Abstract
In chloroplasts of green plants and algae, CO2 is assimilated into triose-phosphates (TPs); a large part of these TPs is exported to the cytosol by a TP/phosphate translocator (TPT), whereas some is stored in the plastid as starch. Plastidial phosphate translocators have evolved from transport proteins of the host endomembrane system shortly after the origin of chloroplasts by endosymbiosis. The red microalga Galdieria sulphuraria shares three conserved putative orthologous transport proteins with the distantly related seed plants and green algae. However, red algae, in contrast to green plants, store starch in their cytosol, not inside plastids. Hence, due to the lack of a plastidic starch pool, a larger share of recently assimilated CO2 needs to be exported to the cytosol. We thus hypothesized that red algal transporters have distinct substrate specificity in comparison to their green orthologs. This hypothesis was tested by expression of the red algal genes in yeast (Saccharomyces cerevisiae) and assessment of their substrate specificities and kinetic constants. Indeed, two of the three red algal phosphate translocator candidate orthologs have clearly distinct substrate specificities when compared to their green homologs. GsTPT (for G. sulphuraria TPT) displays very narrow substrate specificity and high affinity; in contrast to green plant TPTs, 3-phosphoglyceric acid is poorly transported and thus not able to serve as a TP/3-phosphoglyceric acid redox shuttle in vivo. Apparently, the specific features of red algal primary carbon metabolism promoted the evolution of a highly efficient export system with high affinities for its substrates. The low-affinity TPT of plants maintains TP levels sufficient for starch biosynthesis inside of chloroplasts, whereas the red algal TPT is optimized for efficient export of TP from the chloroplast.
In plants, the photosynthetic light reactions provide the energy for major plastid localized pathways, such as CO2 assimilation, the synthesis of starch, fatty acids, several amino acids, nucleic acids, and the reductive assimilation of inorganic ions like nitrate and sulfate (Weber et al., 2005; Zrenner et al., 2006). To supply the cell and the organism with these primary metabolites, a large number of precursors, end products, and intermediates have to be transported across the organelle envelope membrane and therefore present-day plastids are extensively connected to the cytoplasm by metabolite transporters that reside in the envelope membranes (Tegeder and Weber, 2006; Weber and Fischer, 2007).
Chloroplasts originated approximately 1.6 billion years ago through a single primary endosymbiosis between a nonphotosynthetic primitive mitochondriate eukaryote and a cyanobacterium (Yoon et al., 2004; Bhattacharya et al., 2007; Reyes-Prieto et al., 2007). Within a period of 0.15 billion years, establishment of the plastid and divergence of the three major lineages of the Archaeplastida (Adl et al., 2005), that is the red algae (Rhodophyceae), green algae/land plants (Chloroplastida), and glaucophytes (Glaucophyta), began (Bhattacharya et al., 2004; Yoon et al., 2004). Establishment of the chloroplast within the host cell required massive remodeling of its membrane proteome; novel transport proteins to connect its metabolism with the metabolic network of the host cell had to be acquired (Bhattacharya et al., 2007; Weber and Fischer, 2007). Phylogenetic and phylogenomics analyses recently revealed that a large portion of these plastid-resident transporters is host derived, indicating that integration of the chloroplast with host metabolism was predominantly a host-driven process (Tyra et al., 2007). Genes encoding these transporters are not present in extant cyanobacterial genomes but most are conserved throughout the Archaeplastida, indicating they have been established at an early stage during formation of endosymbiosis, likely already at the stage of the protoalga (Tyra et al., 2007).
Of particular importance for establishment of chloroplasts was an efficient and controlled export of photoassimilates from the endosymbiont to its host cell (Weber et al., 2006). In the Chloroplastida, a triose-P (TP) translocator (TPT) exports a significant amount of the dihydroxy-acetonephosphate (DHAP) that is generated by the Calvin-Benson cycle to the cytosol; there it mainly serves as precursor for Suc and cell wall biosynthesis (Flügge, 1999; Reiter, 2002). However, part of the TP is retained in the plastid stroma and used to fuel starch biosynthesis and other intraplastidial biosynthetic pathways (Zeeman et al., 2007). TPT candidate orthologs are highly conserved throughout the photosynthetic eukaryotes (Weber et al., 2006), and transport experiments with isolated organelles or reconstituted organellar membranes demonstrated TP transport activity in the red algae and glaucophytes lineages, respectively (Schlichting and Bothe, 1993; Weber et al., 2004). Apparently, TP already served as the main export photoassimilate at the stage of the proto-alga (Weber et al., 2006). In the plant model organism Arabidopsis (Arabidopsis thaliana), additional sugar phosphate transporters with specialized functions have been characterized (Flügge, 1999). Import of phosphoenolpyruvic acid (PEP) from the cytosol by the PEP/phosphate transporter (PPT) drives fatty acid biosynthesis and synthesis of compounds by the shikimate pathway (i.e. aromatic amino acids; Fischer et al., 1997; Knappe et al., 2003b; Voll et al., 2003) and a Glc-6-P/phosphate transporter (GPT) provides Glc-6-P for starch synthesis in heterotrophic plastids (Kammerer et al., 1998; Niewiadomski et al., 2005). Last, a protein closely related to the GPT, the pentose-P/phosphate transporter (XPT), connects the oxidative pentose-P pathways (OPPPs) in cytosol and plastid (Eicks et al., 2002; Flügge and Gao, 2005).
Phylogenetic analysis showed that candidate orthologs for the PPT and the GPT/XPT type of translocators are present in the genome of the ancient red microalga Galdieria sulphuraria, which is separated from green plants by an evolutionary distance of at least 1 billion years (Yoon et al., 2004). However, reconstituted membrane fractions from G. sulphuraria showed neither significant PEP nor Glc-6-P transport activity (Weber et al., 2004). This raises the intriguing question as to whether the genetic repertoire present in the last common ancestor of Rhodophyceae and Chloroplastida was functionally maintained or whether it diverged after separation of the two phyla. To address this question, we used G. sulphuraria as a model for the Rhodophyceae. The draft genome of G. sulphuraria is publicly available and the unicellular organism shares core features of carbon metabolism with other red algae (Viola et al., 2001; Barbier et al., 2005a). In contrast to higher plants, starch in red algae is produced in the cytosolic compartment, using UDP-Glc as precursor (Coppin et al., 2005; Patron and Keeling, 2005). Cell wall polysaccharides as well as the major soluble carbohydrate floridoside (α-d-galactopyranosyl-1-2′-glycerol) are produced in the same compartment. Notably, in addition to photoautotrophic growth, G. sulphuraria also is able to grow mixo- or heterotrophically on more than 50 different carbon sources (Gross and Schnarrenberger, 1995; Barbier et al., 2005a). Carbon partitioning thus has to be coordinated not only during alternating light and dark periods but also under continuous heterotrophic growth conditions. It is not known in detail how the carbon allocation between the plastid and the cytosol is accomplished in red algae. Based on phylogenetic data, the plastid phosphate translocators presumably are the major routes for metabolite exchange. They have to balance a high demand for photo-assimilates in the cytosol with maintaining sufficient levels of TPs for rhodoplast-localized pathways. Here we report the heterologous expression and biochemical characterization of the plastid phosphate transporter family from G. sulphuraria. With regard to higher plants we will discuss an alternative strategy to fine-tune carbon flux across the plastid membrane in photosynthetic eukaryotes.
RESULTS
Molecular Features of the Plastidic Phosphate Translocator Homologs from G. sulphuraria
Three genes encoding proteins with significant similarity to higher plant plastidic phosphate translocators (pPTs) have previously been identified in the genome of the red alga G. sulphuraria (Weber et al., 2006). Based on detailed phylogenetic analysis, GsHET39C12 is the candidate ortholog to the plastid TPT and will thus be called GsTPT. GsA14H8 and GsA16F5 cluster with the functionally characterized green plant plastid Glc-6-P/phosphate transport proteins (GPTs) and PEP/phosphate translocator proteins (PPTs), respectively. They were thus assigned with the acronyms GsGPT and GsPPT, respectively. The coding sequences of GsTPT and GsPPT contain 1,224 nucleotides, corresponding to 407 amino acid residues and computed molecular masses of 45.83 kD (TPT) and 44.98 kD (PPT). GsGPT has a calculated molecular mass of 45.45 kD, consisting of 410 amino acid residues that are encoded by 1,233 nucleotides. The N-terminal regions of all three proteins represent putative plastid target sequences that display only low sequence similarity with the pPTs from higher plants (Supplemental Fig. S1) and the red alga Cyanidioschyzon merolae. The G. sulphuraria proteins show an average sequence identity of 37% and a 55% similarity with their Arabidopsis homologs and a slightly higher identity of 48% to the corresponding C. merolae proteins (Supplemental Table S2). Six Lys and two Arg residues that are invariantly embedded in conserved motifs in all functionally characterized pPT proteins to date (Knappe et al., 2003a) are also found in all three G. sulphuraria homologs. Relative to the GsTPT sequence, the positively charged amino acids are at positions K123, K199, R261, K266, K267, K361, R362, and K399 (Supplemental Fig. S1). All three putative transport proteins are highly hydrophobic proteins that are predicted by ConPred II (Arai et al., 2004) to contain nine to 10 membrane spanning α-helices. The GsTPT and GsGPT genes contain one intron that separates a short first exon from the residual coding sequence (Supplemental Table S2). GsPPT has a total of four exons. The last exon covers approximately 50% of the codons. All three sequences have been submitted to GenBank and also can be found at the Michigan State University Galdieria genome database (http://genomics.msu.edu/galdieria). Accession numbers and annotations are listed in Supplemental Table S2.
Heterologous Expression of GsTPT, GsPPT, and GsGPT
To assess the substrate specificities of the G. sulphuraria phosphate translocators (PTs), we cloned the corresponding cDNAs into the yeast (Saccharomyces cerevisiae) expression vector pYES/NT under control of the Gal-inducible GAL4 promoter. The regions of the cDNAs encoding for the putative target sequences of each protein were removed and instead fused to an N-terminal hexa-His tag. After transformation of the corresponding constructs into the yeast strain INVSc1, all three pPT homologs could be successfully expressed and accumulated in the membrane fraction (Supplemental Fig. S2). Immunoblot analysis with an anti-poly-His tag antibody verified the Gal-inducible accumulation of the pPT proteins (Supplemental Fig. S2, lane 2–4) compared to controls, which maintained the empty expression vector (Supplemental Fig. S2, lane 1). The calculated molecular masses of the N-terminal His-tagged proteins were 41.6 kD, 45.2 kD, and 41.2 kD for GsTPT, GsPPT, and GsGPT, respectively. The presence of recombinant protein was verified for all biological replicates by western blot before reconstitution.
Functional Analysis of Three Putative Transport Proteins from G. sulphuraria
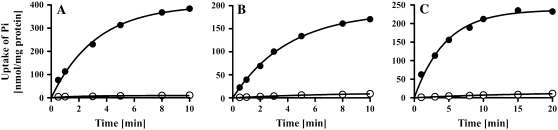
All functionally characterized pPTs of higher plants catalyze, in addition to their characteristic substrates, a strict homo-exchange of orthophosphate (Pi) (Flügge, 1999). To test whether the yeast-expressed, recombinant GsPTs are functional, we thus first examined their ability to catalyze the signature Pi homo-exchange.To this end, they were reconstituted into liposomes that were preloaded with 30 mm Pi (i.e. the liposomes contained 30 mm Pi inside). Then radiolabeled [32P]phosphate was added to the liposomes and the uptake kinetics were recorded. In GsTPT and GsPPT containing proteoliposomes, protein-mediated isotope equilibrium was reached within 10 min, using an external Pi concentration of 0.25 mm (Fig. 1). GsGPT mediated Pi uptake equilibrated after 15 min with a 4 times higher external [32P]Pi concentration of 1 mm. Pi exchange followed a first-order kinetic with an equilibrium plateau (Vmax) of 400 ± 13.74, 187.3 ± 3.31, and 237.1 ± 7.02 nmol mg protein−1 and initial rates of 60.69 ± 2.48, 30.94 ± 0.43, and 23.23 ± 1.27 nmol mg protein−1 min−1 for GsTPT, GsPPT, and GsGPT, respectively. Control membranes from yeast cells transformed with an empty pYES/NT vector or boiled recombinant proteins showed negligible accumulation rates over time (data not shown). Pyridoxal-phosphate and 4,4#-diisothiocyanostilbene-2,2#-disulfonic acid also effectively inhibited GspPT-mediated import of [32P]phosphate (data not shown). Marginal uptake of radiolabeled [32P]phosphate compared to control experiments was measured when vesicles did not contain Pi as a countersubstrate (Fig. 1). In summary, all putative phosphate transport proteins from G. sulphuraria were functionally expressed in yeast and are able to catalyze the signature Pi homo-exchange activity, similar to their higher plant homologs.
Figure 1.
Time kinetics of pPT homologs from G. sulphuraria. A to C, Liposomes were reconstituted with membrane proteins from yeast cells expressing GsTPT (A), GsPPT (B), or GsGPT (C). Uptake of Pi was measured in the presence (•) or absence (○) of intraliposomal Pi (30 mm). Transport was initiated with a final concentration of 0.25 mm [32P]Pi (A and B) or 1 mm [32P]Pi (C). Diagrams represent a typical result of at least three independent studies.
Transport Properties of GsTPT, GsPPT, and GsGPT
To assess the substrate specificity of the recombinant GspPT proteins, vesicles were preloaded with saturating concentrations (i.e. 30 mm) of various counter-exchange substrates and initial rates of [32P]Pi uptake were determined.
GsTPT
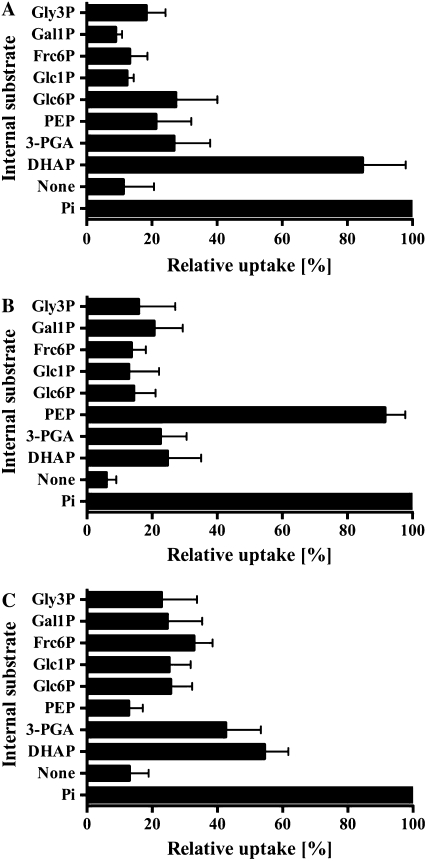
Relative to the Pi/Pi homo-exchange experiment, the uptake rate of [32P]Pi into proteoliposomes reconstituted with GsTPT was 86.7% ± 13.3% when proteoliposomes were preloaded with the TP DHAP. Uptake rates into liposomes preloaded with 3-phosphoglyceric acid (3-PGA), PEP, or Glc-6-P, respectively, were low (Fig. 2A). Uptake was also negligible when GsTPT liposomes were preloaded with Glc-1-P, Fru-6-P, Gal-1-P, glycerol-3-P (Gly-3-P; Fig. 2A), AMP, UMP, gluconate-6-P, ribulose-1,5-bisP, ADP-Glc, UDP-Glc, UDP-Gal, α-ketoglutaric acid, malic acid, pyruvic acid, or oxaloacetic acid (data not shown). GsTPT has an apparent KM value of 0.33 ± 0.07 mm for Pi and DHAP competitively inhibited Pi uptake with a Ki value of 0.5 ± 0.04 mm (Table I). 3-PGA and PEP inhibited [32P]Pi uptake only at nonphysiological concentrations of 8 and 9 mm, respectively, and Glc-6-P did not show significant inhibition of Pi transport at any of the tested concentrations.
Figure 2.
Internal substrate dependence on the transport activities of GsTPT, GsPPT, and GsGPT. A to C, Liposomes were preloaded with various substrates (30 mm) and reconstituted with membrane proteins from yeast cells expressing GsTPT (A), GsPPT (B), or GsGPT (C). Uptake experiments were initiated with 0.25 mm [32P]Pi (A and B) or 1 mm [32P]Pi (C) and terminated after 1.5 min (GsTPT), 2 min (GsPPT), or 4 min (GsGPT). As negative control, proteoliposomes were preincubated (2 min) with inhibitor stop solution to calculate net uptake activity. Relative uptake activities were compared to the Pi/Pi counter-exchange experiment, which was set to 100%. Data are summarized as the arithmetic mean ± se of at least three independent experiments. Glc6P, Glc-6-P; Glc1P, Glc-1-P; Frc6P, Fru-6-P; Gal1P, Gal-1-P; Gly3P, Gly-3-P.
Table I.
Comparison of kinetic constants from G. sulphuraria and higher plant pPT homologs
The Michaelis constant (KM) for Pi was determined using various external [32P]Pi concentrations (0.05–10 mm). The competitive inhibition constant (Ki) of [32P]Pi uptake was assayed at two different external Pi concentrations with increasing inhibitor concentrations (0.05–20 mm; Dixon, 1953). All proteoliposomes were preloaded with 30 mm Pi. Data represent the arithmetic mean ± se of at least three independent experiments. Recombinant higher plant pPT data are taken from Flügge (1999). None, No competitive inhibitory constant could be measured under the given experimental conditions.
| Kinetic Constants | TPT Homolog
|
PPT Homolog
|
GPT Homolog
|
|||
|---|---|---|---|---|---|---|
| Plants | G. sulphuraria | Plants | G. sulphuraria | Plants | G. sulphuraria | |
| mm | mm | mm | ||||
| KM (Pi) | 1.0 | 0.33 ± 0.07 | 0.8 | 0.76 ± 0.08 | 1.1 | 5.07 ± 0.4 |
| Ki (DHAP) | 1.0 | 0.5 ± 0.04 | 8.0 | 7.93 ± 0.23 | 0.6 | None |
| Ki (3-PGA) | 1.0 | 7.71 ± 0.47 | 4.6 | 3.95 ± 0.1 | 1.8 | None |
| Ki (PEP) | 3.3 | 8.75 ± 0.40 | 0.3 | 0.36 ± 0.04 | 2.9 | None |
| Ki (Glc-6-P) | None | None | None | None | 1.1 | None |
GsPPT
Reconstituted GsPPT protein efficiently used Pi and PEP as a countersubstrate for the import of radiolabeled [32P]phosphate (Fig. 2B). Eighteen additional substrates, as listed above for the GsTPT, showed marginal initial Pi uptake rates of less than 30% of the Pi/Pi homo-exchange rate (Fig. 2B; data not shown). GsPPT has an apparent KM value of 0.76 ± 0.075 mm for Pi and a Ki value of 0.36 ± 0.04 mm for PEP. Compared to PEP, 11 times higher 3-PGA and 22 times higher DHAP levels, respectively, were needed to inhibit Pi uptake by 50% (Table I). Glc-6-P had no affinity for the Pi binding site.
GsGPT
Vesicles reconstituted with the GPT homolog from G. sulphuraria were preloaded with the same 20 substrates as given above; in all cases the [32P]Pi uptake was much lower than for liposomes preloaded with 30 mm Pi (Fig. 2C). GsGPT has rather low affinity to Pi (KM, 5.07 ± 0.4 mm; Table I). Compared to nonpreloaded proteoliposomes (13.03% ± 5.9%), approximately 3- to 4-fold higher uptake rates were measured when liposomes were preloaded with either 3-PGA or DHAP. Importantly, however, even high concentrations of 3-PGA and DHAP did not significantly inhibit Pi homo-exchange (Table I). Glc-6-P and PEP are nonrelevant substrates for the GsGPT protein.
Transcript Levels and Protein Activity of the pPT Homologs in G. sulphuraria Cells
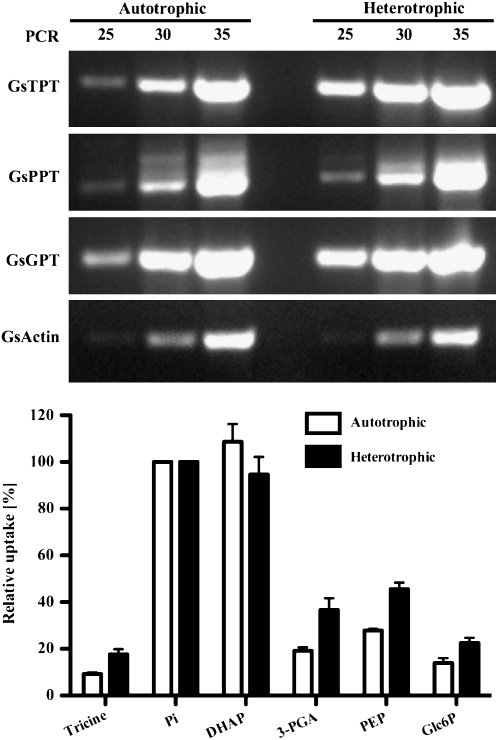
In land plants, TPT expression is confined to photosynthetic tissues whereas GPT expression is highest in heterotrophic tissues (Kammerer et al., 1998; Flügge, 1999; Niewiadomski et al., 2005). PPT genes are expressed at low levels in various tissues (Knappe et al., 2003b). To determine the expression pattern of GspPTs, semiquantitative reverse transcription-PCR was used. Figure 3 shows that GsTPT, GsPPT, and GsGPT transcripts are clearly detectable and abundant under both autotrophic and heterotrophic growth conditions. The steady-state transcript levels of all three genes were higher than those of the reference gene actin (Gs15600; Galdieria Genome Browser Build 3.0). Liposomes reconstituted with total membrane fractions exhibit high [32P]Pi import using DHAP as a countersubstrate under both culturing regimes. Exchange rates with PEP and 3-PGA as counter-exchange substrates were much lower as compared to DHAP. Exchange of Glc-6-P with Pi was close to the background values of unloaded vesicles.
Figure 3.
Transcript abundance and protein activity of GsTPT, GsPPT, and GsGPT in G. sulphuraria cells. Top, Total RNA from auto- and heterotrophically cultured G. sulphuraria cells was isolated, treated with DNase, and used for first-strand cDNA synthesis. Expression of GsTPT, GsPPT, GsGPT, and actin transcripts was amplified by PCR with 25, 30, or 35 elongation cycles and visualized on an ethidium bromide-stained 1.25% (w/v) agarose gel. Bottom, Relative [32P]Pi uptake rates into liposomes reconstituted with total G. sulphuraria membranes isolated from heterotrophically (black bars) or autotrophically (white bars) grown cells, respectively. Membrane fractions were reconstituted into liposomes preloaded with 30 mm various substrates. Experiment was performed as in Figure 2.
DISCUSSION
In this study, we report the molecular and biochemical characteristics of the pPT family from the unicellular red alga G. sulphuraria. All three members of the GsPT gene family were heterologously expressed in yeast, functionally reconstituted into liposomes (Supplemental Fig. S1; Fig. 1), and their substrate specificities and kinetic constants were assessed.
The G. sulphuraria Genome Encodes a High-Affinity TPT with Narrow Substrate Specificity
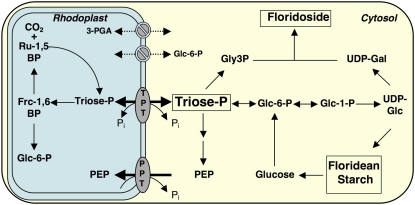
Recombinant G. sulphuraria TPT mediates a strict counterexchange of the TP DHAP with Pi (Fig. 2). Interestingly, the kinetic constants of GsTPT considerably differ from those of the green plant TPT (Table I). The red algal transporter exhibits a 3 times higher affinity for Pi and, as indicated by its low Ki value, the substrate DHAP has a 2-fold higher affinity to the Pi binding site. Further, GsTPT is highly specific for DHAP. In contrast to the green plant ortholog, 3-PGA poorly acts as a countersubstrate, even under saturating conditions (Figs. 2A and 3). In addition, 3-PGA is a weak competitive inhibitor of Pi import with an 8-fold and 15-fold higher Ki value as compared to the TPT from plants and the Ki (DHAP) constant from GsTPT, respectively. These results argue strongly against 3-PGA as a physiological relevant substrate of GsTPT. PEP and Glc-6-P were even less efficient exchange substrates and inhibitors of Pi transport (Fig. 2; Table I). Very likely, the distinct kinetic properties of GsTPT represent an important adaptation to red algal carbon metabolism. In contrast to the Chloroplastida, red algae store an insoluble starch-like polymer called floridean starch and synthesize cell wall building blocks and the main soluble carbohydrate floridoside, composed of UDP-Gal and Gly-3-P moieties in their cytosol (Viola et al., 2001; Collen et al., 2004; Barbier et al., 2005a). Hence, primary carbon partitioning in red algae is predominantly organized within the cytosol and GsTPT enables solely the export of TPs, even at low stromal concentrations, to cope with the massive cytosolic demand of recently fixed carbon (Fig. 4).
Figure 4.
Flexible transport of TP across the plastid membrane is central for the cellular carbon metabolism in the red alga G. sulphuraria. In photosynthetically active cells, TP is exported via the GsTPT from rhodoplasts to sustain soluble and insoluble carbohydrate synthesis (floridoside and floridean starch) in the cytosol. Under heterotrophic growth conditions the same transporter supplies the rhodoplasts with TP for, e.g. hexose-P synthesis. Rhodoplasts are not capable of producing PEP via partial glycolysis and are thus dependent on the import of PEP from the cytosol via the PPT protein for fatty acid and aromatic amino acid biosynthesis. The red algal plastid envelope does not contain any 3-PGA and Glc-6-P transport activities, which is a specific trait of green plants. Frc-1,6BP, Fru-1,6-bisP; Ru-1,5BP, ribulose-1,5-bisP; Gly3P, Gly-3-P.
On the other hand, GsTPT is highly expressed and active in heterotrophically cultured G. sulphuraria cells (Fig. 3). Under these conditions, GsTPT presumably mediates TP import into the plastid, thus mediating carbon flux between cytosol and nonphotosynthetic rhodoplasts. In contrast, land plants export TP from photosynthetic chloroplasts via TPT and import Glc-6-P into heterotrophic plastids via GPT. The TPT is an integral part of photoassimilate partitioning in plants (Häusler et al., 2000; Smith and Stitt, 2007). Accumulation of Suc and hexose-Ps in the cytosol sequester cytosolic Pi and thus slow down the export of TP. A resulting increased stromal 3-PGA/Pi ratio allosterically activates ADP-Glc pyrophosphorylase, which is the committing step of plastidic starch biosynthesis (Ballicora et al., 2004). Plants have to maintain sufficiently high levels of TP in the plastid stroma to enable both the regeneration of the CO2 acceptor ribulose-1,5-bisP and starch biosynthesis during the light period (Zeeman et al., 2007). The apparent lower affinity of the plant transporter for Pi and TP thus ensures higher steady-state levels of TP in the stroma to drive these reactions.
G. sulphuraria Has a PPT with Similar Properties as Its Green Plant Ortholog
PEP uptake from the cytosol is required for stroma-localized fatty acid and shikimate biosynthesis (Flügge, 1999). Previously published results have been ambiguous about the existence of a putative PPT in G. sulphuraria. Phylogenetic analysis revealed an orthologous candidate gene (Weber et al., 2006), although isolated membrane fractions did not exhibit pronounced PEP uptake activity (Weber et al., 2004). Here, we show that EST GsA16F5 (Weber et al., 2006) represents a highly conserved PPT with almost identical kinetic constants in both the green and red lineages (Table I). Both transporters are highly specific for Pi and PEP and poorly accept 3-PGA, DHAP, or any other of the 16 tested metabolites as a substrate (Fig. 2; data not shown). In addition, we employed an improved reconstitution method for total membranes from autotrophic and heterotrophic cell cultures that yielded lower background and a weak but clearly detectable PEP/Pi counter-exchange activity (Fig. 3). The low PEP transport capacity (Fig. 3) mirrors published data from chloroplasts of C3 land plants (Flügge and Weber, 1994). TPT is by far the most abundant transport protein in C3 chloroplasts and accounts for a high DHAP exchange capacity (Flügge, 1999). Fatty acid biosynthesis and the shikimate pathway in red algae are predicted to be similar to those of seed plants and are presumably localized in the plastid, based on phylogenetic analyses and N-terminal target sequence predictions (Weber et al., 2004; Richards et al., 2006).
A question that cannot be conclusively answered at the moment is whether rhodoplasts, analogous to chloroplasts of green plants, also have negligible activities of plastid phosphoglyceromutase and enolase to produce PEP from plastidial 3-PGA (Trimming and Emes, 1993). The G. sulphuraria genome encodes for two phosphoglyceromutase isozymes (Gs04140 and Gs52680) and a single enolase (Gs21490), which are all three computationally predicted as cytosolic enzymes (Nielsen et al., 1997) without any N-terminal targeting sequences, when compared with their homologous genes from Arabidopsis (phosphoglyceromutase: At4g09520, At3g08590, At1g09780; enolase: At2g36530, At2g29560, At1g74030; Friso et al., 2004; Larkin et al., 2007). Alternatively, PEP could be generated from pyruvic acid via the pyruvate Pi dikinase (PPDK) reaction. The annotated PPDK gene Gs42070 is computationally predicted to be localized in the cytosol, exhibiting a high similarity with cyanobacterial PPDK genes, and is not phylogenetically related to the dual targeted enzyme from Arabidopsis (At4g15530; Parsley and Hibberd, 2006). Although computational targeting predictions have to be taken with a grain of salt, bioinformatics analysis indicates that conversion of TP to PEP is not possible in G. sulphuraria, and rhodoplasts thus depend on PEP import from the cytosol to drive PEP-depended reactions in the stroma (Fig. 4).
G. sulphuraria Does Not Possess a Plastidic Hexose-P Importer
Reconstituted recombinant putative GsGPT mediated a strict Pi/Pi homo-exchange that followed a first-order rate kinetics (Fig. 1). An unusually high KM value of 5 mm for Pi (Table I) indicates that GsGPT under physiological conditions is not able to compete with GsTPT and GsPPT for the common substrate Pi. Although liposomes preloaded with high (i.e. saturating) concentrations of DHAP or 3-PGA (i.e. 30 mm internal substrate concentration) showed Pi uptake rates of approximately 50% of Pi/Pi homo-exchange, it is important to notice that neither 3-PGA nor DHAP significantly inhibited Pi/Pi homo-exchange either at physiological concentrations or at 100-fold excess (Table I). The prime substrate candidate Glc-6-P was poorly accepted under saturating conditions (Fig. 2) and it did not inhibit Pi import in the Pi/Pi homo-exchange setup (Table I). These results strongly indicate that 3-PGA, DHAP, and Glc-6-P are not physiologically relevant substrates of the putative GsGPT. We also tested various additional metabolites, such as nucleotide sugars, mononucleotides, hexose-Ps (Glc-1-P, Fru-6-P, and Gal-1-P), pentose-Ps, and a precursor of floridoside biosynthesis, Gly-3-P. All of these are poorly exchanged with [32P]Pi and the physiological substrate of GsGPT thus remains unidentified.
In nongreen plastids of plants, Glc-6-P is the preferred precursor for starch synthesis and NADPH generation via the OPPP due to the absence of a Fru-1,6-bisphosphatase (FBPase) activity (Flügge, 1999). In photosynthetic tissues, plastidic FBPase is inactivated at night and thus hexose-Ps cannot be generated from TPs (and vice versa). In contrast, red algal starch biosynthesis is cytosolic and plastidic FBPase from G. sulphuraria is not subject to a strict redox regulation (Reichert et al., 2003; Oesterhelt et al., 2007). Importing TPs via the GsTPT could thus sustain the production of hexose-P for carbon and NADPH supply in the rhodoplast during the night or prolonged heterotrophic growth conditions, thus bypassing the requirement for a plastidic hexose-P translocator (Fig. 4).
An Operative DHAP/3-PGA Reduction Shuttle Is Unlikely in G. sulphuraria
3-PGA is a major substrate for the TPT in higher plants (Flügge, 1999). In spinach (Spinacia oleracea) and other model plants, 3-PGA levels in the plastid and the cytosol frequently exceed DHAP levels by severalfold (Gerhardt et al., 1987). However, the affinity of the spinach TPT is identical for both metabolites. It has been proposed that in vivo a significant amount of 3-PGA transport occurs across the envelope membrane; the TPT could thus operate as an NADPH reduction equivalent shuttle between stroma and cytosol (Heineke et al., 1991). The substrate specificity and the kinetic constants of recombinant proteins, as discussed in detail earlier, and total membrane fractions from G. sulphuraria presumably cannot sustain a physiological relevant exchange rate of 3-PGA across the rhodoplasts envelope membrane. Still, DHAP offers a possibility to export reduction equivalent to the cytosol. The nonphosphorylating glyceraldehyde-3-P dehydrogenase (Rius et al., 2006), which is also present in the draft genome sequence of the red alga (Gs00840) or the combined glycolytic reaction sequence of glyceraldehyde-3-P dehydrogenase and 3-PGA kinase (Plaxton, 1996) generates NADPH, or NADH and ATP, respectively. However, in contrast to green plants, not 3-PGA but Pi must be postulated as the alternative countersubstrate for DHAP.
Impact of Subtle Differences of TPT in Chloroplastida and Rhodophyta
TP is the main entry and branching point in the glycolytic network in photosynthetically active cells (Plaxton, 1996). Metabolic flux has to be regulated between generating hexose-Ps via gluconeogenesis and glycolytic “downstream” products, such as 3-PGA, PEP, and pyruvic acid. In higher plants, the redox transfer by a TP/3-PGA shuttle via nonphosphorylating glyceraldehyde-3-P dehydrogenase and 3-PGA kinase is proposed to be marginal and glyceraldehyde-3-P dehydrogenase expression is mainly induced under stress conditions, such as anaerobiosis and heat shock or Suc feeding (Yang et al., 1993; Bustos et al., 2008; Holtgrefe et al., 2008). On the other hand, glycolysis has to provide adequate amounts of PEP to replenish TCA intermediates used for biosynthesis of, e.g. amino acids, to produce pyruvic acid for mitochondrial metabolism, or to fuel plastid fatty acid biosynthesis (Plaxton, 1996). Because 3-PGA transport is seemingly a specific adaptation in plants, we hypothesize that 3-PGA transport across the chloroplast envelope membrane might provide a bypass of the complex regulation of the cytosolic TP pool that permits supporting cytosolic PEP biosynthesis and serves under stress conditions as an additional redox shuttle system.
CONCLUSION
Although the primary structures of pPTs are highly conserved between green plants and red algae, these proteins have evolved quite distinct biochemical characteristics in the different lineages of the Archaeplastida. Comparative biochemical analysis of candidate orthologs from distantly related organisms thus provides novel insights into alternative modes for regulating “ancient” metabolic pathways. While the TP/phosphate antiport activity across the plastid envelope membrane is highly conserved across all photosynthetic eukaryotes analyzed to date, the red algal transporters catalyze neither 3-PGA nor Glc-6-P transport (Fig. 4).
G. sulphuraria must have evolved alternative mechanisms to distribute TP into various pools (i.e. floridean starch, floridoside, and fatty acid and amino acid synthesis), such as less stringent redox control of Calvin cycle, OPPP, and glycolysis (Oesterhelt et al., 2007). Plastidial phosphate translocators thus represent crucial components of primary carbon portioning in higher plants and red algae that have evolved to the specific requirements of each lineage through modulation of substrate specificities and kinetic constants.
MATERIALS AND METHODS
Growth and Sampling of Galdieria sulphuraria Cells
Galdieria sulphuraria strain 074W (Gross et al., 1999) was cultured autotrophically in a minimal salt medium at 37°C with a light intensity of 80 μmol photons m−2 s−1. The culture was grown at pH 2.0 in Erlenmeyer flasks under vigorous shaking at ambient air conditions.
Heterotrophic growth of the algal cells was performed at 37°C in culture flasks in complete darkness in the identical salt medium containing 25 mm Glc as sole carbon source. Cells were harvested at the late logarithmic phase by centrifugation (3,000g, 5 min, 4°C), washed with 1×TE buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA), frozen in liquid nitrogen, and stored at −80°C or directly used for nucleic acid extraction (Barbier et al., 2005b) or total membrane enrichment.
Isolation of Genomic DNA and Total RNA from G. sulphuraria
Isolation of genomic DNA was performed as previously described (Barbier et al., 2005b). Briefly, cells were ground in liquid nitrogen and proteins were removed by phenol/chloroform/isoamylalcohol (PCI) extraction. Nucleic acids were precipitated by ethanol and RNA was removed by a DNAse-free RNAse treatment and purified by an additional PCI and ethanol precipitation step. Total RNA was isolated with an acid guanidium isothiocyanate-phenol/chloroform solution as described by Chomczynski and Sacchi (1987).
Enrichment of Total Membrane Fractions from G. sulphuraria
Harvested cells were resuspended in breaking buffer (100 mm HEPES/KOH, pH 7.6, 1 mm EDTA, 5% glycerol, 5 mm ascorbic acid, 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride) to yield an OD600 of approximately 100 (0.5 mL) and an equal volume of acid-washed glass beads (0.4–0.6 mm size; Sigma Aldrich) were added. Cells were lysed with a mixer mill (MM301; Retsch GmbH) for 4 min. Broken cells were diluted with 10 mL breaking buffer and centrifuged (2,000g, 1 min, 4°C). The supernatant contained the total membrane fractions. Membranes were pelleted from the supernatant by ultracentrifugation (100,000g, 50 min, 4°C), resuspended in 0.4 mL of 10 mm HEPES/KOH, pH 7.6, 1 mm MgCl2, and 0.05-mL aliquots reconstituted into liposomes.
Expression of GsPTs in Yeast
The coding sequences of all three GsPT cDNAs were amplified from G. sulphuraria cDNA by PCR (Platinum Pfx polymerase; Invitrogen) using the primer combinations listed in Supplemental Table S1. PCR products were subcloned into the pGEM-T Easy vector system (Promega) and sequenced. Forward and reverse primers for the genes GsHET39C12 and GsA16F5 were designed with BamHI and XhoI restriction recognition sites, respectively. The GsA14H8 specific forward primer had a KpnI and the reverse primer an XhoI restriction site. Each cDNA was ligated in frame with an N-terminal poly-His tag into the yeast (Saccharomyces cerevisiae) expression vector pYES2/NT (Invitrogen). Standard molecular methods were applied for DNA restriction and cloning (Sambrook et al., 1995).
The resulting constructs were transformed into competent yeast INVSc1 cells (Invitrogen). Selection, maintenance of the transformants, and Gal-inducible expression of the recombinant proteins were done according to the manufacturer's instructions (pYES2/NT expression system; Invitrogen). Preparation of yeast membranes containing heterologously expressed GsHET3912, GsA16F5, and GsA14H8 proteins, respectively, was done as described previously (Bouvier et al., 2006), except that the expression was induced at an OD600 of 0.6 in the presence of 2% Gal and cells had been cultured for 8 h at 30°C. Presence of recombinant proteins was verified by standard SDS-PAGE and immunoblot analysis (Sambrook et al., 1995) using an anti-penta-His antibody (Qiagen) and a secondary anti-mouse IgG antibody conjugated with an alkaline phosphatase (Promega).
Reconstitution into Liposomes and Transport Activity Assays
Acetone-washed l-α-phosphatidylcholine (Sigma-Aldrich) had been sonicated (5 min on ice, Branson Sonicator 250, output 2, duty cycle 30%) to a final concentration of 2% (w/v) in 120 mm Tricine-KOH (pH 7.5) and 30 mm internal substrate. Yeast membrane fraction (50 μL) was reconstituted with 950 μL liposome suspension by the freeze-thaw procedure (Kasahara and Hinkle, 1977). After thawing, the proteoliposomes were pulsed 20 times on ice to yield unilamellar vesicles. PD10-desalting columns (GE Healthcare) were preequilibrated with 150 mm Tricine-KOH (pH 7.5) and used to remove nonincorporated substrate from the external medium. If not stated otherwise, all transport studies were initiated with radiolabeled [32P]Pi (GE Healthcare) at a final concentration of 0.25 mm. For each data point, 190 μL proteoliposomes were terminated with 10 μL inhibitor stop solution (200 mm pyridoxal-phosphate, 20 mm 4,4#-diisothiocyanostilbene-2,2#-disulfonic acid, 100 mm Tricine-KOH, pH 8.0). Control experiments with membranes from yeast cells harboring the empty vector were performed in parallel. The addition of inhibitor mix before adding radiolabeled substrate was used to monitor unspecific binding. External phosphate was removed by strong anion-exchange chromatography with AG-1 X8 resin (acetate form, 200–400 mesh, preequilibrated with 150 mm sodium acetate; Bio-Rad). The flow-through of the anion-exchange columns containing the proteoliposomes with the imported radiolabeled phosphate was quantified by a liquid scintillation counting. All substrates used for uptake studies were purchased from Sigma-Aldrich. Kinetic constants were determined by measuring the initial velocity of each experiment. Michaelis-Menten constant (KM) has been analyzed with at least six external phosphate concentrations ranging between 0.05 and 10 mm and competitive inhibition of Pi transport was assessed by the inhibitor constant Ki as described by Dixon (1953). GraphPad-Prism software was used for nonlinear regression fitting of all enzyme kinetic data.
Reverse Transcription-PCR Gene Expression Study
DNAse-treated RNA was used for first-strand cDNA synthesis (SuperScript II first-strand synthesis system; Invitrogen). Oligonucleotide primers for expression studies of the G. sulphuraria genes are summarized in Supplemental Table S1.
Other Methods
Protein concentrations were determined with a standard Bradford assay (Bio-Rad). Proteins in the membrane fractions were delipidated by TCA-acetone extraction before determination (Shultz et al., 2005).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: GsTPT, EU853171; GsPPT, EU853172; and GsGPT, EU853173.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Protein alignment of pPTs from Arabidopsis (At) and G. sulphuraria (Gs).
Supplemental Figure S2. Expression of pPT homologs from G. sulphuraria in yeast.
Supplemental Table S1. Oligonucleotide primers designed in this study.
Supplemental Table S2. Molecular characteristics of the plastidic phosphate transporters GsTPT, GsPPT, and GsGPT from G. sulphuraria.
Supplementary Material
Acknowledgments
We thank Guillaume Barbier and David Gagneul for support with cultivation of G. sulphuraria and the nucleic acid isolation and Prof. Dr. Gerald Schönknecht (Oklahoma State University, Stillwater) for critical discussion.
This work was supported by a grant from the National Science Foundation (award EF–0332882 to A.P.M.W.) and by the Deutsche Forschungsgemeinschaft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Andreas P.M. Weber (andreas.weber@uni-duesseldorf.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adl SM, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, et al (2005) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol 52 399–451 [DOI] [PubMed] [Google Scholar]
- Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T (2004) ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res 32 W390–W393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballicora M, Iglesias A, Preiss J (2004) ADP-glucose pyrophosphorylase: a regulatory enzyme for plant starch synthesis. Photosynth Res 79 1–24 [DOI] [PubMed] [Google Scholar]
- Barbier G, Oesterhelt C, Larson MD, Halgren RG, Wilkerson C, Garavito RM, Benning C, Weber APM (2005. a) Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol 137 460–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier G, Zimmermann M, Weber APM (2005. b) Genomics of the thermo-acidophilic red alga Galdieria sulphuraria. In BH Richard, VL Gilbert, YR Alexei, GR Gladstone, eds, Astrobiology and Planetary Missions, Vol 5906. SPIE, San Diego, pp 590–609
- Bhattacharya D, Archibald JM, Weber APM, Reyes-Prieto A (2007) How do endosymbionts become organelles? Understanding early events in plastid evolution. Bioessays 29 1239–1246 [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Yoon HS, Hackett JD (2004) Photosynthetic eukaryotes unite: Endosymbiosis connects the dots. Bioessays 26 50–60 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Linka N, Isner JC, Mutterer J, Weber APM, Camara B (2006) Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 18 3088–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos DM, Bustamante CA, Iglesias AA (2008) Involvement of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase in response to oxidative stress. J Plant Physiol 165 456–461 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162 156–159 [DOI] [PubMed] [Google Scholar]
- Collen PN, Camitz A, Hancock RD, Viola R, Pedersen M (2004) Effect of nutrient deprivation and resupply of metabolites and enzymes related to carbon allocation in Gracilaria tenuistipitata (Rhodophyta). J Phycol 40 305–314 [Google Scholar]
- Coppin A, Varré JS, Lienard L, Dauvillée D, Guérardel Y, Soyer-Gobillard M-O, Buléon A, Ball S, Tomavo S (2005) Evolution of plant-like crystalline storage polysaccharide in the protozoan parasite Toxoplasma gondii argues for a red alga ancestry. J Mol Evol 60 257–267 [DOI] [PubMed] [Google Scholar]
- Dixon M (1953) The determination of enzyme inhibitor constants. Biochem J 55 170–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicks M, Maurino V, Knappe S, Flugge UI, Fischer K (2002) The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol 128 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K, Kammerer B, Gutensohn M, Arbinger B, Weber A, Hausler RE, Flugge UI (1997) A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 9 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge UI (1999) Phosphate translocators in plastids. Annu Rev Plant Physiol Plant Mol Biol 50 27–45 [DOI] [PubMed] [Google Scholar]
- Flügge UI, Gao W (2005) Transport of isoprenoid intermediates across chloroplast envelope membranes. Plant Biol 7 91–97 [DOI] [PubMed] [Google Scholar]
- Flügge UI, Weber A (1994) A rapid method for measuring organelle-specific substrate transport in homogenates from plant tissues. Planta 194 181–185 [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt R, Stitt M, Heldt HW (1987) Subcellular metabolite levels in spinach leaves: regulation of sucrose synthesis during diurnal alterations in photosynthetic partitioning. Plant Physiol 83 399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross W, Lenze D, Nowitzki U, Weiske J, Schnarrenberger C (1999) Characterization, cloning, and evolutionary history of the chloroplast and cytosolic class I aldolases of the red alga Galdieria sulphuraria. Gene 230 7–14 [DOI] [PubMed] [Google Scholar]
- Gross W, Schnarrenberger C (1995) Heterotrophic growth of two strains of the acido-thermophilic red alga Galdieria sulphuraria. Plant Cell Physiol 36 633–638 [Google Scholar]
- Häusler RE, Schlieben NH, Nicolay P, Fischer K, Fischer KL, Flügge UI (2000) Control of carbon partitioning and photosynthesis by the triose phosphate/phosphate translocator in transgenic tobacco plants (Nicotiana tabacum L.). I. Comparative physiological analysis of tobacco plants with antisense repression and overexpression of the triose phosphate/phosphate translocator. Planta 210 371–382 [DOI] [PubMed] [Google Scholar]
- Heineke D, Riens B, Grosse H, Hoferichter P, Peter U, Flügge UI, Heldt HW (1991) Redox transfer across the inner chloroplast envelope membrane. Plant Physiol 95 1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtgrefe S, Gohlke J, Starmann J, Druce S, Klocke S, Altmann B, Wojtera J, Lindermayr C, Scheibe R (2008) Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol Plant 133 211–228 [DOI] [PubMed] [Google Scholar]
- Kammerer B, Fischer K, Hilpert B, Schubert S, Gutensohn M, Weber A, Flügge UI (1998) Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6-phosphate/phosphate antiporter. Plant Cell 10 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Hinkle PC (1977) Reconstitution and purification of the D-glucose transporter from human erythrocytes. J Biol Chem 252 7384–7390 [PubMed] [Google Scholar]
- Knappe S, Flugge UI, Fischer K (2003. a) Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol 131 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S, Lottgert T, Schneider A, Voll L, Flugge UI, Fischer K (2003. b) Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis-AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J 36 411–420 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10 1–6 [DOI] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge UI, Schneider A (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt C, Klocke S, Holtgrefe S, Linke V, Weber APM, Scheibe R (2007) Redox regulation of chloroplast enzymes in Galdieria sulphuraria in view of eukaryotic evolution. Plant Cell Physiol 48 1359–1373 [DOI] [PubMed] [Google Scholar]
- Parsley K, Hibberd J (2006) The Arabidopsis PPDK gene is transcribed from two promoters to produce differentially expressed transcripts responsible for cytosolic and plastidic proteins. Plant Mol Biol 62 339–349 [DOI] [PubMed] [Google Scholar]
- Patron NJ, Keeling PJ (2005) Common evolutionary origin of starch biosynthetic enzymes in green and red algae. J Phycol 41 1131–1141 [Google Scholar]
- Plaxton WC (1996) The organization and regulation of plant glycolysis. Annu Rev Plant Physiol Plant Mol Biol 47 185–214 [DOI] [PubMed] [Google Scholar]
- Reichert A, Dennes A, Vetter S, Scheibe R (2003) Chloroplast fructose 1,6-bisphosphatase with changed redox modulation: comparison of the Galdieria enzyme with cysteine mutants from spinach. Biochim Biophys Acta 1645 212–217 [DOI] [PubMed] [Google Scholar]
- Reiter WD (2002) Biosynthesis and properties of the plant cell wall. Curr Opin Plant Biol 5 536–542 [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber APM, Bhattacharya D (2007) The origin and establishment of the plastid in algae and plants. Annu Rev Genet 41 147–168 [DOI] [PubMed] [Google Scholar]
- Richards T, Dacks JB, Campbell SA, Blanchard JL, Foster PG, McLeod R, Roberts CW (2006) Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot Cell 5 1517–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius S, Casati P, Iglesias A, Gomez-Casati D (2006) Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol Biol 61 945–957 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1995) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- Schlichting R, Bothe H (1993) The cyanelles (organelles of a low evolutionary scale) possess a phosphate-translocator and a glucose-carrier in Cyanophora paradoxa. Bot Acta 106 428–434 [Google Scholar]
- Shultz RW, Settlage SB, Hanley-Bowdoin L, Thompson WF (2005) A trichloroacetic acid-acetone method greatly reduces infrared autofluorescence of protein extracts from plant tissue. Plant Mol Biol Rep 23 405–409 [Google Scholar]
- Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30 1126–1149 [DOI] [PubMed] [Google Scholar]
- Tegeder M, Weber APM (2006) Metabolite transporters in the control of plant primary metabolism. In WC Plaxton, MT McManus, eds, Control of Primary Metabolism in Plants. Blackwell Publishing Ltd, Oxford, pp 85–120
- Trimming BA, Emes MJ (1993) Glycolytic enzymes in non-photosynthetic plastids of pea (Pisum sativum L.) roots. Planta 190 439–445 [Google Scholar]
- Tyra H, Linka M, Weber APM, Bhattacharya D (2007) Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol 8 R212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola R, Nyvall P, Pedersén M (2001) The unique features of starch metabolism in red algae. Proc R Soc Lond B Biol Sci 268 1417–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll L, Hausler RE, Hecker R, Weber A, Weissenbock G, Fiene G, Waffenschmidt S, Flugge UI (2003) The phenotype of the Arabidopsis cue1 mutant is not simply caused by a general restriction of the shikimate pathway. Plant J 36 301–317 [DOI] [PubMed] [Google Scholar]
- Weber APM, Fischer K (2007) Making the connections—the crucial role of metabolite transporters at the interface between chloroplast and cytosol. FEBS Lett 581 2215–2222 [DOI] [PubMed] [Google Scholar]
- Weber APM, Linka M, Bhattacharya D (2006) Single, ancient origin of a plastid metabolite translocator family in plantae from an endomembrane-derived ancestor. Eukaryot Cell 5 609–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber APM, Oesterhelt C, Gross W, Bräutigam A, Imboden L, Krassovskaya I, Linka N, Truchina J, Schneidereit J, Voll H, et al (2004) EST-analysis of the thermo-acidophilic red microalga Galdieria sulphuraria reveals potential for lipid A biosynthesis and unveils the pathway of carbon export from rhodoplasts. Plant Mol Biol 55 17–32 [DOI] [PubMed] [Google Scholar]
- Weber APM, Schwacke R, Flügge UI (2005) Solute transporters of the plastid envelope membrane. Annu Rev Plant Biol 56 133–164 [DOI] [PubMed] [Google Scholar]
- Yang Y, Kwon HB, Peng HP, Shih MC (1993) Stress responses and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiol 101 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D (2004) A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol 21 809–818 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM (2007) The diurnal metabolism of leaf starch. Biochem J 401 13–28 [DOI] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57 805–836 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.