whole-cell extracts with antibodies against Cox-2 as described below.
Cell biological assays
Analyzing the effects of altered Cox-2 expression on normal and malignant MEC response to TGF-β was determined as follows: (i) EMT induced by TGF-β1 (5 ng/ml) administration as described previously (4,5,7); (ii) Smad3 immunofluorescence using 50 000 cells per well as described previously (17); (iii) cell invasion induced by 2% serum using 25 000 cells per well in a modified Boyden chamber coated with Matrigel matrices (Cell Biolabs, San Diego, CA) as described previously (7) and (iv) synthetic pSBE- (Smad-binding element), pCox-2-, p3TP- and TOPFlash-luciferase reporter gene assays with or without 100 ng per well of either Cox-2 or GSK-3β cDNA using 25 000–30 000 cells per well as described previously (7).
Western blotting analyses
Control and TGF-β-stimulated NMuMG and 4T1 cells were lysed and solubilized in buffer H/Triton X-100 (18) for 30 min on ice. Clarified cell extracts were resolved on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels, transferred electrophoretically to nitrocellulose membranes and blocked in 5% milk prior to incubation with the following primary antibodies (dilutions): (i) anti-Cox-2 (1:2000; Cayman Chemical); (ii) anti-phospho-Smad3 (1:250; Cell Signaling, Danvers, MA); (iii) anti-Smad4 (1:500; Cell Signaling); (iv) phospho-p38 MAPK (1:500; Cell Signaling) and (v) p38 MAPK (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA). The resulting immunocomplexes were visualized by enhanced chemiluminescence. Differences in protein loading were monitored by reprobing stripped membranes with anti-β-actin antibodies (1:5000; Sigma, St Louis, MO).
Semiquantitative real-time PCR analyses
Total RNA from control and Cox-2-deficient NMuMG and 4T1 cells was purified using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations. Afterward, cDNAs were synthesized by iScript reverse transcription (Bio-Rad, Hercules, CA), which then were diluted 10-fold in H2O and employed in semiquantitative real-time PCRs (25 μl) that used the SYBR Green system (Bio-Rad) supplemented with 5 μl of diluted cDNA and 0.1 μM of oligonucleotide pairs listed below. PCRs were performed and analyzed on a Bio-Rad Mini-Opticon detection system, and differences in RNA concentrations were controlled by normalizing individual gene signals to their corresponding glyceraldehyde 3-phosphate dehydrogenase RNA signals. The oligonucleotide primer pairs used were as follows: (i) plasminogen activator inhibitor-1 (NM_008871), forward 5’ GGTGAAACAGGTGGACTTCTCA and reverse 5’ GCATTCAC CAGCACCAGGCGTG (amplicon 144 bp); (ii) Cox-2 (NM_011198), forward 5’ TGGGGTGATGA GCAACTATTCC and reverse 5’ AGGCAATGCGGTTCTGATACTG (amplicon 169 bp); (iii) p15/INK4b (NM_007670), forward 5’ TGCCACCCTTACCAGACCTGTG and reverse 5’ GCAGATAC CTCGCAATGTCACG (amplicon 167 bp) and (iv) glyceraldehyde 3-phosphate dehydrogenase (NM_008084), forward 5’ CAACTTT GGCATTGTGGAAGGGCTC and reverse 5’ GCAGGGATGATGTTCTGGGCAGC (amplicon 129 bp).
PGE2 enzyme-linked immunosorbent assay
Analyzing the production of PGE2 by normal and malignant MECs was carried out as described previously (19). Briefly, control- and Cox-2-deficient NMuMG and 4T1 cells were allowed to adhere overnight onto six-well plates (350 000 cells per well). Adherent cells were washed twice in phosphate-buffered saline the following morning and subsequently were incubated in 1 ml of serum-free media for 60 min at 37°C. Afterward, the resulting conditioned media was collected, diluted in serum-free media (1:4–1:10) and analyzed by PGE2 enzyme-linked immunosorbent assay (Cayman Chemical) according to the manufacturer's instructions. Synthesized PGE2 concentrations were normalized against protein concentrations measured in the corresponding whole-cell extracts.
Results
TGF-β induces Cox-2 expression during EMT and breast cancer progression
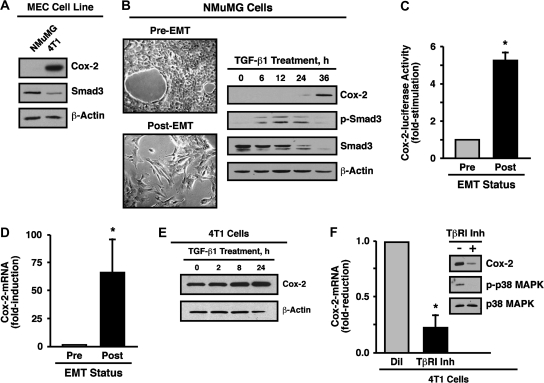
Findings from a variety of epidemiological and experimental studies have established that mammary tumorigenesis upregulates the expression of Cox-2, an event associated with poor patient prognosis and the acquisition of breast cancer metastasis and angiogenesis (8–10). These pathophysiological events mediated by Cox-2 in human breast cancers in many respects mirror those observed for oncogenic TGF-β signaling in these same malignant cells; however, the impact of Cox-2 expression on normal and malignant MEC response to TGF-β remains unknown. To determine how Cox-2 potentially alters MEC response to TGF-β, we studied the expression and activity of Cox-2 in normal NMuMG cells, which routinely are employed to study EMT induced by TGF-β (4–7,20–22), and in malignant, metastatic 4T1 cells, which we (6,7) and others (23–25) recently established as being an important late-stage model of TGF-β-responsive breast cancer. As such, Cox-2 immunoblot analysis of normal NMuMG cells revealed that they express little-to-no Cox-2 protein, whereas malignant, metastatic 4T1 cells exhibited abundant Cox-2 expression (Figure 1A). It is interesting to note that elevated Cox-2 expression in 4T1 cells was associated with a significant reduction in Smad3 expression as compared with normal NMuMG cells (Figure 1A). We demonstrated previously that TGF-β treatment of 4T1 cells induced their expression of luciferase driven by the Cox-2 promoter through a TAB1:TAK1:IKKβ:NF-κB-dependent pathway (7); however, the effect of TGF-β on Cox-2 expression in normal MECs remains unknown. To address this question, we stimulated NMuMG cells with TGF-β1 and subsequently monitored their acquisition of a fibroblastoid morphology as well as changes in their expression of Cox-2 and Smad3 at various times over a period of 36 h. Figure 1B shows that TGF-β stimulation of EMT in NMuMG cells induced their expression of Cox-2 as well as promoted their loss of Smad3. Moreover, EMT induced by TGF-β significantly enhanced NMuMG cell expression of luciferase driven by the Cox-2 promoter (Figure 1C) and transcription of Cox-2 messenger RNA (mRNA) (Figure 1D). Although endogenous Cox-2 expression is robust in untreated 4T1 cells, those stimulated with TGF-β further upregulated their expression of Cox-2 (Figure 1E). Interestingly, administering a TβR-I antagonist to quiescent 4T1 cells significantly reduced their expression of Cox-2 mRNA and protein (Figure 1F). Collectively, these findings demonstrate that the ability of TGF-β induce Cox-2 expression in normal MECs transpires through a transcriptional- and EMT-dependent process. Our findings also suggest that activation of the TGF-β signaling systems represents a major route leading to Cox-2 expression in normal MECs following their induction of EMT and in resting malignant MECs via autocrine TGF-β signaling.
Fig. 1.
TGF-β induces Cox-2 expression during EMT and breast cancer progression. (A) Detergent-solubilized whole-cell extracts prepared from NMuMG and 4T1 cells were immunoblotted with antibodies against Cox-2, Smad3 and β-actin as indicated. Images are from a representative experiment that was performed three times with identical results. (B) NMuMG cells were incubated for 36 h in the absence or presence of TGF-β1 (5 ng/ml) to induce EMT (left panel) and subsequently were immunoblotted for Cox-2, phospho-Smad3, Smad3 and β-actin as indicated (right panel). Images are from a representative experiment that was performed three times with identical results. (C) NMuMG cells were incubated overnight in the absence (pre) or presence (post) of TGF-β1 (5 ng/ml) to induce EMT. Afterward, the cells were transiently transfected with Cox-2-luciferase and β-gal cDNAs and subsequently were incubated for an additional 48 h prior to measuring the luciferase and β-gal activities contained in detergent-solubilized whole-cell extracts. Data are the mean (±SE; n = 3) luciferase activities relative to pre-EMT NMuMG cells (*P < 0.05; Student's t-test). (D) NMuMG cells were incubated for 36 h in the absence (pre) or presence (post) of TGF-β1 (5 ng/ml) to induce EMT. Total RNA was isolated and subjected to semiquantitative real-time PCR to monitor the expression of Cox-2. Data are the mean (±SE; n = 8) fold changes of Cox-2 gene expression relative to pre-EMT NMuMG cells (*P < 0.05; Student's t-test). (E) 4T1 cells were stimulated with TGF-β1 (5 ng/ml) for varying times as indicated and subsequently were immunoblotted for Cox-2 and β-actin as shown. Images are from a representative experiment that was performed twice with identical results. (F) 4T1 cells were treated for 24 h with either diluent (i.e. DMSO, Dil) or the TβR-I type II inhibitor (100 ng/ml, Inh). Total RNA was isolated and subjected to semiquantitative real-time PCR to monitor the expression of Cox-2. Data are the mean (±SE; n = 3) fold changes in Cox-2 gene expression relative to diluent-treated cells (left panel). Detergent-solubilized whole-cell extracts were prepared from 4T1 cell subjected to identical experimental treatments and subsequently were immunoblotted with antibodies against Cox-2, phospho-p38 and p38 as indicated (right panel) (*P < 0.05; Student's t-test).
Cox-2 enhances oncogenic TGF-β signaling by inhibiting Smad2/3 activity
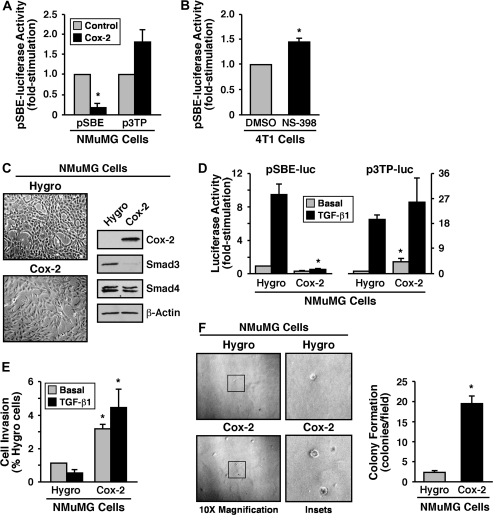
Our findings thus far suggest that Cox-2 expression and its enzymatic products may function in impacting the response of normal and malignant MECs to TGF-β, a supposition supported by several recent findings in the literature (26–28). To further address this question, we transiently transfected NMuMG cells with Cox-2 cDNA and monitored its effects on luciferase activity driven by the synthetic pSBE promoter, which measures the transcriptional activity of Smad3/4, and by the synthetic p3TP promoter, which measures the transcriptional activity of Smad2/3/4 and AP-1. As shown in Figure 2A, elevated Cox-2 expression significantly suppressed SBE-driven luciferase expression. Interestingly, this same experimental condition elicited only a modest increase in basal luciferase expression driven by the synthetic p3TP promoter (Figure 2A) and failed to affect the extent of luciferase expression stimulated by TGF-β (data not shown), suggesting that Cox-2 preferentially inhibits canonical Smad2/3 signaling stimulated by TGF-β. Along these lines, treating quiescent 4T1 cells with the selective Cox-2 antagonist, NS-398, significantly increased the activity of the pSBE-luciferase reporter gene construct (Figure 2B). Although this increase in Smad3/4 signaling appears relatively minor, it is nonetheless statistically significant and likely to be biologically significant, given the extent of autocrine TGF-β signaling experienced by 4T1 cells (24). Thus, upregulated Cox-2 expression may play a role in promoting oncogenic signaling by TGF-β by altering its ability to couple to Smad2/3 in normal and malignant MECs. To further investigate this possibility, we stably expressed Cox-2 in NMuMG cells, which was sufficient to (i) induce their EMT and specific downregulation of Smad3, not Smad4 (Figure 2C); (ii) abrogate the ability of TGF-β to stimulate the expression of luciferase driven by the synthetic pSBE promoter, not the p3TP promoter (Figure 2D); (iii) enhance their invasion through synthetic basement membranes in response to TGF-β (Figure 2E) and (iv) promote their anchorage-independent growth in soft agar (Figure 2F). Taken together, these findings demonstrate that the inappropriate expression of Cox-2 in MECs facilitates oncogenic signaling by TGF-β in part via altered coupling to and expression of Smad3.
Fig. 2.
Cox-2 enhances oncogenic TGF-β signaling by inhibiting Smad2/3 activity. (A) NMuMG cells were transiently transfected with the cDNAs for Cox-2, β-gal and pSBE- or p3TP-luciferase for 48 h as indicated. Afterward, luciferase and β-gal activities contained in detergent-solubilized whole-cell extracts were measured. Data are the mean (±SE; n = 2) luciferase activities relative to control cells (*P < 0.05; Student's t-test). (B) 4T1 cells were transiently transfected with the pSBE-luciferase and β-gal and subsequently were treated with either DMSO (i.e. diluent) or NS-398 (10 μM) for 24 h. Afterward, the luciferase and β-gal activities contained in detergent-solubilized whole-cell extracts were measured. Data are the mean (±SE; n = 3) luciferase activities relative to DMSO-treated cells (*P < 0.05; Student's t-test). (C) Detergent-solubilized whole-cell extracts prepared from control (i.e. empty vector, Hygro) and Cox-2-expressing NMuMG cells (left panel) were immunoblotted with antibodies against COX-2, Smad3, Smad4 and β-actin as indicated. Images are from a representative experiment that was performed three times with identical results. (D) Control (i.e. Hygro) and Cox-2-expressing NMuMG cells were transiently transfected with either pSBE- or p3TP-luciferase and β-gal and subsequently were stimulated overnight with TGF-β1 (5 ng/ml) prior to measuring the luciferase and β-gal activities contained in detergent-solubilized whole-cell extracts. Data are the mean (±SE; n = 3 for SBE; n = 4 for p3TP) luciferase activities relative to control (i.e. Hygro) cells (*P < 0.05; Student's t-test). (E) Control (i.e. Hygro) and Cox-2-expressing NMuMG cells were induced to invade through synthetic basement membranes by serum (2%) in the absence or presence of TGF-β1 (5 ng/ml) as indicated. Data are the mean (±SE; n = 3) invasion relative to that induced by serum in Hygro-expressing cells. (F) Control (i.e. Hygro) and Cox-2-expressing NMuMG cells were cultured in soft agar for 14 days, whereupon NMuMG colony formation was quantified by light microscopy (left panel). Magnification of boxed regions also is shown (middle panel). Colony formation per microscope field (means ± SEs) observed in three independent experiments (*P < 0.05; Student's t-test).
Cox-2 expression inhibits Smad2/3 activity through PGE2 signaling
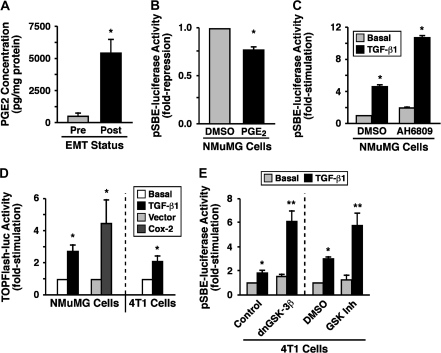
The primary product of Cox-2 activity is PGE2, whose synthesis associates with the development and progression of mammary tumorigenesis through its activation of the EP family (i.e. EPs 1–4) of G protein-coupled receptors (8–10,14,15). Because NMuMG cells undergoing EMT in response to TGF-β upregulate their expression of Cox-2, we evaluated whether these same MECs also exhibit a simultaneous increase in PGE2 production. As expected, TGF-β stimulation of EMT in NMuMG cells did indeed significantly enhance their production of PGE2 as compared with their unstimulated counterparts (Figure 3A). Functionally, administering PGE2 to NMuMG cells resulted in a small, but statistically significant reduction in luciferase expression driven by the synthetic SBE promoter (Figure 3B). The relative difference in the abilities of Cox-2 and PGE2 to repress SBE-luciferase activity probably reflects the continued production of PGE2 by Cox-2 expression versus a single exposure of PGE2, which is converted rapidly to inactive metabolites upon addition to cells (29–31). In support of this supposition, treating NMuMG cells with the non-selective EP receptor antagonist, AH6809, significantly enhanced the coupling of TGF-β to SBE-driven luciferase activity in post-EMT NMuMG cells (Figure 3C).
Fig. 3.
Cox-2 expression inhibits Smad2/3 activity through PGE2 signaling. (A) NMuMG cells were incubated for 36 h in the absence (pre) or presence (post) of TGF-β1 (5 ng/ml) to induce EMT. Afterward, the cells were washed extensively in phosphate-buffered saline and incubated in serum-free media for 1 h prior to measuring PGE2 concentrations by enzyme-linked immunosorbent assay assay. Data are the mean (±SE; n = 3) PGE2 concentrations normalized to corresponding cell protein content (*P < 0.05; Student's t-test). (B) NMuMG cells were transiently transfected with pSBE-luciferase and β-gal and subsequently were treated with DMSO (i.e. diluent) or PGE2 (500 ng/ml) for 24 h. Afterward, the luciferase and β-gal activities contained in detergent-solubilized whole-cell extracts were measured. Data are the mean (±SE; n = 2) luciferase activities relative to DMSO-treated cells (*P < 0.05; Student's t-test). (C) NMuMG cells were incubated overnight in the absence or presence of TGF-β1 (5 ng/ml) to induce EMT. Afterward, the cells were transiently transfected with pSBE-promoter-luciferase and β-gal and subsequently were incubated for an additional 24 h with either DMSO or AH6809 (50 μM) prior to measuring the luciferase and β-gal activities contained in whole-cell extracts. Data are the mean (±SE; n = 4) luciferase activities relative to basal NMuMG cells (*P < 0.05; Student's t-test). (D) Control or Cox-2-expressing NMuMG cells or 4T1 cells were transiently transfected with pTOPFlash-luciferase and β-gal and subsequently were incubated for 14 h in the absence or presence of TGF-β1 (5 ng/ml) as indicated. Afterward, the luciferase and β-gal activities contained in detergent-solubilized whole-cell extracts were measured. Data are the mean (±SE; n = 3 for parental NMuMG and 4T1 cells; n = 2 for Cox-2-transfected cells) luciferase activities relative to corresponding untreated cells (*P < 0.05; Student's t-test). (E) NMuMG cells were incubated overnight in the absence or presence of TGF-β1 (5 ng/ml) to induce EMT. Afterward, the cells were transiently transfected with pSBE-promoter-luciferase and β-gal, together with either empty vector (control) or dominant-negative GSK-3β (dnGSK-3β) as indicated (left panel). Afterward, the transfectants were incubated for an additional 24 h in the absence (DMSO) or presence of a GSK-3β inhibitor (50 nM; right panel) prior to measuring the luciferase and β-gal activities contained in the whole-cell extracts. Data are the mean (±SE; n = 3) luciferase activities relative to basal NMuMG cells (*P < 0.05; Student's t-test).
Through its binding to and activation of EP receptors, PGE2 stimulates a number of effector molecules (14,15) that have been implicated in altering Smad2/3 signaling, including ERK1/2, PI3K/AKT and 3′5′-cyclic adenosine monophosphate (2,3,6). PGE2 also regulates GSK-3β activity (32–34), which recently was shown to phosphorylate Smad3 and induce its degradation (35). Figure 3D shows that administering TGF-β to either NMuMG or 4T1 cells induced their expression of luciferase driven by the synthetic TOPFlash promoter, as did expression of Cox-2 in NMuMG cells. Additionally, treating 4T1 cells with the non-selective Cox inhibitor, indomethacin, enhanced the ability of TGF-β to stimulate SBE-luciferase activity (data not shown), whereas NS-398 administration decreased TOPFlash-driven luciferase activity in these same cells (data not shown). Finally, the ability of TGF-β to induce SBE-luciferase activity in 4T1 cells was greatly potentiated by the expression of kinase-dead GSK-3β or by pharmacological inhibition of GSK-3β activity (Figure 3E). Collectively, these findings suggest that the elevation of Cox-2 expression that occurs during mammary tumorigenesis activates an autocrine PGE2/EP receptor signaling system that uncouples Smad2/3 from TGF-β receptors in part via the activity of GSK-3β.
Cox-2 deficiency enhances Smad2/3 signaling and inhibits PGE2 production and EMT stimulated by TGF-β
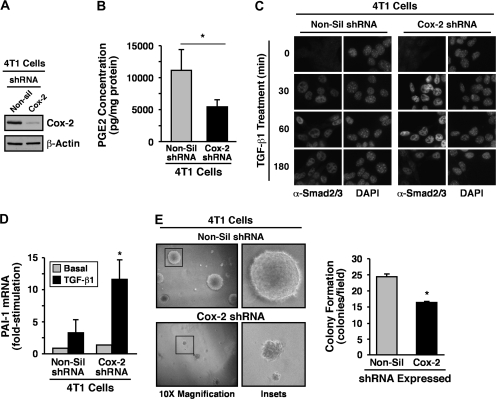
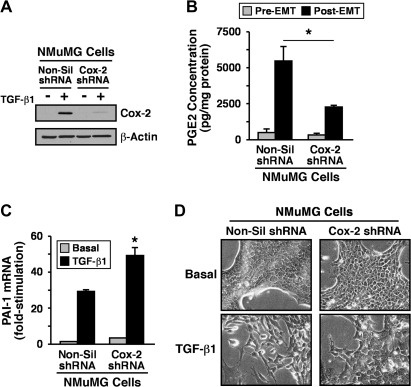
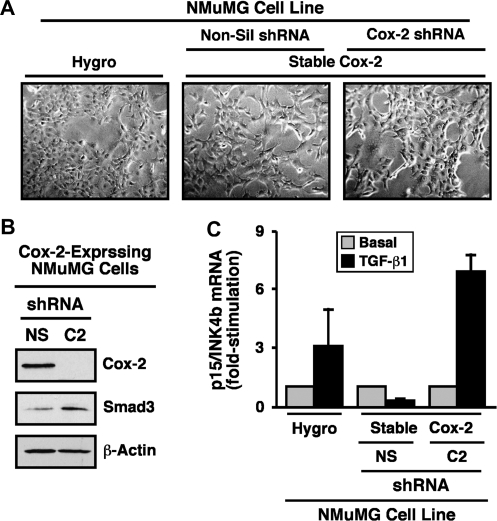
Our findings thus far implicate Cox-2 and its enzymatic product PGE2 as potential inhibitors of Smad2/3 signaling and as mediators of oncogenic signaling stimulated by TGF-β. A corollary states that rendering normal and malignant MECs deficient in Cox-2 will enhance the coupling of Smad2/3 to TGF-β as well as suppress its oncogenic activities. We explored this possibility by infecting normal and malignant MECs with lentivirus encoding for either control (i.e. non-silencing) or Cox-2 shRNA. Consistent with the aforementioned hypothesis, stable expression of Cox-2 shRNA in 4T1 cells significantly reduced their expression of Cox-2 protein (Figure 4A) and, consequently, diminished their synthesis of PGE2 (Figure 4B). Mechanistically, Cox-2 deficiency enhanced the nuclear accumulation of Smad2/3 in response to TGF-β (Figure 4C), an event that resulted in significantly upregulated production of plasminogen activator inhibitor-1 transcripts in Cox-2-deficient 4T1 cells as compared with their Cox-2-expressing counterparts (Figure 4D). Similar outcomes on diminished Cox-2 expression (Figure 5A), on reduced PGE2 production (Figure 5B) and on elevated plasminogen activator inhibitor-1 mRNA transcription (Figure 5C) were observed in TGF-β-treated NMuMG cells engineered to stably express Cox-2 shRNA. Importantly, whereas NMuMG cells expressing non-silencing shRNA readily acquired a fibroblastoid phenotype when stimulated with TGF-β, those rendered Cox-2 deficient failed to undergo EMT in response to TGF-β (Figure 5D). Similarly, the ability of TGF-β to induce EMT and altered actin cytoskeletal architectures in NMuMG cells also was prevented by indomethacin administration (data not shown), suggesting that the expression and activity of Cox-2 is indeed necessary for TGF-β stimulation of EMT in NMuMG cells. Chronic cooperation between TGF-β and cytokine/growth factor receptor signaling systems can stabilize the EMT phenotype in MECs (36,37). Because stable Cox-2 expression was sufficient to induce EMT in NMuMG cells (Figure 2C), we examined whether this cellular and morphological phenotype was metastable and subject to reversion by transducing Cox-2-expressing NMuMG cells with shRNA against this Cox. Figure 6A shows that rendering Cox-2-expressing NMuMG cells deficient in Cox-2 (Figure 6B) did indeed impart an epithelial morphology as compared with their Cox-2-expressing counterparts infected with non-silencing shRNA. Moreover, the loss of Cox-2 also restored NMuMG cell expression of Smad3 (Figure 6B) as well as its ability to mediate p15/INK4b expression in response to TGF-β (Figure 6C). Collectively, our findings reveal an essential function for Cox-2 and PGE2 in suppressing the coupling of TGF-β to Smad2/3 activation and EMT in MECs.
Fig. 4.
Cox-2 deficiency inhibits 4T1 cell PGE2 production and enhances TGF-β stimulation of Smad2/3 signaling. (A) 4T1 cells were rendered Cox-2 deficient by lentiviral-mediated transduction of control (i.e. non-silencing, Non-sil) or Cox-2 shRNA. Following puromycin selection, Cox-2 deficiency was monitored by immunoblotting whole-cell extracts with Cox-2-specific antibodies. Differences in protein loading were monitored using anti-β-actin antibodies. Images are from a representative experiment that was performed three times with identical results. (B) The production of PGE2 by control (i.e. non-silencing, Non-sil) or Cox-2-deficient 4T1 cells was determined by enzyme-linked immunosorbent assay analysis. Data are the mean (±SE; n = 5) PGE2 concentrations normalized to corresponding cell protein content (*P < 0.05; Student's t-test). (C) Control (i.e. non-silencing, Non-sil) or Cox-2-deficient 4T1 cells were incubated in the absence or presence of TGF-β1 (5 ng/ml) for 0–180 min as indicated. Afterward, the cells were fixed in 4% paraformaldehyde and processed for indirect immunofluorescence with anti-Smad2/3 antibodies, following by 4′,6-diamidino-2-phenylindole (DAPI) staining to visualize nuclei. All images were captured on a Nikon Diaphot microscope. Shown are representative images from a single experiment that was performed two times with identical results. (D) Control (i.e. non-silencing, Non-sil) or Cox-2-deficient 4T1 cells were incubated in the absence or presence of TGF-β1 (5 ng/ml) for 24 h. Afterward, total RNA was isolated and subjected to semiquantitative real-time PCR to monitor the expression of plasminogen activator inhibitor-1. Data are the mean (±SE; n = 2) fold changes of plasminogen activator inhibitor-1 gene expression relative to unstimulated control cells (*P < 0.05; Student's t-test). (E) Control (i.e. non-silencing, Non-sil) and Cox-2-deficient (i.e. Cox-2 shRNA) 4T1 cells were cultured in soft agar for 14 days, whereupon 4T1 colony formation was quantified by light microscopy (left panel). Magnification of boxed regions also is shown (middle panel). Colony formation per microscope field (means ± SEs) observed in three independent experiments (*P < 0.05; Student's t-test).
Fig. 5.
Cox-2 deficiency inhibits NMuMG cell PGE2 production and EMT stimulated by TGF-β. (A) NMuMG cells were rendered Cox-2 deficient by lentiviral-mediated transduction of control (i.e. non-silencing, Non-sil) or Cox-2 shRNA. Following puromycin selection, control (i.e. non-silencing, Non-sil) and Cox-2 shRNA-expressing NMuMG cells were incubated in the absence or presence of TGF-β1 (5 ng/ml) for 36 h to induce EMT and subsequently were immunoblotted for Cox-2 and β-actin as indicated. Images are from a representative experiments that was performed three times with identical results. (B) Control (i.e. non-silencing, NS) or Cox-2-deficient (C2) NMuMG cells were incubated for 36 h in the absence (pre) or presence (post) of TGF-β1 (5 ng/ml) to induce EMT. Afterward, the production of PGE2 by control (i.e. non-silencing, Non-sil) or Cox-2-deficient NMuMG cells was determined by enzyme-linked immunosorbent assay analysis. Data are the mean (±SE; n = 2) PGE2 concentrations normalized to corresponding cell protein content (*P < 0.05; Student's t-test). (C) Control (i.e. non-silencing, NS) or Cox-2-deficient (C2) NMuMG cells were incubated in the absence or presence of TGF-β1 (5 ng/ml) for 36 h. Afterward, total RNA was isolated and subjected to semiquantitative real-time PCR to monitor the expression of plasminogen activator inhibitor-1. Data are the mean (±SE; n = 2) fold changes of plasminogen activator inhibitor-1 gene expression relative to unstimulated control cells (*P < 0.05; Student's t-test). (D) Control (i.e. non-silencing, Non-sil) or Cox-2-deficient NMuMG cells were incubated in the absence (pre) or presence (post) of TGF-β1 (5 ng/ml) for 36 h to induce EMT. Bright field images are from a representative experiments that were repeated two times with identical results.
Fig. 6.
Cox-2 deficiency reverts fibroblastoid morphology and restores TGF-β coupling t Smad3 in NMuMG cells. (A) Cox-2-expressing NMuMG cells were subjected to lentiviral-mediated transduction of control (i.e. non-silencing) or Cox-2 shRNA as indicated and subsequently were selected for by resistance to puromycin. Afterward, bright field images of parental (i.e. Hygro), Cox-2-expressing (i.e. non-silencing, Non-sil) or Cox-2-deficient (i.e. Cox-2 shRNA) NMuMG cells were captured from a representative experiment that was performed three times with identical results. (B) Cox-2-expressing (i.e. non-silencing shRNA, NS) or Cox-2-deficient (i.e. Cox-2 shRNA, C2) NMuMG cells were harvested and immunoblotted with antibodies against Cox-2, Smad3 and β-actin as indicated. Images are from a representative experiments that was performed three times with identical results. (C) Parental (i.e. Hygro), Cox-2-expressing (i.e. non-silencing, NS) or Cox-2-deficient (i.e. Cox-2 shRNA, C2) NMuMG cells were incubated for 36 h in the absence or presence of TGF-β1 (5 ng/ml) as indicated. Afterward, total RNA was isolated and subjected to semiquantitative real-time PCR to monitor the expression of p15/INK4b. Data are the mean (±SE; n = 2) fold changes of plasminogen activator inhibitor-1 gene expression relative to unstimulated control cells (*P < 0.05; Student's t-test).
Discussion
Despite nearly three decades of intense research, the molecular mechanisms that enable mammary tumorigenesis to convert TGF-β from a suppressor of breast cancer formation to a promoter of its growth and metastasis remain to be determined definitively. The inability of science and medicine to solve the ‘TGF-β paradox’ as it relates to mammary tumorigenesis probably reflects the heterogeneous nature of human breast cancers, as well as the pathophysiological inputs and outcomes that arise in response to dysregulated TGF-β signaling in multiple tumor components, including malignant MECs and their activated stromal constituents [e.g. fibroblasts, endothelial cells and infiltrating adaptive and innate immune cells (1,2)]. Equally daunting is our failure to fully comprehend how normal MECs integrate canonical Smad2/3 signaling inputs with those arising from their non-canonical counterparts (e.g. integrins/PTKs, MAP kinases, PI3K/AKT and NF-κB) during cytostasis induced by TGF-β and conversely how aberrant and/or amplified activation of these non-canonical signaling inputs conspire with Smad2/3 to facilitate oncogenic signaling stimulated by TGF-β in developing and progressing mammary tumors (1–3). Thus, identifying, characterizing and targeting novel non-canonical pathways activated by TGF-β may provide new inroads to circumvent the TGF-β paradox and restore the tumor-suppressing function of TGF-β in human breast cancers (38).
With this goal in mind, we recently identified and established the importance of two novel pathways that mediate oncogenic signaling by TGF-β—i.e. one involving αvβ3 integrin and Src (4–6) and a second involving the formation of TAB1:TAK1:IKKβ complexes which is essential for NF-κB activation and breast cancer growth stimulated by TGF-β (7). Activation of the latter pathway also was essential for coupling TGF-β to Cox-2 expression in breast cancer cells (7); however, the role of Cox-2 in promoting oncogenic signaling by TGF-β in MECs remained to be clarified. We show here that the ability of TGF-β to stimulate Cox-2 expression in normal MECs transpires through an EMT-dependent mechanism and that this event coincided with upregulated PGE2 production and its ability to blunt of Smad2/3 signaling by attenuating Smad3 expression and nuclear accumulation in normal and malignant MECs. More importantly, we show that upregulated Cox-2 expression is essential for oncogenic signaling stimulated by TGF-β, particularly its ability to induce EMT, invasion and anchorage-independent growth in MECs (Figures 2 and 5). Indeed, similar to aberrant TGF-β signaling, inappropriate Cox-2 activity promotes mammary tumor growth and metastasis to lung and bone as well as predicts for poor clinical outcomes (8–12); it also stimulates the synthesis of proinflammatory molecules [e.g. PGE2, IL-11 and IL-8; (39,40)], which enhance genetic instability (41,42) and create tumor microenvironments that promote malignant progression. It should be noted that we have yet to ascertain whether EMT enables TGF-β to couple to Cox-2 expression or whether Cox-2 expression promotes EMT stimulated by TGF-β. Our bias is that Cox-2 expression is essential for EMT induced by TGF-β, particularly since Cox-2 deficiency prevented NMuMG cells from undergoing EMT in response to TGF-β (Figure 5). A number of studies have implicated activated Ras and ERK1/2 in facilitating EMT stimulated by TGF-β (36,43–45). Whether elevated Cox-2 expression induced by TGF-β can compensate for a lack of activated Ras in NMuMG cells remains to be determined. If so, then it would appear that the differentiation state and/or pre-existing gene expression profile of an individual epithelial cell may in large part determine its relative dependence on Cox-2 when acquiring fibroblastoid phenotypes in response to TGF-β. Although the validity of this hypothesis awaits future investigation, our findings do suggest that the chemotherapeutic targeting of Cox-2 may suppress the formation and progression of mammary tumors not only by alleviating their production of cancer-promoting inflammatory and angiogenic factors but also by inactivating the oncogenic activities of TGF-β in developing and progressing mammary tumors.
A wealth of epidemiological studies have identified Cox-2 as a tumor-promoting molecule, particularly in colorectal cancers whose incidence can be reduced by ∼50% with long-term non-steroidal anti-inflammatory drugs administration (46). Similar population-based studies have revealed that ∼40% of breast cancers exhibit upregulated expression of Cox-2 (8,10), which correlates with decreased disease-free survival (47); these studies also found that regular non-steroidal anti-inflammatory drugs usage significantly reduced the risk and development of breast cancers (48–50). Furthermore, aberrantly elevated Cox-2 expression has been associated with the progression of ductal carcinoma in situ (DCIS) to basal-like mammary tumors, leading to the hypothesis that Cox-2 expression in DCIS may serve as a novel biomarker capable of predicting which DCIS patients probably will progress and develop this most lethal form of breast cancer (51). Along these lines, elevated Cox-2 expression in DCIS has been linked to elevated p38 MAPK activation (52), which we previously identified as an essential mediator of breast cancer growth and metastasis stimulated by TGF-β (6). Thus, it is tempting to speculate that p38 MAPK couples TGF-β to Cox-2 expression in normal and malignant MECs. These epidemiological findings have been bolstered and recapitulated by numerous in vitro analyses of Cox-2 expression and function in human breast cancer cells (47,53) and by preclinical in vivo analyses of Cox-2 activity in mice (54–57), which collectively have established Cox-2 as an essential mediator of mammary tumor formation and their acquisition of invasive, metastatic, angiogenic and high-grade phenotypes (8–10). As mentioned above, aberrant Cox-2 expression probably occurs early during mammary tumorigenesis where its expression strongly associates with the development of DCIS (8–10,51,52,58) and with the upregulated expression of HER2/neu (59,60), which can induce the synthesis of Cox-2 in malignant MECs (61,62). Moreover, Richards et al. (61) found that TGF-β, as well as EGF and phorbol ester, readily induces the expression of Cox-2 and aromatase in human breast cancer cells, although the functional significance of these TGF-β-dependent events in mediating the development and progression of mammary tumors remains unknown. Despite this limitation, these observations do suggest a casual relationship between HER2/neu and TGF-β signaling in mediating inappropriate Cox-2 expression, a supposition supported by the finding that Cox-2 deficiency in mice suppresses the oncogenicity of MMTV-driven HER2/neu overexpression (55). Despite this apparent association, our findings identify the TGF-β signaling system as a major and important route leading to Cox-2 expression in normal and malignant MECs. Indeed, pharmacological inactivation of TβR-I in 4T1 breast cancer cells reduced Cox-2 mRNA and protein content by ∼80% (Figure 1), suggesting that autocrine TGF-β signaling in these malignant MECs largely accounts for their inappropriate expression of Cox-2. Along these lines, Wakefield et al. (24) recently showed that 4T1 cells produce copious amounts of TGF-β1 and, to a lesser extent, TGF-β3, both in vitro and in vivo. Thus, upregulated autocrine TGF-β signaling may represent a novel step in the progression of mammary tumorigenesis that results in elevated Cox-2 expression, findings consistent with those presented herein. In addition, several studies have shown HER2/neu to cooperate with TGF-β in mediating mammary tumorigenesis (63,64), whereas others have established integrins as being essential for the oncogenic activity of epidermal growth factor receptor (65). We too observed αvβ3 integrin to cooperate with and be essential for oncogenic signaling by TGF-β (4,5), particularly its ability to stimulate the growth and pulmonary metastasis of mammary tumors in mice (6). More recently, we found the activity of αvβ3 integrin to mediate Cox-2 expression in response to TGF-β (J.R.Neil, A.J.Galliher-Beckley and W.P.Schiemann, unpublished observation), which together with NF-κB probably represents a major node whereby TGF-β induces EMT in normal and malignant MECs. Future studies clearly need to determine the role of integrins in mediating the coupling of TGF-β to Cox-2 expression as well as examine the potential involvement of HER2/neu signaling in mediating Cox-2 expression and EMT stimulated by TGF-β.
A particularly interesting finding of the present study was the ability of Cox-2 to inhibit Smad2/3 signaling in normal and malignant MECs, an event initiated in part by an autocrine PGE2:EP receptor signaling axis that reduced the expression, nuclear accumulation and transcriptional activity of Smad3 (Figures 3–5). This response appears to be specific for Smad3 as no changes in the cellular levels of Smad4 were detected in Cox-2-manipulated cells. A similar loss of Smad2/3 expression was observed in the mammary glands of mice engineered to express a constitutively active TβR-I (i.e. T204D-TβR-I) under control of the MMTV promoter (66). More recently, GSK-3β-mediated phosphorylation of Thr66 in inactive Smad3 led to its ubiquitination and degradation and consequently to altered sensitivity of cells to TGF-β (35). Along these lines, we found that measures capable of inhibiting GSK-3β activity significantly potentiated the transcriptional activity of Smad2/3 (Figure 3), suggesting that the Cox-2:PGE2:EP receptor signaling axis requires GSK-3β activity to antagonize Smad2/3 signaling stimulated by TGF-β. Although we do not yet know whether GSK-3β directly phosphorylates and inhibits Smad3 signaling in normal and malignant MECs, our findings do show that the amplified activation of the Cox-2:PGE2:EP receptor signaling axis reduces the cellular pool of Smad3 as well as impairs its ability to translocate into the nucleus in response to TGF-β. In addition to activating GSK-3β, EP receptors also activate ERK1/2, PI3K/AKT and 3′5′-cyclic adenosine monophosphate (14,15), all of which regulate Smad2/3 signaling in response to TGF-β (1,3). Thus, the relative contribution of these pathways to Cox-2- and PGE2-mediated regulation of TGF-β signaling in normal and malignant MECs needs to be addressed.
A major drawback associated with the administration of Cox-2 inhibitors in clinical settings is their propensity for eliciting severe cardiovascular reactions (8–10). Because PGE2 primarily mediates the oncogenic activities of aberrant Cox-2 expression, it therefore stands to reason that chemotherapeutic targeting of PGE2 and its EP receptors may mitigate mammary tumorigenesis driven by Cox-2 without engendering life-threatening cardiovascular reactions (14,67). Moreover, the expression of various EP receptor isoforms has been associated with tumorigenesis (14). For instance, increased EP4 receptor expression is observed in colorectal carcinoma cells and contributes to their enhanced proliferation and anchorage-independent growth (68). Additionally, EP1, EP2 and EP4 receptors all have been implicated in promoting mammary tumorigenesis (10,14). Moreover, we show that pan-antagonism of EP receptor activity significantly enhances the coupling of TGF-β to Smad2/3 signaling in normal MECs (Figure 3), suggesting that specific targeting of EP receptors may mimic the tumor-suppressing activities of Cox-2 inhibitors, while simultaneously circumventing their development of untoward cardiovascular episodes. This notion awaits preclinical evaluation, as does the performance of studies aimed at determining which EP receptors alter the response of normal and malignant MECs to TGF-β.
Funding
National Institutes of Health (CA095519, CA114039, CA129359); Komen Foundation to W.P.S.
Acknowledgments
Members of the Schiemann Laboratory are thanked for critical reading of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- Cox
cyclooxygenase
- DCIS
ductal carcinoma in situ
- EMT
epithelial–mesenchymal transition
- ERK
extracellular signal-regulated kinase
- GSK
glycogen synthase kinase
- MAPK
mitogen-activated protein kinase
- MEC
mammary epithelial cell
- mRNA
messenger RNA
- NF-κB
nuclear factor kappa B
- PCR
polymerase chain reaction
- PGE
prostaglandin E2
- PI3K
phosphoinositide 3-kinase
- SBE
smad-binding element
- TGF-β
transforming growth factor-β
- TβR-I
transforming growth factor-β type I
References
- 1.Galliher AJ, et al. Role of TGF-β in cancer progression. Future Oncol. 2006;2:743–763. doi: 10.2217/14796694.2.6.743. [DOI] [PubMed] [Google Scholar]
- 2.Gordon KJ, et al. Role of TGF-β superfamily signaling pathways in human disease. Biochim. Biophys. Acta. 2008;1782:197–228. doi: 10.1016/j.bbadis.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Siegel PM, et al. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 4.Galliher AJ, et al. β3 integrin and Src facilitate TGF-β mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Res. 2006;8:R42. doi: 10.1186/bcr1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galliher AJ, et al. Src phosphorylates Tyr284 in TGF-β type II receptor and regulates TGF-β stimulation of p38 MAPK during breast cancer cell proliferation and invasion. Cancer Res. 2007;67:3752–3758. doi: 10.1158/0008-5472.CAN-06-3851. [DOI] [PubMed] [Google Scholar]
- 6.Galliher-Beckley AJ, et al. Grb2 binding to Tyr284 in TβR-II is essential for mammary tumor growth and metastasis stimulated by TGF-β. Carcinogenesis. 2008;29:244–251. doi: 10.1093/carcin/bgm245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neil JR, et al. Altered TAB1:IκB kinase interaction promotes TGF-β-mediated NF-κB activation during breast cancer progression. Cancer Res. 2008;68:1462–1470. doi: 10.1158/0008-5472.CAN-07-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazhar D, et al. COX inhibitors and breast cancer. Br. J. Cancer. 2006;94:346–350. doi: 10.1038/sj.bjc.6602942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh-Ranger G, et al. The role of Cox-2 in breast cancer: review. Breast Cancer Res. Treat. 2008;109:189–198. doi: 10.1007/s10549-007-9641-5. [DOI] [PubMed] [Google Scholar]
- 10.Howe LR. Inflammation and breast cancer. Cyclooxygenase/prostaglandin signaling and breast cancer. Breast Cancer Res. 2007;9:210. doi: 10.1186/bcr1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh B, et al. COX-2 involvement in breast cancer metastasis to bone. Oncogene. 2007;26:3789–3796. doi: 10.1038/sj.onc.1210154. [DOI] [PubMed] [Google Scholar]
- 13.Langenbach R, et al. Cyclooxygenase-deficient mice. A summary of their characteristics and susceptibilities to inflammation and carcinogenesis. Ann. N. Y. Acad. Sci. 1999;889:52–61. doi: 10.1111/j.1749-6632.1999.tb08723.x. [DOI] [PubMed] [Google Scholar]
- 14.Fulton AM, et al. Targeting prostaglandin E EP receptors to inhibit metastasis. Cancer Res. 2006;66:9794–9797. doi: 10.1158/0008-5472.CAN-06-2067. [DOI] [PubMed] [Google Scholar]
- 15.Narumiya S, et al. Prostanoid receptors: structures, properties, and functions. Physiol. Rev. 1999;79:1193–1226. doi: 10.1152/physrev.1999.79.4.1193. [DOI] [PubMed] [Google Scholar]
- 16.Zufferey R, et al. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 17.Schiemann BJ, et al. SPARC inhibits epithelial cell proliferation in part through stimulation of the TGF-β-signaling system. Mol. Biol. Cell. 2003;14:3977–3988. doi: 10.1091/mbc.E03-01-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiemann WP, et al. A deletion in the gene for TGF-β type I receptor abolishes growth regulation by TGF-β in a cutaneous T-cell lymphoma. Blood. 1999;94:2854–2861. [PubMed] [Google Scholar]
- 19.Bren-Mattison Y, et al. Antitumorigenic effects of PPAR-γ in non-small-cell lung cancer cells are mediated by suppression of Cox-2 via inhibition of NF-κB. Mol. Pharmacol. 2008;73:709–717. doi: 10.1124/mol.107.042002. [DOI] [PubMed] [Google Scholar]
- 20.Miettinen PJ, et al. TGF-β induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piek E, et al. TGF-β type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J. Cell Sci. 1999;112:4557–4568. doi: 10.1242/jcs.112.24.4557. [DOI] [PubMed] [Google Scholar]
- 22.Sokol JP, et al. The use of cystatin C to inhibit epithelial-mesenchymal transition and morphological transformation stimulated by TGF-β. Breast Cancer Res. 2005;7:R844–R853. doi: 10.1186/bcr1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L, et al. Abrogation of TGF-β signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nam JS, et al. An anti-TGF-β antibody suppresses metastasis via cooperative effects on multiple cell compartments. Cancer Res. 2008;68:3835–3843. doi: 10.1158/0008-5472.CAN-08-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge R, et al. Inhibition of growth and metastasis of mouse mammary carcinoma by selective inhibitor of TGF-β type I receptor kinase in vivo. Clin. Cancer Res. 2006;12:4315–4330. doi: 10.1158/1078-0432.CCR-06-0162. [DOI] [PubMed] [Google Scholar]
- 26.Enders GA. Cox-2 overexpression abrogates the antiproliferative effects of TGF-β. Br. J. Cancer. 2007;97:1388–1392. doi: 10.1038/sj.bjc.6604048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang A, et al. Prostaglandin E2 is a potent inhibitor of epithelial-to-mesenchymal transition: interaction with HGF. Am. J. Physiol. Renal Physiol. 2006;291:F1323–F1331. doi: 10.1152/ajprenal.00480.2005. [DOI] [PubMed] [Google Scholar]
- 28.Schiller M, et al. Cyclic adenosine 3′,5′-monophosphate-elevating agents inhibit TGF-β-induced SMAD3/4-dependent transcription via a protein kinase A-dependent mechanism. Oncogene. 2003;22:8881–8890. doi: 10.1038/sj.onc.1206871. [DOI] [PubMed] [Google Scholar]
- 29.Granstrom E, et al. Chemical instability of 15-keto-13,14-dihydro-PGE2: the reason for low assay reliability. Prostaglandins. 1980;19:933–957. doi: 10.1016/0090-6980(80)90127-6. [DOI] [PubMed] [Google Scholar]
- 30.Hamberg M, et al. On the metabolism of prostaglandins E 1 and E 2 in man. J. Biol. Chem. 1971;246:6713–6721. [PubMed] [Google Scholar]
- 31.Fitzpatrick FA, et al. The stability of 13,14-dihydro-15 keto-PGE2. Prostaglandins. 1980;19:917–931. doi: 10.1016/0090-6980(80)90126-4. [DOI] [PubMed] [Google Scholar]
- 32.Takadera T, et al. Prevention of rat cortical neurons from prostaglandin E2-induced apoptosis by GSK-3 inhibitors. Neurosci. Lett. 2006;400:105–109. doi: 10.1016/j.neulet.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 33.Fujino H, et al. Prostaglandin E2 induced functional expression of early growth response factor-1 by EP4, but not EP2, prostanoid receptors via the PI-3K and ERKs. J. Biol. Chem. 2003;278:12151–12156. doi: 10.1074/jbc.M212665200. [DOI] [PubMed] [Google Scholar]
- 34.Lim K, et al. Cox-2-derived prostaglandin E2 activates β-catenin in human cholangiocarcinoma cells: evidence for inhibition of these signaling pathways by omega 3 polyunsaturated fatty acids. Cancer Res. 2008;68:553–560. doi: 10.1158/0008-5472.CAN-07-2295. [DOI] [PubMed] [Google Scholar]
- 35.Guo X, et al. Axin and GSK3- control Smad3 protein stability and modulate TGF-β signaling. Genes Dev. 2008;22:106–120. doi: 10.1101/gad.1590908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grunert S, et al. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat. Rev. Mol. Cell Biol. 2003;4:657–665. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 37.Gotzmann J, et al. Molecular aspects of epithelial cell plasticity: implications for local tumor invasion and metastasis. Mutat. Res. 2004;566:9–20. doi: 10.1016/s1383-5742(03)00033-4. [DOI] [PubMed] [Google Scholar]
- 38.Schiemann WP. Targeted TGF-β chemotherapies: friend or foe in treating human malignancies? Expert Rev. Anticancer Ther. 2007;7:609–611. doi: 10.1586/14737140.7.5.609. [DOI] [PubMed] [Google Scholar]
- 39.Singh B, et al. COX-2 induces IL-11 production in human breast cancer cells. J. Surg. Res. 2006;131:267–275. doi: 10.1016/j.jss.2005.11.582. [DOI] [PubMed] [Google Scholar]
- 40.Singh B, et al. Involvement of IL-8 in COX-2-mediated bone metastases from breast cancer. J. Surg. Res. 2006;134:44–51. doi: 10.1016/j.jss.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 41.Singh B, et al. Cox-2 expression induces genomic instability in MCF10A breast epithelial cells. J. Surg. Res. 2007;140:220–226. doi: 10.1016/j.jss.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 42.Singh B, et al. Cox-2 induces genomic instability, BCL2 expression, doxorubicin resistance, and altered cancer-initiating cell phenotype in MCF7 breast cancer cells. J. Surg. Res. 2008;147:240–246. doi: 10.1016/j.jss.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 43.Xie L, et al. Activation of the ERK pathway is required for TGF-β1-induced EMT in vitro. Neoplasia. 2004;6:603–610. doi: 10.1593/neo.04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oft M, et al. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat. Cell Biol. 2002;4:487–494. doi: 10.1038/ncb807. [DOI] [PubMed] [Google Scholar]
- 45.Janda E, et al. Ras and TGF-β cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J. Cell Biol. 2002;156:299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia Rodriguez LA, et al. Reduced incidence of colorectal adenoma among long-term users of nonsteroidal antiinflammatory drugs: a pooled analysis of published studies and a new population-based study. Epidemiology. 2000;11:376–381. doi: 10.1097/00001648-200007000-00003. [DOI] [PubMed] [Google Scholar]
- 47.Ristimaki A, et al. Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res. 2002;62:632–635. [PubMed] [Google Scholar]
- 48.Gonzalez-Perez A, et al. Effects of non-steroidal anti-inflammatory drugs on cancer sites other than the colon and rectum: a meta-analysis. BMC Cancer. 2003;3:28. doi: 10.1186/1471-2407-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris RE, et al. Reduction in the risk of human breast cancer by selective COX-2 inhibitors. BMC Cancer. 2006;6:27. doi: 10.1186/1471-2407-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris RE, et al. Breast cancer and nonsteroidal anti-inflammatory drugs: prospective results from the Women's Health Initiative. Cancer Res. 2003;63:6096–6101. [PubMed] [Google Scholar]
- 51.Gauthier ML, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gauthier ML, et al. p38 regulates Cox-2 in human mammary epithelial cells and is activated in premalignant tissue. Cancer Res. 2005;65:1792–1799. doi: 10.1158/0008-5472.CAN-04-3507. [DOI] [PubMed] [Google Scholar]
- 53.Liu XH, et al. Differential expression and regulation of Cox-1 and -2 in two human breast cancer cell lines. Cancer Res. 1996;56:5125–5127. [PubMed] [Google Scholar]
- 54.Abou-Issa HM, et al. Dose-response effects of the COX-2 inhibitor, celecoxib, on the chemoprevention of mammary carcinogenesis. Anticancer Res. 2001;21:3425–3432. [PubMed] [Google Scholar]
- 55.Howe LR, et al. HER2/neu-induced mammary tumorigenesis and angiogenesis are reduced in Cox-2 knockout mice. Cancer Res. 2005;65:10113–10119. doi: 10.1158/0008-5472.CAN-05-1524. [DOI] [PubMed] [Google Scholar]
- 56.Liu CH, et al. Overexpression of Cox-2 is sufficient to induce tumorigenesis in transgenic mice. J. Biol. Chem. 2001;276:18563–18569. doi: 10.1074/jbc.M010787200. [DOI] [PubMed] [Google Scholar]
- 57.Narko K, et al. COX-2 inhibitors and genetic background reduce mammary tumorigenesis in Cox-2 transgenic mice. Prostaglandins Other Lipid Mediat. 2005;76:86–94. doi: 10.1016/j.prostaglandins.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 58.Half E, et al. Cox-2 expression in human breast cancers and adjacent ductal carcinoma in situ. Cancer Res. 2002;62:1676–1681. [PubMed] [Google Scholar]
- 59.Vadlamudi R, et al. Regulation of Cox-2 pathway by HER2 receptor. Oncogene. 1999;18:305–314. doi: 10.1038/sj.onc.1202307. [DOI] [PubMed] [Google Scholar]
- 60.Subbaramaiah K, et al. Cox-2 is overexpressed in HER-2/neu-positive breast cancer: evidence for involvement of AP-1 and PEA3. J. Biol. Chem. 2002;277:18649–18657. doi: 10.1074/jbc.M111415200. [DOI] [PubMed] [Google Scholar]
- 61.Richards JA, et al. Signaling pathways regulating aromatase and cyclooxygenases in normal and malignant breast cells. J. Steroid Biochem. Mol. Biol. 2002;80:203–212. doi: 10.1016/s0960-0760(01)00187-x. [DOI] [PubMed] [Google Scholar]
- 62.Wang SC, et al. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell. 2004;6:251–261. doi: 10.1016/j.ccr.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 63.Seton-Rogers SE, et al. ErbB2 and TGF-β: a cooperative role in mammary tumor progression? Cell Cycle. 2004;3:597–600. [PubMed] [Google Scholar]
- 64.Seton-Rogers SE, et al. Cooperation of the ErbB2 receptor and TGF-β in induction of migration and invasion in mammary epithelial cells. Proc. Natl Acad. Sci. USA. 2004;101:1257–1262. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reginato MJ, et al. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat. Cell Biol. 2003;5:733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- 66.Muraoka-Cook RS, et al. Activated type I TGF-β receptor kinase enhances the survival of mammary epithelial cells and accelerates tumor progression. Oncogene. 2006;25:3408–3423. doi: 10.1038/sj.onc.1208964. [DOI] [PubMed] [Google Scholar]
- 67.Ma X, et al. Prostaglandin E receptor EP4 antagonism inhibits breast cancer metastasis. Cancer Res. 2006;66:2923–2927. doi: 10.1158/0008-5472.CAN-05-4348. [DOI] [PubMed] [Google Scholar]
- 68.Chell SD, et al. Increased EP4 receptor expression in colorectal cancer progression promotes cell growth and anchorage independence. Cancer Res. 2006;66:3106–3113. doi: 10.1158/0008-5472.CAN-05-3702. [DOI] [PubMed] [Google Scholar]