Abstract
Yeast Mot1p, a member of the Snf2 ATPase family of proteins, is a transcriptional regulator that has the unusual ability to both repress and activate mRNA gene transcription. To identify interactions with other proteins that may assist Mot1p in its regulatory processes, Mot1p was purified from replicate yeast cell extracts, and Mot1p-associated proteins were identified by coupled multidimensional liquid chromatography and tandem mass spectrometry. Using this approach we generated a catalog of Mot1p-interacting proteins. Mot1p interacts with a range of transcriptional co-regulators as well as proteins involved in chromatin remodeling. We propose that interaction with such a wide range of proteins may be one mechanism through which Mot1p subserves its roles as a transcriptional activator and repressor.
The TATA-binding protein (TBP)1 is required for expression of nearly every eukaryotic gene (1). Once recruited to the promoter by transcriptional activators (2), TBP, along with TBP-associated factors (TAFs), facilitates the nucleation of the preinitiation complex (PIC), which consists of an RNA polymerase and its associated initiation factors (for a review, see Ref. 3). Polymerase-specific subsets of TAFs associate with TBP, forming the SL1 (RNA polymerase I), TFIID (RNA polymerase II), and TFIIIB (RNA polymerase III) TBP·TAF complexes (4). Given the central importance of TBP to nuclear gene transcription, it is not surprising that other regulatory proteins also associate with TBP to modulate its activity (5). One of these TBP-binding proteins is modifier of transcription 1 (Mot1p), a member of the highly conserved Snf2p/Swi2p ATPase family of proteins (6, 7).
Yeast Mot1p is highly evolutionarily conserved, displaying significant sequence homology to orthologs in humans (BTAF1 (8)), Drosophila melanogaster (HEL89B (9)), and Caenorhabditis elegans (BTF1 (10)) especially within the C-terminal ATPase and N-terminal TBP-binding domains (11–13). Multiple HEAT motifs within its N terminus (14) are believed to be responsible for mediating the regulatory protein-protein interactions of Mot1p (12).
MOT1 was initially defined through a genetics screen designed to identify global repressors of transcription. Cells containing the recessive, temperature-sensitive mot1-1 allele expressed a reporter gene in the absence of the required enhancer/transactivator, a result that provided evidence that Mot1p likely functioned as a transcriptional repressor (6). Shortly thereafter, while characterizing the DNA binding behavior of TBP, Auble and Hahn (15) identified an activity, which they termed ATP-dependent inhibitor of TBP binding (ADI), that was capable of dissociating TBP from TATA DNA in the presence of ATP. Upon subsequent purification, they then demonstrated that ADI was encoded by MOT1 (16). At the same time, we identified a 170-kDa protein, which we termed Taf170p, that co-purified with TBP (17); sequence analysis revealed it to be Mot1p. Further we demonstrated that Mot1p forms a complex with TBP distinct from the TFIID TBP·TAF complex (18, 19). Additional genetics evidence suggesting a repressive function of Mot1p was provided by the finding that both Mot1p and the Leu3p repressor are required to effect complete repression of the LEU2 promoter (20). In addition, overexpression of wild-type Mot1p or several mutants confers a dominant negative growth phenotype that can be rescued by co-overexpression of TBP, indicating that Mot1p performs its repressive function, at least in part, by targeting TBP (11, 16, 21).
Although initial evidence pointed to a primarily repressive role for Mot1p, more recent data have indicated that Mot1p also activates gene transcription. Microarray analyses using yeast strains carrying mot1 temperature-sensitive alleles have shown that 10–15% of yeast genes are Mot1p-dependent, and a subset of these require Mot1p for activation of transcription. Chromatin immunoprecipitation (ChIP) assays have localized Mot1p at the promoters of several of these genes during gene transcription activation (22–24). Under conditions inducing the environmental stress response (ESR), such as exposure of yeast cells to heat shock, excess copper, or high salt, sequential ChIP experiments have shown that Mot1p occupies transcriptionally active promoters along with TFIIB and RNA polymerase II, apparently excluding TFIIA. These data led Geisberg and Struhl (25) to hypothesize that Mot1p may replace TFIIA during PIC formation under environmental stress conditions. Subsequent ChIP analyses revealed alterations in TBP binding to promoters in mot1 mutants. This finding suggests that Mot1p may effect both its positive and negative regulatory actions via its TBP displacement activity, either repressing transcription by removing TBP from a transcriptionally competent promoter or activating transcription by removing TBP from “dead-end” complexes thereby freeing it to bind to competent promoters (26, 27). All of these findings are consistent with previous gene-by-gene genetics and ChIP studies (2, 28–30). Alternatively Mot1p has been demonstrated to play a role in chromatin remodeling. Topalidou et al. (31) have demonstrated that Mot1p, working in concert with the Spt·Ada·Gcn5·acetyltransferase (SAGA) complex, remodeled nucleosomes at the GAL1 promoter. Thus, by creating an environment suitable for TBP binding, such remodeling may indirectly lead to recruitment of TBP and activation of gene transcription.
To gain insights into how Mot1p can both activate and repress gene transcription, we directly analyzed Mot1p protein-protein interactions by utilizing multidimensional protein identification technology (MudPIT) (32, 33). We reasoned that by determining the collection of proteins that interact with Mot1p we could gain insights into how it functions. These proteomics analyses indicated that in addition to subunits of the negative cofactor 2 (NC2) and SAGA complexes, both of which had been shown by either genetics or biochemical means to interact with Mot1p (29, 34, 35), Mot1p interacted with components of multiple transcriptional regulatory complexes, many of which modulate chromatin structure and activity. The significance of these data is discussed in terms of possible mechanisms of action of Mot1p during gene transcription regulation.
EXPERIMENTAL PROCEDURES
Plasmids—
Plasmid pDA5, which expresses Stb1p-FLAG3 under the control of the yeast ADH1 promoter, was constructed to allow expression of an epitope-tagged Stb1p. First oligos (FLAG3 Forward, 5′-CTA GTA TGG ATT ATA AAG ATG ATG ATG ATA AAG GTG GTG ATT ATA AAG ATG ATG ATG ATA AAG GTG GTG AAT ATA AAG ATG ATG ATG ATA AAG, and FLAG3 Reverse, 5′-GGA TCC TTT ATC ATC ATC ATC TTT ATA TTC ACC ACC TTT ATC ATC ATC ATC TTT ATA ATC ACC ACC TTT ATC ATC ATC ATC TTT ATA ATC CAT A) were gel-purified and annealed to create an SpeI-BamHI oligo duplex that was cloned into SpeI-BamHI-digested p413ADH (36) to create p413ADH-FLAG3. The STB1 coding sequence was PCR-amplified from genomic DNA (Invitrogen) using oligos that generated BglII and SalI restriction sites (forward, 5′-GCG GCG AGA TCT TCT CAA CCC CAG ATG TCC CCT GAA AAA G, and reverse, 5′-CGC CGC TGC GAC TCA TTC AGT GAG TTT GTC ATC AAT GGA C). The resulting PCR product was digested with BglII and SalI and cloned into BamHI-SalI-digested p413ADH-FLAG3 to create pDA5.
Yeast Methods—
Yeast strains are listed in Table I. Whole cell extracts (WCEs) prepared (37) from YPH252, DPY107, W303, Z1318, YDA106, and YDA136 were used for MudPIT analysis. For confirmatory co-immunoprecipitations (co-IPs), GFP-tagged strains were obtained from the Research Genetics library (38), and MOT1 in these strains was tagged with the HA3 epitope as described previously (19). Strains expressing either Myc13-tagged Itc1p (39) or FLAG3-tagged Dls1p (40) were generously provided by Toshio Tsukiyama, and MOT1 in these strains was tagged with the HA3 epitope (19). pDA5 was transformed into a strain also bearing an HA3-tagged Mot1p (YDA14) to generate YDA76. Alternatively potential target proteins were tagged with the Myc13 (41) and/or the FLAG5 epitope (42) in YDA14. Standard procedures for yeast cell growth and manipulation were utilized (43).
Table I.
Yeast strains
| Strain | Genotype | Ref. |
|---|---|---|
| SC295 | MATa ura3–52 leu2–3,112 reg1–501 gal1 pep4–3 | 93 |
| YPH252 | MATα ura3–52 trp1-Δ1 his3-Δ200 leu2-Δ1 lys2–801amber ade2–101ochre | 19 |
| DPY107 | YPH252 + HA3-MOT1 | 19 |
| W303 | MATa ura3-1 his3–11,15 ade2-1 leu2–3,112 trp1-1 can1–100 | 94 |
| Z1318 | W303 + RAP1-Myc13::TRP1 | 95 |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 96 |
| YDA20 | BY4741 + HIS3-RFA3-GFP + MOT1-HA3 | This study |
| YDA23 | BY4741 + HIS3-YGR130C-GFP + MOT1-HA3 | This study |
| YDA34 | BY4741 + HIS3-ISW1-GFP + MOT1-HA3 | This study |
| YDA36 | BY4741 + HIS3-HHF1-GFP + MOT1-HA3 | This study |
| YDA14 | W303 + MOT1-HA3 | This study |
| YDA39 | YDA14 + IOC2-Myc13::KANMx6 | This study |
| YDA47 | YDA14 + RCO1-Myc13::KANMx6 | This study |
| YDA48 | YDA14 + SDS3-Myc13::KANMx6 | This study |
| YTT2200 | MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1-1 ura3-1 RAD5+DLS1-FLAGx3::HYG | 40 |
| YDA53 | YTT2200 + MOT1-HA3 | This study |
| Z1608 | W303 + HIR2-Myc9::TRP1 | 95 |
| YDA57 | Z1608 + MOT1-HA3 | This study |
| YTT648 | MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1-1 ura3-1 RAD5+ITC1-Myc13::KANMx6 | 39 |
| YDA58 | YTT648 + MOT1-HA3 | This study |
| YDA76 | YDA14 + pDA5 (p413ADH-FLAGx3-STB1) | This study |
| YDA106 | YDA14 + MED6-FLAG5::KANMx6 + EAF6-Myc13::TRP1 | This study |
| YDA122 | YDA14 + NHP10-FLAG5::KANMx6 + POB3-Myc13::TRP1 | This study |
| YDA136 | YDA14 + NHP10-FLAG5::KANMx6 + MED6-Myc13::TRP1 | This study |
Purification of Mot1p—
Mot1p was overexpressed and purified from yeast cells as described by Adamkewicz et al. (44) with the following modifications. Binding to nickel-nitrilotriacetic acid-agarose (Qiagen) was performed in batch with mixing on a tiltboard for 5 h at 4 °C. Following imidazole elution, fractions containing Mot1p were dialyzed against saturated ammonium sulfate, the precipitate was collected by centrifugation (15 min at 17,000 × g), and pellets were resuspended in 20 mm HEPES (pH 7.9) and dialyzed against several changes of the same buffer. Dialyzed Mot1p was centrifuged for 2 min in a microcentrifuge to remove insoluble material and fractionated by chromatography on a Sephacryl S-300 HR column (16 × 600 mm; Amersham Biosciences). Mot1p was localized by SDS-PAGE, fractions containing Mot1p were pooled and concentrated on an Amicon Ultra 15 Ultracel 30,000 membrane (Millipore), and the buffer was exchanged for BA/300 (20 mm HEPES-KOH (pH 7.6), 10% (v/v) glycerol, 300 mm potassium acetate). Purified Mot1p was aliquoted and stored at −80 °C. The yield was ∼5 mg from 300 g of cells.
Superose 6 Fractionation of WCE—
Yeast strain DPY107 was grown in yeast extract peptone dextrose media supplemented with 0.004% adenine (43) to midlog phase (∼2 × 107 cells/ml), and cells were lysed using 0.5-mm glass beads and a Mini-BeadBeater (BioSpec) in BA/300 plus protease inhibitors (1 mm DTT, 1 mm benzamidine, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 0.5 μg/ml pepstatin A, 60 μg/ml Nα-tosyl-l-lysine chloromethyl ketone, 60 μg/ml N-p-tosyl-l-phenylalanine chloromethyl ketone, 0.1 mm PMSF). Following clarification by repeated centrifugation (3 × 20 min in a microcentrifuge), 200 μl (∼4 mg of total protein) of lysate was incubated for 10 min in the presence or absence of 10 μg/ml ethidium bromide (Bio-Rad) and fractionated on a molecular weight-calibrated Superose 6 column (high molecular weight and low molecular weight gel filtration calibration kits, Amersham Biosciences) at 0.2 ml/min, collecting 500-μl fractions. Purified Mot1p (50 μg) was also run across the same column. 20% of each column fraction from WCE or 1% of each recombinant Mot1p (rMot1p) column fraction was separated on 4–12% polyacrylamide gradient Criterion XT gels (Bio-Rad) and transferred to Immobilon-P (Millipore), and Mot1p was detected by immunoblotting with either affinity-purified rabbit anti-Mot1p IgG (30) or mouse monoclonal anti-hemagglutinin (HA)-horseradish peroxidase (HRP) IgG (Roche Applied Science).
Immunoprecipitations (IPs)—
Preparation of WCEs and immunopurifications were performed as described previously (45) except Buffer A/0 (20 mm HEPES-KOH (pH 7.9), 10% (v/v) glycerol) was added to 1 ml of yeast WCE (protein concentration, ∼100 mg/ml) to give a final conductivity equivalent to BA containing 150 mm potassium acetate (BA/150), and Nonidet P-40 (SurfactAmps grade, Pierce) was added to a final concentration of 0.2% (v/v). Immunopurifications were performed using either Sepharose bead-bound affinity-purified rabbit polyclonal anti-Mot1p IgG (30), mouse monoclonal anti-HA IgG (12CA5, Vanderbilt Monoclonal Antibody Core), or nonimmune rabbit IgG (Sigma). Immunopurifications of Mot1p-interacting proteins were performed as above using Sepharose bead-bound mouse monoclonal anti-HA IgG, anti-Myc IgG (Roche Applied Science), or anti-FLAG IgG (IBI Scientific). Proteins were eluted with a 50-fold mole excess of HA (YPYDVPDYA, SynBioSci), c-myc epitope (EQKLISEEDL, Roche Applied Science), or FLAG peptide (DYKDDDDK, SynBioSci).
MudPIT—
All digested samples were analyzed using MudPIT (32, 33). Briefly a fritless, microcapillary 100-μm-inner diameter column was packed with 9 cm of 5-μm C18 reverse-phase material (Synergi 4μ Hydro RP80a, Phenomenex) followed by 3 cm of 5-μm strong cation exchange material (Partisphere SCX, Whatman) and finally 2 cm of C18 reverse-phase material (Synergi 4μ Hydro RP80a, Phenomenex). Trypsin-digested samples were loaded directly onto triphasic columns equilibrated in 0.1% formic acid, 2% acetonitrile. The triphasic column was placed in line with an LCQ-Deca-XP-Plus ion trap mass spectrometry (Thermo, Inc.). An automated six-cycle multidimensional chromatographic separation was performed using buffer A (0.1% formic acid, 5% acetonitrile), buffer B (0.1% formic acid, 80% acetonitrile), and buffer C (0.1% formic acid, 5% acetonitrile, 500 mm ammonium acetate) at a flow rate of 0.3 μl/min. The first cycle was a 20-min isocratic flow of buffer B. Cycles 2–6 consisted of 3 min of buffer A, 2 min of x% buffer C, 5 min of buffer A, and a 60-min linear gradient to 60% buffer B. Cycles 2–6 used 15, 30, 50, 70, and 100% buffer C, respectively. During the linear gradient, eluting peptides were analyzed by one full MS scan (400–2000 m/z) followed by five MS/MS scans on the five most abundant ions in the full MS scan while operating under dynamic exclusion. Original MS data are available on request.
Data Analysis—
Centroided MS/MS scans from Thermo RAW files were exported to mzData 1.05 extensible markup language (XML) files by a modified version of ReAdW. The MyriMatch database search algorithm version 0.2 (46) identified peptides that matched to these files. The software was configured to allow for variable oxidations of methionine (+16 Da), and fragment ions were required to appear within 0.5 m/z of their monoisotopic m/z values. To be compared with a spectrum, a candidate peptide average mass was required to fall within 1.25 m/z of the observed precursor m/z. Only fully tryptic peptides were considered as candidates, but any number of missed cleavages was allowed within each candidate peptide. The orf_trans_all.fasta database from the Saccharomyces Genome Database (downloaded December 5, 2005) was used for the search with each protein included in both normal and reversed orientation for a total of 13,428 sequences.
Raw peptide identifications were assembled to proteins by the IDPicker algorithm (47). First identifications from each reverse-phase LC separation were read, and IDPicker determined the MyriMatch score thresholds for each charge state that would produce no higher than a 5% false discovery rate (FDR) in the passing peptide identifications. The formula used was FDR = (2r)/(f + r) where r is the number of reversed peptides passing the threshold and f is the number of forward peptides passing the threshold. The IDPicker hierarchy divided into experimental and control cohorts with each of these divided into hemagglutinin and immunoglobulin sets. Each MudPIT was kept as a separate column within the appropriate cohort and set. Proteins were required to be supported by two different peptide sequences to be included in the report, although those peptides could come from different MudPITs in the analysis. IDPicker output is available as part of the supplemental information (supplemental Table 1).
Significance analysis of microarrays (SAM; version 2.23A in Microsoft Excel 2003) was utilized to determine peptide count differences between the control and experimental groups of samples (48). The Wilcoxon two-class unpaired test was performed using the default random seed of 1234567 and filtered at a delta of 0.287. Proteins were deemed significant if they had a q value of 10% or less and at least a 2-fold average enrichment. To verify the significance of these proteins, the Mann-Whitney U test was performed for each individual protein, yielding p values <0.05 (95% significance).
Co-IPs/Immunoblots—
For authentication of the interactions observed by mass spectrometry, mouse monoclonal anti-HA-Sepharose beads (12CA5, Vanderbilt Monoclonal Antibody Core) were mixed overnight at 4 °C with either a 50-fold mole excess of HA or FLAG peptide in BA/150. Immunopurifications were performed as described previously (49) with the following modifications. WCE (10 mg of protein) was added to the peptide-blocked, bead-bound antibodies. Ethidium bromide (Bio-Rad) was added to a final concentration of 10 μg/ml, and samples were mixed on a tiltboard for 2 h at 4 °C, washed, and eluted by a 50-fold mole excess of HA peptide. Eluates were analyzed by immunoblotting, utilizing affinity-purified rabbit polyclonal antibodies described previously (45, 49–54) as well as four antibodies we prepared in the same manner (anti-Sgf73p, anti-Spt3p, anti-Ncb1p, and anti-Msn2p) and two (anti-Pob3p and anti-Spt16p) prepared by the University of Texas Southwestern Antibody Production Core. Alternatively mouse monoclonal anti-HA-HRP IgG (Roche Applied Science), mouse monoclonal anti-FLAG-HRP IgG (Sigma), affinity-purified goat polyclonal anti-Esa1p (Santa Cruz Biotechnology), mouse monoclonal anti-Myc-HRP or anti-GFP-HRP IgG (Santa Cruz Biotechnology), or affinity-purified rabbit polyclonal anti-GFP (Abcam) was used.
RESULTS
Mot1p Exists in High Molecular Weight Complexes
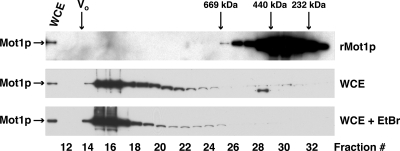
We reasoned that if Mot1p worked through stable associations with transcriptional activators and repressors then WCE Mot1p would size fractionate as heterodisperse, high molecular weight complexes. We tested this hypothesis by fractionating WCE by Superose 6 gel filtration chromatography, monitoring the elution of Mot1p by immunoblotting (Fig. 1). WCE and purified rMot1p were chromatographed sequentially on the same column. rMot1p chromatographed as an asymmetric monomer, molecular mass ∼280 kDa (Fig. 1, top panel), in agreement with previous findings (44). By contrast, the majority of WCE Mot1p chromatographed much more heterogeneously, ranging in size from ∼700 kDa to ≥3 MDa (void volume of the column); ∼70% of Mot1p was present in column fractions 15–17 and exhibited apparent masses in the MDa size range (Fig. 1, middle panel). No change in chromatographic behavior was observed when an aliquot of the same WCE was fractionated in the presence of ethidium bromide (Fig. 1, bottom panel, WCE + EtBr), indicating that the observed fractionation patterns were not mediated by DNA (see Ref. 55); rMot1p fractionated identically in the presence or absence of ethidium bromide (data not shown). Note that rMot1p did not oligomerize even when run at a significantly higher concentration than WCE Mot1p (Fig. 1, compare top panel with middle and lower panels, ∼3,000-fold more rMot1p than WCE Mot1p). We concluded from these data that a large fraction of cellular Mot1p was associated with other proteins in high molecular weight complexes.
Fig. 1.
Yeast Mot1p exists in heterodisperse high molecular weight complexes. Yeast WCE protein (4 mg), prepared from DPY107 cells, was fractionated by fast protein LC on a Superose 6 HR 10/30 column in the absence (middle panel) or presence (lower panel) of ethidium bromide (EtBr). Purified rMot1p (50 μg; top panel) and molecular mass standards (thyroglobulin (669 kDa), ferritin (440 kDa), and Catalase (232 kDa)) were fractionated in parallel. The column void volume (V0) was measured using plasmid pRS305 (molecular mass = 3.4 MDa). Fractions were collected, and aliquots of fractions and WCE were precipitated, separated by SDS-PAGE, and electroblotted to PVDF membranes, and Mot1p was detected with polyclonal anti-Mot1p IgG (rMot1p) or monoclonal anti-HA-HRP IgG (WCE).
Identification of Mot1p-associated Proteins
To rigorously test that Mot1p is indeed associated with other proteins, we performed replicate MudPIT analyses of Mot1p immunopurified from yeast WCE. To comprehensively address this question, we performed seven independent Mot1p immunopurification experiments using WCEs prepared from four yeast strains listed in Table I: YPH252, DPY107, W303, and Z1318. This large number of replicates was performed to maximize our ability to identify Mot1p-interacting proteins, including those protein-protein associations that might be of somewhat lower affinity and/or dynamic. The W303 and Z1318 strains were chosen to examine Mot1p protein-protein interactions in a strain background distinct from YPH252. Mot1p and associated proteins were immunopurified from all four strains using rMot1p affinity-purified, rabbit polyclonal anti-Mot1p IgG, whereas WCE from DPY107, which expresses HA3-tagged Mot1p, was also subjected to affinity purification using mouse monoclonal anti-HA 12CA5 IgG. As controls, either nonimmune rabbit IgG (all strains) or anti-HA 12CA5 IgG (YPH252, W303, and Z1318) was used for mock affinity purification; a total of 12 control immunopurification experiments were performed (Table II). All immunopurifications were conducted in the presence of ethidium bromide to ensure that observed interactions were not DNA-mediated (45, 55).
Table II.
False discovery rate of peptide identification
| Control
|
Experimental
|
||||||
|---|---|---|---|---|---|---|---|
| Samplea | Strain | Abb | FDRc | Sample | Strain | Ab | FDR |
| % | % | ||||||
| HA/1 | W303 | α-HA | 2.6 | HA/1 | DPY107 | α-HA | 1.5 |
| HA/2 | Z1318 | α-HA | 1.7 | HA/2 | DPY107 | α-HA | 1.8 |
| HA/3 | YPH252 | α-HA | 3.6 | Ig/1 | W303 | α-Mot1p | 2.6 |
| HA/4 | Z1318 | α-HA | 1.5 | Ig/2 | Z1318 | α-Mot1p | 1.6 |
| HA/5 | W303 | α-HA | 1.8 | Ig/3 | DPY107 | α-Mot1p | 1.3 |
| HA/6 | W303 | α-HA | 2.4 | Ig/4 | YPH252 | α-Mot1p | 2.0 |
| Ig/1 | W303 | Nonimmune IgG | 2.2 | Ig/5 | DPY107 | α-Mot1p | 2.1 |
| Ig/2 | Z1318 | Nonimmune IgG | 2.3 | ||||
| Ig/3 | DPY107 | Nonimmune IgG | 2.2 | ||||
| Ig/4 | YPH252 | Nonimmune IgG | 2.4 | ||||
| Ig/5 | W303 | Nonimmune IgG | 2.1 | ||||
| Ig/6 | DPY107 | Nonimmune IgG | 2.0 | ||||
Sample numbers correspond to supplemental Tables 1 and 2.
Ab, antibody; indicates the IgG used for immunopurification.
FDR = 2r/(f + r) where r = number of reverse peptide sequence identifications and f = number of forward peptide sequence identifications.
Acquired MS/MS spectra were matched against tryptic peptides derived from the Saccharomyces Genome Database (6714 sequences) using the MyriMatch search algorithm (46). IDPicker (47) assembled lists of peptides identified at a 5% false discovery rate, and the software generated lists showing the number of sequences observed for each protein. Proteins were only considered to be present if represented by at least two independent peptide sequences. To measure the FDR, each database protein sequence was included in both forward and reverse orientations, and the FDR was calculated by dividing twice the number of reverse sequences identified by the total number of sequences identified; the per run FDRs are included in Table II. The aggregate FDR for all 19 runs was 2.0%. The complete data set (1192 proteins, including false positives) is presented in supplemental Table 1.
The data set was analyzed by the SAM method (48) using the parameters described under “Experimental Procedures.” The analysis yielded a list of proteins significantly enriched in the experimental samples as compared with the control samples. The 67 proteins with a SAM-derived q value of 10% or less, a greater than 2-fold average-fold enrichment in the experimental samples as compared with the controls, and a p value of less than 0.05 by Mann-Whitney U test were considered to be significant. Proteins that met these criteria were sorted into complexes or categories using the Munich Information Center for Protein Sequences (MIPS) functional classifications (56).
To validate a subset of the protein-protein interactions observed by MudPIT, confirmatory co-IP reactions were performed. Mot1p was immunopurified from strains expressing an epitope-tagged form of the protein, HA3-Mot1p, using mouse monoclonal anti-HA IgG, and co-purifying proteins were identified by immunoblot using specific, affinity-purified, rabbit polyclonal antibodies where possible (see Figs. 2–6). Alternatively MOT1 was tagged with the HA3 epitope in strains expressing a GFP-, Myc13-, or FLAG3-tagged allele of the putative interacting proteins, and HA3-Mot1p co-purifying proteins were detected by co-IP with monoclonal anti-HA IgG followed by immunoblotting using anti-GFP, anti-Myc, or anti-FLAG IgG. For proteins where the cognate genes were refractory to GFP tagging, Myc13-tagged alleles of the gene of interest were generated in a strain bearing HA3-Mot1p. Co-purifying proteins were then detected by co-IP followed by immunoblotting with mouse monoclonal anti-Myc IgG as described above.
Fig. 2.
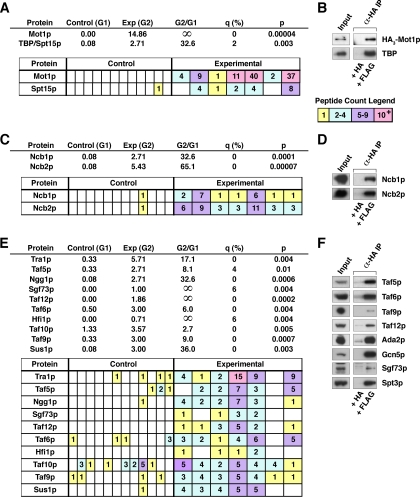
MudPIT identification of previously characterized Mot1p-associated proteins. A, results of MudPIT analysis for Mot1p and TBP/Spt15p. Top, control (Group 1 (G1)) = average number of peptide counts over the 12 control reactions. Experimental (Exp) (Group 2 (G2)) = average number of peptide counts over the seven experimental reactions. Group 2/Group 1 = experimental average/control average. q value, expressed as a percentage, measures the significance of the difference between Group 1 and Group 2 as calculated by SAM. p value was calculated by Mann-Whitney U test. Bottom, also shown are peptide counts, color-coded (see legend, right), for each independent MudPIT run: the first 12 columns are control runs; the last seven columns experimental runs. B, authentication of MS-identified Mot1p protein-protein interactions by co-IP analysis. WCE from DPY107 cells, which express HA3-Mot1p, was immunoprecipitated with bead-bound anti-HA IgG that had been blocked overnight with a 50-fold mole excess of either HA or FLAG peptide. The protein content of the resulting IPs was analyzed by immunoblotting with affinity-purified rabbit polyclonal anti-TBP antibody or anti-HA-HRP IgG in the case of Mot1p. C, results of MudPIT analysis for NC2 ordered by descending molecular weight. D, authentication of Mot1p-NC2 subunit interactions by co-IP. Ncb1p/2p in IPs was scored by immunoblotting with affinity-purified rabbit polyclonal anti-Ncb1p and anti-Ncb2p antibodies. E, SAGA subunits identified by MudPIT analysis. F, authentication of Mot1p-SAGA subunit interactions by co-IP. SAGA subunits in the IPs were identified by immunoblotting with the indicated affinity-purified rabbit polyclonal antibodies.
Fig. 3.
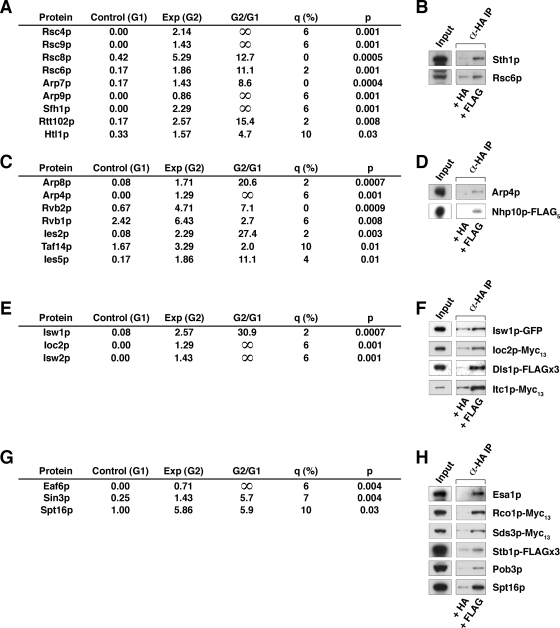
Association of Mot1p with chromatin-remodeling complexes. A, results of MudPIT analysis for RSC. B, authentication of Mot1p-RSC subunit interactions by co-IP performed as in Fig. 2B. C, INO80 subunits identified by MudPIT analysis. D, authentication of Mot1p-INO80 subunit interactions by co-IP. E, ISW1 and ISW2 complex subunits identified by MudPIT analysis. F, authentication of Mot1p-ISW subunit interactions by co-IP. G, NuA4, Sin3, and FACT subunits identified by MudPIT analysis. H, authentication of Mot1p protein-protein interactions by co-IP analysis. Exp, experimental; G1, Group 1; G2, Group 2.
Fig. 4.
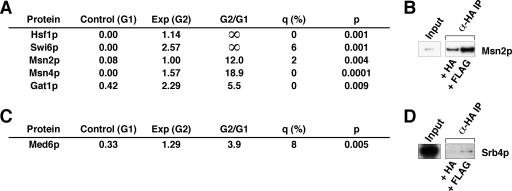
Association of Mot1p with DNA-binding transfactors and the general coactivator, Mediator. A, DNA-binding transfactors identified by MudPIT analysis. B, authentication of Mot1p-Msn2p interaction by co-IP analysis. C, Mediator subunit identified by MudPIT analysis. D, authentication of Mot1p-Srb4p interaction by co-IP analysis. Exp, experimental; G1, Group 1; G2, Group 2.
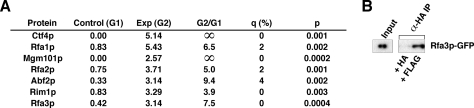
Fig. 5.
Interaction of Mot1p with proteins involved in DNA packaging and maintenance. A, proteins involved in DNA replication, repair, and recombination identified by MudPIT analysis. B, authentication of Mot1p-Rfa3p-GFP interaction by co-IP analysis. Exp, experimental; G1, Group 1; G2, Group 2.
Fig. 6.
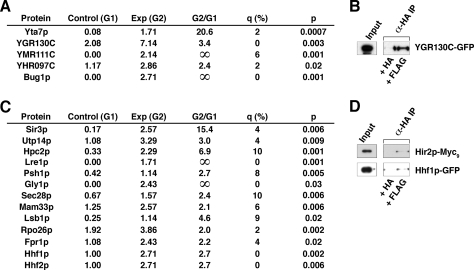
Interaction of Mot1p with other classes of proteins. A, proteins of unknown function identified by MudPIT. B, authentication of Mot1p-YGR130C-GFP interaction by co-IP analysis. C, other Mot1p-interacting proteins identified by MudPIT. D, authentication of Mot1p protein-protein interactions by co-IP analysis. Exp, experimental; G1, Group 1; G2, Group 2.
Identification of Known Mot1p-associated Proteins: Authentication of MS-identified Mot1p-associated Proteins
Mot1p was identified in all seven experimental samples; peptide counts (nonredundant peptides identifying a cognate protein) ranged from 1 to as high as 40 with an average of 15 Mot1p peptide counts per run (Fig. 2A). Note that the conditions used for elution of immunopurified Mot1p (4 m urea) typically were not sufficient to fully disrupt the antibody·antigen complex, which resulted in a reduction of the amount of Mot1p in the eluted fraction (45).
To validate our experimental approach, we first looked for the presence of proteins that have been shown previously, either by genetics or biochemical means, to interact with Mot1p. First, we looked for TBP, also known as Spt15p, a defining biochemical marker of Mot1p (1). TBP was identified in five of seven experimental samples (Fig. 2A). Note that we have shown previously (19) that only a fraction of Mot1p (approximately 50%) is TBP-associated. Similarly both the Ncb1p and Ncb2p subunits of the NC2 complex were detected in all seven experimental samples (Fig. 2C). Although each of the 20 currently described subunits of SAGA were in fact identified in our analyses, only 10 subunits met the statistical cutoff (q ≤ 10%, p < 0.05) and are listed in Fig. 2E (see also supplemental Table 2).
To authenticate the interactions between Mot1p and TBP, NC2, and SAGA, co-IP studies were performed using WCE derived from strain DPY107, which expresses HA3-Mot1p. WCE was incubated with bead-bound anti-HA antibody that had been blocked by incubation with either HA or FLAG peptide prior to and throughout incubation with WCE. IPs were then eluted with HA peptide, and a portion of the eluate was SDS-PAGE-fractionated, blotted, and probed with our affinity-purified anti-TBP, anti-NC2, or anti-SAGA subunit rabbit polyclonal antibodies. The efficiency of immunoprecipitation was scored by probing with mouse monoclonal anti-HA IgG to assess HA3-Mot1p in the IP as compared with the non-bead-bound fraction (data not shown); IP efficiency was >90%. Immunoprecipitation of Mot1p coprecipitated TBP (Fig. 2B) and NC2 (Fig. 2D) as well as all eight subunits tested from the 20-subunit SAGA complex: Ada2p, Gcn5p, Sgf73p, Spt3p, Taf5p, Taf6p, Taf9p, and Taf12p (Fig. 2F). Note that although not deemed statistically significant in the proteomics analysis, Gcn5p and Spt3p (only one peptide sequence identified for each) and Ada2p (q = 36%, p = 0.06) did indeed co-purify with Mot1p as scored by co-IP. The identification of multiple proteins and protein complexes previously shown to interact with Mot1p demonstrated the ability of our proteomics analyses to detect Mot1p-associated proteins.
Identification of Novel Mot1p-associated Proteins
Association of Mot1p with Chromatin-remodeling Complexes: ATP-dependent Remodelers—
Our proteomics analysis revealed interactions between Mot1p and several chromatin-remodeling complexes (Fig. 3). Subunits of the RSC (remodel the structure of chromatin) complex were highly and specifically enriched in the immunopurifications analyzed by MudPIT (Fig. 3A). Of the 17 subunits defining the complex, 14 were identified in the affinity-purified samples; nine were above our statistical cutoff (i.e. q ≤ 10%, p < 0.05). Both Rsc6p, which was identified in our proteomics data, and Sth1p, which fell outside of the statistical cutoff (q = 36%, p = 0.06), were demonstrated to co-purify with Mot1p by co-IP analysis (Fig. 3B). A recent high throughput proteomics analysis also identified interactions between Mot1p and the Rsc3p and Sth1p subunits of RSC (57); although these subunits were present in our samples as well, they fell outside of our statistical cutoff (Rsc3p: q = 24%, p = 0.08; Sth1p: q = 36%, p = 0.06).
The INO80 chromatin-remodeling complex was also well represented in the MudPIT analysis (Fig. 3C). Seven of the 13 known subunits of INO80 were identified (58, 59). By co-IP analysis we demonstrated that both Arp4p, a subunit shared with the NuA4 (nucleosome acetyltransferase of H4) complex, as well as the INO80-specific subunit Nhp10p, which was not identified during our proteomics analysis, co-purified with Mot1p (Fig. 3D).
Additionally two of the four subunits of the Isw1a and Isw1b (imitation switch) remodeling complexes, Isw1p and Ioc2p, were detected (Fig. 3E), a finding consistent with the identification of Isw1p and Ioc3p in the high throughput proteomics analyses noted above (57). Isw1p-GFP and Ioc2p-Myc13 were shown to co-purify with Mot1p by co-IP (Fig. 3F). Another member of the highly conserved ISW1 family, Isw2p, a subunit of the ISW2 remodeling complex that acts in concert with the Rpd3·Sin3 complex to effect repression of early meiotic genes (39), was also detected (Fig. 3E). Two of the four subunits of the ISW2 complex, Itc1p-Myc13 and Dls1p-FLAG3, were successfully co-purified with Mot1p by co-IP (Fig. 3F). Unlike Dls1p, which was not present in our proteomics analysis, Itc1p was identified in our samples; however, it did not meet the statistical cutoff (q = 36%, p = 0.06).
Association of Mot1p with Chromatin-remodeling Complexes: Chromatin Covalent Modifiers—
In addition to the ATP-dependent RSC, ISW1 and ISW2, and INO80 chromatin remodelers, our analysis also revealed the presence of complexes that covalently modify chromatin. As mentioned above, the histone acetyltransferase (HAT) activity-containing SAGA complex was well represented (Fig. 2E). The presence of a second complex with HAT activity, NuA4, was indicated by the identification of Eaf6p (Fig. 3G). Although Eaf6p is itself present in both the NuA3 and NuA4 complexes, further analysis by co-IP revealed an association between Mot1p and Esa1p (Fig. 3H), the essential catalytic subunit of the NuA4 complex. Other NuA4 subunits identified include Tra1p (shared with SAGA) and Arp4p (shared with INO80; Fig. 3D).
Mot1p also interacts with chromatin modifiers with histone deacetylase (HDAC) activity. Sin3p, resident in several different chromatin-active complexes with HDAC activity, namely the Rpd3L, Rpd3S, and Sin3 complexes, was also identified (Fig. 3G). Co-IP analyses demonstrated that Mot1p interacts with all three Sin3p-containing complexes. Although not present in our proteomics analysis, the complex-specific subunits Rco1p-Myc13 (Rpd3S), Sds3p-Myc13 (Rpd3L), and Stb1p-FLAG3 (Sin3) all co-purified with Mot1p (Fig. 3H).
Association of Mot1p with Chromatin-remodeling Complexes: Other Chromatin Modifiers—
Although not a chromatin modifier in the sense of RSC/INO80 and SAGA, the heterodimeric Spt16p·Pob3p chromatin-modulating FACT (facilitates chromatin transcription) complex was first identified as a factor required for efficient transcription elongation on a nucleosomal template (60) by a mechanism later shown to result in destabilization of the nucleosome (61). More recently, FACT has also been shown to play a role in the initiation of transcription both by directly facilitating PIC formation (62) and by suppressing initiation from cryptic promoters (63). In our proteomics analysis, the Spt16p subunit of the FACT complex was identified (Fig. 3G). However, the Pob3p subunit, although present in these samples and successfully authenticated by co-IP (Fig. 3H), did not meet the statistical cutoff (q = 23%, p = 0.1).
Association of Mot1p with DNA-binding Transfactors and the General Coactivator, Mediator—
Our data revealed that five known RNA polymerase II-specific DNA binding transfactors were Mot1p-associated: Gat1p, Hsf1p, Msn2p, Msn4p, and Swi6p (Fig. 4A). Interestingly four of these, Gat1p, Hsf1p, Msn2p, and Msn4p, are involved in activation of transcription supporting the environmental stress response. As noted earlier, Mot1p has been implicated as playing a key role in mediating aspects of the ESR. The interaction between Mot1p and Msn2p was validated by co-IP (Fig. 4B).
Also identified as a potential Mot1p-interacting complex was the general coactivator, Mediator. By mass spectrometry, we demonstrated the presence of Med6p (Fig. 4C), a subunit of the head module of Mediator. To authenticate an interaction between Mot1p and the Mediator head module, HA3-Mot1p was immunoprecipitated from DPY107 WCE, and immunoblots were performed utilizing affinity-purified rabbit polyclonal anti-Srb4p IgG (Fig. 4D).
Interaction of Mot1p with Proteins Involved in DNA Packaging and Maintenance—
Mot1p was also found to be associated with several proteins involved in aspects of the packaging and maintenance of DNA (Fig. 5A). Most significantly, all three subunits of replication factor A (RFA), a complex required for DNA replication, repair, and recombination (64), were associated with Mot1p. This finding was authenticated by co-IP, which showed that Rfa3p-GFP co-purified with Mot1p (Fig. 5B). Ctf4p, which is involved in mitotic DNA replication (65), was also identified as a putative interacting protein. Finally three proteins involved in replication and repair of mitochondrial DNA, Abf2p, Rim1p, and Mgm101p, were identified (66).
Interaction of Mot1p with Other Classes of Proteins—
MudPIT identified five proteins with unknown or poorly characterized functions (Fig. 6A), including Yta7p, a protein with an ATPase motif; Bug1p, which is believed to be involved in trafficking to the Golgi (67); and the putative proteins encoded by YHR097C, YGR130C (authenticated by co-IP; Fig. 6B), and YMR111C.
Finally we also identified several proteins that fall outside easily definable groups (Fig. 6C). Of these, four play roles in regulating transcription. Psh1p, a putative transcription elongation factor, has been reported in a high throughput proteomics analysis to associate with the two subunits of the FACT complex, Pob3p and Spt16p (68). Lre1p, which contributes to laminase resistance, is also involved in heat stress resistance via inhibition of the protein kinase Cbk1p (69), whereas a silencing protein, Sir3p, represses gene transcription by modulating chromatin structure (70). Lastly Hpc2p, a subunit of the recently characterized HIR complex, a corepressor that represses transcription of histone genes by promoting histone deposition (71), was observed as Mot1p-associated. Additionally although not itself identified in our proteomics analysis, Hir2p, a subunit of the HIR complex, did co-purify with Mot1p by co-IP (Fig. 6D).
As with the recent high throughput MS analyses (57), our data also demonstrated the presence of histone proteins in the immunopurified fractions, namely Hhf1p and Hhf2p, which was authenticated by co-IP (Fig. 6D). Of the remaining proteins (Fig. 6C), Utp14p, a component of the pre-18 S rRNA processing small subunit processome (72), is also Mot1p-associated, a finding made more interesting in light of recent evidence demonstrating a role for Mot1p in the regulation of transcription of rRNA (73). Finally Gly1p is involved in threonine and glycine metabolism (74), Sec28p is part of the coatomer complex (75), Mam33p is a mitochondrial matrix protein involved in oxidative phosphorylation (76), Lsb1p is involved in the actin cytoskeleton functions (77), Rpo26p is a subunit common to all three RNA polymerases (78), and Fpr1p is a putative regulator of the nonhistone chromatin-binding protein Hmo1p (79).
Mot1p Is Present in Multiple, Discrete Subcomplexes in Yeast WCE
The large numbers of transcriptionally related proteins identified in association with Mot1p could be explained in two ways. One possible explanation could be that nuclear proteins aggregate into a single, massive complex and that Mot1p serves to nucleate the formation of this “megacomplex” of transcriptionally active proteins. Alternatively Mot1p could be interacting with discrete subsets of protein complexes, resulting in multiple, physically distinct, Mot1p-containing complexes.
To distinguish between these possibilities, we performed MudPIT analyses of three protein complexes we had identified previously as being Mot1p-associated: Mediator (using Med6p as bait), Ino80 (Nhp10p as bait), and NuA4/3 (Eaf6p as bait). We constructed two yeast strains bearing multiple tagged proteins (YDA106 (HA3-Mot1p, Med6p-FLAG5, and Eaf6p-Myc13) or YDA136 (HA3-Mot1p, Nhp10p-FLAG5, and Med6p-Myc13)). WCEs were prepared as before, and immunopurifications were performed using Sepharose bead-bound monoclonal anti-HA, anti-Myc, or anti-FLAG IgGs. The immunopurified proteins were specifically eluted with the appropriate epitope peptide, trypsinized, and analyzed by MudPIT as described above. As controls, the same purifications were performed on the untagged cognate strain (W303). Data processing was performed by an approach similar to that used previously. We reasoned that, in the case of a Mot1p-nucleated megacomplex (Fig. 7A), each of the protein complexes identified as Mot1p-interacting would be linked to one another via association with Mot1p. In this case, the purification of any one of these complexes should also result in purification of the other complexes, assuming co-purification of Mot1p. However, if Mot1p is present in multiple, discrete functional complexes, purification of any one Mot1p-interacting complex could instead result in the purification of at most a subset of these complexes.
Fig. 7.
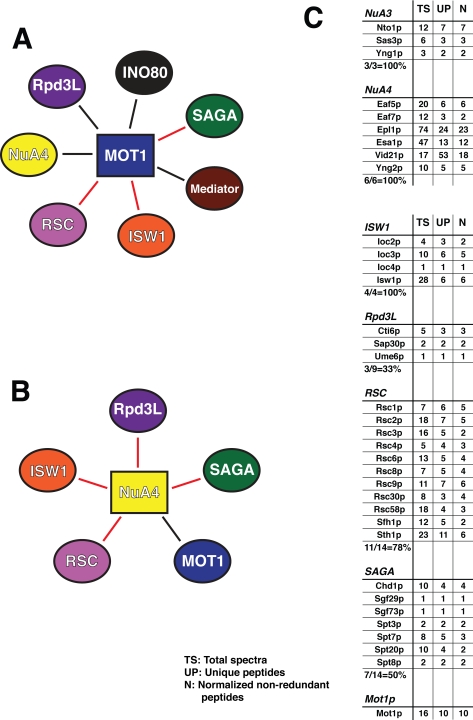
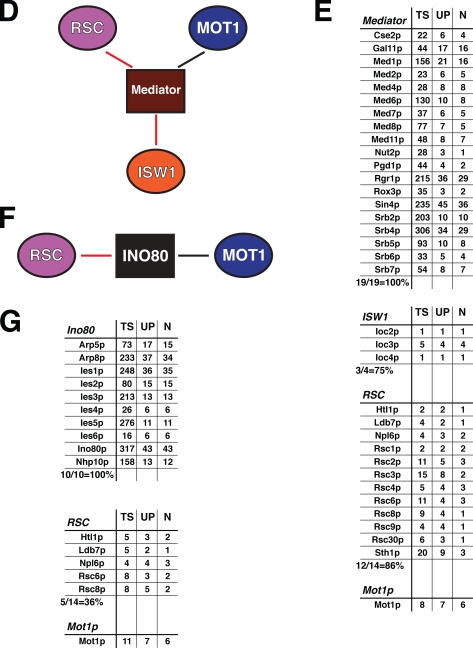
Mot1p participates in multiple, distinct functional complexes. A, schematic illustration of a hypothetical, Mot1p-nucleated megacomplex. In A, B, D, and F, the protein (complex) used as bait in the immunopurification experiment is indicated by a rectangle, associated proteins are indicated by ovals, red lines are used throughout to indicate protein-protein interactions that have been described previously either via genetics or high throughput MS analyses, and black lines demarcate novel interactions observed in our analyses. B, schematic of identified interactions between protein complexes revealed by MudPIT analysis of NuA4/3 (via Eaf6p). C, MudPIT results for NuA4/3 immunopurification. TS, total spectra; UP, unique peptides; N, non-redundant peptides normalized to the control immunopurification. Results for the bait complexes are shown first followed by those for the identified interacting complexes. D, schematic representation of protein complexes found to interact with Mediator. E, MudPIT results for Mediator (Med6p) immunopurification. F, schematic representation of Ino80-associated protein complexes. G, MudPIT results for Ino80 (Nhp10p) immunopurification.
The resulting protein lists obtained from these analyses (the complete data set listing non-redundant peptides is presented in supplemental Table 3) were analyzed using the Clusterer application of the Bioinformatic Graphical Comparative Analysis Tools (BIGCAT) analysis software suite (80, 81). Data were normalized by subtracting the appropriate background from the number of non-redundant peptides identifying each protein. These results were then clustered by supervised clustering using the Enrichment clustering algorithm (Clusterer output included in supplemental Table 3). Lists of known protein complexes were compiled, and subunits of a given complex were classified as being unique if they were not present in another protein complex as defined by the Saccharomyces Genome Database (accessed May 9, 2008). To assess the efficacy of a given immunopurification, we verified that 100% of the subunits unique to the bait complex(es) were present. A protein complex was identified as interacting with the immunopurified complex if both of the following criteria were met: (a) at least 30% of the subunits unique to that complex were identified, and (b) where the reverse IP was performed (e.g. Mediator-NuA4 and NuA4-Mediator), the complexes were identified in both directions.
As indicated in Fig. 7 (B–G), Mot1p co-purified with each of the three complexes examined. Fig. 7B schematically illustrates the protein complexes found to interact with NuA4/3 when the Eaf6p subunit was used as bait: namely the RSC, ISW1, Rpd3L, and SAGA complexes in addition to Mot1p. As indicated by the red connecting lines, with the exception of the interaction between NuA4 and Mot1p, shown in black, each of these protein complex interactions has been described previously in the literature either through a high throughput MS screen (82) or a large scale genetics assay (83, 84). MudPIT results are shown (Fig. 7C), indicating both that the efficacy of immunopurification was high, revealing all of the complex-specific subunits of both the NuA3 and NuA4 complexes, and also that the interacting complexes were well represented, ranging from 33% of the Rpd3L-specific subunits to 100% of ISW1 subunits being identified in our sample.
Similarly when the Mediator immunopurification was analyzed (Fig. 7, D and E), it was found to contain 100% of the Mediator-specific subunits as well as the majority of subunits specific to the ISW1 and RSC complexes. Again the interactions between Mediator and components of the ISW1 and RSC complexes are consistent with the literature (84, 85). Immunopurification of the Ino80 complex via the Nhp10p subunit (Fig. 7, F and G) resulted in co-purification of all 10 Ino80-specific subunits. In addition to Mot1p, 36% of the subunits specific to the RSC complex were also identified. Interactions between the RSC and Ino80 complexes have been shown via both genetics (84) and high throughput MS (57, 86) approaches.
Of the potential interacting complexes, only RSC was identified in all three immunopurifications, although Mot1p was also present in each sample. If a Mot1p-nucleated megacomplex existed, it would have been predicted that immunopurification of any one of these complexes should co-purify a very similar, if not identical, group of interacting proteins. As our data show, this is not the case. Rather immunopurification of each of these complexes co-purified Mot1p as well as different subsets of protein complexes. Thus we conclude that Mot1p likely exists in many different functional complexes; of course, ultimately this question can only be answered by the purification and characterization of the many Mot1p-containing complexes described here.
DISCUSSION
In an effort to more fully understand the mechanism of action of the transcriptional regulator Mot1p, we utilized MudPIT proteomics analysis to assemble a comprehensive list of Mot1p-interacting proteins. In addition to identifying several proteins that had been shown previously by genetics or biochemical means to interact with Mot1p, including TBP and the NC2 and SAGA complexes, our data also revealed many interactions that had not yet been described, such as interactions between Mot1p and the chromatin-active INO80, ISW2, NuA4, Rpd3L·Rpd3S·Sin3, and FACT complexes.
High throughput proteomics analyses have been performed by two other groups (57, 82, 86), and Mot1p was one of the many proteins studied by these investigators. However, because of the large number of distinct proteins studied in their proteomics analyses, neither group has undertaken extensive analysis of a single protein target with as many replicate samples as we have performed. Thus, Krogan et al. (82) reported very few Mot1p protein-protein interactions (Rpn6p, Rpn12p, Met6p, Dug1p, and Ctf3p), and in fact, there is no overlap between either our data set or that of Gavin et al. (57, 86).
In addition to the large number of samples analyzed in our study, our use of affinity-purified polyclonal anti-Mot1p IgG to perform the purifications for mass spectrometric analysis should enable identification of a larger number of interacting proteins. The previously published high throughput analyses utilized only tandem affinity purification with a C-terminal tandem affinity purification tag, leading to possible problems of epitope accessibility should the tag be hidden within a particular protein complex and of interference of such a large tag with the formation of some protein complexes.
Mot1p May Be Recruited to Promoters via Direct or Indirect Protein-Protein Interactions with DNA-binding Transfactors—
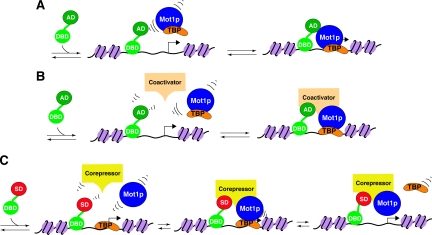
If in fact Mot1p is required to be present at the promoter of specific genes to regulate transcription, one question that remains to be answered is how can Mot1p be recruited to the correct target promoters? One possible means of achieving gene-specific targeting would be for a DNA-binding transfactor to bind an enhancer and, either directly or indirectly, interact with Mot1p to bring it to the correct promoter; such transfactors could be activators or repressors of transcription (Fig. 8). With this in mind, we scanned our proteomics data for the presence of possible DNA-binding transcription factors.
Fig. 8.
Models for the involvement of Mot1p-associated proteins in transcriptional activation and repression. A, direct recruitment of Mot1p by DNA-bound transactivator proteins. Either Mot1p (blue) or Mot1p complexed with TBP (orange) may be recruited to a target promoter by direct interaction with a DNA-bound transactivator (green; DBD, DNA-binding domain; AD, activation domain). Recruitment of a Mot1p-TBP heterodimer is a mechanism by which TBP could be delivered to a target promoter, thereby leading to PIC formation and transcription activation (92). B, indirect recruitment of Mot1p by enhancer-bound coactivators. Mot1p or Mot1p-TBP could be recruited by a coactivator protein or protein complex (tan) that has itself been recruited to the enhancer by interaction with a DNA-bound transactivator. Subsequently Mot1p-TBP recruitment could again result in delivery of TBP to the promoter, or Mot1p may act in concert with coactivators to modulate chromatin structure, thus allowing stable binding of TBP to the promoter, PIC formation, and activation of transcription. C, indirect recruitment of Mot1p by repressor element-bound corepressors. To effect repression of transcription, Mot1p could be recruited by a corepressor protein or protein complex (yellow) that is bound to repressor cis-elements via interaction with a DNA-bound transcription repressor (green/red; SD, silencing domain), leading to alterations in the post-translational modification status of chromatin and/or the removal of TBP from the targeted promoter.
Of the five transcription factors that met our statistical cutoff, four (Gat1p, Hsf1p, Msn2p, and Msn4p) are involved in activation of the ESR with both Msn2p and Msn4p playing important, rather general roles in ESR activation. This finding is especially interesting in light of previous data suggesting a role for Mot1p in the response to stress (25). Association with known DNA-binding transfactors could provide an avenue for direct recruitment of Mot1p to specific target promoters (Fig. 8A).
Mot1p could also be recruited to a target promoter by interaction with a co-regulator protein or protein complex(es). The co-regulators RSC, SAGA, and Mediator (as documented in our work) have all been shown to be recruited to target promoters by enhancer-bound DNA-binding transfactors. Thus, in the case of transcriptional activation involving coactivators, Mot1p could be recruited (as a monomer or in the form of a Mot1p-TBP heterodimer) following interactions with enhancer-bound coactivators (Fig. 8B). Similarly Mot1p could be recruited by transcriptional repressors or corepressors via specific Mot1p-corepressor interactions (Fig. 8C). Such interactions could lead to chromatin remodeling, modulation of histone post-translational modification status, or TBP removal at the target gene(s). It is also important to note that TBP ejection from the PIC could indirectly lead to activation of other genes when TBP levels are limiting.
Transcriptional Outcome of the Interaction of Mot1p with Chromatin-modulating Proteins—
The finding that Mot1p can interact with many different co-regulatory proteins, most of which are known to have chromatin-modifying activities, provides one possible mechanism by which a single protein could play such disparate roles in transcription, functioning on certain genes as a repressor while on other genes as an activator. The accessibility of promoters to the transcriptional machinery is largely dependent on the structure of chromatin, which is altered by two general classes of chromatin remodelers: proteins that covalently modify histones and other transcription proteins and ATP-dependent nucleosome-remodeling factors (87). As such, chromatin modulators play an important role in regulating gene transcription both in terms of activation and repression (88).
Our analysis revealed that Mot1p interacts with members of both classes of chromatin remodelers. ATP-dependent nucleosome-remodeling factors, such as RSC, ISW1 and ISW2, and INO80, can effect either repression or activation of transcription of certain target genes. Additionally members of both classes of covalent histone modifiers, HATs and HDACs, are Mot1p-associated. The SAGA and NuA4 complexes exhibit HAT activity and generally contribute critically to activation of gene transcription, whereas complexes with HDAC activity, such as the Rpd3L·Rpd3S·Sin3 complex(es), generally repress transcription by deacetylating histones. By interacting with complexes with opposing biochemical activities and thus differing transcriptional regulatory functions, Mot1p could itself participate in repression or activation of transcription at individual promoters.
A Possible Role for Mot1p in DNA Replication?—
Our discovery of several proteins involved in the packaging and maintenance of DNA was unexpected. Although high throughput proteomics analyses had also revealed Abf2p as possibly Mot1p-associated (57), the physiological significance of this finding was uncertain: Abf2p is involved in mitochondrial DNA replication, and there is currently no evidence that Mot1p is located in mitochondria. However, in addition to Abf2p and two other mitochondrial proteins, our studies also revealed interactions between Mot1p and several other nuclear proteins involved in DNA maintenance, notably the members of the RFA complex.
Besides its well established function in repair, recombination, and replication of DNA, yeast RFA has been shown to play a possible role in regulating gene transcription. RFA binds to the upstream repressing sequence of several genes, including genes involved in DNA metabolism as well as glucose-regulated genes (for a review, see Ref. 89). Thus, the finding that Mot1p interacts with proteins important in the maintenance of DNA could indicate either a role for Mot1p in DNA replication, an activity that has been ascribed to a growing number of chromatin modifiers, including Rpd3p and the RSC complex (90, 91), or may provide further evidence suggestive of a role in transcription regulation for these replication proteins.
Summary—
Based on the results of our proteomics analysis of Mot1p-associated proteins, we propose that interaction of Mot1p with a wide range of proteins and protein complexes involved in the regulation of transcription, both at the level of chromatin remodeling and of recruitment of transcription machinery to the DNA, represents a likely mechanism through which Mot1p performs the seemingly incongruous task of acting both as an activator and repressor of transcription. Future work will examine the mechanisms through which interaction of Mot1p with these many regulatory proteins turns gene transcription on and off.
Acknowledgments
We thank Toshio Tsukiyama for yeast strains, Yuichiro Takagi for Mediator antibody, and Justin Layer for plasmid p413ADH-FLAG3. We also thank our laboratory colleagues for thoughtful comments and assistance.
Footnotes
Published, MCP Papers in Press, July 2, 2008, DOI 10.1074/mcp.M800221-MCP200
The abbreviations used are: TBP, TATA-binding protein; ADI, ATP-dependent inhibitor; ChIP, chromatin immunoprecipitation; co-IP, co-immunoprecipitation; ESR, environmental stress response; FACT, facilitates chromatin transcription; FDR, false discovery rate; HA, hemagglutinin; HAT, histone acetyltransferase; HDAC, histone deacetylase; HIR, histone regulatory; HRP, horseradish peroxidase; IP, immunoprecipitation; ISW, imitation switch; MudPIT, multidimensional protein identification technology; NC2, negative cofactor 2; NuA, nucleosome acetyltransferase; PIC, preinitiation complex; RFA, replication factor A; RSC, remodel the structure of chromatin; SAGA, Spt·Ada·Gcn5·acetyltransferase; SAM, significance analysis of microarrays; TAF, TBP-associated factor; WCE, whole cell extract; TF, transcription factor; GFP, green fluorescent protein; Mot1p, modifier of transcription 1; rMot1p, recombinant Mot1p; BA, Buffer A.
This work was supported, in whole or in part, by National Institutes of Health Grants GM52461 (to P. A. W.), GM64779, HL68744, ES11993, and CA098131 (to A. J. L.), and CA126218 (to D. L. T.) and Contract HHSN266200400079C/N01-AI-40079 from NIAID and the United States Department of Health and Human Services. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Pugh, B. F. ( 2000) Control of gene expression through regulation of the TATA-binding protein. Gene (Amst.) 255, 1–14 [DOI] [PubMed] [Google Scholar]
- 2.Li, X. Y., Virbasius, A., Zhu, X., and Green, M. R. ( 1999) Enhancement of TBP binding by activators and general transcription factors. Nature 399, 605–609 [DOI] [PubMed] [Google Scholar]
- 3.Roeder, R. G. ( 2005) Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579, 909–915 [DOI] [PubMed] [Google Scholar]
- 4.Albright, S. R., and Tjian, R. ( 2000) TAFs revisited: more data reveal new twists and confirm old ideas. Gene (Amst.) 242, 1–13 [DOI] [PubMed] [Google Scholar]
- 5.Lee, T. I., and Young, R. A. ( 1998) Regulation of gene expression by TBP-associated proteins. Genes Dev. 12, 1398–1408 [DOI] [PubMed] [Google Scholar]
- 6.Davis, J. L., Kunisawa, R., and Thorner, J. (1992) A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 1879–1892 [DOI] [PMC free article] [PubMed]
- 7.Eisen, J. A., Sweder, K. S., and Hanawalt, P. C. ( 1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23, 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chicca, J. J., II, Auble, D. T., and Pugh, B. F. ( 1998) Cloning and biochemical characterization of TAF-172, a human homolog of yeast Mot1. Mol. Cell. Biol. 18, 1701–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman-Levi, R., Miller, C., Bogoch, J., and Zak, N. B. ( 1996) Expanding the Mot1 subfamily: 89B helicase encodes a new Drosophila melanogaster SNF2-related protein which binds to multiple sites on polytene chromosomes. Nucleic Acids Res. 24, 3121–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. elegans Sequencing Consortium ( 1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 [DOI] [PubMed]
- 11.Adamkewicz, J. I., Hansen, K. E., Prud’homme, W. A., Davis, J. L., and Thorner, J. ( 2001) High affinity interaction of yeast transcriptional regulator, Mot1, with TATA box-binding protein (TBP). J. Biol. Chem. 276, 11883–11894 [DOI] [PubMed] [Google Scholar]
- 12.Pereira, L. A., van der Knaap, J. A., van den Boom, V., van den Heuvel, F. A., and Timmers, H. T. ( 2001) TAF(II)170 interacts with the concave surface of TATA-binding protein to inhibit its DNA binding activity. Mol. Cell. Biol. 21, 7523–7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Knaap, J. A., Borst, J. W., van der Vliet, P. C., Gentz, R., and Timmers, H. T. ( 1997) Cloning of the cDNA for the TATA-binding protein-associated factor(II)170 subunit of transcription factor B-TFIID reveals homology to global transcription regulators in yeast and Drosophila. Proc. Natl. Acad. Sci. U. S. A. 94, 11827–11832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrade, M. A., Petosa, C., O’Donoghue, S. I., Muller, C. W., and Bork, P. ( 2001) Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 [DOI] [PubMed] [Google Scholar]
- 15.Auble, D. T., and Hahn, S. ( 1993) An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 7, 844–856 [DOI] [PubMed] [Google Scholar]
- 16.Auble, D. T., Hansen, K. E., Mueller, C. G., Lane, W. S., Thorner, J., and Hahn, S. ( 1994) Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8, 1920–1934 [DOI] [PubMed] [Google Scholar]
- 17.Poon, D., and Weil, P. A. ( 1993) Immunopurification of yeast TATA-binding protein and associated factors. Presence of transcription factor IIIB transcriptional activity. J. Biol. Chem. 268, 15325–15328 [PubMed] [Google Scholar]
- 18.Poon, D., Bai, Y., Campbell, A. M., Bjorklund, S., Kim, Y. J., Zhou, S., Kornberg, R. D., and Weil, P. A. ( 1995) Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 92, 8224–8228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poon, D., Campbell, A. M., Bai, Y., and Weil, P. A. ( 1994) Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-TBP-associated factor complex distinct from transcription factor IID. J. Biol. Chem. 269, 23135–23140 [PubMed] [Google Scholar]
- 20.Wade, P. A., and Jaehning, J. A. ( 1996) Transcriptional corepression in vitro: a Mot1p-associated form of TATA-binding protein is required for repression by Leu3p. Mol. Cell. Biol. 16, 1641–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auble, D. T., Wang, D., Post, K. W., and Hahn, S. ( 1997) Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol. Cell. Biol. 17, 4842–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrau, J. C., Van Oevelen, C. J., Van Teeffelen, H. A., Weil, P. A., Holstege, F. C., and Timmers, H. T. ( 2002) Mot1p is essential for TBP recruitment to selected promoters during in vivo gene activation. EMBO J. 21, 5173–5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dasgupta, A., Darst, R. P., Martin, K. J., Afshari, C. A., and Auble, D. T. ( 2002) Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc. Natl. Acad. Sci. U. S. A. 99, 2666–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geisberg, J. V., Moqtaderi, Z., Kuras, L., and Struhl, K. ( 2002) Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 8122–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geisberg, J. V., and Struhl, K. ( 2004) Cellular stress alters the transcriptional properties of promoter-bound Mot1-TBP complexes. Mol. Cell 14, 479–489 [DOI] [PubMed] [Google Scholar]
- 26.Biswas, D., Yu, Y., Mitra, D., and Stillman, D. J. ( 2006) Genetic interactions between Nhp6 and Gcn5 with Mot1 and the Ccr4-Not complex that regulate binding of TATA-binding protein in Saccharomyces cerevisiae. Genetics 172, 837–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dasgupta, A., Juedes, S. A., Sprouse, R. O., and Auble, D. T. ( 2005) Mot1-mediated control of transcription complex assembly and activity. EMBO J. 24, 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collart, M. A. ( 1996) The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol. Cell. Biol. 16, 6668–6676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madison, J. M., and Winston, F. ( 1997) Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muldrow, T. A., Campbell, A. M., Weil, P. A., and Auble, D. T. ( 1999) MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol. Cell. Biol. 19, 2835–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Topalidou, I., Papamichos-Chronakis, M., Thireos, G., and Tzamarias, D. ( 2004) Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment. EMBO J. 23, 1943–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Link, A. J., Eng, J., Schieltz, D. M., Carmack, E., Mize, G. J., Morris, D. R., Garvik, B. M., and Yates, J. R., III ( 1999) Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682 [DOI] [PubMed] [Google Scholar]
- 33.Washburn, M. P., Wolters, D., and Yates, J. R., III ( 2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 34.van Oevelen, C. J., van Teeffelen, H. A., and Timmers, H. T. ( 2005) Differential requirement of SAGA subunits for Mot1p and Taf1p recruitment in gene activation. Mol. Cell. Biol. 25, 4863–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darst, R. P., Dasgupta, A., Zhu, C., Hsu, J. Y., Vroom, A., Muldrow, T., and Auble, D. T. ( 2003) Mot1 regulates the DNA binding activity of free TATA-binding protein in an ATP-dependent manner. J. Biol. Chem. 278, 13216–13226 [DOI] [PubMed] [Google Scholar]
- 36.Mumberg, D., Muller, R., and Funk, M. ( 1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene (Amst.) 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 37.Woontner, M., Wade, P. A., Bonner, J., and Jaehning, J. A. ( 1991) Transcriptional activation in an improved whole-cell extract from Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 4555–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O’Shea, E. K. ( 2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 39.Goldmark, J. P., Fazzio, T. G., Estep, P. W., Church, G. M., and Tsukiyama, T. ( 2000) The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103, 423–433 [DOI] [PubMed] [Google Scholar]
- 40.McConnell, A. D., Gelbart, M. E., and Tsukiyama, T. ( 2004) Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol. Cell. Biol. 24, 2605–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. ( 1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 42.Noguchi, E., Noguchi, C., McDonald, W. H., Yates, J. R., III, and Russell, P. ( 2004) Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol. Cell. Biol. 24, 8342–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guthrie, C., and Fink, G. R. ( 1991) Guide to yeast genetics and molecular biology. Methods Enzymol. 194, 1–933 [PubMed] [Google Scholar]
- 44.Adamkewicz, J. I., Mueller, C. G., Hansen, K. E., Prud’homme, W. A., and Thorner, J. ( 2000) Purification and enzymic properties of Mot1 ATPase, a regulator of basal transcription in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 275, 21158–21168 [DOI] [PubMed] [Google Scholar]
- 45.Sanders, S. L., Jennings, J., Canutescu, A., Link, A. J., and Weil, P. A. ( 2002) Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22, 4723–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tabb, D. L., Fernando, C. G., and Chambers, M. C. ( 2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, B., Chambers, M. C., and Tabb, D. L. ( 2007) Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tusher, V. G., Tibshirani, R., and Chu, G. ( 2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanders, S. L., and Weil, P. A. (2000) Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 275, 13895–13900 [DOI] [PubMed]
- 50.Sanders, S. L., Klebanow, E. R., and Weil, P. A. ( 1999) TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274, 18847–18850 [DOI] [PubMed] [Google Scholar]
- 51.Cairns, B. R., Lorch, Y., Li, Y., Zhang, M., Lacomis, L., Erdjument-Bromage, H., Tempst, P., Du, J., Laurent, B., and Kornberg, R. D. ( 1996) RSC, an essential, abundant chromatin-remodeling complex. Cell 87, 1249–1260 [DOI] [PubMed] [Google Scholar]
- 52.Saha, A., Wittmeyer, J., and Cairns, B. R. ( 2002) Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16, 2120–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cairns, B. R., Erdjument-Bromage, H., Tempst, P., Winston, F., and Kornberg, R. D. ( 1998) Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell 2, 639–651 [DOI] [PubMed] [Google Scholar]
- 54.Takagi, Y., and Kornberg, R. D. ( 2006) Mediator as a general transcription factor. J. Biol. Chem. 281, 80–89 [DOI] [PubMed] [Google Scholar]
- 55.Lai, J. S., and Herr, W. ( 1992) Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. U. S. A. 89, 6958–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guldener, U., Munsterkotter, M., Kastenmuller, G., Strack, N., van Helden, J., Lemer, C., Richelles, J., Wodak, S. J., Garcia-Martinez, J., Perez-Ortin, J. E., Michael, H., Kaps, A., Talla, E., Dujon, B., Andre, B., Souciet, J. L., De Montigny, J., Bon, E., Gaillardin, C., and Mewes, H. W. ( 2005) CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 33, D364–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gavin, A. C., Aloy, P., Grandi, P., Krause, R., Boesche, M., Marzioch, M., Rau, C., Jensen, L. J., Bastuck, S., Dumpelfeld, B., Edelmann, A., Heurtier, M. A., Hoffman, V., Hoefert, C., Klein, K., Hudak, M., Michon, A. M., Schelder, M., Schirle, M., Remor, M., Rudi, T., Hooper, S., Bauer, A., Bouwmeester, T., Casari, G., Drewes, G., Neubauer, G., Rick, J. M., Kuster, B., Bork, P., Russell, R. B., and Superti-Furga, G. ( 2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 58.Shen, X., Mizuguchi, G., Hamiche, A., and Wu, C. ( 2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544 [DOI] [PubMed] [Google Scholar]
- 59.Shen, X., Ranallo, R., Choi, E., and Wu, C. ( 2003) Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12, 147–155 [DOI] [PubMed] [Google Scholar]
- 60.Orphanides, G., LeRoy, G., Chang, C. H., Luse, D. S., and Reinberg, D. ( 1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92, 105–116 [DOI] [PubMed] [Google Scholar]
- 61.Belotserkovskaya, R., Oh, S., Bondarenko, V. A., Orphanides, G., Studitsky, V. M., and Reinberg, D. ( 2003) FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 [DOI] [PubMed] [Google Scholar]
- 62.Biswas, D., Yu, Y., Prall, M., Formosa, T., and Stillman, D. J. ( 2005) The yeast FACT complex has a role in transcriptional initiation. Mol. Cell. Biol. 25, 5812–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mason, P. B., and Struhl, K. ( 2003) The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23, 8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Longhese, M. P., Plevani, P., and Lucchini, G. ( 1994) Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol. Cell. Biol. 14, 7884–7890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanna, J. S., Kroll, E. S., Lundblad, V., and Spencer, F. A. ( 2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 21, 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Contamine, V., and Picard, M. ( 2000) Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 64, 281–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Behnia, R., Barr, F. A., Flanagan, J. J., Barlowe, C., and Munro, S. ( 2007) The yeast orthologue of GRASP65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic. J. Cell Biol. 176, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krogan, N. J., Kim, M., Ahn, S. H., Zhong, G., Kobor, M. S., Cagney, G., Emili, A., Shilatifard, A., Buratowski, S., and Greenblatt, J. F. ( 2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22, 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Versele, M., and Thevelein, J. M. ( 2001) Lre1 affects chitinase expression, trehalose accumulation and heat resistance through inhibition of the Cbk1 protein kinase in Saccharomyces cerevisiae. Mol. Microbiol. 41, 1311–1326 [DOI] [PubMed] [Google Scholar]
- 70.Gasser, S. M., and Cockell, M. M. ( 2001) The molecular biology of the SIR proteins. Gene (Amst.) 279, 1–16 [DOI] [PubMed] [Google Scholar]
- 71.Prochasson, P., Florens, L., Swanson, S. K., Washburn, M. P., and Workman, J. L. ( 2005) The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19, 2534–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dragon, F., Gallagher, J. E., Compagnone-Post, P. A., Mitchell, B. M., Porwancher, K. A., Wehner, K. A., Wormsley, S., Settlage, R. E., Shabanowitz, J., Osheim, Y., Beyer, A. L., Hunt, D. F., and Baserga, S. J. ( 2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417, 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dasgupta, A., Sprouse, R. O., French, S., Aprikian, P., Hontz, R., Juedes, S. A., Smith, J. S., Beyer, A. L., and Auble, D. T. ( 2007) Regulation of rRNA synthesis by TATA-binding protein-associated factor Mot1. Mol. Cell. Biol. 27, 2886–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Monschau, N., Stahmann, K. P., Sahm, H., McNeil, J. B., and Bognar, A. L. ( 1997) Identification of Saccharomyces cerevisiae GLY1 as a threonine aldolase: a key enzyme in glycine biosynthesis. FEMS Microbiol. Lett. 150, 55–60 [DOI] [PubMed] [Google Scholar]
- 75.Duden, R., Kajikawa, L., Wuestehube, L., and Schekman, R. ( 1998) epsilon-COP is a structural component of coatomer that functions to stabilize α-COP. EMBO J. 17, 985–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seytter, T., Lottspeich, F., Neupert, W., and Schwarz, E. ( 1998) Mam33p, an oligomeric, acidic protein in the mitochondrial matrix of Saccharomyces cerevisiae is related to the human complement receptor gC1q-R. Yeast 14, 303–310 [DOI] [PubMed] [Google Scholar]
- 77.Madania, A., Dumoulin, P., Grava, S., Kitamoto, H., Scharer-Brodbeck, C., Soulard, A., Moreau, V., and Winsor, B. ( 1999) The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell 10, 3521–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Archambault, J., and Friesen, J. D. ( 1993) Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol. Rev. 57, 703–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dolinski, K. J., and Heitman, J. ( 1999) Hmo1p, a high mobility group 1/2 homolog, genetically and physically interacts with the yeast FKBP12 prolyl isomerase. Genetics 151, 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fleischer, T. C., Weaver, C. M., McAfee, K. J., Jennings, J. L., and Link, A. J. ( 2006) Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 20, 1294–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McAfee, K. J., Duncan, D. T., Assink, M., and Link, A. J. ( 2006) Analyzing proteomes and protein function using graphical comparative analysis of tandem mass spectrometry results. Mol. Cell. Proteomics 5, 1497–1513 [DOI] [PubMed] [Google Scholar]
- 82.Krogan, N. J., Cagney, G., Yu, H., Zhong, G., Guo, X., Ignatchenko, A., Li, J., Pu, S., Datta, N., Tikuisis, A. P., Punna, T., Peregrin-Alvarez, J. M., Shales, M., Zhang, X., Davey, M., Robinson, M. D., Paccanaro, A., Bray, J. E., Sheung, A., Beattie, B., Richards, D. P., Canadien, V., Lalev, A., Mena, F., Wong, P., Starostine, A., Canete, M. M., Vlasblom, J., Wu, S., Orsi, C., Collins, S. R., Chandran, S., Haw, R., Rilstone, J. J., Gandi, K., Thompson, N. J., Musso, G., St Onge, P., Ghanny, S., Lam, M. H., Butland, G., Altaf-Ul, A. M., Kanaya, S., Shilatifard, A., O’Shea, E., Weissman, J. S., Ingles, C. J., Hughes, T. R., Parkinson, J., Gerstein, M., Wodak, S. J., Emili, A., and Greenblatt, J. F. ( 2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 83.Mitchell, L., Lambert, J. P., Gerdes, M., Al-Madhoun, A. S., Skerjanc, I. S., Figeys, D., and Baetz, K. ( 2008) Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol. Cell. Biol. 28, 2244–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Collins, S. R., Miller, K. M., Maas, N. L., Roguev, A., Fillingham, J., Chu, C. S., Schuldiner, M., Gebbia, M., Recht, J., Shales, M., Ding, H., Xu, H., Han, J., Ingvarsdottir, K., Cheng, B., Andrews, B., Boone, C., Berger, S. L., Hieter, P., Zhang, Z., Brown, G. W., Ingles, C. J., Emili, A., Allis, C. D., Toczyski, D. P., Weissman, J. S., Greenblatt, J. F., and Krogan, N. J. ( 2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810 [DOI] [PubMed] [Google Scholar]
- 85.Wilson, B., Erdjument-Bromage, H., Tempst, P., and Cairns, B. R. ( 2006) The RSC chromatin remodeling complex bears an essential fungal-specific protein module with broad functional roles. Genetics 172, 795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., Remor, M., Hofert, C., Schelder, M., Brajenovic, M., Ruffner, H., Merino, A., Klein, K., Hudak, M., Dickson, D., Rudi, T., Gnau, V., Bauch, A., Bastuck, S., Huhse, B., Leutwein, C., Heurtier, M. A., Copley, R. R., Edelmann, A., Querfurth, E., Rybin, V., Drewes, G., Raida, M., Bouwmeester, T., Bork, P., Seraphin, B., Kuster, B., Neubauer, G., and Superti-Furga, G. ( 2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 87.Shahbazian, M. D., and Grunstein, M. ( 2007) Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 88.Narlikar, G. J., Fan, H. Y., and Kingston, R. E. ( 2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487 [DOI] [PubMed] [Google Scholar]
- 89.Wold, M. S. ( 1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 90.Vogelauer, M., Rubbi, L., Lucas, I., Brewer, B. J., and Grunstein, M. ( 2002) Histone acetylation regulates the time of replication origin firing. Mol. Cell 10, 1223–1233 [DOI] [PubMed] [Google Scholar]
- 91.Koyama, H., Itoh, M., Miyahara, K., and Tsuchiya, E. ( 2002) Abundance of the RSC nucleosome-remodeling complex is important for the cells to tolerate DNA damage in Saccharomyces cerevisiae. FEBS Lett. 531, 215–221 [DOI] [PubMed] [Google Scholar]
- 92.Gumbs, O. H., Campbell, A. M., and Weil, P. A. ( 2003) High-affinity DNA binding by a Mot1p-TBP complex: implications for TAF-independent transcription. EMBO J. 22, 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joshua-Tor, L., Xu, H. E., Johnston, S. A., and Rees, D. C. ( 1995) Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6. Science 269, 945–950 [DOI] [PubMed] [Google Scholar]
- 94.Rothstein, R. J. ( 1983) One-step gene disruption in yeast. Methods Enzymol. 101, 202–211 [DOI] [PubMed] [Google Scholar]
- 95.Simon, I., Barnett, J., Hannett, N., Harbison, C. T., Rinaldi, N. J., Volkert, T. L., Wyrick, J. J., Zeitlinger, J., Gifford, D. K., Jaakkola, T. S., and Young, R. A. ( 2001) Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106, 697–708 [DOI] [PubMed] [Google Scholar]
- 96.Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O’Shea, E. K., and Weissman, J. S. ( 2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]