Abstract
Background:
Cell-cycle checkpoint regulates cell cycle progression and proliferation. Alterations in cell-cycle control mechanisms are linked to tumorigenesis.
Methods:
This case-control study included 147 cases and 147 controls. We used a pathway-based approach to assess the association between 10 potential functional single-nucleotide polymorphisms from seven cell-cycle control genes and the risk of oral premalignant lesions (OPLs). We also used classification and regression tree analysis to examine high-order gene-gene and gene-smoking interactions.
Results:
Compared with the homozygous wild-type GG genotype of CCND1 P241P, individuals with the AG genotype exhibited an increased risk of OPL (odds ratio, 1.58; 95% confidence interval, 0.89–2.83), and carriers of the AA genotype had a significantly increased risk of OPL (odds ratio, 2.75; 95% confidence interval, 1.33–5.71), with risk increasing significantly with the increasing number of variant alleles (P = 0.006). The risk of OPL increased significantly as the number of unfavorable genotypes in the pathway increased (P = 0.002). The final decision tree in the CART analysis contained five terminal nodes. Compared with the never smokers (the lowest risk group), the odds ratios for terminal nodes 2 through 5 ranged from 1.21 to 5.40.
Conclusions:
Our results illustrated the advantage of using a pathway-based approach for analyzing gene-gene and gene-smoking interactions. Specifically, we showed that genetic polymorphisms in cell-cycle control pathway genes may contribute to the risk of OPL.
Keywords: Cell-cycle pathway, SNP, Oral premalignant lesion, CART
INTRODUCTION
The chief clinical manifestations of oral premalignant lesions (OPL) are leukoplakia, erythroplakia, lichen planus, and submucous fibrosis [1]. The risk of developing oral cancer is high in individuals with OPLs. Lee et al. followed 70 OPL patients with a medium follow-up of 7 years. The resulting incidence rate of cancer in upper aerodigestive tract following treatment was 31.4% [2]. The probability of showing evidence of severe epithelial dysplasia or malignancy at diagnosis was 90% for erythroplakia [3]. A few studies suggest that the most common manifestation of OPL, leukoplakia, has a co-incidence rate of up to 60% at the time of diagnosis of oral squamous cell carcinoma [1]. The major risk factors for oral tumorigenesis are tobacco chewing, cigarette smoking, and alcohol consumption, but evidence also supports the role of genetic susceptibility [4-7].
A study of the role of genetic polymorphisms in OPLs can be approached in different ways. A candidate gene study assesses the effects of a single genetic polymorphism at a time, whereas a pathway-based approach examines the joint effects of a panel of genetic polymorphisms in the same pathway. Because cancer is a multistep, multigenetic process, single-gene analyses usually provide limited power. Therefore, a pathway-based approach may provide more insight and enhance the prediction power[8-10].
Chromosomal damage induced during carcinogenesis leads to the activation of cell-cycle checkpoint controls that regulate cell progression and proliferation. Under cell-cycle control, the affected cells experience either slow growth or apoptosis [11]. Alterations in various cell cycle control mechanisms are linked to tumorigenesis [12-14]. Specifically, the alterations in the expression level of cell cycle proteins were observed in both premalignant and malignant lesions arising in the oral cavity [15]. The aim of this case-control analysis was to use a pathway-based approach to assess the association between 10 single-nucleotide polymorphisms (SNPs) from seven cell-cycle checkpoint pathway genes (P53, P21, P27, CDK4, CDK6, CCND1, and STK15) and the risk of OPL. We also used classification and regression tree (CART) analysis to examine high-order gene-gene and gene-smoking interactions.
MATERIALS AND METHODS
Study population
Adult patients (>17yrs) with histologically confirmed OPL (leukoplakia and/or erythroplakia) were recruited between 1997 and 2006 from the patient population at the University of Texas M. D. Anderson Cancer Center in Houston, Texas. Patients who had acute intercurrent illnesses or infections, those who had undergone retinoid or carotenoid therapy within 3 months before study entry, and those with a history of cancer (except nonmelanoma skin cancer) who had received treatment within 2 years prior to enrollment, were excluded from the study. Healthy individuals with no prior history of cancer (except nonmelanoma skin cancer) were selected from a database of control subjects recruited at Kelsey-Seybold clinics, the largest private multispecialty physician group in the Houston metropolitan area. Controls were matched to cases by age (±5 years), sex, and ethnicity. We included a total of 147 patients with OPL and 147 healthy controls in this study.
Epidemiologic data were collected including sociodemographic characteristics, recent and prior tobacco use, and alcohol consumption. Blood was drawn into heparinized tubes and delivered to the laboratory for DNA extraction and other molecular analyses. Each sample was given a study identification number before delivery to the laboratory. All laboratory workers were blinded to the case-control status of the samples. All study participants were provided written informed consent in accordance with the institutional review boards of M. D. Anderson Cancer Center and Kelsey-Seybold Clinic.
Genotyping
We conducted genotyping using the TaqMan method with a 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA), except for the p53 intron 3 polymorphism, which we genotyped using polymerase chain reaction (PCR)–restriction fragment length polymorphism [16]. DNA samples (5 ng) were mixed with 1× TaqMan buffer A, deoxynucleotide triphosphate (200 μM), MgCl2 (5 mM), AmpliTaq Gold (0.65 U), each primer (900 nM), and each probe (200 nM) to produce the amplification mixes (5 μL). The mixes were placed in the GeneAmp PCR system (Applied Biosystems) to carry out the reaction in the following sequence of thermal conditions: 95°C for 10 minutes, followed by 50 cycles of 92°C for 30 seconds and 60°C for 1 minute. The final genotyping result was provided by the built-in software of the system.
Statistical Analysis
The χ2-test was used to test for differences between the patients and the controls subjects in the distributions of sex, ethnicity, and smoking status. We used Pearson's χ2 and Fisher's exact tests to assess the association between each SNP in patients and control subjects. We applied Student's t test (normal distribution variables) or rank sum test (non-normal distribution variables) for continuous variables. Unconditional multivariate logistic regression was performed to compute odds ratios (ORs) as estimates of relative risk for overall and stratified analyses with 95% confidence intervals (CIs) while adjusting for possible confounding by age, sex, ethnicity, alcohol use and smoking status (never, former, or current) when appropriate. An ever smoker was defined as an individual who had smoked at least 100 cigarettes in his/her lifetime. Ever smokers included former smokers, current smokers, and recent quitters (quit within the previous year). A former smoker was defined as an individual who had quit smoking at least 1 year before the interview.
In addition, we used a pathway-based approach to evaluate OPL risk as a function of the number of unfavorable genotypes in the cell-cycle control pathway. We determined the best fitting model for each SNP, i.e. dominant risk model (comparing homozygous wild-type genotype with variant allele carrying genotypes), recessive risk model (comparing homozygous wild-type and heterozygous genotypes with homozygous variant genotype), or additive model on the basis of P value. A SNP is included in our pathway-based analysis if the P-value for the best fitting model is less than 0.5. Without prior knowledge of the genetic variant or OPL risk, we assumed that the minor variant was the risk allele. However, because our assumption may have been inaccurate, we used a more stringent measure to reassign the unfavorable genotype if the OR ≤ 0.75 or OR ≤ 1.0 and the P value was ≤ 0.05. If the genotype frequency for the variant homozygous genotype in both patients and control subjects was <5%, we combined it with that of the heterozygous genotype. All P values were two-sided, and associations were considered statistically significant a P<0.05. We used Stata software (version 8.0; Stata Co., College Station, TX) to complete these analyses.
For high-order gene-gene and gene-environmental interactions, we used CART analysis (HelixTree Genetics Analysis Software, version 4.1.0; Golden Helix, Inc., Bozeman, MT) to build a decision tree via recursive partitioning. Before growing a tree, we defined the measure for goodness of split using multiplicity-adjusted P values and controlled tree growth at 0.05. Starting with the root node that contained all the patient and control subject data, the CART process identified the split with the smallest multiplicity-adjusted P value, i.e., the most optimal split, for the root node. This process continued for each subsequent node until there was no statistically significant split or there were <10 subjects in the terminal node.
Results
The study population consisted of 147 patients with OPL and 147 healthy controls, frequency- matched by age, sex, and ethnicity (Table 1). Although the control subjects were slightly older than the patients (59.1 ± 11.0 years vs. 57.5 ± 13.6 years), the difference was not statistically significant (P = 0.26). As shown in Table 1, there was a significant difference in smoking status between the two groups: 68.7% of patients and 44.9% of controls were ever smokers (P < 0.001). However, control subjects had significantly more alcohol use than cases (68.0% v.s. 53.7%, P = 0.012).
Table 1.
Host Characteristics
| Variable | Control(%) | Case(%) | P-value |
|---|---|---|---|
| Total | 147 | 147 | |
| Gender | |||
| Male | 82(55.8) | 82(55.8) | |
| Female | 65(44.2) | 65(44.2) | 1 |
| Smoking Status | |||
| Never | 81(55.1) | 46(31.3) | |
| Ever | 66(44.9) | 101(68.7) | <0.001 |
| Former | 55(37.4) | 58(39.5) | |
| Current & RQ* | 11(7.5) | 43(29.3) | <0.001 |
| Ethnicity | |||
| Caucasian | 129(87.8) | 129(87.8) | |
| Hispanic | 11(7.5) | 11(7.5) | |
| African American | 7(4.8) | 7(4.8) | 1 |
| Alcohol usage | |||
| Never | 47(32.0) | 68(46.3) | |
| Ever | 100(68.0) | 79(53.7) | 0.012 |
| Age, mean(+/−SD) | 59.1(11.0) | 57.5(13.6) | 0.26 |
RQ – Recent Quitter
There was no significant departure from Hardy-Weinberg equilibrium (HWE) for the 10 SNPs in control subjects. Compared with the homozygous wild-type GG genotype of CCND1 P241P, those with the heterozygous genotype AG exhibited an increased risk of OPL (OR, 1.58; 95% CI, 0.89–2.83), and carriers of the variant homozygous genotype AA had a significantly increased risk of OPL (OR, 2.75; 95% CI, 1.33–5.71), with the risk increasing significantly with the increasing number of variant alleles (P = 0.006) (Table 2). In the stratified analysis (Table 2 and 3), elevated risk conferred by the CCND1 variant allele was more evident among men (OR, 2.55; 95% CI, 1.08–6.04 for AG; OR, 9.89; 95% CI, 3.00–32.68 for AA; P for trend < 0.001), whereas no significant association was found among women. This trend of increasing risk for CCND1 was also seen in ever smokers (OR, 1.41; 95% CI, 0.64–3.14 for AG; OR, 2.96; 95% CI, 1.09–8.05 for AA; P for trend = 0.036) and younger individuals (age ≤ 60 years old) (OR, 2.08; 95% CI, 0.95–4.57 for AG; OR, 2.75; 95% CI, 1.02-7.42; P for trend = 0.03). Overall there was no significant association between P53 R72P or CDK6 and OPL risk. However, never smokers with the homozygous variant CC genotype of P53 R72P exhibited a significantly decreased risk of OPL (OR, 0.16; 95% CI, 0.03–0.99). Younger individuals carrying the same genotype of this SNP showed a decreased risk of OPL (OR, 0.19; 95% CI, 0.03–1.29), although it is not statistically significant at P = 0.05. Younger patients with the heterozygous genotype CT of cyclin-dependent kinase 6 (CDK6) had a significantly reduced risk of OPL compared with the homozygous wild-type genotype CC (OR, 0.44 95% CI, 0.19–0.98). There was a boarder-line significantly decreased risk of OPL for carriers of variant allele of CDK6 among younger individuals (OR, 0.46, 95% CI, 0.21-1.00).
Table 2.
Genetic polymorphisms in cell-cycle pathway for oral premalignant lesion risk
| Overall | Never Smoker | Ever Smoker | ||||||
|---|---|---|---|---|---|---|---|---|
| Subjects, N |
||||||||
| Polymorphism | Controls | Cases | OR(95% CI)* | P value | OR(95% CI) † | P-value | OR(95% CI)⊥ | P-value |
| CCND1 P241P | 147‡ | 144‡ | ||||||
| GG | 56 (38.1) | 37 (25.7) | Ref. | Ref. | Ref. | |||
| AG | 69 (46.9) | 70 (48.6) | 1.58(0.89-2.83) | 0.12 | 1.81(0.76-4.35) | 0.183 | 1.41(0.64-3.14) | 0.394 |
| AA | 22 (15.0) | 37 (25.7) | 2.75(1.33-5.71) | 0.006 | 2.66(0.85-8.31) | 0.092 | 2.96(1.09-8.05) | 0.033 |
| dominant | 1.86(1.08-3.21) | 0.025 | 2.00(0.87-4.60) | 0.102 | 1.78(0.84-3.75) | 0.13 | ||
| recessive | 2.09(1.11-3.95) | 0.023 | 1.85(0.69-4.99) | 0.223 | 2.41(1.00-5.76) | 0.049 | ||
| P for trend | 0.006 | 0.077 | 0.036 | |||||
| P53 intron 3 | 147 | 111 | ||||||
| No insert | 103 (70.1) | 76 (68.5) | Ref. | Ref. | Ref. | |||
| 1 insert | 41 (27.9) | 33 (29.7) | 1.17(0.65-2.11) | 0.604 | 1.22(0.49-3.05) | 0.669 | 1.11(0.51-2.45) | 0.79 |
| 2 insert | 3 (2.0) | 2 (1.80) | 1.15(0.16-8.48) | 0.891 | 1.44(0.18-11.83) | 0.732 | ||
| dominant | 1.17(0.66-2.08) | 0.599 | 1.25(0.52-2.99) | 0.620 | ||||
| P53 intron 6 | 146 | 102 | ||||||
| GG | 104 (71.2) | 73 (71.6) | Ref. | Ref. | Ref. | |||
| GA | 40 (27.4) | 27 (26.4) | 1.04(0.56-1.91) | 0.9061 | 1.06(0.43-2.63) | 0.899 | 1.06(0.45-2.45) | 0.9 |
| AA | 2 (1.4) | 2 (2.0) | 2.18(0.27-17.68) | 0.465 | 3.21(0.36-28.60) | 0.295 | ||
| dominant | 1.08(0.60-1.97) | 0.795 | 1.20(0.50-2.84) | 0.685 | ||||
| P53 R72P | 137 | 110 | ||||||
| GG | 68 (49.6) | 62 (56.4) | Ref. | Ref. | Ref. | |||
| GC | 53 (38.7) | 42 (38.2) | 0.83(0.47-1.46) | 0.508 | 0.87(0.35-2.16) | 0.762 | 0.80(0.38-1.68) | 0.555 |
| CC | 16 (11.7) | 6 (5.4) | 0.47(0.15-1.44) | 0.185 | 0.16(0.03-0.99) | 0.049 | 1.11(0.21-5.90) | 0.898 |
| dominant | 0.76(0.44-1.30) | 0.314 | 0.62(0.26-1.46) | 0.277 | 0.83(0.40-1.71) | 0.611 | ||
| recessive | 0.51(0.17-1.52) | 0.224 | 0.17(0.03-1.00) | 0.051 | 1.23(0.24-6.30) | 0.805 | ||
| P for trend | 0.192 | 0.082 | 0.737 | |||||
| CDK4 3′UTR | 143 | 136 | ||||||
| AA | 73 (51.1) | 64 (47.1) | Ref. | Ref. | Ref. | |||
| AC | 56 (39.2) | 64 (47.1) | 1.29(0.76-2.19) | 0.343 | 2.01(0.88-4.56) | 0.097 | 0.96(0.47-1.97) | 0.921 |
| CC | 14 (9.8) | 8 (5.9) | 0.64(0.24-1.72) | 0.381 | 0.95(0.21-4.27) | 0.950 | 0.44(0.11-1.68) | 0.229 |
| dominant | 1.15(0.70-1.91) | 0.576 | 1.77(0.81-3.89) | 0.152 | 0.85(0.43-1.69) | 0.65 | ||
| recessive | 0.57(0.22-1.49) | 0.256 | 0.68(0.16-2.91) | 0.608 | 0.45(0.12-1.64) | 0.224 | ||
| P for trend | 0.959 | 0.373 | 0.381 | |||||
| P27 5′UTR | 145 | 123 | ||||||
| CC | 82 (56.6) | 65 (52.9) | Ref. | Ref. | Ref. | |||
| CT | 55 (37.9) | 48 (39.0) | 0.99(0.57-1.71) | 0.967 | 0.56(0.23-1.35) | 0.195 | 1.28(0.61-2.71) | 0.51 |
| TT | 8 (5.5) | 10 (8.1) | 1.45(0.48-4.36) | 0.51 | 1.02(0.23-4.48) | 0.981 | 2.29(0.38-13.85) | 0.366 |
| dominant | 1.04(0.62-1.77) | 0.875 | 0.63(0.28-1.42) | 0.264 | 1.37(0.67-2.81) | 0.394 | ||
| recessive | 1.46(0.49-4.28) | 0.496 | 1.27(0.30-5.40) | 0.749 | 2.07(0.35-12.20) | 0.42 | ||
| P for trend | 0.69 | 0.464 | 0.311 | |||||
| P21 3′UTR | 143 | 137 | ||||||
| CC | 121 (84.6) | 116 (84.7) | Ref. | Ref. | Ref. | |||
| CT | 21 (14.7) | 19 (13.9) | 0.92(0.44-1.91) | 0.819 | 1.30(0.39-4.36) | 0.668 | 0.75(0.28-1.99) | 0.566 |
| TT | 1 (0.7) | 2 (1.5) | 2.36(0.16-35.96) | 0.535 | 3.69(0.13- 104.80) |
0.444 | ||
| dominant | 0.97(0.47-1.98) | 0.924 | 1.38(0.42-4.52) | 0.597 | 0.81(0.31-2.11) | 0.669 | ||
| CDK6 3′UTR | 142 | 129 | ||||||
| CC | 81 (57.0) | 86 (66.7) | Ref. | Ref. | Ref. | |||
| CT | 55 (38.7) | 35 (27.1) | 0.64(0.37-1.13) | 0.123 | 0.82(0.35-1.96) | 0.659 | 0.48(0.22-1.02) | 0.057 |
| TT | 6 (4.2) | 8 (6.2) | 1.62(0.50-5.17) | 0.419 | 1.36(0.27-6.75) | 0.707 | 1.90(0.30-11.90) | 0.495 |
| dominant | 0.73(0.43-1.24) | 0.248 | 0.90(0.40-2.02) | 0.790 | 0.55(0.27-1.15) | 0.112 | ||
| recessive | 1.90(0.60-5.99) | 0.272 | 1.46(0.30-7.03) | 0.639 | 2.60(0.43-15.90) | 0.301 | ||
| P for the trend |
0.596 | 0.980 | 0.336 | |||||
| STK15 F31I | 144 | 136 | ||||||
| TT | 88 (61.1) | 78 (57.4) | Ref. | Ref. | Ref. | |||
| TA | 53 (36.8) | 49 (36.0) | 1.08(0.64-1.83) | 0.77 | 1.36(0.61-3.02) | 0.455 | 0.77(0.37-1.59) | 0.476 |
| AA | 3 (2.1) | 9 (6.6) | 2.49(0.59-10.46) | 0.213 | 1.05(0.08-14.51) | 0.971 | 4.72(0.54-41.63) | 0.162 |
| dominant | 1.17(0.70-1.94) | 0.556 | 1.34(0.61-2.92) | 0.469 | 0.93(0.46-1.86) | 0.829 | ||
| recessive | 2.41(0.58-10.00) | 0.224 | 0.95(0.07-13.01) | 0.968 | 5.24(0.61-45.21) | 0.132 | ||
| P for the trend |
0.352 | 0.518 | 0.647 | |||||
| STK15 I57V | 143 | 137 | ||||||
| GG | 104 (72.7) | 104 (75.9) | Ref. | Ref. | Ref. | |||
| GA | 36 (25.2) | 30 (21.9) | 0.81(0.45-1.48) | 0.498 | 0.72(0.27-1.87) | 0.494 | 0.85(0.38-1.88) | 0.687 |
| AA | 3 (2.1) | 3 (2.2) | 1.17(0.22-6.29) | 0.854 | 2.94(0.28-31.37) | 0.372 | ||
| dominant | 0.84(0.47-1.50) | 0.553 | 0.62(0.24-1.59) | 0.317 | 0.95(0.44-2.04) | 0.894 | ||
Adjusted by age, gender, ethnicity, smoking status(never, former, and current), and alcohol use
Adjusted by age, gender, ethnicity, and alcohol use
Adjusted by age, gender, ethnicity, smoking status(former and current), and alcohol use
Abbreviations: CI = confidence interval; OR = odds ratio.
Due to missing genotyping information, the number which indicated the number of individuals with genotyping data varies for control and case subjects
Table 3.
Stratified analysis for CCND1, P53 R72P, and CDK6 by gender and age
| Male | Female | Young Individual(Age ≤60) | Old Individual (Age>60) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | Case | OR(95% CI) * | Control | Case | OR(95% CI) * | Control | Case | OR(95% CI) ‡ | Control | Case | OR(95% CI) ‡ | |
| CCND1 P241P | 82 | 81 | 65 | 63 | 79 | 83 | 68 | 61 | ||||
| GG | 33 | 15 | 1 | 23 | 22 | 1 | 36 | 23 | 1 | 20 | 14 | 1 |
| AG | 39 | 43 | 2.55(1.08-6.04) | 30 | 27 | 0.85(0.36-2.04) | 31 | 40 | 2.08(0.95-4.57) | 38 | 30 | 0.97(0.38-2.49) |
| AA | 10 | 23 | 9.89(3.00-32.68) | 12 | 14 | 0.84(0.29-2.48) | 12 | 20 | 2.75(1.02-7.42) | 10 | 17 | 2.67(0.83-8.58) |
| Dominant | 3.51(1.56-7.93) | 0.85(0.38-1.92) | 2.26(1.09-4.71) | 1.30(0.54-3.14) | ||||||||
| Recessive | 5.37(1.94-14.88) | 0.92(0.36-2.40) | 1.83(0.76-4.44) | 2.72(1.00-7.36) | ||||||||
| P for trend | <0.0001 | 0.729 | 0.030 | 0.127 | ||||||||
| P53 R72P | 77 | 61 | 60 | 49 | 73 | 64 | 64 | 46 | ||||
| GG | 37 | 36 | 1 | 31 | 26 | 1 | 33 | 36 | 1 | 35 | 26 | 1 |
| GC | 30 | 20 | 0.65(0.28-1.50) | 23 | 22 | 1.15(0.50-2.64) | 28 | 26 | 1.06(0.48-2.33) | 25 | 16 | 0.59(0.24-1.46) |
| CC | 10 | 5 | 0.63(0.17-2.42) | 6 | 1 | 12 | 2 | 0.19(0.03-1.29) | 4 | 4 | 0.83(0.13-5.42) | |
| Dominant | 0.65(0.30-1.41) | 0.92(0.41-2.08) | 0.85(0.40-1.81) | 0.62(0.26-1.46) | ||||||||
| Recessive | 0.75(0.21-2.76) | 0.19(0.03-1.22) | 0.99(0.16-6.29) | |||||||||
| P for trend | 0.31 | 0.434 | 0.268 | 0.368 | ||||||||
| CDK6 3′UTR | 80 | 77 | 62 | 52 | 75 | 75 | 67 | 54 | ||||
| CC | 46 | 54 | 1 | 35 | 32 | 1 | 43 | 57 | 1 | 38 | 29 | 1 |
| CT | 29 | 20 | 0.64(0.29-1.43) | 26 | 15 | 0.57(0.24-1.38) | 29 | 15 | 0.44(0.19-0.98) | 26 | 20 | 1.06(0.45-2.49) |
| TT | 5 | 3 | 0.72(0.12-4.13) | 1 | 5 | 7.75(0.76-79.39) | 3 | 3 | 0.68(0.12-3.95) | 3 | 5 | 3.17(0.58-17.32) |
| Dominant | 0.65(0.30-1.41) | 0.78(0.34-1.78) | 0.46(0.21-1.00) | 1.23(0.54-2.78) | ||||||||
| Recessive | 0.84(0.15-4.77) | 9.50(0.96-94.52) | 0.89(0.16-5.05) | 3.09(0.59-16.07) | ||||||||
| P for trend | 0.33 | 0.743 | 0.095 | 0.344 | ||||||||
Adjusted by age, ethnicity, smoking status(never, former, and current), and alcohol use
Adjusted by age, gender, ethnicity, smoking status(never, former, and current), and alcohol use
In this pathway-based analysis, we excluded P53 intron3, P53 intron6, P21 3′UTR, and STK15 I57V because P-values for the best fitting model were greater than 0.5. For the other SNPs, the unfavorable genotypes were AG/AA for CCND1 P241P, GG/GC for P53 R72P, AA/AC for CDK4 3′UTR, TT for P27 5′UTR, CC for CDK6 3′UTR, and AA for STK15 F31I, As shown in Table 3, we categorized the subjects into three risk groups according to the number of unfavorable genotypes: low-risk group (1∼2 unfavorable genotypes), medium-risk group (3 unfavorable genotypes), and high-risk group (4∼5 unfavorable genotypes). The OR for the medium-risk group was 5.01 (95% CI, 1.74–14.40) and for the high-risk group was 6.18 (95% CI, 2.14–17.86). The risk of OPL increased significantly as the number of unfavorable genotypes increased (P = 0.002).
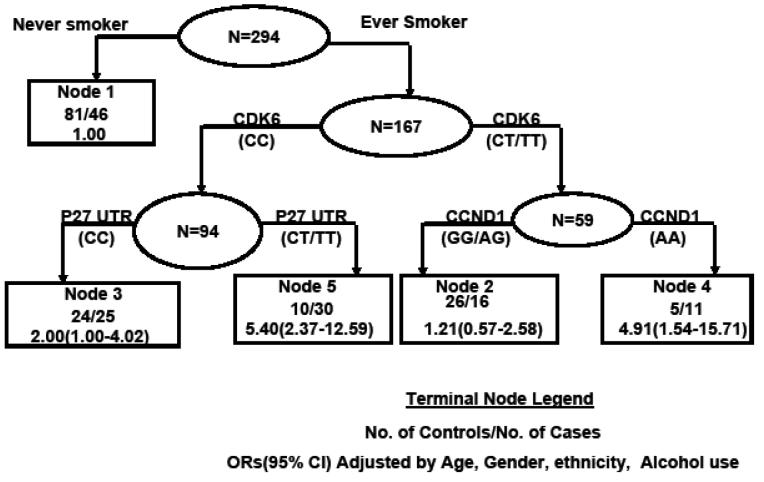
Ten genetic polymorphisms and smoking status were included in the CART analysis. The initial split of the root node was smoking status, showing the strongest association with OPL risk and supporting its critical role in the risk of OPL (Figure 1). A total of five terminal nodes were constructed in the final tree. Never smokers, the lowest risk group, were in terminal node 1. Table 4 shows the association between terminal node and OPL risk. The ORs for terminal nodes 2 to 5 ranged from 1.21 to 5.40 compared with terminal node 1, adjusting for age, sex, ethnicity, and alcohol use. The highest-risk group, terminal node 5, contained ever smokers carrying the common homozygous genotype of CDK6 3′UTR and the variant allele P27 5′UTR. The five terminal nodes were categorized into three risk groups on the basis of estimated OR: low (OR ≤1.21), medium (1.21 <OR ≤ 2), and high (OR > 2). The risk of OPL was significantly elevated in the three risk groups (P for trend< 0.001).
Figure 1.
Classification and regression tree analysis of genetic polymorphisms in cell cycle pathways and smoking status with OPL risk.
Table 4.
Combined analysis for the number of unfavorable genotypes* in cell-cycle control pathways
| Risk (No. of unfavorable genotypes) |
No. of controls |
No. of patients |
OR (95% CI) | P value |
|---|---|---|---|---|
| Low(1∼2) | 37 | 5 | Ref. | |
| Medium (3) | 56 | 39 | 5.01(1.74-14.40) | 0.0028 |
| High (4∼5) | 39 | 43 | 6.18(2.14-17.86) | 0.0008 |
| P for the trend | 0.002 |
Unfavorable genotypes: CCND1 P241P(AG/AA), P53 R72P (GG/GC), CDK4 3′UTR (AA/AC), P27 5′UTR (TT), CDK6 3′UTR(CC), STK15 F31I (AA),
Adjusted by age, gender, ethnicity, smoking status, and alcohol use
Abbreviations: CI = confidence interval; OR = odds ratio.
Discussion
In this study, we used a pathway-based approach to increase our understanding of the complex interaction between genes in the cell-cycle control pathway, smoking status and the overall risk of OPL.
Overall, in individual SNP analysis only the CCND1 P241P polymorphism was significantly associated with an increased risk of OPL for variant homozygous. Compared with the common homozygous genotype (GG), the rare homozygous genotype (AA) of CCND1 was associated with a 2.5-fold increased risk of OPL. The per-allele OPL risk for the rare allele A of CCND1 was 1.65 (P = 0.006). The CCND1 gene, located at 11q13, encodes protein cyclin D1 and regulates the G1 phase of the cell cycle. The rare allele A of CCND1 has been associated with an increased risk of numerous malignancies, including cancers of the lungs, skin, head and neck, prostate, bladder, kidneys, esophagus, colorectum, endometrium, and larynx [4,17-20]. The alternative splicing of CCND1 mRNA into two transcripts, a and b, has been observed to be modulated by the AG alleles at nucleotide 870 within the splicer donor region of exon 4 [21]. Transcript b, which is preferentially spliced by allele A, exhibits a greater potential to become an oncogene in human malignancies. In the stratified analysis, an increased risk associated with the variant allele of CCND1 was observed in men, younger individuals, and ever smokers. The increased risk of the CCND1 variant allele with colorectal cancer among younger individuals and males was also observed by Huang et al. [20] in the Taiwanese population.
Due to the complex nature of cancer, single SNP analysis usually provides undetectable low or minor risk associated with cancer as shown by this study. Setting threshold P-value to 0.5, we will be able to include the minor, low risks in our pathway-based analysis while excluding SNPs without association with risk of OPL. The pathway based analysis using selected genetic polymorphisms in the cell-cycle pathway showed a promising advantage over the single-candidate gene method. We found that individuals with three unfavorable genotypes exhibited a 5.01-fold increased risk of OPL, and those with more than three unfavorable genotypes showed a 6.18-fold increased risk, compared to individuals with one or two unfavorable genotypes. Each additional number of unfavorable genotypes was associated with a 1.79-fold (P for trend = 0.002) significantly increased risk.
Traditional logistic regression analysis cannot handle the sparseness caused by high-order interaction, i.e., empty cells in the contingency table. Unlike logistic regression, CART analysis is an explorative, nonparametric approach that requires no assumption of a genetic model. Zhang and Bonney [22] illustrated the applicability of tree-based analyses for genetic association studies. In our study, when compared with genetic polymorphisms, smoking was the most dominant factor in OPL risk. Among ever smokers, CART analysis identified CDK6-P27 and CDK6-CCND1 interactions. Ever smokers with a common homozygote CDK6 genotype and a variant allele-carrying genotype of P27 5′UTR had the highest percentage of OPL cases (75%). Individuals who smoked and carried at least one variant allele of CDK6 3′UTR and common homozygous and heterozygous genotypes of CCND1 P241P had the second-lowest percentage of OPL cases (38%). Both CDK6-P27 and CDK6-CCND1 interactions are biologically feasible. Cyclin-dependent kinase 6, CDK6, is located in chromosome region 7q21-q22 and starts its activity in the mid-G1 phase. It is crucial for progression from G1 phase and G1/S phase transition through interaction with CCND1. The complex mix, made up of CDK6/CDK4 with CCND1, regulates the tumor-suppressor protein pRB. Deregulated CDK6 expression has been associated with the dysfunction of cell-cycle control [23]. P27 belongs to the KIP family of CDK inhibitors, which negatively regulate the interaction between CCND1 and CDK6/CDK4. Low expression of P27 has been linked to many tumor types and tumor progression [24]. The polymorphism of P27 5′UTR is within a U-rich element and necessary for the transcription of P27 mRNA [25]. Of note, the high-order gene-gene interactions identified above were found only in ever smokers, and no significant SNPs was identified for never smokers. Therefore, the risk of OPL may be regulated by potential higher-order gene-gene and gene-smoking interactions.
In conclusion, this study assessed a panel of genetic polymorphisms in cell-cycle control pathway and the OPL risk. We performed a pathway based approach and a multifaceted approach (CART) to analyze high-order gene-gene and gene-smoking interactions. Our results revealed that the alteration of risk of OPL by SNPs in the cell-cycle control pathway can be attributed to smoking status. Therefore, this pathway-based approach has the advantage of identifying the complex relationship between genetic polymorphisms and cancer susceptibility involving multiple factors.
Table 5.
Association of terminal nodes from CART analysis and risk of oral premalignant lesion
| Terminal | No. of controls |
No. of patients |
OR | 95% CI | P value |
|---|---|---|---|---|---|
| 1 | 81 | 46 | Ref. | ||
| 2 | 26 | 16 | 1.21 | 0.57-2.58 | 0.625 |
| 3 | 24 | 25 | 2.00 | 1.00-4.02 | 0.050 |
| 4 | 5 | 11 | 4.91 | 1.54-15.71 | 0.007 |
| 5 | 10 | 30 | 5.40 | 2.37-12.29 | <0.001 |
| P for the trend | <0.001 | ||||
| Risk group | |||||
| Low (OR<1.2) | 107 | 62 | Ref. | ||
| Medium (1.2 ≤ OR <2) | 24 | 25 | 1.90 | 0.98-3.68 | 0.058 |
| High (OR ≥ 2) | 15 | 41 | 4.99 | 2.51-9.93 | <0.001 |
| P for the trend | <0.001 | ||||
Adjusted by age, gender, ethnicity, and alcohol use
Abbreviations: CART = classification and regression tree; CI = confidence interval; OR = odds ratio.
Acknowledgments
Grant support: National Cancer Institute grants CA106451 and CA097007
References
- 1.Mithani SK, Mydlarz WK, Grumbine FL, Smith IM, Califano JA. Molecular genetics of premalignant oral lesions. Oral Dis. 2007;13:126–33. doi: 10.1111/j.1601-0825.2006.01349.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee JJ, Hong WK, Hittelman WN, Mao L, Lotan R, Shin DM, Benner SE, Xu XC, Lee JS, Papadimitrakopoulou VM, Geyer C, Perez C, Martin JW, El-Naggar AK, Lippman SM. Predicting cancer development in oral leukoplakia: ten years of translational research. Clin Cancer Res. 2000;6:1702–10. [PubMed] [Google Scholar]
- 3.Shafer WG, Waldron CA. Erythroplakia of the oral cavity. Cancer. 1975;36:1021–8. doi: 10.1002/1097-0142(197509)36:3<1021::aid-cncr2820360327>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 4.Huang M, Spitz MR, Gu J, Lee JJ, Lin J, Lippman SM, Wu X. Cyclin D1 gene polymorphism as a risk factor for oral premalignant lesions. Carcinogenesis. 2006;27:2034–7. doi: 10.1093/carcin/bgl048. [DOI] [PubMed] [Google Scholar]
- 5.Raju B, Mehrotra R, Oijordsbakken G, Al-Sharabi AK, Vasstrand EN, Ibrahim SO. Expression of p53, cyclin D1 and Ki-67 in pre-malignant and malignant oral lesions: association with clinicopathological parameters. Anticancer Res. 2005;25:4699–706. [PubMed] [Google Scholar]
- 6.Rosin MP, Cheng X, Poh C, Lam WL, Huang Y, Lovas J, Berean K, Epstein JB, Priddy R, Le ND, Zhang L. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin Cancer Res. 2000;6:357–62. [PubMed] [Google Scholar]
- 7.Wu X, Lippman SM, Lee JJ, Zhu Y, Wei QV, Thomas M, Hong WK, Spitz MR. Chromosome instability in lymphocytes: a potential indicator of predisposition to oral premalignant lesions. Cancer Res. 2002;62:2813–8. [PubMed] [Google Scholar]
- 8.Wu X, Gu J, Grossman HB, Amos CI, Etzel C, Huang M, Zhang Q, Millikan RE, Lerner S, Dinney CP, Spitz MR. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006;78:464–79. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu J, Zhao H, Dinney CP, Zhu Y, Leibovici D, Bermejo CE, Grossman HB, Wu X. Nucleotide excision repair gene polymorphisms and recurrence after treatment for superficial bladder cancer. Clin Cancer Res. 2005;11:1408–15. doi: 10.1158/1078-0432.CCR-04-1101. [DOI] [PubMed] [Google Scholar]
- 10.Han J, Colditz GA, Samson LD, Hunter DJ. Polymorphisms in DNA double-strand break repair genes and skin cancer risk. Cancer Res. 2004;64:3009–13. doi: 10.1158/0008-5472.can-04-0246. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Shin DM, El-Naggar A, Lee JS, Corrales C, Lippman SM, Hong WK, Hittelman WN. Chromosome polysomy and histological characteristics in oral premalignant lesions. Cancer Epidemiol Biomarkers Prev. 2001;10:319–25. [PubMed] [Google Scholar]
- 12.Collins K, Jacks T, Pavletich NP. The cell cycle and cancer. Proc Natl Acad Sci U S A. 1997;94:2776–8. doi: 10.1073/pnas.94.7.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–7. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 14.Shintani S, Mihara M, Nakahara Y, Kiyota A, Ueyama Y, Matsumura T, Wong DT. Expression of cell cycle control proteins in normal epithelium, premalignant and malignant lesions of oral cavity. Oral Oncol. 2002;38:235–43. doi: 10.1016/s1368-8375(01)00048-3. [DOI] [PubMed] [Google Scholar]
- 15.Schoelch ML, Regezi JA, Dekker NP, Ng IO, McMillan A, Ziober BL, Le QT, Silverman S, Fu KK. Cell cycle proteins and the development of oral squamous cell carcinoma. Oral Oncol. 1999;35:333–42. doi: 10.1016/s1368-8375(98)00098-0. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, Zhao H, Amos CI, Shete S, Makan N, Hong WK, Kadlubar FF, Spitz MR. p53 Genotypes and Haplotypes Associated With Lung Cancer Susceptibility and Ethnicity. J Natl Cancer Inst. 2002;94:681–90. doi: 10.1093/jnci/94.9.681. [DOI] [PubMed] [Google Scholar]
- 17.Le Marchand L, Seifried A, Lum-Jones A, Donlon T, Wilkens LR. Association of the cyclin D1 A870G polymorphism with advanced colorectal cancer. Jama. 2003;290:2843–8. doi: 10.1001/jama.290.21.2843. [DOI] [PubMed] [Google Scholar]
- 18.Kang S, Kim JW, Park NH, Song YS, Kang SB, Lee HP. Cyclin D1 polymorphism and the risk of endometrial cancer. Gynecol Oncol. 2005;97:431–5. doi: 10.1016/j.ygyno.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 19.Rydzanicz M, Golusinski P, Mielcarek-Kuchta D, Golusinski W, Szyfter K. Cyclin D1 gene (CCND1) polymorphism and the risk of squamous cell carcinoma of the larynx. Eur Arch Otorhinolaryngol. 2006;263:43–8. doi: 10.1007/s00405-005-0957-7. [DOI] [PubMed] [Google Scholar]
- 20.Huang WS, Tang R, Lin PY, Changchien CR, Chen JS, Chiang JM, Yeh CY, Wang JY, Hsieh LL. Impact of the cyclin D1 A870G polymorphism on susceptibility to sporadic colorectal cancer in Taiwan. Dis Colon Rectum. 2006;49:602–8. doi: 10.1007/s10350-005-0311-6. [DOI] [PubMed] [Google Scholar]
- 21.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–8. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Bonney G. Use of classification trees for association studies. Genet Epidemiol. 2000;19:323–32. doi: 10.1002/1098-2272(200012)19:4<323::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 23.Ch'ng S, Sullivan M, Yuan L, Davis P, Tan ST. Mast cells dysregulate apoptotic and cell cycle genes in mucosal squamous cell carcinoma. Cancer Cell Int. 2006;6:28. doi: 10.1186/1475-2867-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clurman BE, Porter P. New insights into the tumor suppression function of P27(kip1) Proc Natl Acad Sci U S A. 1998;95:15158–60. doi: 10.1073/pnas.95.26.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez P, Diez-Juan A, Coto E, Alvarez V, Reguero JR, Batalla A, Andres V. A single-nucleotide polymorphism in the human p27kip1 gene (−838C>A) affects basal promoter activity and the risk of myocardial infarction. BMC Biol. 2004;2:5. doi: 10.1186/1741-7007-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]