Abstract
TBP functions in transcription initiation in all eukaryotes and in Archaebacteria. Although the 181–amino acid (aa) carboxyl (C-) terminal core of the protein is highly conserved, TBP proteins from different phyla exhibit diverse sequences in their amino (N-) terminal region. In mice, the TBP N-terminus plays a role in protecting the placenta from maternal rejection; however the presence of similar TBP N-termini in nontherian tetrapods suggests that this domain also has more primitive functions. To gain insights into the pretherian functions of the N-terminus, we investigated its phylogenetic distribution. TBP cDNAs were isolated from representative nontetrapod jawed vertebrates (zebrafish and shark), from more primitive jawless vertebrates (lamprey and hagfish), and from a prevertebrate cephalochordate (amphioxus). Results showed that the tetrapod N-terminus likely arose coincident with the earliest vertebrates. The primary structures of vertebrate N-termini indicates that, historically, this domain has undergone events involving intragenic duplication and modification of short oligopeptide-encoding DNA sequences, which might have provided a mechanism of de novo evolution of this polypeptide.
Keywords: transcription, TFIID, cyclostome, minisatellite duplication, polypeptide genesis
Introduction
The C-terminal core of the TATA-binding protein (TBP) is one of the most evolutionarily conserved protein domains known; however the TBP N-terminal region varies greatly between taxa (Hernandez 1993). Published sequences indicate that all tetrapods (amphibians, reptiles, birds, and mammals) have similar TBP N-termini (Hernandez 1993; Sumita et al. 1993; Nakashima et al. 1995; Shimada et al. 1999). Conversely, TBP sequences from nonvertebrate metazoans, including both protostomes (e.g., arthropods) and lower deuterostomes (e.g., echinoderms), lack nonrepetitive sequences resembling the tetrapod TBP N-terminus (NCBI/GenBank) (Lichtsteiner and Tjian 1993; Muhich et al. 1990).
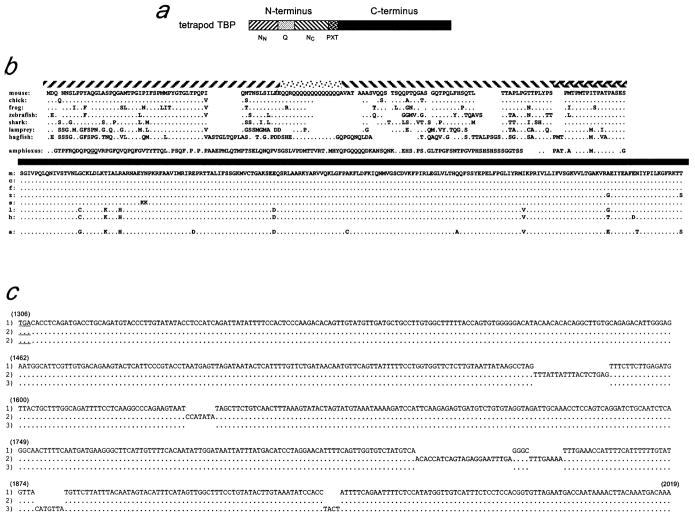
The tetrapod TBP N-terminus can be divided into four subdomains (fig. 1a). Two of these, entitled NN and NC, which do not resemble each other or other known proteins, flank a central glutamine-rich repeat region, called Q. Near the junction with the conserved C-terminus is an imperfect repeat region of sequence (PXT)n, where X is generally M, A, or I. This repeat exists in most or all metazoan TBPs, and we suspect it serves as a connector that integrates phyla-specific properties of the various metazoan N termini with the universal functions of the C-terminus.
Fig. 1.
Chordate TBPs. (a) Linear depiction of vertebrate TBP with domains indicated by different shading. (b) a.a. sequences of TBP proteins, with shaded bars above corresponding to the regions indicated in (a). Dots represent amino acids that are identical to the mouse sequence; gaps in alignment are exhibited by gaps in the presentation. The longer amphioxus N terminus is shown with the two extra a.a. underlined. For hagfish, variants containing 11, 12, and 13 Qs in the Q domain were isolated; 12 Qs are shown. (c) Oligonucleotide insertion-based polymorphisms in the aTBP 3′ UTR. Sequences for the three 3′-UTR mRNA isoforms that we identified are shown beginning at the stop codon (underlined). Nucleic acid positions for isoform number 1 are shown above the sequence in parentheses.
We hypothesized that the N-terminus functions as a “signaling port” by which the basal transcription machinery receives specific regulatory signals and that variation within this region represents commitment to gene-regulation processes that are unique to each phylum. We previously showed that mice lacking most of this domain exhibit no defects in basal cellular/metabolic functions nor in any other functions shared between mammals and nonvertebrate eukaryotes (Hobbs et al. 2002; Schmidt et al. 2003), as one might predict based on the absence of related sequences from lower eukaryotes. Rather, homozygous mutants exhibit a lethal defect in the function of the midgestational placenta, which can be rescued by providing mutant fetuses with a wild-type tetraploid placenta. Genetic rescue studies suggest mutant failure results from perturbation of a β2-microglobulin–dependent process that the placenta uses to evade maternal immune rejection (Hobbs et al. 2002).
The physiological site of the defect in these mice, the chorionic placenta, is found only in therian mammals. However, the presence of a similar TBP N-terminus among all tetrapods suggests this domain confers a fitness advantage on nonplacental as well as placental tetrapods. To gain insights into the preplacental functions of this domain, we isolated TBP cDNAs from strategically placed representatives of nontetrapod vertebrate lineages (teleost, shark, lamprey, and hagfish) and from an advanced non-vertebrate deuterostome (amphioxus). Within the resolution afforded by analyses of extant species, this domain was precisely restricted to vertebrates. Comparisons between species revealed that multiple events involving intragenic duplication of oligopeptide-encoding DNA sequences have occurred and persisted in this domain.
Materials and Methods
Isolation of cDNA and Genomic tbp Clones
Sources cited in the acknowledgments provided cDNA libraries from amphioxus (Branchiostoma floridae) (Langeland et al. 1998), Atlantic sea lamprey (Petromyzon marinus) (Tomsa and Langeland 1999), nurse shark (Ginglymostoma cirratum) (Kandil et al. 1996), Pacific hagfish (Eptatretus stoutii) (Nagata et al. 2002), and zebrafish (Danio rerio). A cDNA λ library was produced in a ZAP II vector using poly-A+=mRNA isolated from spawning Atlantic sea lamprey testes. The library contained 2.25 × 106 primary clones; 84% contained inserts of approximately 1.5 kb average length.
For screening zebrafish and shark libraries, a cDNA probe containing most of the C- terminus and 222 bp of 3′ untranslated region (UTR) of mouse (m) TBP was used. For screening lamprey and amphioxus libraries (8 × 105 to 10 × 105 phage screened per library), we used a mixed-species probe containing the same mTBP cDNA fragment and (1) an 840-bp Hinc II/Sca I fragment of shark TBP cDNA (144 bp of N-terminus, entire C-terminus, and 149 bp of 3′ UTR); and (2) a 697-bp fragment of zebrafish TBP cDNA (start codon to a position midway through the C-terminus). TBP-containing inserts were recombined into SK- plasmid vectors and inserts were sequenced in both directions. Five amphioxus, two lamprey, two shark, and five zebrafish TBP cDNA clones (designated aTBP, lTBP, sTBP, and zfTBP, respectively) were sequenced.
We isolated 23 TBP clones from 1.8 × 106 plaques of the hagfish library (designated hfTBP); however none were full length. The two longest cDNAs encoded the entire C-terminus, but only 91 aa of the N-terminal region. The sequence revealed Pst I, Hind III, and Eco RI sites in the 5′ end of the C-terminal protein-coding DNA. Genomic Southern blots suggested that the hagfish gene also contained Pst I, Eco RI, and Hind III sites at approximately 2.2 kb, approximately 3.5 kb, and approximately 6.7 kb upstream of the TBP N-terminal/C-terminal junction, respectively. Using Hind III ligation-mediated PCR (hfTBP-specific primer: 5′-TAT GGA TCC TGA CGT TGT GCT TCC ACT G-3′), we isolated and cloned a 1,359-bp genomic fragment, including more of the TBP N-terminal region. This fragment contained two exons, which, together, encoded a protein domain homologous to the entire exon 3–encoded N-terminal region of mTBP. It did not, however, include the region homologous to mTBP exon 2, (start codon and subsequent 17 aa) (Sumita et al. 1993; Ohbayashi et al. 1996). A primer to sequences from the 5′ protein-coding region of this clone (5′-TAT GGA TCC AGC ATC TAG TTG GTT CTG CC-3′) was used in combination with the vector-encoded T3 primer to screen 24 portions (1 × 106 phage/portion) of the hagfish library by PCR. Two portions contained full-length hfTBP cDNAs, including 200 bp of 5′ UTR.
Sequence Alignments, dN and dS Estimations, and Relatedness Predictions
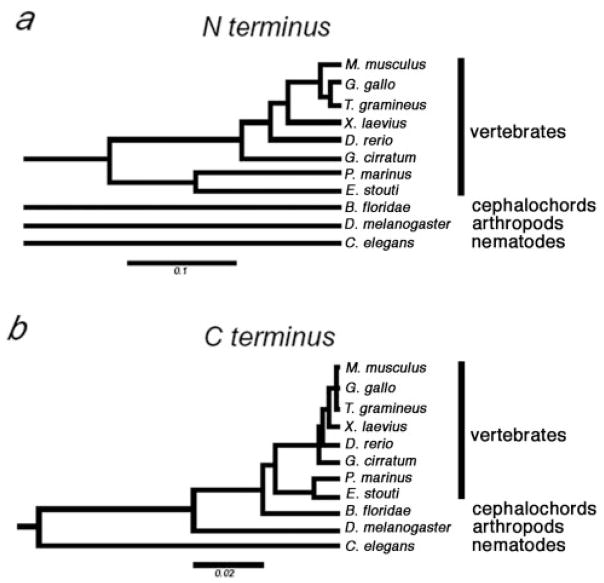
Initial pair-wise alignments were performed using Blast (NCBI/GenBank). Progressive multiple sequence alignments were performed using ClaustalW software (Thompson, Higgins, and Gibson 1994) and best-fit/gap-placement was confirmed manually (fig. 1) (Thompson, Higgins, and Gibson 1994). dN/dS estimations were performed with the MEGA version 2.1 software package (Kumar et al. 2001) using five methods (Li, Wu, and Luo 1985; Nei and Gojobori 1986; Li 1993; Pamilo and Bianchi 1993; Comeron 1995) that differ in their treatment of transition versus transversion substitutions and their treatment of different-fold degenerate sites (Nei and Kumar 2000), as described in table 1. Relatedness was predicted on nucleotide sequences derived from gap-free polypeptide alignments as the Jukes-Cantor correction of p-distance for nonsynonymous substitutions using the modified Nei-Gojobori method (Nei and Kumar 2000) within MEGA 2.1. Relatedness is presented in figure 2 as distance trees built using the UPGMA Tree Making method (Kumar et al. 2001) within MEGA 2.1. Best-fit, gap-free N-terminal alignments between vertebrate and lower metazoan species were nearly arbitrary because no regions of homology are identifiable outside the PXT region (e.g., compare vertebrate and amphioxus sequences in figure 1b). Therefore, although the modified Nei-Gojobori method within MEGA 2.1 (Kumar et al. 2001) gave p-distance values, the tree in figure 2a is not drawn this deep because we did not wish to imply a common ancestry for these polypeptide domains.
Table 1.
Estimates of Nonsynonymous/Synonymous Substitution Frequencies
|
dN/dSa |
|||||||
|---|---|---|---|---|---|---|---|
| Polypeptide | Codons | Accession Numbersb | Method 1 | Method 2 | Method 3 | Method 4 | Method 5 |
| TBP N-terminus | 114 | D01034, D83127, D31776, X66033 | 0.03745 | 0.05385 | 0.04535 | 0.04742 | 0.05039 |
| TBP C-terminus | 181 | D01034, D83127, D31776, X66033 | 0.00588 | 0.00741 | 0.00696 | 0.00791 | 0.00936 |
| Histone H2B | 125 | NM_030082, D70896, X03018 | 0.07043 | 0.08456 | 0.06683 | 0.06594 | 0.07875 |
| Translation eEF-2 | 857 | BC007152, U46663, BC044327 | 0.02910 | 0.02086 | 0.02952 | 0.02274 | 0.02692 |
| α-Actin | 376 | NM_007392, M10607, AF416707, X12525 | 0.01839 | 0.02392 | 0.02085 | 0.02183 | 0.02557 |
Five different methods of estimating dN/dS were used within MEGA version 2.1, which differ in their treatment of different-fold degenerate codons and in their treatment of transition versus transversion substitutions. Although absolute values differ between methods, conclusions concerning the relative intensity of purifying selection between polypeptides were consistent with all methods. Method 1, Nei-Gojobori; Method 2, Modified Nei-Gojobori; Method 3, Li-Wu-Luo; Method 4, Pamilo-Bianchi-Li; Method 5, Kumar (Kumar et al. 2001).
For H2B and eEF-2, reptile sequences were not available in NCBI/GenBank. Analyses excluding reptiles from all comparisons qualitatively matched to those including reptiles (data not shown).
Fig. 2.
Predicted phylogeny of TBP N-termini (a) and C-termini (b). Relatedness is depicted as trees, with the p-distance scale indicated below each tree. The N-terminal sequences for B. floridae, D. melanogaster, and C. elegans cannot be reliably aligned with each other or with vertebrate TBP sequences, and thus these branches do not connect to the vertebrate tree or with each other (a) (see Materials and Methods). We find no evidence that the N-termini from these four groups are ancestrally related. Conversely, the C-terminal sequences are all homologous and an interconnected rooted tree is depicted (b).
Phyla-Specific Intron Analysis
Primers that would amplify TBP sequences from all vertebrates were designed to protein-coding sequences on either side of the extra intron identified in hagfish (upstream primer: 5′-TAT AAG CTT CAG TCC CAT GAT GAT GCC NTA YGG SAC-3′; downstream primer: 5′-TAT CTC GAG CTG TGG AAC GAT DCC HGA RCT CTC-3′; D =A/G/T, H =A/C/T, N =A/C/G/T, R =A/G, S =C/G, Y =C/T). Genomic PCR products were inserted into plasmid vectors and clones were sequenced.
GenBank accession numbers are AY168624 to AY168633. Ten sequence files were deposited.
Results
Tetrapod TBP
TBP cDNAs have been sequenced from multiple representatives of each tetrapod group, and all are highly related, especially within the C-terminal region (Tamura et al. 1991; Hernandez 1993). Indeed, our comparisons of the C-terminus from one representative of each tetrapod group revealed only 0.14 substitutions per 100 aa per class representative (NCBI/GenBank accession numbers: mammals, M. musculus [D01034]; birds, G. gallo [D83127]; reptiles, T. gramineus [D31776]; and amphibians X. laevius [X66033]). Moreover, estimation of nonsynonymous/synonymous substitutions (dN/dS) from published tetrapod TBP C-termini gave values between 0.006 and 0.009, which is roughly threefold lower than those calculated for α-actin, fivefold lower than those for translation elongation factor 2 (eEF-2), and 10-fold lower than those for histone H2B (table 1). In the absence of selection, a dN/dS of approximately 1.0 would be expected (reviewed in Nei and Kumar 2000, Yang 2001, and Yang 2002). The low dN/dS values for the tetrapod TBP C-termini suggest that this domain has been subjected to extraordinarily strong purifying selection (Nei and Kumar 2000; Yang 2001; Yang 2002). However, greater variation is seen within the N-terminus (Tamura et al. 1991). Thus, six substitutions per 100 aa per species were counted between the same four tetrapods (data not shown), and dN/dS values of 0.04 to 0.06 were obtained (table 1), which were comparable to those of H2B.
Although these results suggested that the TBP N-terminus was as conserved between tetrapods groups as was H2B, published data indicated that nonvertebrate species, including nematodes, insects, and echinoderms, lacked sequences resembling the tetrapod N-terminus (NCBI/GenBank) (Muhich et al. 1990; Tamura et al. 1991; Lichtsteiner and Tjian 1993). Therefore, we initiated a study aimed at determining which species between echinoderms and tetrapods contained sequences related to the tetrapod N-terminus.
Phylogenetic Distribution of the N-Terminus
TBP cDNAs were isolated from a cephalochordate (amphioxus), from two primitive jawless vertebrates (hagfish and lamprey), from a jawed cartilaginous fish (nurse shark), and from a teleost (zebrafish). Three TBP mRNA isoforms, encoding two protein isoforms, were isolated from amphioxus. The two proteins differed only by a GQ insertion/deletion within an XQ heterodipeptide repeat region (fig. 1b). In addition, three different 3′ UTR isoforms were found, which differed by the presence or absence of oligonucleotide insertions (fig. 1c). Thus, as compared with the shortest isoform (number 1), isoform number 2 had four insertions of lengths 7, 8, 18, and 23 bp, respectively. Isoform number 3 lacked these insertions, but had two other insertions of lengths 4 and 7 bp. The 8-bp insertion in the former isoform, and the 7-bp insertion in the latter, are direct repeats of adjacent sequences; other oligonucleotide insertions resemble nearby sequences, suggesting that they might be diverged repeats.
The amphioxus N-terminus, like those of all metazoans, had a PXT repeat; however, outside of this region, it lacked sequences suggesting homology with either the tetrapod N terminus (fig. 1b) or with TBP domains in any more primitive metazoans (NCBI/GenBank [data not shown]). Conversely, zebrafish, shark, lamprey, and hagfish TBP N-termini resembled those of tetrapods (fig. 1b). Both within the C-terminal domain and within the NN region, hagfish and lamprey TBP proteins had shared amino acids that differed from those in higher vertebrates (fig. 1b). Phylogenetic relationship predictions based on either the N-terminus or the C-terminus grouped lamprey and hagfish within a clade and separated vertebrates from all lower metazoans (fig. 2). Nevertheless, the hagfish N-terminus was longer than those of other vertebrates, including lamprey, largely as a result of being interspersed with several additional oligopeptide domains (fig. 1b). A recent report of TBP cDNA sequences from a Pacific lamprey (Lethenteron reissneri) and medaka (Oryzias latipes) (Hoshiyama, Kuma, and Miyata 2001) is consistent with the lamprey and teleost sequences described here.
It was interesting that, although no single-nucleotide polymorphisms were found between the open reading frames of different hagfish clones, individual cDNAs encoded 11, 12, or 13 Q residues within the Q region. TBP Q repeat-length variation also occurs between individual humans, and this has been attributed to possible expansion/deletion of trinucleotide microsatellites (Rubinsztein et al. 1996; Koide et al. 1999; Yamada, Tsuji, and Takahashi 2000). Such repeat-length variation between individuals within a species, in the absence of any silent single-base polymorphisms, indicates that these Q-region expansions/contractions persist more frequently than do point mutations within the N-terminus. However, unlike some other Q-repeat regions, for example, in Huntingtin protein (Yamada, Tsuji, and Takahashi 2000), both Q-encoding codons (CAA and CAG) are found in the Q regions of tbp genes from most species, including hagfish (fig. 1b) (NBCI/GenBank). In the presence of frequent expansion/contraction of Q codons, the apparently low rate of point mutation is unlikely to account for Q codon heterogeneity. This suggests that the mechanism that alters the length of this domain does not tend toward fixation of one codon or the other (see Discussion).
The PXT repeat also varies in length between metazoan phyla, and since the repeat unit contains three different amino acids, repeat length variation cannot be easily explained by trinucleotide microsatellite duplications/deletions. Rather, it requires reiteration of larger (≥9 bp) minisatellites. Interestingly, hagfish exhibited one more PXT repeat than did other vertebrates, including lamprey (fig. 1b). Thus, the mechanism that alters the length of the TBP PXT repeat has been active at least once since divergence of hagfishes from lampreys.
Within the NN and NC regions of hagfish TBP, short motifs were found that were absent from other vertebrates (fig. 1b, amino acids 44 to 52 and 102 to 109). These domains each appear to be diverged tandem duplications of sequences immediately adjacent to them. Thus, the NC motif, TTALPSG, exists in two additional imperfect repeats in all species, and the NN motif is an apparent diverged duplication of the adjacent oligopeptide sequence (amino acids 53 to 61; consensus: ) (fig. 1b). At least one other region in the hagfish NN region, beginning at amino acid 36 (TGLTPQP), resembles the core of this repeat. This region is a part of the most highly conserved portion of the NN region, suggesting that this sequence is ancient and that its integrity is important.
Phylogenetic Distribution of Introns Within Sequences Encoding the N-Terminus
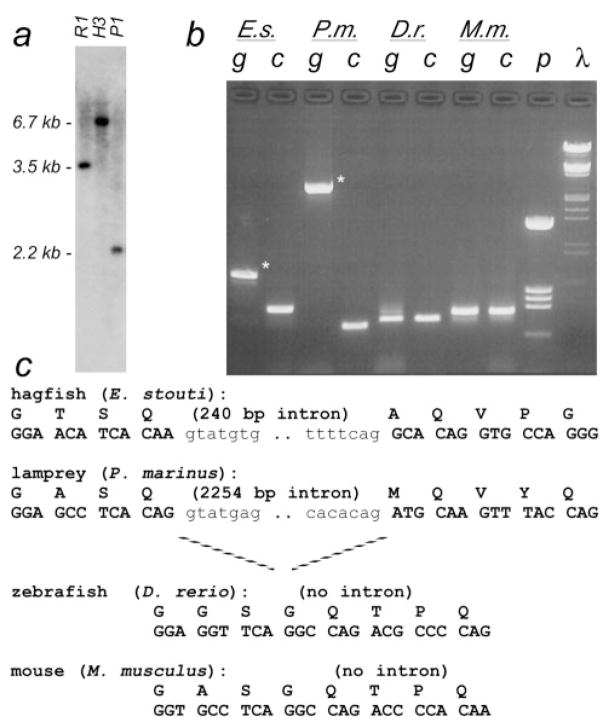
The positions of introns within genes are highly conserved during evolution (Long, de Souza, and Gilbert 1995; de Souza, Long, and Gilbert 1996; Gilbert, de Souza, and Long 1997). Although the TBP N-terminus in tetrapods is encoded almost entirely by a single large exon (Nakashima et al. 1995; Ohbayashi et al. 1996; Shimada et al. 1999), our analyses of tbp genomic sequences revealed an unexpected “extra” intron in this region of the hagfish tbp gene (see Materials and Methods). Analysis of genomic DNAs from the other vertebrates in our study indicated that lamprey also had an intron at this position; however, zebrafish and shark did not (fig. 3). This intron forms a phylogenetic marker that distinguishes hagfishes and lampreys, as a group, from all other vertebrates.
Fig. 3.
Intron/exon structure of TBP N-terminus. (a) Southern blot of hagfish genomic DNA using a probe to the Q/NC region. Sequence analysis of partial cDNA clones indicated the presence of Eco RI (R1), Hind III (H3), and Pst I (P1) sites in the 5′end of C-terminus–encoding sequences. The upstream portion of the 6.7-kb Hind III fragment (second lane) was cloned by ligation-mediated PCR, revealing an unexpected intron interrupting the NC region of the gene. (b) Distribution of intron C. Primers were designed to amplify across this novel splice junction from all vertebrates (E.s., hagfish; P.m., lamprey; D.r., zebrafish; M.m., mouse). Amplification of cDNA samples (lanes labeled “c”) confirmed that the primers worked in all species. Amplification of genomic DNA (“g”) showed a larger product in hagfish and lamprey (asterisks), indicating the region contained an intron in these species. λ, HindIII/EcoRI-cut λ-DNA markers; p, HinfI-cut pBS+ markers. (c) Genomic DNA and amino acid sequence of the junction region. Intron sequences in lowercase font.
Discussion
The C-terminal 181-aa core of TBP is found from Archaea to man and may be the most highly conserved protein motif in the living world. The fusion of this evolutionarily static protein motif to the phyla-specific N-terminus provided a convenient baseline control for analyses of evolutionary changes within the N-terminus. Unexpectedly, the sequences of hfTBP and lTBP revealed the apparently repetitive nature of the ancestral N-terminal domain (fig. 1b) and the presence of the cyclostome-specific intron (fig. 3). The findings presented here lend insights into our understanding of early vertebrate phylogeny, the mechanisms by which a novel polypeptide motif like the TBP N-terminus might evolve, and the physiological functions of this domain.
Vertebrate Phylogeny and the TBP N-Terminus
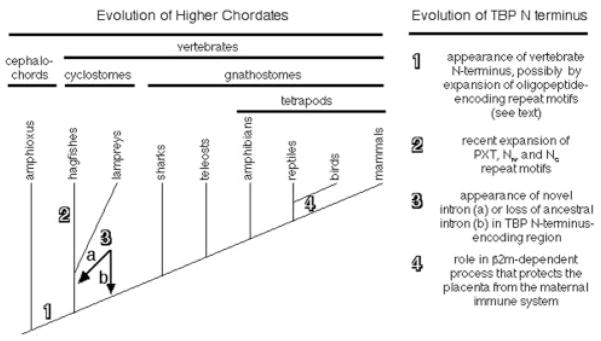
Although the phylogenetic position of hagfishes has been controversial (Janvier 1999; Neidert et al. 2001; Delarbre et al. 2002), a recent report based on mitochondrial genomes concluded that hagfishes and lampreys share a clade, “cyclostomata,” within vertebrata (fig. 4) (Delarbre et al. 2002). Relatedness prediction algorithms for both the N-terminal and the C-terminal regions of TBP group the hagfish and lamprey polypeptides within a separate vertebrate clade (fig. 2). Moreover, hagfishes and lampreys, but not jawed vertebrates, have an intron at an identical position within the region encoding the N-terminus (fig. 3). Because the entire protein-coding regions of these genes are homologous to those of higher vertebrates, the designations for conserved (i.e., homologous) exons and introns should be retained. Therefore, we have designated this new intron “C” for “cyclostomata,” the clade to which this intron is restricted. The exons on either side of intron C are designated “3-left” and “3-right” to indicate that they are homologous to the left and right sides of exon 3 from higher vertebrates, respectively. We cannot determine whether intron C arose immediately after divergence of the lineage leading to cyclostomes or was more primitive but was lost from the lineage leading to jawed vertebrates (fig. 4). Nevertheless, in combination, our TBP sequence data and the presence of the cyclostome-specific intron C support the proposed monophyletic origin of cyclostomes (Delarbre et al. 2002).
Fig. 4.
Transitions in the tbp gene during vertebrate phylogeny. At left is a diagram of vertebrate evolution based on published reports (Janvier 1999; Neidert et al. 2001; Delarbre et al. 2002). At right is a list of major modifications found in the structure or function of TBP, with the numbers corresponding to the indicated points on the diagram at left.
We show that all vertebrates share homologous TBP N-termini, whereas the domain from amphioxus is dissimilar (fig. 1b). Thus, the vertebrate TBP N-terminus either arose coincident with the first vertebrates, or it immediately predated their appearance. Although studies on mice lacking this domain have not yet provided evidence that N-terminus–dependent processes played a causal role in the genesis of vertebrates (Hobbs et al. 2002; Schmidt et al. 2003), the precise vertebrate distribution and strong amino acid conservation of this domain suggest it might perform critical vertebrate-specific functions (see below).
Evolution of the Vertebrate TBP N-Terminus
The results presented here suggest the TBP N-terminus has undergone multiple events involving duplication of oligopeptide-encoding domains (fig. 1b). Expansion of heterodipeptide or larger repeats would likely require duplications of 6-bp, 9-bp, or larger minisatellite sequences. The 3′ UTR sequences of individual TBP cDNAs from a polyclonal amphioxus population suggested that insertion/deletion of small minisatellite sequences occurs frequently in the tbp gene (fig. 1c). We also documented duplications in sequences encoding homooligopeptides (Q region), heterodipeptides (GQ polymorphism in amphioxus), heterotripeptides (PXT repeats), and larger heterooligopeptides (NN and NC repeats). A previous study noted the presence of several other 6-aa to 9-aa oligopeptide repeats within the TBP N-termini of C. elegans, Drosophila, and humans, and suggested that these repeats might function in transcription (Lichtsteiner and Tjian 1993). Here we propose a model by which duplication and divergence of oligopeptide units might have allowed de novo evolution of this vertebrate-specific polypeptide domain.
It is generally accepted that two major routes by which novel genes appear is by duplication and divergence of existing genes or by duplication, “shuffling,” and divergence of existing exons (reviewed in Gilbert, de Souza, and Long 1997). Much less is known about the evolution of novel proteins or protein domains by other mechanisms. Our data suggest that, at or about the time of the appearance of the first vertebrates, the TBP N-terminus might have evolved de novo by duplications and divergence of minisatellite-encoded oligopeptide domains. Minisatellites are known to be major contributors to repetitive genomic regions (Heringa 1998). They have also been implicated in creating the repetitive polymorphisms within in the mucin genes (Fowler, Vinall, and Swallow 2001). We theorize that duplication and divergence of minisatellite repeats can also contribute to the genesis of novel polypeptide domains.
Regions of arbitrary length might be duplicated during replication, as has been proposed as a general mechanism for minisatellite expansion (Levinson and Gutman 1987). Indeed, only one of the six minisatellite insertions that we found in the amphioxus 3′ UTRs (noncoding) was a multiple of 3 nt in length (fig. 1c). In protein-coding regions, modifications that altered the reading frame would disrupt the downstream TBP C-terminus and therefore would not persist. As a result, neither the nucleic acid sequence nor the amino acid sequence would need to have a deterministic effect on the reiterative mechanism, either in terms of reading-frame maintenance or in terms of the positions or lengths of the duplications. Rather, disruptive duplications or deletions would reduce fitness and would be lost from the lineage by natural selection. Modeling reveals that intragenic mini-satellite duplications tend to tandemly reiterate amino acid motifs rather than introduce novel amino acids (fig. 5a). Once dimerized, subsequent duplications of the same size will add another repeat of the exact same unit even if they initiate out-of-step with the existing repeats (fig. 5b). The propensity of this mechanism to duplicate short oligopeptide-encoding domains suggests it could play an important role in creating many reiterative secondary structures, such as the B-ZIP domain (7-aa diverged-repeat helix) (Landschultz, Johnson, and McKnight 1988), the ankyrin repeat (33-aa diverged fold structure) (Bork 1993; Mosavi, Minor, and Peng 2002), the collagen repeat (GXP repeat), the C-terminal domain (CTD) repeat on the large subunit of RNA pol II (7-aa repeat) (Ahearn et al. 1987), and others.
Fig. 5.
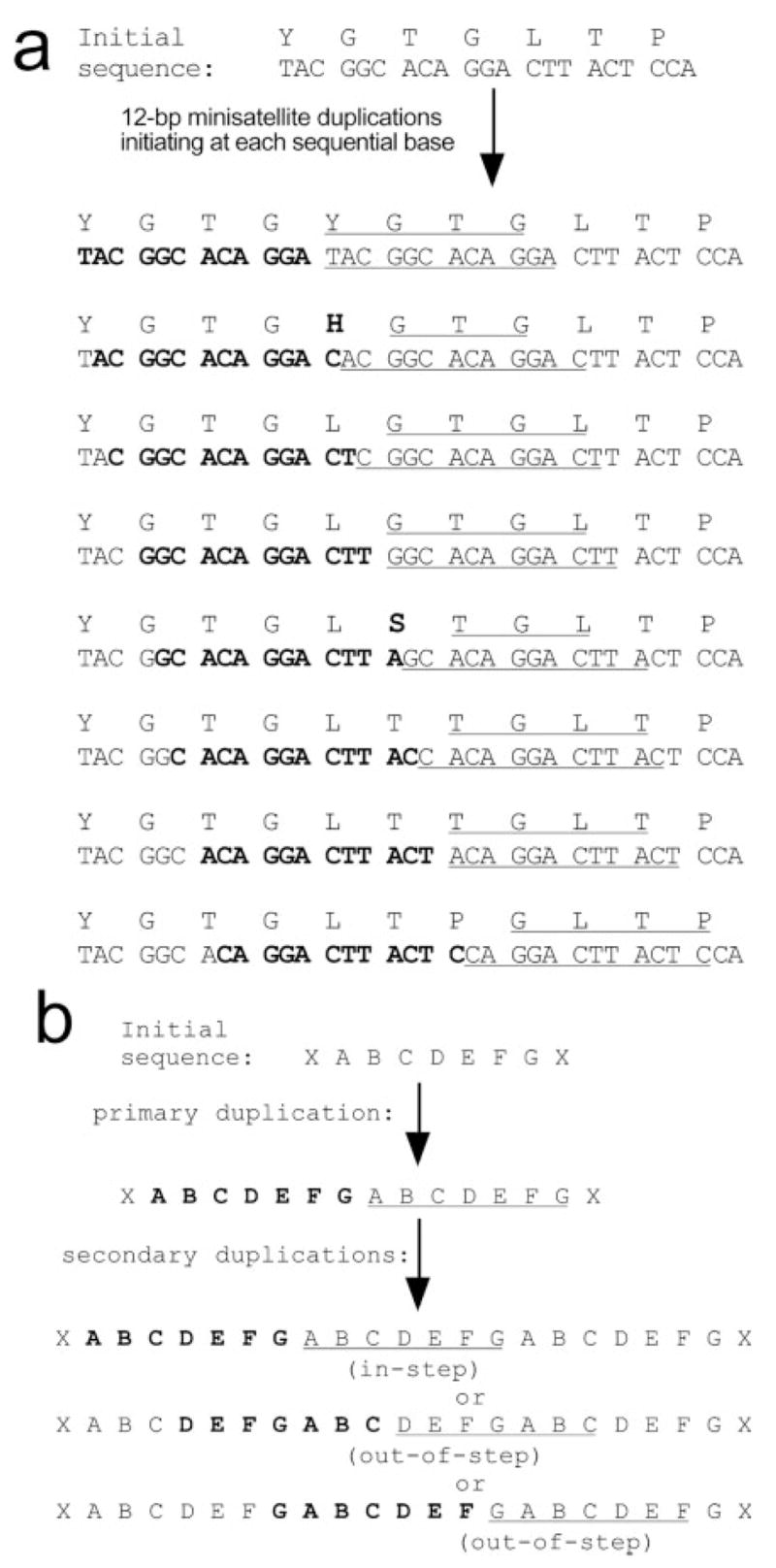
Models of minisatellite-dependent, oligopeptide-encoding sequence duplications. (a) Reading-frame independent preservation of amino acid sequences. A 7-aa region of the mouse NN region (top) was chosen as a starting sequence; however similar results are obtained with most arbitrary sequences (not shown). Twelve–base pair minisatellite duplications were modeled starting at each of eight consecutive nucleotides. Duplicated nucleotides are designated by bold font and underlining. Duplicated amino acids are underlined; novel amino acids are in bold. Few duplications (two of eight depicted) yield a novel amino acid, and this only occurs at the template/copy junction. (b) Within repeats, duplications preserve repeat periodicity. Model shows duplication of a 7-aa motif, “ABCDEFG” (21-bp minisatellite). Bold and underline indicate duplicated regions. Secondary duplications that initiate either at the first amino acid within the repeat (residue A, “in-step”) or at any other amino acid within the repeat (residues D and G shown, “out-of-step”) lead to the same trimeric repeat: ABCDEFGABCDEFGABCDEFG. The models in (a) and (b) show that imprecise or arbitrary duplications, as long as they occur in multiples of 3 bp, will tend to reiterate existing amino acid sequences and repeat motifs, rather than to insert novel amino acids or disrupt repeat units.
Minisatellite duplications might also help explain why in TBP, unlike in Huntingtin, both CAA and CAG codons are present in the Q region of most vertebrate species. Previously, length polymorphisms within the TBP Q region, like that within the Huntingtin Q region, were proposed to result from trinucleotide microsatellite expansions (Rubinsztein et al. 1996; Koide et al. 1999). If, instead, sequences within this region expanded as multi-codon minisatellites, as would be the case for the larger repeats within the N-terminus, then both Q codons might cotemplate individual duplication events. This could help account for the persistence of both codons in the Q region of most species.
Functions of the TBP N-Terminus
Because all vertebrates, but no nonvertebrates, possess a TBP N-terminus that is globally homologous to that of tetrapods, this domain likely first appeared in an ancestor shared by all extant vertebrates but not by amphioxus (fig. 2). This suggests the vertebrate TBP N-terminus likely participates in functions that arose coincident with the earliest vertebrates and that it has imparted a fitness advantage on all subsequent vertebrate lineages.
Anatomically, the transition to vertebrates is correlated with the appearance of characteristics such as vertebrae, neural crest tissue, placodes, and others (Neidert et al. 2001; Delarbre et al. 2002). This transition was also associated with gene duplications leading to novel members of some developmental transcription factor families, including the Otx and Dlx families (Tomsa and Langeland 1999; Neidert et al. 2001), which participate in neural crest formation (Dolle, Price, and Duboule 1992; Vignali et al. 2000). However, the pathways in which these factors function (Simeone, Puelles, and Acampora 2002), as well as all of the known regulators and pathways that define neural crest (Baker and Bronner-Fraser 1997; Langeland et al. 1998; Neidert et al. 2001; Garcia-Castro, Marcelle, and Bronner-Fraser 2002; Knecht and Bronner-Fraser 2002), are far more ancient than is neural crest, per se. In the absence of a truly “vertebrate-specific” pathway regulating neural crest formation, it has been theorized that vertebrate-specific regulation of preexisting pathways allowed formation of this tissue uniquely in the vertebrate lineage (Baker and Bronner-Fraser 1997). In this light, it is interesting to consider that our work suggests the promoters of all genes in all vertebrates, but not in nonvertebrate phyla, are bound by TBP molecules that contain a unique N-terminal domain. In other words, all vertebrate transcription initiation complexes are “vertebrate-specific,” not because of the DNA sequence of their promoters, but rather, because of this novel polypeptide domain within the basal transcription machinery.
The physiological functions for which natural selection has conserved this domain may differ between vertebrate phyla. Thus, although disruption of the TBP N-terminus in mice leads to a defect in therian pregnancy (Hobbs et al. 2002), no other vertebrates share this process, and, by inference, natural selection must have conserved this domain for other functions in pretherian vertebrates. These observations are consistent with model for evolution of this protein domain that we proposed based on functional studies in the mutant mice in which existing motifs can acquire novel activities that might later become “backed-up” to make the system robust (Schmidt et al. 2003). Since mice lacking the N-terminus exhibit only a defect in an evolutionary recent function, one might hypothesize that, if the N-terminus retains any of its more ancestral functions in mice, these can be accomplished by redundant mechanisms.
Acknowledgments
We thank K. Daughenbaugh, J. Prigge, D. Schwart-zenberger, and M. Pratt in our group for technical assistance; A. Rzhetsky at Columbia University for consultation and advice; W. Swink at the Hammond Bay Biological Station for providing lampreys; J. Seaborne and D. Headlee at the Oregon Department of Fish and Wildlife for providing hagfish; and J. Langeland at Kalamzoo College, M. Flajnik at the University of Maryland, and D. Grunwald at the University of Utah for libraries and reagents. This work was supported by a Basil O’Connor Award from the March of Dimes Foundation, a Montana Agricultural Experimental Station appointment, and a grant from the National Science Foundation to EES. MAES/MSU/COA manuscript #2002-01.
Literature Cited
- Ahearn JMJ, Bartolomei MS, West ML, Cisek LJ, Corden JL. Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. J Biol Chem. 1987;262:10695–10705. [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. The origins of the neural crest. Part II: an evolutionary perspective. Mech Dev. 1997;69:13–29. doi: 10.1016/s0925-4773(97)00129-9. [DOI] [PubMed] [Google Scholar]
- Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- Comeron JM. A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J Mol Evol. 1995;41:1152–1159. doi: 10.1007/BF00173196. [DOI] [PubMed] [Google Scholar]
- Delarbre C, Gallut C, Barriel V, Janvier P, Gachelin G. Complete mitochondrial DNA of the hagfish, Eptatretus burgeri: the comparative analysis of mitochondrial DNA sequences strongly supports the cyclostome monophyly. Mol Phylogenet Evol. 2002;22:184–192. doi: 10.1006/mpev.2001.1045. [DOI] [PubMed] [Google Scholar]
- de Souza S, Long M, Gilbert W. Introns and gene evolution. Genes Cells. 1996;1:493–505. doi: 10.1046/j.1365-2443.1996.d01-264.x. [DOI] [PubMed] [Google Scholar]
- Dolle P, Price M, Duboule D. Expression of murine Dlx-1 homeobox gene during facial, ocular, and limb development. Differentiation. 1992;49:93–99. doi: 10.1111/j.1432-0436.1992.tb00773.x. [DOI] [PubMed] [Google Scholar]
- Fowler J, Vinall L, Swallow D. Polymorphism of the human muc genes. Front Biosci. 2001;6:1207–1215. doi: 10.2741/A674. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- Gilbert W, de Souza SJ, Long M. Origin of genes. Proc Natl Acad Sci USA. 1997;94:7698–7703. doi: 10.1073/pnas.94.15.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heringa J. Detection of internal repeats: how common are they? Curr Opin Struct Biol. 1998;8:338–345. doi: 10.1016/s0959-440x(98)80068-7. [DOI] [PubMed] [Google Scholar]
- Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- Hobbs NK, Bondareva AA, Barnett S, Capecchi MR, Schmidt EE. Removing the TBP N-terminus disrupts placental β2-microgolbulin-dependent interactions with the maternal immune system. Cell. 2002;110:43–54. doi: 10.1016/s0092-8674(02)00806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshiyama D, Kuma K, Miyata T. Extremely reduced evolutionary rate of TATA-box binding protein in higher vertebrates and its evolutionary implications. Gene. 2001;280:169–173. doi: 10.1016/s0378-1119(01)00766-1. [DOI] [PubMed] [Google Scholar]
- Janvier P. Catching the first fish. Nature. 1999;402:21–22. [Google Scholar]
- Kandil E, Namikawa C, Nonaka M, Greenberg AS, Flajnik MF, Ishibashi T, Kasahara M. Isolation of low molecular mass polypeptide complementary DNA clones from primitive vertebrates: implications for the origin of MHC class I- restricted antigen presentation. J Immunol. 1996;156:4245–4253. [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Koide R, Kobayashi S, Shimohata T, Ikeuchi T, Maruyama M, Saito M, Yamada M, Takahashi H, Tsuji S. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum Mol Genet. 1999;8:2047–2053. doi: 10.1093/hmg/8.11.2047. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Landschultz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Langeland JA, Tomsa JM, Jackman WRJ, Kimmel CB. An amphioxus snail gene: expression in paraxial mesoderm and neural plate suggests a conserved role in patterning the chordate embryo. Dev Genes Evol. 1998;208:569–577. doi: 10.1007/s004270050216. [DOI] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Li WH. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Lichtsteiner S, Tjian R. Cloning and properties of the Caenorhabditis elegans TATA-box-binding protein. Proc Natl Acad Sci USA. 1993;90:9673–9677. doi: 10.1073/pnas.90.20.9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, de Souza SJ, Gilbert W. Evolution of the intron-exon structure of eukaryotic genes. Curr Opin Genet Dev. 1995;5:774–778. doi: 10.1016/0959-437x(95)80010-3. [DOI] [PubMed] [Google Scholar]
- Mosavi LK, Minor DLJ, Peng ZY. Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci USA. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhich ML, Iida CT, Horikoshi M, Roeder RG, Parker CS. cDNA clone encoding Drosophila transcription factor TFIID. Proc Natl Acad Sci USA. 1990;87:9148–9152. doi: 10.1073/pnas.87.23.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Suzuki T, Ohta Y, Flajnik MF, Kasahara M. The leukocyte common antigen (CD45) of the Pacific hagfish, Eptatretus stoutii: implications for the primordial function of CD45. Immunogenetics. 2002;54:286–291. doi: 10.1007/s00251-002-0469-1. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Nobuhisa I, Deshimaru M, Ogawa T, Shimohigashi Y, Fukumaki Y, Hattori M, Sakaki Y, Hattori S, Ohno M. Structures of genes encoding TATA box-binding proteins from Trimeresurus gramineus and T. flavoviridis snakes. Gene. 1995;152:209–213. doi: 10.1016/0378-1119(94)00681-h. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- Neidert AH, Virupannavar V, Hooker GW, Langeland JA. Lamprey Dlx genes and early vertebrate evolution. Proc Natl Acad Sci USA. 2001;98:1665–1670. doi: 10.1073/pnas.98.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi T, Schmidt EE, Makino Y, Kishimoto T, Nabeshima Y, Muramatsu M, Tamura T. Promoter structure of the mouse TATA-binding protein (TBP) gene. Biochem Biophys Res Commun. 1996;225:275–280. doi: 10.1006/bbrc.1996.1166. [DOI] [PubMed] [Google Scholar]
- Pamilo P, Bianchi NO. Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol Biol Evol. 1993;10:271–281. doi: 10.1093/oxfordjournals.molbev.a040003. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Leggo J, Crow TJ, DeLisi LE, Walsh C, Jain S, Paykel ES. Analysis of poly-glutamine-coding repeats in the TATA-binding protein in different human populations and in patients with schizophrenia and bipolar affective disorder. Am J Med Genet. 1996;67:495–498. doi: 10.1002/(SICI)1096-8628(19960920)67:5<495::AID-AJMG12>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Bondareva AA, Radke JR, Capecchi MR. Fundamental cellular processes do not require vertebrate-specific sequences in the TATA-binding protein. J Biol Chem. 2003;278:6168–6174. doi: 10.1074/jbc.M211205200. [DOI] [PubMed] [Google Scholar]
- Shimada M, Ohbayashi T, Ishida M, Nakadai T, Makino Y, Aoki T, Kawata T, Suzuki T, Matsuda Y, Tamura T. Analysis of the chicken TBP-like protein (tlp) gene: evidence for a striking conservation of vertebrate TLPs and for a close relationship between vertebrate tbp and tlp genes. Nucleic Acids Res. 1999;27:3146–3152. doi: 10.1093/nar/27.15.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Puelles E, Acampora D. The Otx family. Curr Opin Genet Dev. 2002;12:409–415. doi: 10.1016/s0959-437x(02)00318-0. [DOI] [PubMed] [Google Scholar]
- Sumita K, Makino Y, Katoh K, Kishimoto T, Muramatsu M, Mikoshiba K, Tamura T. Structure of a mammalian TBP (TATA-binding protein) gene: isolation of the mouse TBP genome. Nucleic Acids Res. 1993;21:2769. doi: 10.1093/nar/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Sumita K, Fujino I, Aoyama A, Horikoshi M, Hoffmann A, Roeder RG, Muramatsu M, Mikoshiba K. Striking homology of the ‘variable’ N-terminal as well as the ‘conserved core’ domains of the mouse and human TATA-factors (TFIID) Nucleic Acids Res. 1991;19:3861–3865. doi: 10.1093/nar/19.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsa JM, Langeland JA. Otx expression during lamprey embryogenesis provides insights into the evolution of the vertebrate head and jaw. Dev Biol. 1999;207:26–37. doi: 10.1006/dbio.1998.9163. [DOI] [PubMed] [Google Scholar]
- Vignali R, Colombetti S, Lupo G, Zhang W, Stachel S, Harland RM, Barsacchi G. XOtx5b, a new member of the Otx gene family, may be involved in anterior and eye development in Xenopus laevius. Mech Dev. 2000;96:3–13. doi: 10.1016/s0925-4773(00)00367-1. [DOI] [PubMed] [Google Scholar]
- Yamada M, Tsuji S, Takahashi H. Pathology of CAG repeat diseases. Neuropathology. 2000;20:319–325. doi: 10.1046/j.1440-1789.2000.00354.x. [DOI] [PubMed] [Google Scholar]
- Yang Z. Adaptive molecular evolution. In: Balding D, Bishop M, Cannings C, editors. Handbook of statistical genetics. Wiley; New York: 2001. pp. 327–350. [Google Scholar]
- Yang Z. Inference of selection from multiple species alignments. Curr Opin Genet Dev. 2002;12:688–694. doi: 10.1016/s0959-437x(02)00348-9. [DOI] [PubMed] [Google Scholar]