Abstract
Glucosinolates (GSLs) are amino acid-derived secondary metabolites with diverse biological activities dependent on chemical modifications of the side chain. We previously identified the flavin-monooxygenase FMOGS-OX1 as an enzyme in the biosynthesis of aliphatic GSLs in Arabidopsis (Arabidopsis thaliana) that catalyzes the S-oxygenation of methylthioalkyl to methylsulfinylalkyl GSLs. Here, we report the fine mapping of a quantitative trait locus for the S-oxygenating activity in Arabidopsis. In this region, there are three FMOs that, together with FMOGS-OX1 and a fifth FMO, form what appears to be a crucifer-specific subclade. We report the identification of these four uncharacterized FMOs, designated FMOGS-OX2 to FMOGS-OX5. Biochemical characterization of the recombinant protein combined with the analysis of GSL content in knockout mutants and overexpression lines show that FMOGS-OX2, FMOGS-OX3, and FMOGS-OX4 have broad substrate specificity and catalyze the conversion from methylthioalkyl GSL to the corresponding methylsulfinylalkyl GSL independent of chain length. In contrast, FMOGS-OX5 shows substrate specificity toward the long-chain 8-methylthiooctyl GSL. Identification of the FMOGS-OX subclade will generate better understanding of the evolution of biosynthetic activities and specificities in secondary metabolism and provides an important tool for breeding plants with improved cancer prevention characteristics as provided by the methylsulfinylalkyl GSL.
Glucosinolates (GSLs) are amino acid-derived secondary metabolites present in the order Brassicales. Upon disruption of plant tissue by, for example, wounding or mastication, GSLs are hydrolyzed by the thioglucosidases, myrosinases, which produce a range of breakdown products, primarily isothiocyanates and nitriles, with diverse biological activities (Halkier and Gershenzon, 2006; Zhang et al., 2006b). GSLs (typically their hydrolysis products) act as antimicrobials against pathogens, feeding deterrents toward generalist herbivores, and as attractants for specialist herbivores (Kliebenstein et al., 2001a; Tierens et al., 2001). For humans, the well-studied sulforaphane, which is derived from 4-methylsulfinylbutyl (4-MSB) GSL and the isothiocyanates derived from 7-methylsulfinylheptyl (7-MSH) GSL and 8-methylsulfinyloctyl (8-MSO) GSL, are considered very potent cancer-preventive agents because of their strong induction of xenobiotic phase II detoxification enzymes in animals (Zhang et al., 1992; Rose et al., 2000). These isothiocyanates can decrease the risk of developing different cancers, such as breast cancer (Rose et al., 2005), gastric cancer (Fahey et al., 2002), and skin cancer (Talalay et al., 2007). The potent cancer-preventive property of the hydrolysis product of methylsulfinylalkyl (MS) GSLs makes it desirable to characterize the genes that encode for the enzymes that catalyze the S-oxygenation reaction from methylthioalkyl (MT) to MS GSLs.
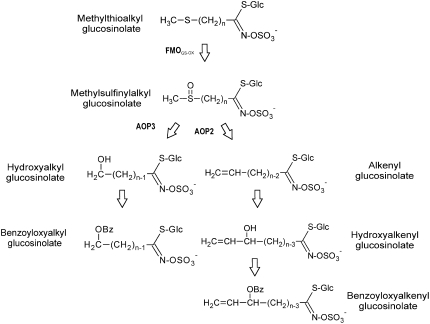
Biosynthesis of MS GSLs can be divided into three separate phases (i.e. Met chain elongation, GSL core structure formation [Halkier and Gershenzon 2006], and, finally, an S-oxygenating reaction [Fig. 1]). An Arabidopsis (Arabidopsis thaliana) flavin-monooxygenase FMOGS-OX1 was recently shown to catalyze the S-oxygenation of MT to MS GSLs (Hansen et al., 2007). The presence of MS GSLs in FMOGS-OX1 T-DNA knockout mutants indicated that additional genes catalyzing this reaction are present (Hansen et al., 2007).
Figure 1.
Potential secondary modifications of Met-derived GSLs in Arabidopsis. AOP2 and AOP3, 2-oxoglutarate-dependent dioxygenase. For MT and MS, n = 3 to 8. For OH and benzoyloxyalkyl, n = 2 to 3. For alkenyl, n = 2 to 5. For hydroxyalkenyl and benzoyloxyalkenyl, n = 3.
The conversion of MT to MS GSLs was originally studied via genetic means using natural variation in Arabidopsis and Brassica napus as defined by the GSL S-oxygenation (GS-OX) quantitative trait loci (QTLs; Giamoustaris and Mithen, 1996; Kliebenstein et al., 2001a, 2001b, 2002). These studies identified several independent GS-OX loci that mapped near FMOGS-OX1, although they did not overlap with the physical position of FMOGS-OX1. These GS-OX loci even showed structural specificity such that some affected all aliphatic GSLs, whereas others were specific to subsets of substrates. The QTL study suggests that the GS-OX loci near the physical position of FMOGS-OX1 may contain the candidate genes for the additional MT to MS S-oxygenating enzyme activity previously suggested (Hansen et al., 2007).
In this article, we identify four FMOGS-OX1-related genes encoding for enzymes that catalyze the MT to MS S-oxygenation reaction. By fine genetic mapping analysis, we found that three FMOGS-OX1 homologs, At1g62540, At1g62570, and At1g62560, mapped to a 0.2-Mb area containing a GS-OX QTL in multiple Arabidopsis populations. The five FMO genes At1g65860 (FMOGS-OX1), At1g62540 (FMOGS-OX2), At1g62560 (FMOGS-OX3), At1g62570 (FMOGS-OX4), and At1g12140 (FMOGS-OX5) were found within a subclade of the FMO phylogeny that (at least presently) consists of genes from only cruciferous species. Furthermore, we characterize the Arabidopsis enzymes in this subclade and show that FMOGS-OX2 to FMOGS-OX4 are able to catalyze the S-oxygenation independent of chain length, as was observed for FMOGS-OX1, and that FMOGS-OX5 is specific for 8-methylthiooctyl (8-MTO) GSL.
RESULTS
Phylogenetic Tree of Plant FMOs in Proposed Clade III
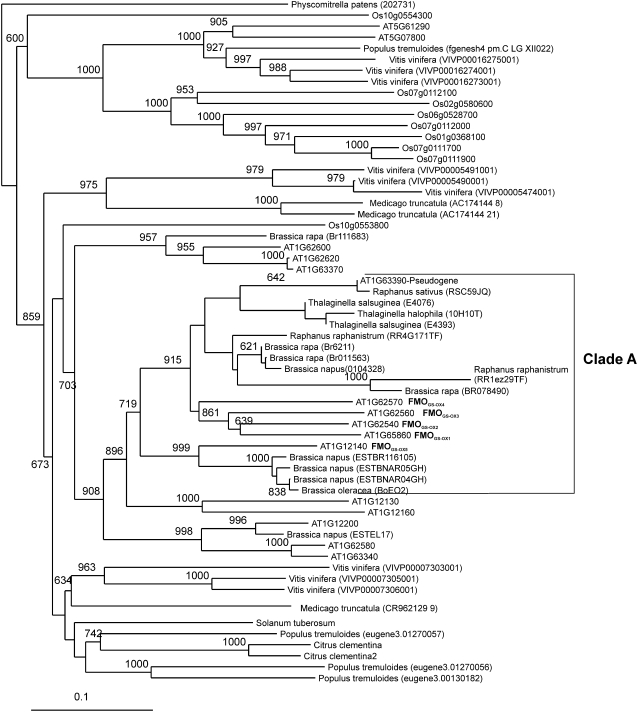
Previous phylogenetic analysis had shown three plant FMO clades from the genomic sequence of rice (Oryza sativa), Arabidopsis, and poplar (Populus tremuloides; Hansen et al., 2007). Clade III is the putative S-oxygenation clade and contains what appears to be crucifer-specific radiation of FMOs, which includes the biochemically characterized FMOGS-OX1 (Hansen et al., 2007). In the intervening time, several additional plant genomic sequences became available and allowed for retesting of this FMO clade's specificity to crucifers. To do this, we obtained all of the FMOs from the genomic sequences of poplar, rice, Medicago truncatula, grape (Vitis vinifera), and Physcomitrella patens and created a complete phylogeny to identify those FMOs present in clade III, which has a single ancestor present in P. patens. To further extend this analysis, we utilized the FMOGS-OX and related Arabidopsis sequences to identify all ESTs that were at least 80% identical at the amino acid level. These ESTs identified one potato (Solanum tuberosum) FMO, two Citrus clementine FMOs, and a set of crucifer FMOs from Raphanus, Brassica, and Thalaginella ssp. (Fig. 2; Supplemental Table S1). Phylogenetic analysis showed that only genomic and EST sequences from the Cruciferae clustered within the FMOGS-OX subclade, leading us to hypothesize that this is a crucifer-specific clade of FMOs (Fig. 2). Because GSLs are also unique to these plants, these crucifer-specific FMOs provide candidate genes for the residual MT to MS S-oxygenating enzyme activity present in the FMOGS-OX1 knockout (Hansen et al., 2007). Interestingly, this clade is marked by numerous species-specific duplications as shown by the grape and rice radiations (Fig. 2).
Figure 2.
Phylogenetic analysis of FMOGS-OX homologous sequences. At1g12140, At1g62580, and At1g65860 were used to search the available Genomic and EST databases for homologous FMO sequences in clade III (Hansen et al., 2007). The genomic sequences were utilized as full-length proteins. The EST sequences were aligned to identify the minimum unigene set. Clade A, Subclade that appears to be crucifer specific.
Fine-Scale Mapping of GS-OX
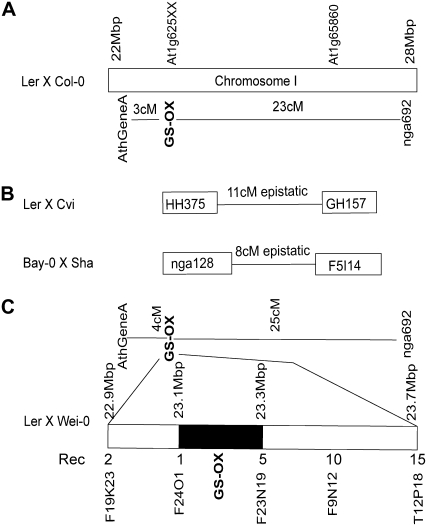
Previous quantitative genetics analysis mapped a GS-OX locus on Arabidopsis chromosome I in crude proximity to the characterized FMOGS-OX1 (Kliebenstein et al., 2001b; Hansen et al., 2007). This GS-OX locus controlled differences in the ratio of MT to MS GSLs in the Landsberg erecta (Ler) × Columbia-0 (Col-0) recombinant inbred line (RIL) population (Kliebenstein et al., 2001a). We have conducted a detailed analysis of the existing data in the larger Ler × Cape Verde Islands (Cvi) and Bay-0 × Shahdara (Sha) RIL populations. This showed that the GS-OX locus was in fact two closely linked loci that epistatically control the ratio of MT to MS GSLs (Supplemental Tables S2 and S3; Kliebenstein et al., 2001a; Wentzell et al., 2007). One of these two loci, as shown by the GH157 and F5I14 markers in the Ler × Cvi and Bay-0 × Sha populations, respectively, was tightly linked to FMOGS-OX1 (At1g65860; Fig. 3). Further, the At1g65860 transcript had a significant cis-expression QTL in the Bay-0 × Sha population that agreed with FMOGS-OX1 likely being the functional basis of one of the two GS-OX loci in this region (Wentzell et al., 2007; West et al., 2007). However, this FMOGS-OX1 QTL was not at the genetic position of the originally identified GS-OX locus (Kliebenstein et al., 2001b; Fig. 3).
Figure 3.
Genetic mapping of GS-OX in multiple Arabidopsis RIL populations. The position of the GS-OX loci [controlling MT:(MT + MS) ratio] on chromosome I in four different Arabidopsis RIL populations is presented. The genomic position of At1g65860 (FMOGS-OX1) and At1g62540 to At1g62580 (At1g625XX) is presented for reference. A, For the Ler × Col-0 population, GS-OX was mapped as a qualitative locus and the position is presented in relation to the AthGeneA and nga692 markers. B, In Ler × Cvi and Bay-0 × Sha, GS-OX was mapped as a quantitative trait and the one LOD intervals for the identified loci on chromosome I presented with the genetic distance between them. The tested marker for each QTL is positioned relative to its physical position on chromosome I. Epistatic shows that there was a significant interaction between these two QTLs. C, For the Ler × Wei-0 F2 population, GS-OX was mapped as a qualitative locus and the position is presented in relation to the AthGeneA and nga692 markers. The Ler × Wei-0 population was used to fine map the major GS-OX locus and the fine-mapping information is presented at the bottom with the genomic position of the markers indicated. Rec, Number of recombinations between the given marker and the GS-OX phenotype in 400 F2 individuals. No markers were identified that could further resolve the position of the GS-OX locus as indicated by the black box.
To better resolve the molecular genetic basis of the originally identified GS-OX locus, we generated a F2 population by crossing the Ler and Wei-0 accessions, which have a strong difference in the GS-OX phenotype, but have the same allelic status at the epistatic GS-Elong and GS-AOP QTLs (Kliebenstein et al., 2001b; Wentzell et al., 2007). This population thereby allowed us to focus on GS-OX variation. We genotyped and HPLC phenotyped 400 F2 individuals and fine-scale mapped the GS-OX QTL in this population to a 200-kb region between 23.1 and 23.3 Mb on Arabidopsis chromosome I. This is in the same position as the original GS-OX locus from the Ler × Col-0 population, as well as proximal to the nga128 and HH375 markers from the Ler × Cvi and Bay-0 × Sha populations (Fig. 3; Loudet et al., 2002). This fine-scale map position did not include the FMOGS-OX1 gene, but did include three tandem duplicates that are the closest homologs of FMOGS-OX1, At1g62540, At1g62560, and At1g62570 (Fig. 2). Additionally, all three genes had cis-expression QTLs in the available Bay × Sha QTL data, suggesting that gene expression rather than enzyme activity variation in this tandem gene family underlies the GS-OX QTL linked to the nga128 and HH375 markers in multiple Arabidopsis populations (West et al., 2007). No recombination events between these genes could be identified. The phylogenetic relationship of these FMOs to a gene encoding an enzyme that converts MT to MS GSLs, their physical proximity to a locus controlling the conversion of MT to MS GSLs, as well as the presence of expression diversity, led us to hypothesize that these genes are FMOGS-OXs within Arabidopsis. We then proceeded to directly test this biochemical hypothesis on these FMOs and on At1g12140, which belongs to the proposed FMOGS-OX clade (Fig. 2).
Recombinant FMOGS-OXs Catalyze the Conversion of MT to MS GSLs
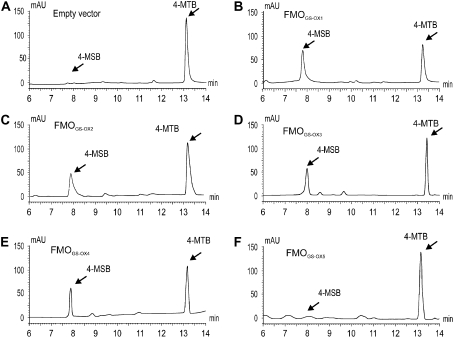
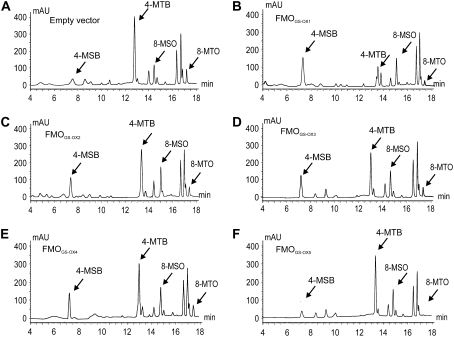
We previously showed that FMOGS-OX1 S-oxygenates desulfo and intact MT GSLs, but not other precursors in the Met-derived GSL biosynthesis pathway in Arabidopsis (Hansen et al., 2007). Therefore, we utilized desulfo MT GSLs as substrate to test whether the other Arabidopsis proteins in the FMOGS-OX subclade catalyzed the S-oxygenation of MT to MS GSLs. FMOGS-OX1, FMOGS-OX2, FMOGS-OX3, FMOGS-OX4, and FMOGS-OX5 were individually expressed in Escherichia coli. Spheroplasts of E. coli expressing these FMOGS-OX proteins were incubated with desulfo 4-methylthiobutyl (4-MTB) GSL for 1 h, and desulfo GSLs were analyzed by HPLC. Recombinant protein of FMOGS-OX2, FMOGS-OX3, and FMOGS-OX4 all catalyzed the production of desulfo 4-MSB GSL (Fig. 4, C–E) in levels comparable to the FMOGS-OX1 (Fig. 4B). Control spheroplasts transformed with empty vector did not show a significant production of desulfo 4-MSB (Fig. 4A). Recombinant protein of the most distant member of this subclade, FMOGS-OX5, did not convert 4-MTB into 4-MSB (Figs. 2 and 4F). This indicated that either FMOGS-OX5 cannot catalyze the S-oxygenation of MT GSLs or it may have substrate specificity for other MT GSLs than 4-MTB.
Figure 4.
Analysis of S-oxygenation activity of FMOGS-OX1 to FMOGS-OX5 for 4-MTB GSL. Spheroplasts of E. coli expressing empty vector were used as negative control. A, Negative control; B, assay of FMOGS-OX1; C, assay of FMOGS-OX2; D, assay of FMOGS-OX3; E, assay of FMOGS-OX4; F, assay of FMOGS-OX5. Ratio of 4-MTB:(4-MTB + 4-MSB) was calculated to represent the S-oxygenating activity. Significance of the difference between the ratio of 4-MTB:(4-MTB + 4-MSB) for FMOGS-OX versus the negative control was determined by ANOVA. For FMOGS-OX1 to FMOGS-OX4, P < 0.001; for FMOGS-OX5, P = 0.16.
The aliphatic, Met-derived GSLs are divided into classes of different side chain lengths: short chain, propyl (C3) and butyl (C4); middle chain, pentyl (C5) and hexyl (C6); and long chain, heptyl (C7) and octyl (C8). Because 4-MTB is the only commercially available MT GSL, we extracted desulfo GSLs from seeds of the Arabidopsis accession Col-0 to use as substrates in the FMOGS-OX enzyme assay. 4-MTB and 8-MTO GSLs are the dominant GSLs in these seeds. This allowed us to test whether any of the FMOGS-OXs catalyzes the S-oxygenation of MT GSLs with chain lengths other than 4-MTB. Spheroplasts of E. coli expressing FMOGS-OX1, FMOGS-OX2, FMOGS-OX3, FMOGS-OX4, and FMOGS-OX5 were incubated with the desulfo GSLs derived from seeds, followed by HPLC analyses. Consistent with the previous work, FMOGS-OX1, FMOGS-OX2, FMOGS-OX3, and FMOGS-OX4, but not FMOGS-OX5, catalyzed the conversion of 4-MTB to 4-MSB (Fig. 5, B–F). Interestingly, all five recombinant proteins converted 8-MTO to 8-MSO (Fig. 5, B–F). Thus, FMOGS-OX1, FMOGS-OX2, FMOGS-OX3, and FMOGS-OX4 catalyze the S-oxygenating reaction for both short-chain and long-chain MT GSL, whereas FMOGS-OX5 has a more limited substrate specificity as indicated by its specificity for 8-MTO. This suggests that this cluster of FMOs is involved in GSL biosynthesis and that the enzymes have evolved different substrate specificities.
Figure 5.
Analysis of S-oxygenation activity of FMOGS-OX1 to FMOGS-OX5 for methythioalkyl GSL. The activity of each recombinant FMOGS-OX was detected in assays using desulfo GSL extracts from Arabidopsis seed with a final concentration of 2 mm total desulfo GSL as substrate. Spheroplasts of E. coli expressing empty vector were used as negative control. A, Negative control; B, assay of FMOGS-OX1; C, assay of FMOGS-OX2; D, assay of FMOGS-OX3; E, assay of FMOGS-OX4; F, assay of FMOGS-OX5. Ratio of 4-MTB:(4-MTB + 4-MSB) was calculated to represent the S-oxygenating activity for short-chain GSL. The significance of the difference between the ratio 4-MTB:(4-MTB + 4-MSB) of the respective FMOGS-OX and the negative control was determined by ANOVA. For FMOGS-OX1 to FMOGS-OX4, P < 0.001; for FMOGS-OX5, P = 0.304. Ratio of 8-MTO:(8-MTO + 8-MSO) was calculated to represent the S-oxygenating activity for long-chain GSL. The significance of the difference between the ratio 8-MTO:(8-MTO + 8-MSO) of the respective FMOGS-OX and the negative control was determined by ANOVA. For FMOGS-OX1 to FMOGS-OX5, P < 0.001.
GSL Analyses of FMOGS-OX Knockout Mutants
To validate the S-oxygenation activities of these FMOs in planta, we obtained two independent T-DNA knockout mutants for, respectively, FMOGS-OX2 and FMOGS-OX4, and one T-DNA knockout mutant for, respectively, FMOGS-OX3 and FMOGS-OX5. These T-DNA mutants were confirmed as having no detectable transcript for the corresponding FMOGS-OX by reverse transcription (RT)-PCR (Supplemental Fig. S1). For each FMOGS-OX knockout mutant, we measured GSL content in leaves and seeds of segregating progeny obtained from a heterozygous parent (Supplemental Table S4). By analyzing the GSL content in wild-type and homozygous knockout plants in a segregating population derived from a single heterozygous parent, we minimize the influence of potential maternal effects. From the HPLC data, the ratio of MT GSL to the sum of MT and MS GSL, which represents the S-oxygenation activity for the conversion from MT GSL to MS GSL, was calculated for each chain length. For FMOGS-OX2 and FMOGS-OX4, there was no statistically significant difference between the MT:(MT + MS) ratios of the two independent T-DNA knockout mutants of the same gene, and the data from the mutants were pooled.
In agreement with predicted biochemistry for FMOGS-OX2, its homozygous knockout mutants showed an increased ratio of MT:(MT + MS) for the butyl, pentyl, heptyl, and octyl Met-derived GSLs in both leaves and seeds in comparison with wild-type plants (Table I). A homozygous knockout mutant in the proposed long-chain-specific FMOGS-OX5 had an increased MT:(MT + MS) for C8 GSLs in seeds, but also for other chain lengths (Table II). This agrees with the observation that only 8-MTO was found to be a substrate for recombinant FMOGS-OX5. The observed changes in the knockout mutants are likely not absolute due to compensatory function present in the other functioning FMOGS-OXs. In contrast, the T-DNA knockout mutant of FMOGS-OX4 did not show an increase in MT:(MT + MS) in either leaves or seeds (Table III). This may be due to low expression or low in planta activity in the Col-0 accession, in which case a mutant phenotype will be hidden by functional redundancy with the other FMOGS-OXs.
Table I.
Altered GS-OX activity in the FMOGS-OX2 T-DNA mutant
GSL content in seeds and leaves was analyzed. All plants were segregants derived from a parental line heterozygous for the T-DNA knockout allele. MT:(MS + MT) represents the S-oxygenation activity for the conversion from MT GSL to MS GSL. This T-DNA mutant is in a wild-type Col-0 background.
| MT:(MS + MT) | Leaf Tissue
|
Seed Tissue
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Homozygous Knockout (na = 52)
|
Homozygous Wild Type (n = 36)
|
Pd | Homozygous Knockout (n = 24)
|
Homozygous Wild Type (n = 24)
|
P | |||||
| Meanb | sec | Mean | se | Mean | se | Mean | se | |||
| Propyl GSL (C3) | 0.12 | 0.008 | 0.13 | 0.011 | NSe | ND | NDf | |||
| Butyl GSL (C4) | 0.33 | 0.011 | 0.24 | 0.013 | <0.001 | 0.70 | 0.008 | 0.60 | 0.023 | 0.001 |
| Pentyl GSL (C5) | 0.24 | 0.013 | 0.17 | 0.015 | <0.001 | 0.79 | 0.024 | 0.73 | 0.040 | <0.001 |
| Hexyl GSL (C6) | ND | ND | ND | ND | ||||||
| Heptyl GSL (C7) | 0.24 | 0.009 | 0.22 | 0.016 | <0.001 | 0.54 | 0.034 | 0.51 | 0.030 | 0.001 |
| Octyl GSL (C8) | 0.10 | 0.005 | 0.09 | 0.008 | 0.001 | 0.32 | 0.014 | 0.29 | 0.022 | 0.003 |
Number of individual lines measured per genotype class.
Mean is the mean value of MT:(MS + MT).
Standard error for the mean value.
P value for GSL differences between the two genotypes as determined by ANOVA.
Nonsignificant P values (P > 0.05).
Given GSL was not detectable; therefore, no statistical analyses were conducted.
Table II.
Altered GS-OX activity in the FMOGS-OX5 T-DNA mutant
GSL content in seeds and leaves was analyzed. All plants were segregants derived from a parental line heterozygous for the T-DNA knockout allele. MT:(MS + MT) represents the S-oxygenation activity for the conversion from MT GSL to MS GSL. This T-DNA mutant is in a wild-type Col-0 background.
| MT:(MS + MT) | Leaf Tissue
|
Seed Tissue
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Homozygous Knockout (na = 35)
|
Homozygous Wild Type (n = 29)
|
Pd | Homozygous Knockout (n = 29)
|
Homozygous Wild Type (n = 26)
|
P | |||||
| Meanb | sec | Mean | se | Mean | se | Mean | se | |||
| Propyl GSL (C3) | NDe | ND | 0.03 | 0.000 | 0.03 | 0.000 | NS | |||
| Butyl GSL (C4) | 0.20 | 0.000 | 0.20 | 0.000 | NSf | 0.80 | 0.010 | 0.79 | 0.010 | NS |
| Pentyl GSL (C5) | ND | ND | 0.91 | 0.010 | 0.90 | 0.010 | NS | |||
| Hexyl GSL (C6) | ND | ND | ND | ND | ||||||
| Heptyl GSL (C7) | 0.20 | 0.020 | 0.20 | 0.030 | NS | ND | ND | |||
| Octyl GSL (C8) | 0.27 | 0.040 | 0.28 | 0.050 | NS | 0.39 | 0.010 | 0.37 | 0.010 | <0.001 |
Number of individual lines measured per genotype class.
Mean value of MT:(MS + MT).
Standard error for the mean value.
P value for GSL differences between the two genotypes as determined by ANOVA.
Given GSL was not detectable; therefore, no statistical analyses were conducted.
Nonsignificant P values (P > 0.05).
Table III.
Altered GS-OX activity in the FMOGS-OX4 T-DNA mutant
GSL content in seeds and leaves was analyzed. All plants were segregants derived from a parental line heterozygous for the T-DNA knockout allele. MT:(MS + MT) represents the S-oxygenation activity for the conversion from MT GSL to MS GSL. This T-DNA mutant is in a wild-type Col-0 background.
| MT:(MS + MT) | Leaf Tissue
|
Seed Tissue
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Homozygous Knockout (na = 40)
|
Homozygous Wild Type (n = 37)
|
Pd | Homozygous Knockout (n = 23)
|
Homozygous Wild Type (n = 24)
|
P | |||||
| Meanb | sec | Mean | se | Mean | se | Mean | se | |||
| Propyl GSL (C3) | 0.26 | 0.020 | 0.24 | 0.020 | NSe | NDf | ND | |||
| Butyl GSL (C4) | 0.41 | 0.020 | 0.39 | 0.020 | NS | 0.72 | 0.010 | 0.71 | 0.010 | NS |
| Pentyl GSL (C5) | 0.23 | 0.020 | 0.22 | 0.020 | NS | 0.79 | 0.020 | 0.76 | 0.020 | NS |
| Hexyl GSL (C6) | ND | ND | ND | ND | ||||||
| Heptyl GSL (C7) | 0.25 | 0.010 | 0.23 | 0.010 | NS | 0.62 | 0.020 | 0.58 | 0.030 | NS |
| Octyl GSL (C8) | 0.11 | 0.010 | 0.12 | 0.010 | NS | 0.32 | 0.010 | 0.31 | 0.010 | NS |
Number of individual lines measured per genotype class.
Mean value of MT:(MS + MT).
Standard error for the mean value.
P value for GSL differences between the two genotypes as determined by ANOVA.
Nonsignificant P values (P > 0.05).
Given GSL was not detectable; therefore, no statistical analyses were conducted.
The only available T-DNA knockout mutant for FMOGS-OX3 is in the Ler accession. In contrast to the Col-0 accession, Ler has predominantly propyl C3 instead of butyl C4 Met-derived GSLs (Kliebenstein et al., 2001b). Another difference between the two ecotypes is that Ler expresses a functional AOP3 (2-oxoglutarate-dependent dioxygenase; Kliebenstein et al., 2001c), which catalyzes the conversion from MS GSL to hydroxyalkyl (OH) GSL (Fig. 1). This results in accumulation of OH GSL instead of MS GSL in Ler. Therefore, the ratio of MT GSL to the sum of MT and OH GSLs was used as a measure for GS-OX activity in Ler. Significant increase of 3-MT:(3-MT + 3-OH) was observed in both leaves and seeds in homozygous knockout mutants compared to wild-type plants (Table IV). No significant difference was found for other MT GSLs in this knockout mutant, which suggests a preference of FMOGS-OX3 for short-chain MT GSLs in Ler. In our data, there is agreement between the in vitro and in planta activities for the majority of FMOGS-OXs.
Table IV.
Altered GS-OX activity in the FMOGS-OX3 T-DNA mutant
GSL content in seeds and leaves was analyzed. All plants were segregants derived from a parental line heterozygous for the T-DNA knockout allele. MT:(OH + MT) represents the S-oxygenation activity for the conversion from MT GSL to MS GSL. This T-DNA mutant is in a wild-type Ler background.
| MT:(OH + MT) | Leaf Tissue
|
Seed Tissue
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Homozygous Knockout (na = 21)
|
Homozygous Wild Type (n = 26)
|
Pd | Homozygous Knockout (n = 17)
|
Homozygous Wild Type (n = 19)
|
P | |||||
| Meanb | sec | Mean | se | Mean | se | Mean | se | |||
| Propyl GSL (C3) | 0.38 | 0.020 | 0.02 | 0.020 | <0.001 | 0.72 | 0.030 | 0.21 | 0.020 | <0.001 |
| Butyl GSL (C4) | NDe | ND | ND | ND | ||||||
| Pentyl GSL (C5) | ND | ND | ND | ND | ||||||
| Hexyl GSL (C6) | ND | ND | ND | ND | ||||||
| Heptyl GSL (C7) | ND | ND | 0.74 | 0.040 | 0.72 | 0.040 | NS | |||
| Octyl GSL (C8) | 0.11 | 0.010 | 0.09 | 0.010 | NSf | 0.64 | 0.030 | 0.56 | 0.030 | NS |
Number of individual lines measured per genotype class.
Mean value of MT:(MS + MT).
Standard error for the mean value.
P value for GSL differences between the two genotypes as determined by ANOVA.
ND, GSL was not detectable; therefore, no statistical analyses were conducted.
NS, Nonsignificant P values (P > 0.05).
Ratio of MT:(MT + MS) Decreased in FMOGS-OX Overexpressers
To further complement the in vitro data, we individually overexpressed all four FMOs in Col-0. For each 35S∷FMOGS-OX construct, two independent T1 lines were identified, segregants from these lines were genotyped, and GSLs measured from leaves and seeds of homozygotes and wild-type offspring (Supplemental Table S5). The ratio of MT:(MT + MS) was calculated for each chain length to estimate the GS-OX activity. There was no statistically significant difference between the two independent transgenic lines for any 35S∷FMOGS-OX construct, and therefore data from the two lines were pooled.
In leaves, MT GSLs were extensively converted into MS GSLs in the 35S∷FMOGS-OX2 and 35S∷FMOGS-OX3 lines, resulting in a significant decrease of the MT:(MT + MS) ratio in comparison to the wild type (Tables V and VI). In seeds of the 35S∷FMOGS-OX3, all MT GSLs were converted to MS GSLs, whereas only the short-chain butyl C4 Met-derived GSL showed a slight decrease in MT:(MT + MS) ratio in the seeds of 35S∷FMOGS-OX2 (Tables V and VI). These data, combined with the T-DNA and in vitro analysis, suggest that, whereas these two genes are both FMOGS-OX enzymes, they have slightly different specificities.
Table V.
Altered GS-OX activity in the FMOGS-OX2 overexpression lines
GSL content in seeds and leaves was analyzed. All plants were segregants derived from two independent T1 generation 35S∷FMOGS-OX2 lines. MT:(MS + MT) represents the S-oxygenation activity for the conversion from MT GSL to MS GSL.
| MT:(MS + MT) | Leaf Tissue
|
Seed Tissue
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
35S∷FMOGS-OX2 (na = 46)
|
Col-0 (n = 24)
|
Pdgene |
35S∷FMOGS-OX2 (n = 23)
|
Col-0 (n = 18)
|
Pgene | |||||
| Meanb | sec | Mean | se | Mean | se | Mean | se | |||
| Propyl GSL (C3) | 0.07 | 0.002 | 0.08 | 0.006 | <0.001 | NDe | ND | |||
| Butyl GSL (C4) | 0.18 | 0.004 | 0.34 | 0.013 | <0.001 | 0.67 | 0.013 | 0.71 | 0.011 | 0.046 |
| Pentyl GSL (C5) | 0.03 | 0.014 | 0.47 | 0.015 | <0.001 | 0.73 | 0.024 | 0.76 | 0.017 | NSf |
| Hexyl GSL (C6) | ND | ND | ND | ND | ||||||
| Heptyl GSL (C7) | 0.08 | 0.011 | 0.37 | 0.016 | <0.001 | 0.52 | 0.031 | 0.54 | 0.031 | NS |
| Octyl GSL (C8) | 0.03 | 0.006 | 0.19 | 0.009 | <0.002 | 0.32 | 0.015 | 0.32 | 0.010 | NS |
Number of individual lines measured per genotype class.
Mean value of MT:(MS + MT).
Standard error for the mean value.
P value for GSL differences between the two genotypes as determined by ANOVA.
Given GSL was not detectable; therefore, no statistical analyses were conducted.
Nonsignificant P values (P > 0.05).
Table VI.
Altered GS-OX activity in the FMOGS-OX3 overexpression lines
GSL content in seeds and leaves was analyzed. All plants were segregants derived from two independent T1 generation 35S∷FMOGS-OX3 lines. MT:(MS + MT) represents the S-oxygenation activity for the conversion from MT GSL to MS GSL.
| MT:(MS + MT) | Leaf Tissue
|
Seed Tissue
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
35S∷FMOGS-OX3 (na = 19)
|
Col-0 (n = 13)
|
Pdgene |
35S∷FMOGS-OX3 (n = 12)
|
Col-0 (n = 8)
|
Pgene | |||||
| Meanb | sec | Mean | se | Mean | se | Mean | se | |||
| Propyl GSL (C3) | 0.05 | 0.000 | 0.12 | 0.010 | <0.001 | 0.01 | 0.000 | 0.03 | 0.000 | <0.001 |
| Butyl GSL (C4) | 0.02 | 0.000 | 0.22 | 0.020 | <0.001 | 0.35 | 0.020 | 0.74 | 0.020 | <0.001 |
| Pentyl GSL (C5) | 0.03 | 0.010 | 0.31 | 0.030 | <0.001 | 0.48 | 0.030 | 0.83 | 0.020 | <0.001 |
| Hexyl GSL (C6) | NDe | ND | 0.13 | 0.020 | 0.33 | 0.030 | <0.001 | |||
| Heptyl GSL (C7) | 0.07 | 0.020 | 0.29 | 0.040 | <0.001 | 0.35 | 0.020 | 0.63 | 0.020 | <0.001 |
| Octyl GSL (C8) | 0.04 | 0.010 | 0.18 | 0.020 | <0.001 | 0.15 | 0.010 | 0.33 | 0.010 | <0.001 |
Number of individual lines measured per genotype class.
Mean value of MT:(MS + MT).
Standard error for the mean value.
P value for GSL differences between the two genotypes as determined by ANOVA.
Given GSL was not detectable; therefore, no statistical analyses were conducted.
Table VII.
Altered GS-OX activity in the FMOGS-OX4 overexpression lines
GSL content in seeds and leaves was analyzed. All plants were segregants derived from two independent T1 generation 35S∷FMOGS-OX4 lines. MT:(MS + MT) represents the S-oxygenation activity for the conversion from MT GSL to MS GSL.
| MT:(MS + MT) | Leaf Tissue
|
Seed Tissue
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
35S∷FMOGS-OX4 (na = 66)
|
Col-0 (n = 23)
|
Pdgene |
35S∷FMOGS-OX4 (n = 13)
|
Col-0 (n = 6)
|
Pgene | |||||
| Meanb | sec | Mean | se | Mean | se | Mean | se | |||
| Propyl GSL (C3) | 0.13 | 0.010 | 0.13 | 0.010 | NSe | ND | ND | |||
| Butyl GSL (C4) | 0.26 | 0.010 | 0.25 | 0.010 | 0.001 | 0.75 | 0.040 | 0.74 | 0.050 | 0.030 |
| Pentyl GSL (C5) | 0.22 | 0.010 | 0.22 | 0.010 | NS | ND | ND | |||
| Hexyl GSL (C6) | NDf | ND | ND | ND | ||||||
| Heptyl GSL (C7) | 0.26 | 0.010 | 0.25 | 0.010 | NS | 0.37 | 0.040 | 0.30 | 0.080 | NS |
| Octyl GSL (C8) | 0.14 | 0.010 | 0.14 | 0.010 | NS | 0.21 | 0.030 | 0.18 | 0.050 | NS |
Number of individual lines measured per genotype class.
Mean value of MT:(MS + MT).
Standard error for the mean value.
P value for GSL differences between the two genotypes as determined by ANOVA.
Nonsignificant P values (P > 0.05).
Given GSL was not detectable; therefore, no statistical analyses were conducted.
35S∷FMOGS-OX4 did not show a decrease in the ratio of MT:(MT + MS) either in leaves or in seeds (Table VII). For C4 GSLs, a very slight, but statistically significant, increase of MT:(MT + MS) was detected, indicating a possible repression of the conversion from 4-MTB to 4-MSB. As with the other FMOGS-OX overexpresser data, the 35S∷FMOGS-OX4 data were derived from two independent, segregating T1 transgenic overexpression lines that exhibited a significant decrease of the MT:(MT + MS) ratio in comparison to wild-type plants (Supplemental Fig. S2; Supplemental Table S6). This suggests that FMOGS-OX4 catalyzes the S-oxygenation reaction and that the 35S∷FMOGS-OX4 transgene was silenced in the T2 generation.
In agreement with its predicted substrate specificity, 35S∷FMOGS-OX5 lines had a significant decrease of MT:(MT + MS) for only octyl C8 GSL in seeds compared to wild type (Table VIII). Interestingly, 7-methylthioheptyl (7-MTH) GSL, another long-chain MT GSL, had a similar concentration as 8-MTO in seeds of the wild-type plant, but we did not detect any 7-MSH GSL, indicating that no significant conversion from 7-MTH to 7-MSH occurred in 35S∷FMOGS-OX5 (Supplemental Table S5). This confirmed that FMOGS-OX5 is specific for the 8-MTO GSL substrate.
Table VIII.
Altered GS-OX activity in the FMOGS-OX5 overexpression lines
GSL content in seeds and leaves was analyzed. All plants were segregants derived from two independent T1 generation 35S∷FMOGS-OX5 lines. MT:(MS + MT) represents the S-oxygenation activity for the conversion from MT GSL to MS GSL.
| MT:(MS + MT) | Leaf Tissue
|
Seed Tissue
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
35S∷FMOGS-OX5 (na = 40)
|
Col-0 (n = 36)
|
Pdgene |
35S∷FMOGS-OX5 (n = 28)
|
Col-0 (n = 30)
|
Pgene | |||||
| Meanb | sec | Mean | se | Mean | se | Mean | se | |||
| Propyl GSL (C3) | NDe | ND | 0.03 | 0.000 | 0.03 | 0.000 | NS | |||
| Butyl GSL (C4) | 0.22 | 0.000 | 0.30 | 0.000 | 0.017 | 0.82 | 0.010 | 0.72 | 0.040 | NS |
| Pentyl GSL (C5) | ND | ND | 0.90 | 0.010 | 0.94 | 0.010 | NS | |||
| Hexyl GSL (C6) | ND | ND | ND | ND | ||||||
| Heptyl GSL (C7) | 0.13 | 0.030 | 0.14 | 0.020 | NSf | ND | ND | |||
| Octyl GSL (C8) | 0.11 | 0.000 | 0.11 | 0.000 | NS | 0.14 | 0.020 | 0.34 | 0.020 | <0.001 |
Number of individual lines measured per genotype class.
Mean value of MT:(MS + MT).
Standard error for the mean value.
P value for GSL differences between the two genotypes as determined by ANOVA.
Given GSL was not detectable; therefore, no statistical analyses were conducted.
Nonsignificant P values (P > 0.05).
DISCUSSION
Modifications of the GSL side chain are of particular importance because the biological activity of the GSL hydrolysis products is determined to a large extent by the structure of the side chain. We identified four new FMO genes encoding for enzymes capable of S-oxygenating aliphatic GSLs. Three of these FMOs, FMOGS-OX2 to FMOGS-OX4, were identified by fine mapping of a 200-kb region containing a GS-OX QTL on chromosome I in a Ler × Wei-0 F2 population. The three FMOs clustered together with FMOGS-OX5 and the previously characterized FMOGS-OX1 in a subclade that appears to be crucifer specific. FMOGS-OX2, FMOGS-OX3, and FMOGS-OX4 S-oxygenated all available MT GSLs as did FMOGS-OX1, whereas FMOGS-OX5 showed high substrate specificity to 8-MTO GSL.
Substrate Specificity of FMOGS-OXs
In animals, there are five functionally expressed FMO genes that detoxify a vast spectrum of xenobiotics. This vast spectrum is due to broad substrate specificity that makes the mammalian FMOs capable of oxidizing thousands of plant natural products as well as thousands of synthetic therapeutic drugs (Krueger and Williams, 2005; Cashman and Zhang, 2006). Plants, on the other hand, have many more FMO genes, possibly with more restricted substrate specificity. In Arabidopsis, there are 29 genes with homology to known FMOs (Schlaich, 2007). The enzyme FMOGS-OX1 was shown to have substrate specificity in that it required an S-Glc group on its substrates (Hansen et al., 2007). FMOGS-OX1 S-oxygenated desulfo and intact GSLs, but not the structurally related Met and aldoxime that do not contain an S-Glc group. In this article, we show additional substrate specificity in plant FMOs because we found that FMOGS-OX5 is specific for long-chain aliphatic GSLs. This indicates that the S-Glc group is not the only structural requirement for the substrate of FMOGS-OX5. Size restriction might be another factor determining substrate specificity whereby FMOGS-OX5 excludes the short-chain GSLs.
It is reported that when there is no available substrate, the FMO proteins exist as 4α-hydroperoxy flavin, which is a precharged complex containing a reduced form of fatty acid dehydrogenase and NADPH that is ready to act on substrates (Eswaramoorthy et al., 2006). The 4α-hydroperoxy flavin complex of FMOGS-OX5 may have a tertiary structure in the binding site that only fits with 8-MTO and prevents shorter chain-length MT GSLs from either entering or being properly held by the enzyme. However, the mechanism of the conformational and chemical complementarities between the MT GSLs and FMOGS-OX enzymes remains to be shown.
The phylogenetic tree of plant FMOs in rice, poplar, and Arabidopsis contains three clades (Hansen et al., 2007). Our phylogenetic analysis of FMOGS-OX homologs (Fig. 2) is based on the genomic sequences present in clade III and EST sequences with more than 80% identity to at least one of the three FMOs used for database searching. In this proposed S-oxygenation clade III (Fig. 2), FMOGS-OX1 to FMOGS-OX5 are the only characterized genes. Given that poplar, rice, alfalfa (Medicago sativa), grape, tomato (Solanum lycopersicum), lemon (Citrus limon), and moss do not produce GSLs, other FMOs in this clade may be involved in S-oxygenation for a diversity of sulfur-containing compounds and therefore contribute to a diversity of biological functions (Fig. 2).
Possible Biological Function of FMOGS-OXs
We have shown that the biochemical function of the FMOGS-OXs is to catalyze the S-oxygenation of the endogenous substrate MT GSL to MS GSL. The physiological function of FMOGS-OXs depends on the biological activity of the hydrolysis products of the MS GSLs and of further modified GSLs. For humans, the isothiocyanate hydrolysis products of 4-MSB, 7-MSH, and 8-MSO GSLs have been shown to be strong inducers of phase II enzymes and thereby function as cancer-preventing agents (Rose et al., 2000; Fahey et al., 2002). In planta, MS GSLs have been shown to play a role in protection against insects (Rohr et al., 2006), and the isothiocyanate derived from 4-MSB GSL has been shown to play a role in protecting Arabidopsis against pathogens (Tierens et al., 2001). This indicates that FMOGS-OXs are important for plant defense responses. In some Arabidopsis accessions, MS GSLs can be converted to OH, alkenyl, hydroxylalkenyl, and benzoyloxy GSLs (Fig. 1), adding more layers of complexity to the warfare between Arabidopsis and its enemies (Kliebenstein et al., 2001c; Tierens et al., 2001). The plants overexpressing FMOGS-OXs showed significant increases in the accumulation of MS GSLs at the expense of the MT GSLs. This indicates that the production of MS GSLs and the further modified GSLs downstream in the pathway (Fig. 1) require the FMOGS-OX proteins. This is an important consideration when FMOGS-OXs are utilized in breeding and genetic engineering toward plants with a high level of cancer-preventive methylsulfinylalkyl isothiocyanates, increased pathogen resistance, or decreased level of the deleterious downstream GSLs, such as progoitrin, 2-hydroxy-but-3-enyl GSL (Kliebenstein et al., 2001a; Halkier and Gershenzon 2006). In addition, when altering plants via breeding or engineering approaches toward isothiocyanates of MS GSLs, it is important to also incorporate genes affecting the breakdown of GSLs into the scheme to ensure that isothiocyanates are produced.
Gene Duplication and Evolution of FMOGS-OXs
The Arabidopsis FMOGS-OXs are present in three gene clusters that appear to have evolved through a combination of local tandem duplication, whole-genome duplications, and a distal duplication (Fig. 2; Vision et al., 2000; Blanc and Wolfe 2004; Rizzon et al., 2006). Interestingly, this group of FMOs may represent crucifer-specific radiation because we have to date only identified cruciferous genes to reside within this FMOGS-OX subclade. Whereas we utilized all available sequence databases, further work is required to fully validate whether this is in fact crucifer-specific radiation associated with the evolution of GSL biosynthesis. Thus, this FMO subclade begins to allow potential detailed analysis of how gene duplicates can either partition the original function (subfunctionalization) or derive entirely new functions (neofunctionalization; Lynch and Conery, 2000). In the Arabidopsis FMOGS-OX part of this FMO subclade, there is evidence for subfunctionalization; however, the true direction of this process will require the identification and biochemical characterization of FMOGS-OX ancestral genes in species basal to the Cruciferae. Phylogenetic analysis suggests that there might be a precursor FMOGS-OX, which first duplicated into FMOGS-OX5 and a FMOGS-OX1 to FMOGS-OX4 precursor. The different substrate specificity between FMOGS-OX5 and FMOGS-OX1 to FMOGS-OX4 is potentially an instance of biochemical subfunctionalization whereby FMOGS-OX5 is likely subfunctionalized for long chain-length substrate, whereas FMOGS-OX1 to FMOGS-OX4 retained broad chain-length specificity from the precursor protein. Further subfunctionalization, potentially from tissue-specific expression patterns, can also be found within FMOGS-OX1 to FMOGS-OX4 and is evidenced by the knockout mutant analysis where both FMOGS-OX1 and FMOGS-OX2 control 8-MSO production in planta, whereas FMOGS-OX3, which is capable of catalyzing this reaction in vitro, does not control this reaction in planta (Tables I and II; Hansen et al., 2007). These slight differences between FMOGS-OX genes might be explained by analysis of microarray data (http://www.genevestigator.ethz.ch; Zimmermann et al., 2004) showing that these genes have different expression patterns across organs and growth stages. Specific identification and validation of the tissue expression patterns of these FMOGS-OXs and the substrates for the other members of this FMO clade could help our understanding of how and when duplicates undergo either biochemical or expression-based subfunctionalization.
Gene Duplication and Quantitative Genetic Variation
The association of the duplicated FMOGS-OXs with independent QTLs for the S-oxygenation reaction in Arabidopsis raises another possible role for duplicated gene families (Fig. 2). Duplicated gene families, while providing redundancy to a system, may also enhance the potential for quantitative variation within a trait. This is illustrated by the fact that each of the FMOGS-OXs has a large expression polymorphism, yet there is only quantitative variation for this trait rather than qualitative. As such, the large polymorphisms in each independent gene are dampened by the presence of the other genes. While duplicated genes do show enhanced levels of genetic variation as would be expected under this model (Gu et al., 2004; Kliebenstein, 2008), it remains to be seen whether duplicated gene families show any bias in controlling quantitative trait variation.
In summary, identification of S-oxygenating activity of the FMOGS-OXs may impact both applied and basic research fields. These genes can potentially be applied in genetic engineering for the production of 4-MSB, 7-MSH, and 8-MSO GSLs, or removal of 2-hydroxy-but-3-enyl GSL. In addition, the characterization of the FMOGS-OXs will help to gain more biochemical insight into plant FMO proteins, which we have just begun to learn about, and it may also bring clues for the functions of the noncharacterized plant FMOs. Finally, this well-defined gene family may provide an optimal model for studying neo- versus subfunctionalization following gene duplication.
MATERIALS AND METHODS
Generation of Phylogenetic Tree
The entire FMO complement from the genomic sequence for Medicago truncatula, grape (Vitis vinifera), Physcomitrella patens, rice (Oryza sativa), Arabidopsis (Arabidopsis thaliana), and poplar (Populus tremuloides) were obtained and translated into their corresponding amino acid sequence. Gene abbreviations are per genome consortium convention or previous publication (Hansen et al., 2007; Supplemental Fig. S1). All FMO amino acid sequences were used to construct a complete neighbor-joining tree with 1,000 bootstraps in ClustalX. The P. patens sequence most closely associated with FMOGS-OX1 was used to define the S-oxygenation subclade for further analysis (Fig. 2). This subclade includes all previous poplar and rice sequences predicted to be within the S-oxygenation subclade. We then identified all EST sequences showing at least 80% amino acid identity with one of three FMOs and included these in the phylogenetic analysis. All of these EST sequences clustered within the P. patens sequence on an unrooted cladogram containing all sequences and, as such, we utilized the P. patens sequence as a root for the presented cladogram focused on the S-oxygenation subclade (Fig. 2). Only those branches with at least 600 of 1,000 bootstrap support are labeled.
Genetic Mapping of GS-OX
The accessions Ler and Wei-0 of Arabidopsis were crossed and the resulting F1 selfed to generate an F2 mapping population. These accessions were chosen because they have the same alleles at the AOP and Elong loci and thereby the only aliphatic GSL structural polymorphism between these accessions is a GS-OX polymorphism (Kliebenstein et al., 2001b; Burow et al., 2008). Four hundred F2 plants were screened for recombination events between the AthGeneA and nga692 markers on chromosome I using a previously described high-throughput DNA extraction and PCR protocol (Kliebenstein et al., 2001b, 2001c). GSLs were analyzed via HPLC for all 400 lines. All recombinant F2 were selfed and eight F3 progeny tested from each parent to assess segregation of the GS-OX phenotype to confirm the parental genotype at GS-OX. Microsatellite markers were made using available genomic sequence for fine-scale mapping. These markers were F19K23 (F, GGTCTAATTGCCGTTGTTGC; R, GAATTCTGTAACATCCCATTTCC), F23N19 (F, TTCCACACTTTCACCGATCA; R, GCTTTTGTTTCTCCCCTTCC), F9N12 (F, CGAGTCTAGAAACTCCGGTGA; R, TCAGCGTAAACCGTCTTTCC); T12P18 (F, GAGAGTCTTCTATTAGAAG; R, CAAATTTGTTAATGTGCGTG); and F24O1 (F, TGCTTCAAGCCCAAGCTTAT; R, AGCGGCAAGAGGAAGTATGA). All recombinant F2 progeny were genotyped with these markers to resolve the position of GS-OX.
Previous HPLC data on GSL accumulation in the leaves and seeds of the Ler × Cvi RILs was reanalyzed to measure the GS-OX phenotype for the short-chain aliphatic GSLs (Supplemental Tables S2 and S3; Kliebenstein et al., 2001a). QTL cartographer was used to map QTLs for this trait as previously described and ANOVA utilized to test the significant QTLs for an impact on the short-chain GS-OX phenotype (Kliebenstein et al., 2001a). The same ANOVA was used to test for epistatic interactions between significant QTLs using the previously published Bay-0 × Sha GSL data (Wentzell et al., 2007).
Heterologous Expression of FMOGS-OX Proteins in Escherichia coli
The coding sequences of the four FMOGS-OXs were amplified by RT-PCR. Total plant RNA was isolated with TRIzol (Invitrogen) according to the manufacturer's recommendations. First-strand cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). The coding sequence of FMOGS-OX2 (At1g62540) was amplified from first-strand cDNA with the following primers: 5′-ATGGCACCAGCTCAAAACCC-3′ and 5′-GAGGATATGGGAAGGATG GAAACTAAT-3′. The coding sequence of FMOGS-OX3 (At1g62560) was amplified with the following primers: 5′-ATGGCACCAGCTCAAAACCAAATC-3′ and 5′-TCTTCCATTTTCGAGGTAATAAG-3′. The coding sequence of FMOGS-OX4 (At1g62570) was amplified with the following primers: 5′-ATGGCACCAGCTCCTAGTCCAAT-3′ and 5′-TCTTCCGGATTCGAGAAAACGA-3′. The coding sequence of FMOGS-OX5 (At1g12140) was amplified with the following primers: 5′-ATGGCACCAGCACGAACCCGA-3′ and 5′-AGATTCCAATAACTGAGAAGGAAG-3′. The amplified PCR products were cloned into pBAD-TOPO vector using pBAD-TOPO TA expression kit (Invitrogen) according to the manufacturer's protocol, resulting in Ara-inducible expression constructs for His-tagged FMOGS-OX proteins. The constructs were confirmed by sequencing. Constructs expressing FMOGS-OX proteins and a negative control (empty pBAD-TOPO vector) were transformed into the E. coli strain TOP10 (Invitrogen). Expression of FMOGS-OXs was performed according to the manufacturer's recommendation with 0.02% Ara followed by growth at 28°C, 250 rpm for 2 h. E. coli spheroplasts were isolated as previously described (Hansen et al., 2007).
Spheroplast Enzymatic Assays
The enzymatic activity of the four FMOGS-OXs was analyzed by spheroplast enzymatic assays. A 100-μL volume of assay solution contained spheroplasts corresponding to 50 μg of total E. coli protein, substrate, 0.1 m Tricine (pH 7.9), and 0.25 mm NADPH. The reaction mixture was incubated for 1 h at 30°C followed by the addition of 100 μL methanol and centrifugation at 5,000g for 2 min. Supernatant (200 μL) was lyophilized and dissolved in 50 μL water. In the assays using desulfo 4-MTB GSL as substrate, final substrate concentration was 0.25 mm. In the assays using desulfo GSL extracts from Arabidopsis seeds as substrate, final concentration was 2 mm total GSLs.
Plant Growth
Plants were grown in a growth chamber at a photosynthetic flux of 100 μE at 20°C and 70% relative humidity with a 16/8-h photoperiod.
Genotyping and RT-PCR of FMOGS-OX T-DNA Insertion Mutants
Two T-DNA insertion mutants for each of FMOGS-OX2 and FMOGS-OX4 and one T-DNA insertion mutant for FMOGS-OX5 in Col-0 background were obtained. One T-DNA insertion mutant in ecotype Ler background was obtained for FMOGS-OX3.
The insertion mutants for FMOGS-OX2 were the Salk_080561 line (FMOGS-OX2-a) and Salk_098896 line (FMOGS-OX2-b; Alonso et al., 2003). Primers for genotyping FMOGS-OX2-a were 5′-TTTCCCACGATGAGATCTTTG-3′, 5′-TGCGTGTCTTTATAACACGTTTC-3′, and the T-DNA-specific primer LBa1 5′-TGGTTCACGTAGTGGGCCATCG-3′. Primers for genotyping FMOGS-OX2-b are 5′-TTTGCCACTGCTGGATTTTAG-3′, 5′-ACCCAGTGCAATACACAATGC-3′, and T-DNA-specific primer LBa1.
The insertion mutant for FMOGS-OX3 was GT13906 line (Martienssen, 1998) in Ler background and genotyping primers were 5′-CCAAGCTTGATTAACTCGCATC-3′, 5′-GCTCTAGACGCAATGTGGACTT-3′, and T-DNA-specific primer DS5-2, 5′-GCGTTTTGTATATCCCGTTTCCGT-3′.
The insertion mutants for FMOGS-OX4 were Salk-059185 line (FMOGS-OX4-a) and Salk_078861 line (FMOGS-OX4-b; Alonso et al., 2003). Genotyping primers for FMOGS-OX4-a were 5′-CATTTCGCAGAACCAAACATC-3′, 5′-GACGTTTTTCGAAAGTGTTGG-3′, and T-DNA-specific primer LBa1. Genotyping primers for FMOGS-OX4-b were 5′-TAGCGCAGGTGGAAAAATATG-3′, 5′-AACGTTGGGATACGTGTTGAG-3′, and T-DNA-specific primer LBa1.
The insertion mutants for FMOGS-OX5 were WiscDsLox361H10 line (Woody et al., 2007). Genotyping primers for FMOGS-OX5 were 5′-CCCTGGCCATGAATCTATACC-3′, 5′-AAAATGTCTTCGGTTGGTTCC-3′, and T-DNA-specific primer 5′-AACGTCCGCAATGTGTTATTAAGTTGTC-3′.
Leaves from homozygous FMOGS-OX2-a, FMOGS-OX2-b, FMOGS-OX3, FMOGS-OX4-a, FMOGS-OX4-b, and FMOGS-OX5 were harvested 20 to 25 d after germination. RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). The primers used in cloning of each gene for heterologous expression in E. coli were used as RT-PCR primers.
GSL Extraction and Analysis
GSL extraction was performed as previously described (Hansen et al., 2007). Fifty to 100 mg leaves and 10 to 20 mg seeds materials were used for the extraction. HPLC analysis was described previously (Hansen et al., 2007).
Analysis of GSL Content in FMOGS-OX Knockout Mutants
For each FMOGS-OX T-DNA insertion mutant line, a single heterozygous plant was allowed to self-pollinate. Plants from segregating seeds were grown in two independent replicates. At 20 to 25 d, individual leaves from each plant were harvested for GSL analysis and for PCR genotyping. GSL contents in wild-type, heterozygous, and homozygous plants were compared. Nested ANOVA was used to test the impact of the T-DNA insertion within each of FMOGS-OX genes on all individual GSLs and resultant variables as described previously (Hansen et al., 2007). Seeds were analyzed the same way as leaves.
FMOGS-OX Overexpression in Arabidopsis
The coding sequences of the four FMOGS-OXs were amplified from the constructs for the heterologous expression in E. coli (described above) with Pfu Turbo CX Hotstart DNA polymerase (Stratagene). The overexpression constructs driven by the cauliflower mosaic virus 35S promoter were created by cloning the above PCR product into pCAMBIA230035Su using the USER method as described (Nour-Eldin et al., 2006).
Primers for amplification of FMOGS-OX2 were 5′-GCTTAAUATGGCACCAGCTCAAAACC-3′ and 5′-GGTTTAAUTTAGAGGATATGGGAAGG-3′. Primers for FMOGS-OX3 were 5′-GGCTTAAUATGGCACCAGCTCAAAACCA-3′ and 5′-GGTTTAAUCATCTTCCATTTTCGAGGTAATAA-3′.
Primers for FMOGS-OX4 were 5′-GGCTTAAUATGGCACCAGCTC-3′ and 5′-GGTTTAAUCGTAGTCAAACTTCATCTTCCG-3′. Primers for FMOGS-OX5 were 5′-GGCTTAAUATGGCACCAGCACGAACCCGA-3′ and 5′-GGTTTAAUTCAAGATTCCAATAACTGAGAAGG-3′.
Plant Transformation
The overexpression constructs with FMOGS-OXs driven by the 35S promoter were transformed into Agrobacterium tumefaciens strain C58 (Zambryski et al., 1983), and then into Arabidopsis (Col-0) via the floral-dip method (Zhang et al., 2006a). Transgenic plants were selected on 0.5× Murashige and Skoog medium with 50 μg·mL−1 kanamycin.
Analysis of GSL Content in FMOGS-OX Overexpression Lines
For each FMOGS-OX gene, two independent T1 transgenic lines with high FMOGS-OX activity were selected for further analysis. Plants from each transgenic line were grown in two independent biological replicates. Leaf material from each plant was harvested for GSL analysis and for genotyping for the presence or absence of the NptII transgene using the primers 5′-CAGCAATATCACGGGTAGCCA-3′ and 5′-GGCTATTCGGCTATGACTGGG-3′. GSL contents in transgenic and wild-type Arabidopsis plants were compared. Nested ANOVA was used to test the impact of FMOGS-OX overexpression on all individual GSLs and resultant variables as previously described (Hansen et al., 2007). Seeds were harvested and analyzed the same way as described above.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Confirmation of FMOGS-OX T-DNA mutants by RT-PCR.
Supplemental Figure S2. GSL profile of seed in T1 generation of 35S∷FMOGS-OX4 and wild-type plants.
Supplemental Table S1. FMOGS-OX homologous sequences utilized in Figure 2.
Supplemental Table S2. Ler × Cvi short-chain aliphatic seed FMOGS-OX analysis.
Supplemental Table S3. Ler × Cvi short-chain aliphatic leaf FMOGS-OX analysis.
Supplemental Table S4. GSL profile of FMOGS-OX T-DNA knockout mutants.
Supplemental Table S5. GSL profiles of 35S∷FMOGS-OX4 overexpression lines and wild-type plants.
Supplemental Table S6. Altered FMOGS-OX activity in the T1 generation of FMOGS-OX4 overexpression lines.
Supplementary Material
This work was supported by the Villum Kann Rasmussen (VKR) Foundation (grant to VKR Research Centre for Pro-Active Plants); the National Science Foundation (grant nos. DBI–0642481 and MCB–0323759 to D.J.K.); Research School for Biotechnology graduate school (Ph.D. stipend to B.G.H.); and a Marie Curie IIF fellowship (contract no. MIF1–CT–2006–022344 to J.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Barbara Ann Halkier (bah@life.ku.dk).
The online version of this article contains Web-only data.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burow M, Zhang ZY, Ober JA, Lambrix VM, Wittstock U, Gershenzon J, Kliebenstein DJ (2008) ESP and ESM1 mediate indol-3-acetonitrile production from indol-3-ylmethyl glucosinolate in Arabidopsis. Phytochemistry 69 663–671 [DOI] [PubMed] [Google Scholar]
- Cashman JR, Zhang J (2006) Human flavin-containing monooxygenases. Annu Rev Pharmacol Toxicol 46 65–100 [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy S, Bonanno JB, Burley SK, Swaminathan S (2006) Mechanism of action of a flavin-containing monooxygenase. Proc Natl Acad Sci USA 103 9832–9837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A (2002) Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc Natl Acad Sci USA 99 7610–7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamoustaris A, Mithen R (1996) Genetics of aliphatic glucosinolates. IV. Side-chain modification in Brassica oleracea. Theor Appl Genet 93 1006–1010 [DOI] [PubMed] [Google Scholar]
- Gu Z, Rifkin SA, White KP, Li WH (2004) Duplicate genes increase gene expression diversity within and between species. Nat Genet 36 577–579 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57 303–333 [DOI] [PubMed] [Google Scholar]
- Hansen BG, Kliebenstein DJ, Halkier BA (2007) Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J 50 902–910 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ (2008) A role for gene duplication and natural variation of gene expression in the evolution of metabolism. PLoS ONE 3 e1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Figuth A, Mitchell-Olds T (2002) Genetic architecture of plastic methyl jasmonate responses in Arabidopsis thaliana. Genetics 161 1685–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Gershenzon J, Mitchell-Olds T (2001. a) Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T (2001. b) Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol 126 811–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T (2001. c) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger SK, Williams DE (2005) Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther 106 357–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104 1173–1184 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155 [DOI] [PubMed] [Google Scholar]
- Martienssen RA (1998) Functional genomics: probing plant gene function and expression with transposons. Proc Natl Acad Sci USA 95 2021–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Norholm MHH, Jensen JK, Halkier BA (2006) Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34 e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS (2006) Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol 2 e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr F, Ulrichs C, Mucha-Pelzer T, Mewis I (2006) Variability of aliphatic glucosinolates in Arabidopsis and their influence on insect resistance. Commun Agric Appl Biol Sci 71 507–515 [PubMed] [Google Scholar]
- Rose P, Faulkner K, Williamson G, Mithen R (2000) 7-Methylsulfinylheptyl and 8-methylsulfinyloctyl isothiocyanates from watercress are potent inducers of phase II enzymes. Carcinogenesis 21 1983–1988 [DOI] [PubMed] [Google Scholar]
- Rose P, Huang Q, Ong CN, Whiteman M (2005) Broccoli and watercress suppress matrix metalloproteinase-9 activity and invasiveness of human MDA-MB-231 breast cancer cells. Toxicol Appl Pharmacol 209 105–113 [DOI] [PubMed] [Google Scholar]
- Schlaich NL (2007) Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci 12 412–418 [DOI] [PubMed] [Google Scholar]
- Talalay P, Fahey JW, Healy ZR, Wehage SL, Benedict AL, Min C, Dinkova-Kostova AT (2007) Sulforaphane mobilizes cellular defenses that protect skin against damage by UV radiation. Proc Natl Acad Sci USA 104 17500–17505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierens KFM, Thomma BPHJ, Brouwer M, Schmidt J, Kistner K, Porzel A, Mauch-Mani B, Cammue BPA, Broekaert WF (2001) Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol 125 1688–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD (2000) The origins of genomic duplications in Arabidopsis. Science 290 2114–2117 [DOI] [PubMed] [Google Scholar]
- Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ (2007) Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet 3 e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MAL, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, Clair DA (2007) Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody S, Austin-Phillips S, Amasino R, Krysan P (2007) The WiscDsLox T-DNA collection: an arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120 157–165 [DOI] [PubMed] [Google Scholar]
- Zambryski P, Joos H, Genetello C, Leemans J, Vanmontagu M, Schell J (1983) Ti-plasmid vector for the introduction of DNA into plant-cells without alteration of their normal regeneration capacity. EMBO J 2 2143–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006. a) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protocols 1 641–646 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Talalay P, Cho CG, Posner GH (1992) A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc Natl Acad Sci USA 6 2399–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ober JA, Kliebenstein DJ (2006. b) The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 18 1524–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.