Abstract
The degradation pathway of glutathione (GSH) in plants is not well understood. In mammals, GSH is predominantly metabolized through the γ-glutamyl cycle, where GSH is degraded by the sequential reaction of γ-glutamyl transpeptidase (GGT), γ-glutamyl cyclotransferase, and 5-oxoprolinase to yield glutamate (Glu) and dipeptides that are subject to peptidase action. In this study, we examined if GSH is degraded through the same pathway in Arabidopsis (Arabidopsis thaliana) as occurs in mammals. In Arabidopsis, the oxoprolinase knockout mutants (oxp1-1 and oxp1-2) accumulate more 5-oxoproline (5OP) and less Glu than wild-type plants, suggesting substantial metabolite flux though 5OP and that 5OP is a major contributor to Glu steady-state levels. In the ggt1-1/ggt4-1/oxp1-1 triple mutant with no GGT activity in any organs except young siliques, the 5OP concentration in leaves was not different from that in oxp1-1, suggesting that GGTs are not major contributors to 5OP production in Arabidopsis. 5OP formation strongly tracked the level of GSH in Arabidopsis plants, suggesting that GSH is the precursor of 5OP in a GGT-independent reaction. Kinetics analysis suggests that γ-glutamyl cyclotransferase is the major source of GSH degradation and 5OP formation in Arabidopsis. This discovery led us to propose a new pathway for GSH turnover in plants where GSH is converted to 5OP and then to Glu by the combined action of γ-glutamyl cyclotransferase and 5-oxoprolinase in the cytoplasm.
Glutathione (GSH), the tripeptide γ-Glu-Cys-Gly, plays various important roles in plants. Through its thiol residue, it performs redox reaction, enabling it to be a regulator of redox homeostasis (Foyer and Noctor, 2005). It detoxifies photosynthetically generated hydrogen peroxide as a component of the ascorbate-GSH cycle (Noctor and Foyer, 1998). GSH is polymerized to form phytochelatins, (γ-Glu-Cys)2-11-Gly, that chelate heavy metals and then transport them into the vacuole, where they are metabolically inactive (Cobbett and Goldsbrough, 2002). Toxic xenobiotics, including herbicides, are conjugated with GSH by glutathione S-transferases and sequestered into the vacuole (Marrs, 1996). GSH is also involved in controlling cell size and root development by regulating the cell cycle (Vernoux et al., 2000; Xiang et al., 2001). GSH exists in high concentration in plant tissues and acts as a substrate for central biochemicals, particularly Cys, and Cys availability for protein synthesis and as a precursor for metabolites may be related to the rate of GSH degradation (Leustek et al., 2000).
GSH is synthesized from Glu, Cys, and Gly in a two-step, ATP-dependent reaction. First, γ-glutamyl-Cys (γ-EC) is synthesized by γ-EC synthetase (Hell and Bergmann, 1990), and then Gly is incorporated by GSH synthetase (Wang and Oliver, 1996). The activity of γ-EC synthetase is regulated by GSH levels (Hell and Bergmann, 1990; Jez et al., 2004). In Arabidopsis (Arabidopsis thaliana), γ-EC synthetase and GSH synthetase are encoded by single genes, GSH1 (May and Leaver, 1994) and GSH2 (Wang and Oliver, 1996), respectively, and expression of both genes is regulated transcriptionally and translationally by heavy metals, jasmonic acid, and oxidative stress (Xiang and Oliver, 1998). γ-EC synthetase is exclusively localized in the plastids, whereas GSH synthetase, albeit also present in the chloroplasts, is, to a large extent, a cytosolic protein (Wachter et al., 2005). Recently, it was reported that restricting glutathione synthesis to the cytosol is sufficient for normal plant development (Pasternak et al., 2008).
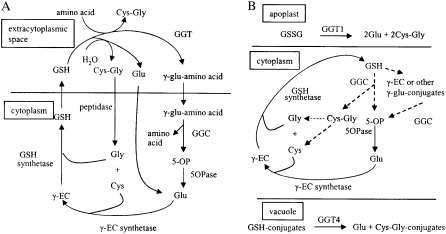
Compared with GSH synthesis, its metabolism is less understood in plants. In animals, the γ-glutamyl cycle is responsible for GSH metabolism (Meister and Larsson, 1995; Fig. 1A). In this cycle, extracellular GSH is broken down and constituent amino acids are absorbed into the cell and used to reform GSH. γ-Glutamyl transpeptidase (GGT) initiates GSH breakdown and is on the plasma membrane, with its active site in the extracellular space. GGT transfers γ-linked Glu from GSH to water or another amino acid, producing either Glu or a γ-glutamyl amino acid, respectively. The resulting Glu and γ-glutamyl amino acids are transported back into the cell. As originally proposed, the movement of this γ-glutamyl amino acid into the cell was viewed as an important route of amino acid uptake (Meister, 1973), but some authors have presented experimental data suggesting that the most common product of the GGT reaction is free Glu (Hanigan and Pitot, 1985), thus decreasing the role in amino acid transport without affecting its importance in GSH movement. Inside the cell, γ-glutamyl cyclotransferase (GGC) converts any γ-glutamyl amino acids formed into 5-oxoproline (5OP; synonyms: pyroglutamate and pyrrolidone carboxylate) and the free amino acid. 5-Oxoprolinase (5OPase) hydrolyzes 5OP to Glu in an ATP-dependent reaction. Cys-Gly produced from GSH by GGT is hydrolyzed at the cell surface to Cys and Gly by a dipeptidase before these components reenter the cell. Glu, Cys, and Gly released from GSH are recycled for GSH synthesis in the cell by γ-EC synthetase and GSH synthetase. Activities of GGT, GGC, and 5OPase are highest in the kidney along with other secretory tissues. In the kidney, GSH is filtered from the blood into the excretory stream. The γ-glutamyl cycle is responsible for reabsorbing the GSH by first triggering its breakdown and then, after reabsorption of the component amino acids, catalyzing their conversion back to GSH. This idea is supported in GGT knockout mice that showed glutathionuria, Cys deficiency, and growth and reproductive defects that can be reversed by feeding acetylcysteine (Lieberman et al., 1996; Harding et al., 1997; Kumar et al., 2000).
Figure 1.
Models for GSH degradation in animals and plants. The solid lines are experimentally confirmed pathways, and the dashed lines are proposed pathways. A, The γ-glutamyl cycle as proposed for animals. GSH is degraded in the extracellular space by GGT, and the constituent amino acids are transported back to the cytoplasm to resynthesize GSH. The GSH hydrolase activity of GGT predominates over the transpeptidase reaction. B, GSH metabolism proposed in Arabidopsis. GSH degradation occurs in the cytoplasm by GGC, with only minor GGT involvement in total GSH turnover. Roles of the GGTs are limited to the extracellular space, such as the degradation of oxidized GSH (GSSG) by GGT1 in the apoplast and the degradation of GSH conjugates by GGT4 in the vacuole.
In plants that lack a formal excretory system, the presence of a γ-glutamyl cycle has yet to be determined. All of the enzymes in this cycle are reported in plants except the Cys-Gly dipeptidase. Soluble and bound GGT activity has been detected in tomato (Solanum lycopersicum), onion (Allium cepa), radish (Raphanus sativus), and Arabidopsis (Martin and Slovin, 2000; Storozhenko et al., 2002; Nakano et al., 2004; Shaw et al., 2005; Grzam et al., 2007; Martin et al., 2007; Ohkama-Ohtsu et al., 2007a, 2007b). Recently, we analyzed the functions of Arabidopsis GGT proteins using knockout mutants (Ohkama-Ohtsu et al., 2007a, 2007b). There are three functional GGT proteins in Arabidopsis, GGT1, GGT2, and GGT4. GGT4 (At4g29210) was originally called GGT 3 in our earlier papers (Ohkama-Ohtsu et al., 2007a, 2007b) but was renamed here to agree with the nomenclature of Martin et al. (2007), who simultaneously discovered it function (Grzam et al., 2007). GGT1 mitigates oxidative stress by degrading oxidized GSH in the extracellular space, and GGT2 (also found in the apoplast) may be involved in GSH transport into siliques (Ohkama-Ohtsu et al., 2007a). GGT4 is located in the vacuole, where it is responsible for the degradation of GSH conjugates formed by glutathione S-transferase reactions (Ohkama-Ohtsu et al., 2007b).
Less is known about the function of 5OPase and GGC in plants. 5OPase activity was first detected in higher plants by Mazelis and Pratt (1976). They showed conversion of isotope-labeled 5OP to Glu in both monocots and dicots. Plant 5OPase was first purified from wheat germ (Triticum aestivum) by Mazelis and Creveling (1978), who also observed 5OPase activity in various organs, including roots and seeds, from a range of species. Subcellular localization of 5OPase was analyzed in cultured tobacco (Nicotiana tabacum) cells by Rennenberg et al. (1981). Almost all of the activity was reported to be in the soluble cytoplasmic fraction. Feeding isotope-labeled GSH to cultured tobacco cells demonstrated that 5OP is a degradation product of GSH (Rennenberg et al., 1980). GGC was studied in tobacco suspension cultures by Steinkamp and Rennenberg (1985, 1987), who suggested that this soluble enzyme is also localized in the cytoplasm.
Here, we studied 5OP synthesis and degradation using knockout mutants of the Arabidopsis 5OPase. Surprisingly, millimolar concentrations of 5OP accumulate in these knockout mutants, suggesting substantial carbon and nitrogen flow through 5OP to Glu in Arabidopsis. Combining the 5OPase with GGT knockouts demonstrated that while 5OP is made from GSH and γ-EC, it is not derived via the action of GGT. Furthermore, GGC enzyme kinetic analyses suggested that GSH is a more important source of 5OP than γ-EC under physiological conditions. These results raise questions about the importance of the traditional γ-glutamyl cycle in plants and identify a new pathway that may also be important in other organisms.
RESULTS
The Arabidopsis 5OPase Gene
In Arabidopsis, a single gene (At5g37830) shows high homology to 5OPase from animals and was named OXOPROLINASE1 (OXP1). The translated sequence of Arabidopsis OXP1 is 57% identical and 71% to 72% similar to the protein from the sequences of Rattus norvegicus, Mus musculus, Homo sapiens, and Bos taurus. High similarity to animal 5OPases is observed throughout the Arabidopsis protein, including the N terminus (Supplemental Fig. S1), suggesting that Arabidopsis 5OPase, like the animal proteins (Meister and Larsson, 1995), localizes to the cytoplasm. This was supported by computer prediction using PSORT (http://psort.hgc.jp/) and TargetP (http://www.cbs.dtu.dk/services/TargetP/) and agrees with the localization in tobacco (Rennenberg et al., 1981).
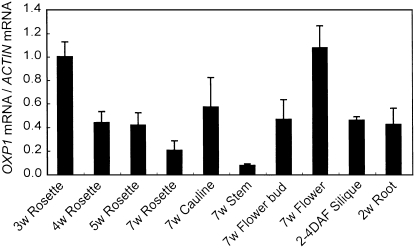
Expression of the OXP1 gene was analyzed using real-time PCR. OXP1 transcript was observed in all organs examined, with the highest levels in flowers and 3-week-old rosette leaves and the lowest in stems (Fig. 2).
Figure 2.
Organ-specific expression pattern for OXP1. The accumulation of OXP1 mRNAs in Columbia wild-type plants is shown. Rosette leaves from soil-grown 3-, 4-, 5-, and 7-week-old plants, cauline leaves, stems, and flowers from soil-grown 7-week-old plants, and siliques (2–4 d after flowering [DAF]) and roots of liquid-cultured 12-d-old plants were harvested and total RNA was extracted. RNA was reverse transcribed and subjected to real-time PCR analysis to monitor the amplification of OXP1 cDNAs. The accumulation of OXP1 mRNAs relative to that of ACTIN was determined. Values are normalized with the means of rosette leaves from 3-week-old plants. Means and sd values of three independent biological replications are shown.
The oxp1 Mutants
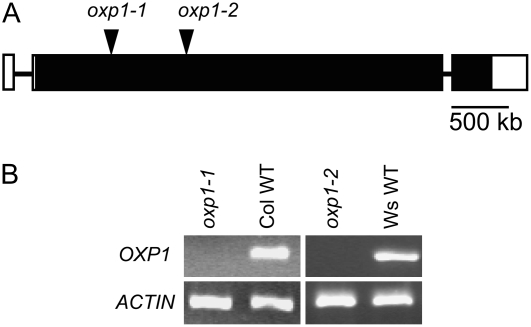
Knockout mutants are effective tools for analyzing the function of genes in planta. Two allelic mutants disrupting OXP1 were obtained and designated oxp1-1 and oxp1-2 (Fig. 3A). oxp1-1 is in the Columbia background with a T-DNA insertion in the second exon of OXP1. oxp1-2 is in the Wassilewskija background and also has a T-DNA insertion in the second exon. OXP1 mRNA was not detected by reverse transcription (RT)-PCR analysis either in the oxp1-1 or oxp1-2 plants (Fig. 3B). In order to verify that oxp1 is the only gene encoding 5OPase activity, we measured the ability of dialyzed protein extracts from wild-type and oxp1-1 plants to convert 5OP into Glu (Table I). Both wild-type and oxp1-1 extracts formed a small amount of Glu (possibly released from proteins) in the absence of added 5OP, but only the wild-type line was able to convert added 5OP to Glu. The lack of detectable 5OPase activity in the oxp1-1 mutant plants verifies that OXP1 is the only gene encoding 5OPase in Arabidopsis and that no other enzyme catalyzes this reaction.
Figure 3.
The oxp1-1 and oxp1-2 knockout mutants. A, Structure of the OXP1 gene in Arabidopsis. The white boxes represent untranslated regions, the black boxes represent coding regions, and the black lines represent introns. The T-DNA insertions in oxp1-1 and oxp1-2 are shown. B, OXP1 transcript accumulation. RNA was isolated from wild-type (WT) and knockout mutant plants and subjected RT-PCR.
Table I.
5OPase activity in wild-type and oxp1-1 plants
Glu production from 5OP was measured in an in vitro 5OPase activity assay using extracts from 20-d-old leaves. The means ± sd of three biological replicates are shown.
| Line | With Substrate | Without Substrate |
|---|---|---|
| nmol mg−1 protein min−1 | ||
| Columbia wild type | 0.39 ± 0.04a | 0.04 ± 0.02 |
| oxp1-1 | 0.04 ± 0.02 | 0.03 ± 0.01 |
Significant difference between with and without substrate (one-tailed Student's t test, P < 0.05).
Both the oxp1-1 and oxp1-2 plants were grown on soil until they set seeds. The oxp1-2 mutant flowered approximately 5 d earlier than Wassilewskija wild-type plants, but the early-flowering phenotype is unlikely to be due to disruption of the OXP1 gene because this phenotype was not observed in oxp1-1. Root growth was not significantly different from that of wild-type plants in both oxp1-1 and oxp1-2 when they were grown vertically on agar plates for 14 d (data not shown). No morphological phenotypes were observed in oxp1-1, so this allele was used in most metabolism studies. Neither mutant line showed differences in thiol levels and composition from wild-type plants, which is not unexpected given that this disruption occurred relatively late in the GSH catabolic pathway.
5OP Accumulates in the oxp1 Mutants
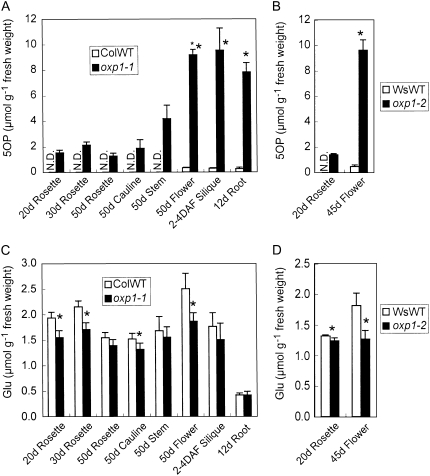
5OP was measured by HPLC (Nishimura et al., 2001) after ethanolic plant extracts were passed through a Dowex 50 (H+) column (Orlowski et al., 1969). 5OP distributions in both Columbia wild-type and oxp1-1 plants are shown in Figure 4A. In leaves, 5OP was not detected in wild-type plants, while it accumulated to approximately 2 μmol g−1 fresh weight in the oxp1-1 plants, where its further metabolism was blocked. In stems, 5OP was undetectable in wild-type plants but accumulated to 4 μmol g−1 fresh weight in the mutant. In sink tissues, roots, flowers, and siliques, 5OP was barely detectable in wild-type plants but accumulated to nearly 10 μmol g−1 fresh weight in the oxp1-1 plants. Very similar results were seen in leaves and flowers of the oxp1-2 plants (Fig. 4B). Given the lack of apparent phenotype, the accumulation of high 5OP levels in oxp1-1 suggests that this intermediate is not toxic. The identity and amount of 5OP measured by HPLC was confirmed using capillary electrophoresis time-of-flight mass spectrometry (CE-TOF/MS). The 5OP concentration in 30-d-old leaves determined by CE-TOF/MS was 3.1 ± 0.2 μmol g−1 fresh weight (n = 6) in the oxp1-1 plants. While 5-OP was undetectable in 30-d-old leaves of Columbia wild-type plants by HPLC, the more sensitive CE-TOF/MS method confirmed a background level of 0.5 ± 0.1 μmol g−1 fresh weight (n = 6).
Figure 4.
5OP and Glu concentrations in wild-type (WT) and oxp1 plants. A, 5OP in Columbia (Col) wild-type and oxp1-1 plants. B, 5OP in Wassilewskija (Ws) wild-type and oxp1-2 plants. C, Glu in Col wild-type and oxp1-1 plants. D, Glu in Ws wild-type and oxp1-2 plants. Roots of liquid-cultured 12-d-old plants, rosette leaves from soil-grown 20-, 30-, and 50-d-old plants, and cauline leaves, stems, and flowers from soil-grown 50-d-old plants and siliques (2–4 d after flowering [DAF]) were harvested from Col wild-type and oxp1-1 plants and assayed for 5OP and Glu. Rosette leaves from soil-grown 20-d-old plants and flowers from soil-grown 45-d-old plants were harvested from Ws wild-type and oxp1-2 plants and assayed for 5OP and Glu. The means ± sd of three (5OP) or four (Glu) biological replicates are shown. Asterisks indicate significant differences between Col wild-type and oxp1-1 plants or between Ws wild-type and oxp1-2 plants. (P < 0.05, one-tailed Student's t test). N.D., Not detected.
Glu Concentrations Decrease in the oxp1 Mutants
5OPase hydrolyzes 5OP to Glu; thus, analysis of Glu levels in the oxp1 plants would provide some insights into the contribution of this reaction to Glu formation. Glu concentrations in 20- and 30-d-old rosette leaves, 50-d-old cauline leaves, and flowers were significantly (14%–30%) lower in the oxp1-1 plants compared with wild-type plants (Fig. 4C). In all other organs except roots, Glu concentrations were also lower in oxp1-1 plants, but the differences were not statistically significant. The same results were observed in leaves and flowers from oxp1-2 plants (Fig. 4D). These results suggest that 5OPase contributes up to 30% of the steady-state Glu level in Arabidopsis.
Growth studies showed that despite the decreased Glu levels, the oxp1 plants were no more sensitive to nitrogen deficiency than wild-type plants and that the levels of the key amino acids Asp, Ser, Gln, Gly, Thr, Arg, and Ala were unchanged (data not shown).
5OP Concentrations Are Not Changed by ggt Mutations
In order to determine the relative contributions of extracellular GGT1 and vacuolar GGT4 to 5OP production, the ggt1-1/oxp1-1 (double), ggt4-1/oxp1-1 (double), and ggt1-1/ggt4-1/oxp1-1 (triple) mutants were constructed by crossing mutant plants. Since GGT2 is only expressed in young siliques (Ohkama-Ohtsu et al., 2007a), it was not included in this study. As shown in Supplemental Table S1 and our previous reports (Ohkama-Ohtsu et al., 2007a, 2007b), GGT activity in the leaves from ggt1 and ggt4 plants was about 10% and 90% of wild-type GGT activity, respectively, while the ggt1/ggt4 double mutant had no detectable GGT activity. As the ggt1-1 and ggt4-1 plants are in the Landsberg background (Ohkama-Ohtsu et al., 2007a, 2007b) and oxp1-1 is in Columbia, oxp1-1 was crossed to Landsberg wild-type plants, and a mixture of F3 seeds derived from five F2 plants carrying the homozygous oxp1-1 mutation was used as a control. 5OP concentrations in leaves of the 20-d-old ggt1-1/oxp1-1, ggt4-1/oxp1-1, and ggt1-1/ggt4-1/oxp1-1 plants (Table II) were not significantly different from those in the oxp1-1 plants. These results clearly demonstrate that GGTs are not in the major pathway for 5OP production in Arabidopsis.
Table II.
The effect of the ggt1 and ggt4 mutations on 5OP concentrations in oxp1-1 Arabidopsis leaves
5OP concentrations in leaves of 20-d-old plants were measured by HPLC. The means ± sd of five biological replicates are shown. None of these values was significantly different by a one-tailed Student's t test (P > 0.05).
| Line | 5OP |
|---|---|
| oxp1-1 (Landsberg) | 1,370 ± 30 |
| ggt1-1/oxp1-1 | 1,440 ± 100 |
| ggt4-1/oxp1-1 | 1,390 ± 60 |
| ggt1-1/ggt4-1/oxp1-1 | 1,340 ± 50 |
5OP Decreases in Plants with Less GSH or γ-EC
Having demonstrated that GGTs were not significant contributors to 5OP formation, this means that the 5OP was not being produced from the γ-glutamyl amino acid that can be produced by the GGT reaction in the γ-glutamyl cycle (Meister and Larsson, 1995). The roles of the major endogenous γ-glutamyl amino acids, GSH and γ-EC, in 5OP production were then determined. To know whether 5OP came from GSH or γ-EC in Arabidopsis, an inhibitor of γ-EC synthetase, buthionine sulfoximine (BSO), was supplied to oxp1-1 mutant plants grown in liquid culture. GSH and 5OP concentrations were determined in 14-d-old seedlings that were treated with BSO for 1 to 4 d. Control plants were cultured without BSO for 14 d. In a preliminary experiment, GSH concentrations were higher in plants treated with BSO for 3 or 4 d compared with plants treated for 1 or 2 d, so fresh BSO was added every 2 d. Compared with nontreated plants, GSH and γ-EC concentrations were substantially decreased in plants treated with BSO. The 5OP level was lowered in all plants treated with BSO, and the decrease was significant after 2 d (Table III). The decrease in the percentage of 5OP is smaller than that for GSH or γ-EC (6% for 5OP versus 81% for GSH and 54% for γ-EC at day 1) because of the high background 5OP concentration in oxp1-1 plants at the time of BSO addition and the fact that no further 5OP metabolism occurs in this mutant. Although the ratio is different, the absolute decreases in 5OP and GSH levels at day 1 are very similar.
Table III.
5OP and thiol concentrations in BSO-treated oxp1-1 seedlings
BSO was added to liquid culture at 1 mm for the indicated days. All plants were 14 d old when harvested. Where indicated, BSO was added 1 to 4 d before the plants were harvested and the metabolites were measured. The 40% decrease in the 5OP concentration in the oxp1-1 mutant was caused by the 2.4-fold increase in plant biomass (1.19 ± 0.07 g per 30 plants at 10 d, 2.86 ± 0.10 g per 30 plants at 14 d; n = 6) during the of BSO treatment but not by 5OP metabolism in the oxp1-1 mutant. The means ± sd of five biological replicates are shown. N.D., Not detected.
| Treatment | 5OP | GSH | γ-EC | Cys |
|---|---|---|---|---|
| nmol g−1 fresh weight | ||||
| No BSO | 5,280 ± 450 | 369 ± 55 | 12.2 ± 2.4 | 37 ± 5 |
| 1 d | 4,990 ± 570 | 71 ± 26a | 5.6 ± 1.9a | 57 ± 4a |
| 2 d | 4,740 ± 240a | 36 ± 5a | N.D. | 52 ± 10a |
| 3 d | 3,970 ± 390a | 27 ± 3a | N.D. | 62 ± 9a |
| 4 d | 3,160 ± 280a | 31 ± 4a | N.D. | 64 ± 5a |
Significant difference from nontreated plants (one-tailed Student's t test, P < 0.05).
This correlation between GSH/γ-EC and 5OP levels was further confirmed in the cad2-1/oxp1-1 double mutants. cad2-1 (Columbia background) has a defect in the γ-EC synthetase gene, and both GSH and γ-EC levels were less than half compared with wild-type plants (Cobbett et al., 1998). In agreement with the previous report, GSH and γ-EC concentrations in leaves of the cad2-1/oxp1-1 double mutant were 21% and 39% of those in the oxp1-1 single mutant (Table IV). 5OP concentration in the cad2-1/oxp1-1 double mutants was decreased to 46% of that in the oxp1-1 single mutant, again demonstrating a tight coupling between GSH/γ-EC and 5OP levels.
Table IV.
The effect of the cad2 mutation on 5OP and thiol concentrations
5OP and thiol concentrations in leaves of 20-d-old plants were determined. The means ± sd of four biological replicates are shown.
| Line | 5OP | GSH | γ-EC | Cys |
|---|---|---|---|---|
| nmol g−1 fresh weight | ||||
| oxp1-1 | 1,470 ± 110 | 293 ± 29 | 12.4 ± 2.2 | 13 ± 2 |
| cad2-1/oxp1-1 | 670 ± 40a | 61 ± 6a | 4.8 ± 1.7a | 60 ± 15a |
Significant difference between oxp1-1 and cad2-1/oxp1-1 plants (one-tailed Student's t test, P < 0.05).
Both the results with BSO and with the cad2-1/oxp1-1 plants suggested that 5OP is produced from GSH and/or γ-EC, although the relative contribution of each could not be determined. To address this issue, an experiment was designed where GSH levels could be increased without increasing γ-EC. To do this, liquid-cultured oxp1-1 plants were supplemented with 2 mm GSH and incubated for 20 h. Compared with plants without GSH supplementation, the 5OP concentration was significantly increased in the GSH-fed oxp1-1 mutants (Table V). In the GSH-treated oxp1-1 plants, GSH, Cys-Gly, and Cys were also increased, but γ-EC was not changed. GSH was also provided to the ggt1-1/ggt4-1/oxp1-1 triple mutant to eliminate the possibility that GSH was converted to γ-glutamyl amino acids by GGTs that then acted as 5OP precursors. In the ggt1-1/ggt4-1/oxp1-1 triple mutant, 5OP was also increased following GSH treatment, while the γ-EC levels were not (Table V). The increases in GSH and 5OP were similar in magnitude.
Table V.
The effect of GSH application on 5OP concentrations
Nine-day-old liquid-cultured plants were supplemented with 2 mm GSH and incubated for 20 h. The means ± sd of five biological replicates are shown. N.D., Not detected.
| Line | GSH Added | 5OP | GSH | γ-EC | Cys-Gly | Cys |
|---|---|---|---|---|---|---|
| nmol g−1 fresh weight | ||||||
| oxp1-1 | 0 mm | 4,880 ± 990 | 354 ± 29 | 14.8 ± 1.8 | N.D. | 47 ± 8 |
| 2 mm | 6,150 ± 590a | 1,519 ± 144a | 14.8 ± 1.9 | 10.4 ± 0.9 | 293 ± 54a | |
| ggt1-1/ggt4-1/oxp1-1 | 0 mm | 4,150 ± 340 | 368 ± 49 | 16.0 ± 2.1 | N.D. | 43 ± 8 |
| 2 mm | 6,060 ± 570a | 1,347 ± 55a | 15.1 ± 0.8 | N.D. | 90 ± 10a | |
Significant difference from plants without GSH (one-tailed Student's t test, P < 0.05).
Finally, the ability of plants to degrade GSH in the absence of GGTs was shown by adding BSO to the ggt1-1/ggt4-1 double knockout and wild-type plants. GSH concentrations in liquid-cultured plants were determined at 8, 12, 16, and 20 h after addition of BSO. As shown in Table VI, the rate of GSH degradation was similar between wild-type and ggt1-1/ggt4-1 plants, suggesting that under these conditions the GGTs were not contributing significantly to GSH turnover.
Table VI.
The rate of GSH degradation in Landsberg wild-type and ggt1-1/ggt4-1 mutant plants
Ten-day-old liquid-cultured plants were supplemented with BSO at 1 mm and incubated for 8, 12, 16, or 20 h, then concentrations of thiols were determined. The means ± sd of five biological replicates are shown.
| Line | BSO | GSH | γ-EC | Cys |
|---|---|---|---|---|
| nmol g−1 fresh weight | ||||
| Landsberg wild type | 0 mm | 443 ± 43 | 10.0 ± 1.0 | 40 ± 8 |
| 1 mm, 8 h | 297 ± 24 | 8.6 ± 1.9 | 42 ± 10 | |
| 1 mm, 12 h | 197 ± 52 | 7.5 ± 2.0 | 69 ± 5 | |
| 1 mm, 16 h | 132 ± 19 | 7.4 ± 2.5 | 80 ± 10 | |
| 1 mm, 20 h | 113 ± 27 | 6.3 ± 0.9 | 105 ± 24 | |
| ggt1-1/ggt4-1 | 0 mm | 404 ± 49 | 13.4 ± 2.9 | 39 ± 9 |
| 1 mm, 8 h | 267 ± 73 | 9.2 ± 0.4 | 48 ± 10 | |
| 1 mm, 12 h | 217 ± 58 | 8.6 ± 1.9 | 71 ± 2 | |
| 1 mm, 16 h | 149 ± 23 | 8.6 ± 0.9 | 86 ± 14 | |
| 1 mm, 20 h | 105 ± 20 | 8.5 ± 1.1 | 116 ± 26 | |
Taken together, these results demonstrate that GSH is the major source for 5OP synthesis, this reaction does not involve GGT, and GGT is not the prominent reaction of GSH turnover.
GGC Enzyme Kinetics with GSH and γ-EC
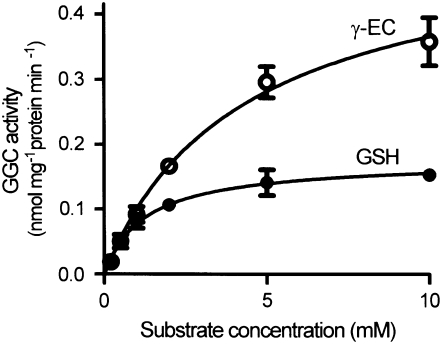
The enzyme kinetics of GGC were studied to determine the relative potential for GSH and γ-EC to serve as sources of 5OP in Arabidopsis tissues. In order to eliminate the possibility that GSH was converted to other γ-glutamyl amino acids by GGTs, the enzyme extract was prepared from liquid-cultured ggt1-1/ggt4-1 double mutant seedlings that do not have detectable GGT activity. Measurements of GGC activities at different substrate concentrations showed that GGC activity exhibited Michaelis-Menten kinetics with either GSH or γ-EC (Fig. 5). The calculated Km and Vmax values were 1.28 mm and 0.18 nmol mg−1 protein min−1 for GSH and 4.24 mm and 0.52 nmol mg−1 protein min−1 for γ-EC.
Figure 5.
GGC enzyme activity. GGC activities in 10-d-old liquid-cultured ggt1-1/ggt4-1 double knockout plants were determined with GSH (black circles) or γ-EC (white circles) as a substrate. The means ± sd of three experiments are shown.
Glutaminyl Cyclotransferase Is Not a Major Contributor of 5OP Production
The gene corresponding to GGC has never been identified from plants. Genes encoding glutaminyl cyclotransferase (QCT), which converts Gln and N-terminal glutaminyl residues in peptides to 5OP and 5OP residues, were identified from papaya (Carica papaya; Dahl et al., 2000) and Arabidopsis (Schilling et al., 2007). The papaya and Arabidopsis QCT were much more active in converting N-terminal Gln than Glu to 5OP (Schilling et al., 2004, 2007). We obtained an Arabidopsis T-DNA knockout mutant for QCT (At4g25720; Supplemental Fig. S2, A and B) to examine if this gene corresponds to GGC activity. GGC activity in the At4g25720 mutant was not different from that in wild-type plants (Supplemental Fig. S2C), suggesting that GGC is not encoded by this gene. Furthermore, we examined if QCT contributes to 5OP production. There was no significant difference in 5OP concentration between oxp1-2 and the At4g25720/oxp1-2 mutant (Supplemental Fig. S2D), indicating that At4g25720 is not involved in the major pathway for 5OP production.
DISCUSSION
In this study, we analyzed the synthesis and metabolism of 5OP in Arabidopsis using knockout mutants for 5OPase, the enzyme responsible for 5OP conversion to Glu. Disruption of OXP1, the gene for 5OPase, resulted in a large increase in 5OP levels and in some organs significant decreases in Glu levels. It was interesting how much impact blocking 5OP metabolism had on steady-state Glu levels. While the lack of effect of the oxp1-1 mutant on Glu levels in roots suggests that this tissue can compensate for the disruption in 5OP metabolism, the 14% to 30% decrease in Glu levels in leaves, flowers, and siliques suggests that 5OP is a major source of Glu formation in these organs. Future flux studies will be necessary to estimate the actual amount of Glu derived from 5OP.
In the oxp1 mutants, 5OP accumulated to millimolar levels in all tissues tested, with the highest levels in roots, flowers, and young siliques. Genetic and metabolic experiments demonstrated that GSH is the predominant source of 5OP. It is possible that γ-EC is also contributing, but since this intermediate normally occurs at concentrations that are 10- to 20-fold lower than GSH and since 5OP synthesis seemed to track GSH level and not γ-EC concentration, its role is likely to be relatively minor. Assuming exponential growth and a constant rate of 5OP synthesis, oxp1 leaves accumulated 5OP at a rate of about 40 nmol g−1 fresh weight h−1 (Fig. 4), a value that agrees reasonably well with observed in vivo rates of GSH breakdown when GSH synthesis is blocked by BSO, about 30 nmol g−1 fresh weight h−1 (Table III). The similarity between rates of GSH degradation and 5OP accumulation suggests that the majority of GSH is metabolized through 5OP and that the major portion of 5OP is derived from GSH. These results also suggest that mechanisms for GSH degradation that do not proceed through 5OP, such as the GSH hydrolase activity of GGT or the newly discovered DUG hydrolase system in yeast (Ganguli et al., 2007), are of limited importance in total GSH turnover in plants.
GGC kinetic analyses supported the idea that GSH is a more important source of 5OP than γ-EC (Fig. 5). Although the Vmax for GGC was 3.3 times higher with γ-EC than with GSH, the Km was 2.9 times lower with GSH than with γ-EC, suggesting that GGC has a higher affinity for GSH than γ-EC. GGC reaction rates were calculated using the best available estimates of cytoplasmic GSH or γ-EC concentrations. Fricker et al. (2000) determined cytoplasmic GSH concentration in the roots of intact Arabidopsis seedlings after labeling with monochlorobimane to give fluorescent GSH S-bimane conjugates and imaging using confocal laser scanning microscopy. The cytoplasmic GSH concentration they measured was 2 to 3 mm. Assuming that γ-EC concentration in the cytoplasm is also proportionately higher than that in whole tissues, the cytoplasmic γ-EC concentration would be about 0.1 mm. Using these values with the kinetic constants we determined in the Michaelis-Menten equation, the GGC reaction rates in the cytoplasm were calculated as 0.11 nmol mg−1 protein min−1 for GSH and 0.02 nmol mg−1 protein min−1 for γ-EC, suggesting that 5.5 times more 5OP is synthesized from GSH than from γ-EC in vivo. The GGC reaction rate with GSH in the cytoplasm is equivalent to 60 nmol g−1 fresh weight h−1, a value that compares extremely well with the rates of GSH breakdown when GSH synthesis is blocked by BSO and of 5OP formation in the oxp1 mutants. These findings all support the suggestion the GGC is responsible for most GSH degradation in vivo.
The protein and gene corresponding to GGC are unidentified in plants, although QCT has been eliminated as a possibility (Supplemental Fig. S2). Specific identification of the Arabidopsis gene encoding GGC and analysis of the mutant are necessary to verify its function and to test our hypothesis that it is the major source of GSH metabolism in vivo. Recently, the gene encoding GGC was identified from humans (Oakley et al., 2008). Homology searches using the human GGC sequence did not identify any similar proteins in plants (Oakley et al., 2008), although GGC activity has been detected in tobacco (Steinkamp and Rennenberg, 1985, 1987) and Arabidopsis (this study). This suggests that the plant and animal enzymes are too highly diverged to be recognized by this method. Human GGC contains a BtrG-like fold that might be a signature feature of GGCs. A small family of Arabidopsis proteins with unknown function also contains this fold (de la Cruz et al., 2008) and may include the Arabidopsis GGC.
The lack of involvement of GGT as a major component of GSH metabolism agrees with the observations in our GGT knockout plants, where no significant changes in GSH levels were measured in any tissues except in the apoplast (Ohkama-Ohtsu et al., 2007a). While some GSH metabolism occurs through the GGT enzymes in plants and this metabolism is physiologically important in plants, it is clearly a minor portion of total GSH degradation.
Therefore, there is a fundamental difference in the ways plants and animals metabolize GSH. In animals, GSH is predominantly metabolized in the nephron tubes of the kidneys. GGT catalyzes this step, most likely releasing Glu and Cys-Gly, with the latter being hydrolyzed outside of the cell. The hydrolysis of GSH into component amino acids initiates its rapid transport into the cell, preventing the loss of these amino acids in the excretory system. In plants, the GGT reaction is a minor component under our experimental conditions. Rather, GSH is rapidly degraded through 5OP by GGC. The differences between the plants and animals may reflect the lack of an excretory system in plants. Having demonstrated this variation of the γ-glutamyl cycle in plants, where 5OP is formed from GSH by GGC and not GGT, it is interesting to ask if GGC fills the same role in animals. We were only able to prove this pathway in plants because of the possibility of creating organisms that lacked both GGT and 5OPase activity. This has not been undertaken in animals.
Although both GGT and GGC hydrolyze γ-glutamyl bonds, the lack of GGT activity in the ggt1/ggt4 double mutant indicates that GGT activity was distinct from GGC activity in our GGT activity assay (Ohkama-Ohtsu et al., 2007b). This was supported by the fact that human GGC did not metabolize γ-glutamyl-p-nitroanilide, the substrate for the GGT assay (Orlowski et al., 1969), and that the product of the reaction is 5OP and not Glu.
Figure 1B shows our current model for GSH degradation in plants, in which cytoplasmic GSH is converted to 5OP in a GGT-independent manner. While we have generally presented the conversion of GSH to 5OP by GGC as a one-step process, it is also possible to postulate an intermediate. The higher Vmax with γ-EC might support the notion that γ-EC or some other γ-glutamyl amino acids could be preferred GGC substrates and that a two-step conversion occurs. High enough levels of γ-EC or other γ-glutamyl amino acids to drive this reaction have not yet been observed in Arabidopsis. Phytochelatins could also act as 5OP precursors. This would require that GSH is first converted to phytochelatins by phytochelatin synthase before GGC removes the N-terminal γ-glutamyl group to make 5OP. Recently, the cytosolic hydrolysis of GSH conjugates to γ-EC conjugates by phytochelatin synthase was reported (Beck et al., 2003; Blum et al., 2007). Since phytochelatin synthase requires a heavy metal, this does not seem likely to be important in our in vitro GGC assay.
While GSH degradation may have a number of physiological functions, one purpose could be to provide Cys for protein synthesis and as a precursor for numerous metabolites. GSH is the major storage and transport form for Cys, and it must be broken down in order to release the amino acid (Leustek et al., 2000). In the future, it will be interesting to know how the futile cycles between GSH synthesis and degradation, which appear to be in the same tissues, are controlled.
MATERIALS AND METHODS
Plant Materials
Unless otherwise indicated, Arabidopsis (Arabidopsis thaliana) plants were grown at 22°C with a 24-h photoperiod (150 μmol m−2 s−1). Liquid culture was performed as described (Xiang and Oliver, 1998). Root length measurement on agar plates and nitrogen-deficient treatment in hydroponics culture were as described by Ohkama-Ohtsu et al. (2007b).
The oxp1-1 mutant (ecotype Columbia) was provided by the Salk Institute (Alonso et al., 2003) and obtained from the Arabidopsis Biological Resource Center (stock no. SALK_078745). The oxp1-1 mutant is a result of a T-DNA insertion into the second exon of the OXP1 gene. The homozygous plants containing the insert were screened by PCR using the gene-specific primers OXP1-1F (5′-CGTTGACGTACCACCCATATCA-3′) and OXP1-1R (5′-GGGAACTACTGTGGCAACGAAT-3′) and the T-DNA left-border primer pROKr3 (5′-CCTTTCGCTTTCTTCCCTTCCTTTCT-3′; Lin and Oliver, 2008).
The oxp1-2 mutant (ecotype Wassilewskija) was provided by INRA (http://dbsgap.versailles.inra.fr/publiclines/; stock no. EYU82). The oxp1-2 mutant is a result of a T-DNA insertion in the second exon of the OXP1 gene. The homozygous plants containing the insert were screened by PCR using the gene-specific primers OXP1-2F (5′-GCATGCGAGTGCATGATTCT-3′) and OXP1-2R (5′-ATCAGGTCCAGCAGGAGGAG-3′) and the T-DNA left-border primer LB4 (5′-CGTGTGCCAGGTGCCCACGGAATAGT-3′; http://www-ijpb.versailles.inra.fr/en/sgap/equipes/fichiers/FST_information.htm).
The At4g25720 mutant (ecotype Wassilewskija) was provided by INRA (stock no. EYH83). The At4g25720 mutant is a result of a T-DNA insertion in the first exon of At4g25720. The homozygous plants containing the insert were screened by PCR using the gene-specific primers 5′-CGAACCTCTTCCGAGAAATG-3′ and 5′-GAGGCGAACAGGAAGTTTTG-3′ and the T-DNA left-border primer LB4 described above.
ggt1-1, ggt4-1 (originally called ggt3-1 [Ohkama-Ohtsu et al., 2007b] but renamed ggt4-1 here), and ggt1-1/ggt4-1 double mutant plants have been described (Ohkama-Ohtsu et al., 2007a, 2007b). ggt1-1/oxp1-1, ggt4-1/oxp1-1, ggt1-1/ggt4-1/oxp1-1, and At4g25720/oxp1-2 double and triple mutants were generated by crossing followed by selection using PCR with the primers described above or by Ohkama-Ohtsu et al. (2007a, 2007b). oxp1-1 (Landsberg) was generated by crossing the oxp1-1 mutant to Landsberg wild-type plants followed by screening with the PCR primers described above.
Metabolite Analyses
HPLC Analyses
As 5OP is known to be chemically generated by heat or weak acid from Glu or Gln, plant tissues were homogenized in ice-cold 80% ethanol. After centrifugation at 16,000g for 10 min, the supernatant was collected and the ethanol was removed by vacuum evaporation at room temperature. The residue was dissolved in 2 mL g−1 tissue of 50 mm Tris-HCl, pH 8.0. After centrifugation at 16,000g for 10 min, 60 μL of the supernatant was passed through a 0.8-mL Dowex 50-X16 column (Orlowski et al., 1969). The column was washed with deionized water until 1.6 mL of effluent was collected. The effluent was dried under vacuum, and the residue dissolved in 60 μL of deionized water.
5OP was determined using HPLC as described (Nishimura et al., 2001) with modifications. The HPLC system used a C18 column (4.6 mm × 150 mm; Alltech) and a mobile phase of 2% (v/v) aqueous perchloric acid at 1.0 mL min−1. 5OP was detected at 210 nm at 5.2 min (Supplemental Fig. S3). 5OP levels were undetectable in leaf extracts from wild-type plants but detected in leaf extracts from the oxp1-1 mutant plants (Supplemental Fig. S3). Internal standards were used to confirm the 5OP peak and the efficiency of the extraction method (Supplemental Fig. S3). No Glu or Gln was converted to 5OP in this method.
Thiols were extracted, reduced with dithiothreitol (DTT), and quantified by HPLC as their monobromobimane derivatives (Ohkama-Ohtsu et al., 2007a).
Amino acid analysis was done following extraction in cold 80% ethanol. After centrifugation at 16,000g for 10 min, the supernatant was removed and dried under vacuum, dissolved in deionized water, and analyzed by reverse-phase HPLC using the AccQ Fluor Reagent Kit (Waters). Glu did not separate from Asn and was measured using an l-Glu assay kit (Seikagaku) and confirmed by CE-TOF/MS.
CE-MS Analyses
Frozen Arabidopsis seedlings were homogenized with Zirconia beads using a Mixer Mill (Retsch) at 27 Hz for 3 min. Twenty volumes of methanol (20 μL mg−1 fresh weight) including 8 μm internal standard, Met sulfone, that was used for compensation of the peak area after CE-MS analysis, was added, and again the mixture was homogenized at 27 Hz for 1 min. The sample solution was then centrifuged at 20,400 g for 3 min at 4°C. Five hundred microliters of chloroform and 200 μL of water were added to the supernatants. This mixture was vortexed for 3 min and centrifuged at 20,400g for 3 min at 4°C. The upper layer was evaporated for 30 min at 45°C by a centrifugal concentrator and then separated into two layers. The upper layer (100–200 μL) was centrifugally filtered through a Millipore 5-kD cutoff filter at 9,100g for 90 min. The filtrate was dried for 120 min by a centrifugal concentrator. The residue was dissolved into 20 μL of water containing a reference compound (3-aminopyrrolidine). The enzyme reaction mixtures were also prepared as described above. The final solution (20 μL) was used to quantify the contents of 5OP, Glu, and Gln by cation analysis using CE-MS. The CE-MS system and conditions were as described by Watanabe et al. (2008). Quantifications were performed using calibration curves of each compound.
The in vivo rates of 5OP synthesis in oxp1-1 and GSH degradation in the presence of BSO were calculated as dC/dt = r − μc, where r = rate of synthesis or degradation (nmol g−1 fresh weight h−1), c = amount of chemical (nmol), dt = change in time, and μ = specific growth rate (h−1).
RT-PCR and Quantitative Real-Time PCR
Total RNA was treated with DNaseI (Invitrogen; http://www.invitrogen.com) and reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) and oligo(dT)20 according to the manufacturer's instructions. Real-time PCR analysis was carried out using the iCycler iQ real-time PCR detection system (Bio-Rad; www.bio-rad.com) and iQ SYBR Green Supermix (Bio-Rad) as recommended by the manufacturer. Sequences of primers used in RT-PCR were 5′-GAAGCAGTTACATCTGCAGCGG-3′ and 5′-CAGAACCGGGTACCTTTGCTC-3′ for OXP1 and 5′-GACGCCTTTACTCAGGGTCTTC-3′ and 5′-CCATCGGTAGCCAGTCCC-3′ for At4g25720. Sequences of primers used in real-time PCR analysis were 5′-TTGGAGGTTTGGCCATTTTCA-3′ and 5′-GCCAAAAGTCTTGGACCAGCA-3′ for OXP1. The ACTIN8 gene (An et al., 1996) was chosen as a control, and its expression was determined using primers as described by Goto and Naito (2002).
5OPase Assay
The 5OPase activity assay was performed as described by Mazelis and Creveling (1978) with modifications. Leaves from 20-d-old plants were homogenized in ice-cold (2 mL g−1 fresh weight) 20 mm HEPES (pH 7.4) containing 5 mm DTT and then centrifuged for 10 min at 16,000g, 4°C. The supernatant solution was dialyzed against 20 mm HEPES (pH 7.4) overnight at 4°C before the 5OPase assay. 5OPase activity was determined at 30°C in an assay mixture of 100 mm Na glycinate, pH 9.5, 5 mm ATP, 2.5 mm MnCl2, 2.5 mm MgCl2, 20 mm (NH4)2SO4, 5 mm DTT, and 2.5 mm 5OP in a total volume of 200 μL. Under these conditions, 5OPase activity was constant for at least 60 min. As a negative control without substrate, deionized water replaced 5OP. The reaction was stopped by adding 20 μL of 1 n acetic acid and heating for 5 min at 100°C. The heated reaction mixture was centrifuged, and an aliquot of the supernatant solution was assayed for Glu by CE-TOF/MS. The conditions of CE-TOF/MS were the same as those used for 5OP determination described above.
GGC Activity Assay
The GGC activity assay was performed as described by Steinkamp and Rennenberg (1985, 1987) with modifications. Ten-day-old liquid-cultured ggt1-1/ggt4-1 seedlings were homogenized to a fine powder in liquid nitrogen, then suspended in 5 mL g−1 tissue of a solution consisting of 50 mm Tris-HCl, pH 8.0, and 5 mm 2-mercaptoethanol. After centrifugation at 13,000g and 4°C for 15 min, the supernatant was brought to 85% saturation with solid ammonium sulfate. The precipitate was collected by centrifugation at 13,000g and 4°C for 15 min and dissolved in a minimum volume of 50 mm Tris-Cl, pH 8.0. The enzyme solution was dialyzed against 50 mm Tris-Cl, pH 8.0, at 4°C overnight before the assay. Protein concentrations were determined as described by Bradford (1976). GSH or γ-EC purchased from Sigma (http://www.sigma-aldrich.com/) was used as a substrate. When purchased, the γ-EC was not pure enough for the assay. It was further purified by HPLC in the same conditions used in the 5OP determination. The γ-EC fraction was collected and concentrated under vacuum. The perchloric acid was removed from the concentrated γ-EC solution by repeating the HPLC with deionized water as the mobile phase. γ-EC was collected, dried under vacuum, and dissolved in 250 mm Tris-HCl, pH 8.0. GGC activity was determined at 30°C in an assay mixture of 100 mm Tris-HCl, pH 8.0, 1 mm DTT, and the substrate in a total volume of 48 μL. The reaction was stopped with 6 μL of 1.5 n HCl. Under this condition, GGC activity was constant for 60 min with up to 0.38 mg of protein in the reaction mixture. 5OP was determined using HPLC as described above. 5OP produced chemically was determined by incubating the substrates in the same conditions except that Tris-Cl, pH 8.0, buffer was added instead of the enzyme solution. 5OP produced from the enzyme solution without added substrates was determined by incubating the enzyme solution in the same conditions except that Tris-Cl, pH 8.0, buffer was added instead of the substrate. 5OP produced by GGC was calculated as follows: (5OP produced in the enzyme solution with substrate) − (5OP produced chemically) − (5OP produced in the enzyme solution without substrate). Km and Vmax were calculated by fitting the first-order kinetic equation Y = Vmax × X/(Km + X) using GraphPad Prism 5 software (GraphPad Software).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the translated sequence of the Arabidopsis OXP1 gene and protein sequences of 5OPase from animals.
Supplemental Figure S2. At4g25720 (QCT) knockout mutants.
Supplemental Figure S3. Detection of 5OP using HPLC.
Supplemental Table S1. γ-Glutamyl transferase activity in the wild type and ggt mutants.
Supplementary Material
Acknowledgments
We thank Ms. Shoko Shinoda (RIKEN Plant Science Center) for excellent technical support of the CE-TOF/MS analyses.
This work was supported by the U.S. Department of Agriculture, the National Research Initiative Program (grant no. 30471038), the Chinese Academy of Science (grant no. KSCX2–SW–3), and the RIKEN Special Postdoctoral Researcher Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: David J. Oliver (doliver@iastate.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- An YQ, McDowell JM, Huang SR, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10 107–121 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Beck A, Lendzian K, Oven M, Christmann A, Grill E (2003) Phytochelatin synthase catalyzes key step in turnover of glutathione conjugates. Phytochemistry 62 423–431 [DOI] [PubMed] [Google Scholar]
- Blum R, Beck A, Korte A, Stenge A, Letzel T, Lendzian K, Grill E (2007) Function of phytochelatin synthase in catabolism of glutathione-conjugates. Plant J 49 740–749 [DOI] [PubMed] [Google Scholar]
- Cobbett C, Goldsbrough P (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53 159–182 [DOI] [PubMed] [Google Scholar]
- Cobbett CS, May MJ, Howden R, Rolls B (1998) The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in γ-glutamylcysteine synthetase. Plant J 16 73–78 [DOI] [PubMed] [Google Scholar]
- Dahl SW, Slaughter C, Lauritzen C, Bateman RC Jr, Connerton I, Pedersen J (2000) Carica papaya glutamine cyclotransferase belongs to a novel plant enzyme subfamily: cloning and characterization of the recombinant enzyme. Protein Expr Purif 20 27–36 [DOI] [PubMed] [Google Scholar]
- de la Cruz NB, Peterson FC, Volkman BF (2008) Solution structure of At3g28950 from Arabidopsis thaliana. Proteins 71 546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker MD, May M, Meyer AJ, Sheard N, White NS (2000) Measurement of glutathione levels in intact roots of Arabidopsis. J Microsc 198 162–171 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli D, Kumar C, Bachhawat AK (2007) The alternative pathway of glutathione degradation is mediated by a novel protein complex involving three new genes in Saccharomyces cerevisiae. Genetics 175 1137–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto DB, Naito S (2002) AtMRD1 and AtMRU1, two novel genes with altered mRNA levels in the methionine over-accumulating mto1-1 mutant of Arabidopsis thaliana. Plant Cell Physiol 43 923–931 [DOI] [PubMed] [Google Scholar]
- Grzam A, Martin MN, Hell R, Meyer AJ (2007) γ-Glutamyl transpeptidase GGT4 initiates vacuolar degradation of glutathione S-conjugates in Arabidopsis. FEBS Lett 581 3131–3138 [DOI] [PubMed] [Google Scholar]
- Hanigan MH, Pitot HC (1985) Gamma-glutamyl transpeptidase: its role in hepatocarcinogenesis. Carcinogenesis 6 165–172 [DOI] [PubMed] [Google Scholar]
- Harding CO, Williams P, Wagner E, Chang DS, Wild K, Colwell RE, Wolff JA (1997) Mice with genetic γ-glutamyl transpeptidase deficiency exhibit glutathionuria, severe growth failure, reduced life spans, and infertility. J Biol Chem 272 12560–12567 [DOI] [PubMed] [Google Scholar]
- Hell R, Bergmann L (1990) γ-Glutamylcysteine synthetase in higher-plants: catalytic properties and subcellular localization. Planta 180 603–612 [DOI] [PubMed] [Google Scholar]
- Jez JM, Cahoon RE, Chen S (2004) Arabidopsis thaliana glutamate-cysteine ligase: functional properties, kinetic mechanism, and regulation of activity. J Biol Chem 279 33463–33470 [DOI] [PubMed] [Google Scholar]
- Kumar TR, Wiseman AL, Kala G, Kala SV, Matzuk MM, Lieberman MW (2000) Reproductive defects in γ-glutamyl transpeptidase-deficient mice. Endocrinology 141 4270–4277 [DOI] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick JA, Davies JP (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51 141–165 [DOI] [PubMed] [Google Scholar]
- Lieberman MW, Wiseman AL, Shi ZZ, Carter BZ, Barrios R, Ou CN, Chévez-Barrios P, Wang Y, Habib GM, Goodman JC, et al (1996) Growth retardation and cysteine deficiency in γ-glutamyl transpeptidase-deficient mice. Proc Natl Acad Sci USA 93 7923–7926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Oliver DJ (2008) The role of acetyl-CoA synthetase in Arabidopsis. Plant Physiol 147 1822–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA (1996) The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Plant Mol Biol 47 127–158 [DOI] [PubMed] [Google Scholar]
- Martin MN, Saladores PH, Lambert E, Hudson AO, Leustek T (2007) Localization of members of the γ-glutamyl transpeptidase family identifies sites of glutathione and glutathione S-conjugate hydrolysis. Plant Physiol 144 1715–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MN, Slovin JP (2000) Purified γ-glutamyl transpeptidases from tomato exhibit high affinity for glutathione and glutathione S-conjugates. Plant Physiol 122 1417–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May MJ, Leaver CJ (1994) Arabidopsis thaliana γ-glutamylcysteine synthetase is structurally unrelated to mammalian, yeast, and Escherichia coli homologs. Proc Natl Acad Sci USA 91 10059–10063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazelis M, Creveling RK (1978) 5-Oxoprolinase (L-pyroglutamate hydrolase) in higher plants. Plant Physiol 62 798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazelis M, Pratt HM (1976) In vivo conversion of 5-oxoproline to glutamate by higher plants. Plant Physiol 57 85–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A (1973) On the enzymology of amino acid transport. Science 180 33–39 [DOI] [PubMed] [Google Scholar]
- Meister A, Larsson A (1995) Glutathione synthetase deficiency and other disorders of the γ-glutamyl cycle. In CR Scriver, AL Beaudet, WS Sly, D Valle, eds, The Metabolic and Molecular Bases of Inherited Disease, Vol 1. McGraw-Hill, New York, pp 1461–1495
- Nakano Y, Okawa S, Yamaguchi T, Koizumi Y, Sekiya J (2004) Occurrence of two forms of γ-glytamyltransferase in radish plant. Plant Biotechnol 3 243–246 [Google Scholar]
- Nishimura A, Itoh H, Oyama H, Murao S, Oda K (2001) A simultaneous assay method for L-glutamate and L-pyroglutamate contents in soy sauce using a 5-oxoprolinase (without ATP hydrolyzing activity). Biosci Biotechnol Biochem 65 477–479 [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49 249–279 [DOI] [PubMed] [Google Scholar]
- Oakley AJ, Yamada T, Liu D, Coggan M, Clark AG, Board P (2008) The identification and structural characterization of C7ORF24 as γ-glutamyl cyclotransferase: an essential enzyme in the γ-glutamyl cycle. J Biol Chem 283 22031–22042 [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Radwan S, Peterson A, Zhao P, Badr AF, Xiang C, Oliver DJ (2007. a) Characterization of the extracellular γ-glutamyl transpeptidases GGT1 and GGT2 in Arabidopsis. Plant J 49 865–877 [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Zhao P, Xiang C, Oliver DJ (2007. b) Glutathione conjugates in the vacuole are degraded by γ-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J 49 878–888 [DOI] [PubMed] [Google Scholar]
- Orlowski M, Richman P, Meister A (1969) Isolation and properties of γ-L-glutamylcyclotransferase from human brain. Biochemistry 8 1048–1055 [DOI] [PubMed] [Google Scholar]
- Pasternak M, Lim B, Wirtz M, Hell R, Cobbett CS, Meyer AJ (2008) Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J 53 999–1012 [DOI] [PubMed] [Google Scholar]
- Rennenberg H, Steinkamp R, Kesselmeier J (1981) 5-Oxoprolinase in Nicotiana tabacum: catalytic properties and subcellular localization. Physiol Plant 62 211–216 [Google Scholar]
- Rennenberg H, Steinkamp R, Polle A (1980) Evidence for the participation of a 5-oxoprolinase in degradation of glutathione in Nicotiana tabacum. Z Naturforsch 35c 708–711 [DOI] [PubMed] [Google Scholar]
- Schilling S, Hoffmann T, Manhart S, Hoffmann M, Demuth HU (2004) Glutaminyl cyclase unfold glutamyl cyclase activity under mild acid conditions. FEBS Lett 563 191–196 [DOI] [PubMed] [Google Scholar]
- Schilling S, Stenzel I, von Bohlen A, Wermann M, Schulz K, Demuth HU, Wasternack C (2007) Isolation and characterization of the glutaminyl cyclases from Solanum tuberosum and Arabidopsis thaliana: implications for physiological functions. Biol Chem 388 145–153 [DOI] [PubMed] [Google Scholar]
- Shaw ML, Pither-Joyce MD, MacCallum JA (2005) Purification and cloning of a γ-glutamyl transpeptidase from onion (Allium cepa). Phytochemistry 66 515–522 [DOI] [PubMed] [Google Scholar]
- Steinkamp R, Rennenberg H (1985) Degradation of glutathione in plant cells: evidence against the participation of a γ-glutamyl transpeptidase. Z Naturforsch 40c 29–33 [Google Scholar]
- Steinkamp R, Rennenberg H (1987) γ-Glutamylcyclotransferase in tobacco suspension cultures: catalytic properties and subcellular localization. Physiol Plant 69 499–503 [Google Scholar]
- Storozhenko S, Belles-Boix E, Babiychuk E, Herouart D, Davey MW, Slooten L, Van Montagu M, Inzé D, Kushnir S (2002) γ-Glutamyl transpeptidase in transgenic tobacco plants: cellular localization, processing, and biochemical properties. Plant Physiol 128 1109–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inzé D, et al (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter A, Wolf S, Steininger H, Bogs J, Rausch T (2005) Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J 41 15–30 [DOI] [PubMed] [Google Scholar]
- Wang CL, Oliver DJ (1996) Cloning of the cDNA and genomic clones for glutathione synthetase from Arabidopsis thaliana and complementation of a gsh2 mutant in fission yeast. Plant Mol Biol 31 1093–1104 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Kusano M, Oikawa A, Fukushima A, Noji N, Saito K (2008) Physiological roles of the β-substituted alanine synthase gene family in Arabidopsis. Plant Physiol 146 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Oliver DJ (1998) Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 10 1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Werner BL, Christensen EM, Oliver DJ (2001) The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiol 126 564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.