Abstract
Topoisomerase IIα interacts with numerous nuclear factors, through which it is engaged in diverse nuclear events such as DNA replication, transcription and the formation or maintenance of heterochromatin. We previously reported that topoisomerase IIα interacts with RNA helicase A (RHA), consistent with a recent view that topoisomerases and helicases function together. Intrigued by our observation that the RHA–topoisomerase IIα interaction is sensitive to ribonuclease A, we explored whether the RHA–topoisomerase IIα interaction can be recapitulated in vitro using purified proteins and a synthetic RNA. This work led us to an unexpected finding that an RNA-binding activity is intrinsically associated with topoisomerase IIα. Topoisomerase IIα stably interacted with RNA harboring a 3′-hydroxyl group but not with RNA possessing a 3′-phosphate group. When measured in decatenation and relaxation assays, RNA binding influenced the catalytic function of topoisomerase IIα to regulate DNA topology. We discuss a possible interaction of topoisomerase IIα with the poly(A) tail and G/U-rich 3′-untranslated region (3′-UTR) of mRNA as a key step in transcription termination.
INTRODUCTION
Topoisomerase IIα is a multifunctional enzyme that catalyzes the relaxation of supercoiled DNA, decatenation of interlinked DNA and unknotting of intramolecularly linked DNA by passing a DNA helix through a transient double-strand break in a second helix (1,2). Consistent with its role in chromosome segregation, topoisomerase II was identified as ScI (scaffold-1), a major component of mitotic chromosomes together with ScII (3). By employing a temperature-sensitive top2 mutant in yeast (4–6) or utilizing topoisomerase II-specific inhibitors (7,8), it was demonstrated that topoisomerase II is involved in chromosome condensation. In addition, when topoisomerase II function is blocked after chromosome condensation, cells are arrested at metaphase and chromatids fail to separate (5,9,10). Thus, it is likely that a topoisomerase II-mediated decatenation reaction not only facilitates chromosome condensation but also plays an important role in chromosome segregation.
Topoisomerases are also implicated in RNA metabolism. In 1987, Liu and Wang (11) proposed ‘the twin supercoil model’ to explain the generation of negative supercoils and positive supercoils during transcription and their subsequent removal by topoisomerase I and gyrase, respectively. Since then, accumulating experimental evidence in both prokaryotes and yeast support critical roles for topoisomerases in transcription (12–16). In Drosophila, topoisomerase II interacts with at least three factors, including SCF (supercoiling factor) (17), CHRAC (chromatin accessibility complex) (18) and HDAC1/2 (histone deacetylase 1 and 2) in NuRD (nucleosome remodeling and deacetylase) complex (19). SCF and topoisomerase II function jointly to introduce negative superhelical turns into DNA in vitro and co-localize to puffs on polytene chromosomes (17), implying a role for the SCF–topoisomerase II complex in transcription. Although it is dispensable for the nucleosome spacing activity of CHRAC in vitro, topoisomerase II is critical for the in vivo function of CHRAC (18). The involvement of topoisomerase II in transcription has also been suggested by the repression of genes containing Polycomb group (PcG) target sequences in Drosophila. (20). A co-repressor function of topoisomerase II is further suggested by an observation that it interacts with histone deacetylases HDAC1/2 constituting the NuRD complex that exhibits ATP-dependent nucleosome remodeling activity (19).
A role for topoisomerase IIα in RNA metabolism has also been implied from its physical association with nucleolus-derived cytoplasmic foci (NDF) in telophase (21) and from its co-immunoprecipitation with nucleophosmin/B23 (22), which is a known component of NDF (23). In addition, Tani and co-workers (24) isolated five yeast mutants (ptr7 to ptr11) that accumulate poly(A)+ RNA at a nonpermissive temperature. Among them, ptr11 exhibited the ‘cut’ (cell untimely torn) phenotype and generated anucleolate nuclei. Nuclear accumulation of poly(A)+ mRNA was observed only in anucleolate nuclei and only for intron-containing transcripts. Unexpectedly, ptr11+ gene was identified as top2+ that encodes topoisomerase II, implying a role for topoisomerase II in nucleolus-dependent mRNA export.
Consistent with the postulated roles for topoisomerase II in RNA metabolism summarized above, we previously reported the interaction of topoisomerase IIα with RNA helicase A (RHA) in an RNase-sensitive manner (25,26). Being a member of the DEAH family of ATPase proteins (27–32), RHA plays a critical role in mammalian development (33) and mediates the interaction between CBP/p300 and RNA polymerase II (34). Similarly, lethality of mouse embryos lacking topoisomerase IIα but not its isoenzyme topoisomerase IIβ testifies to the essential roles played by topoisomerase IIα during development (35,36). Like topoisomerase IIα, RHA also participates in diverse nuclear processes, suggested by its interaction with various nuclear factors, including the tumor suppressor BRCA1 (37), HAP95 highly homologous to AKAP95 (A-kinase anchoring protein 95) (38), the SMN (survival motor neuron) complex (39), an E2-type ligase Ubc9 (40), the RISC (RNA-induced silencing complex) (41) and a NF90/45 (nuclear factor 90/45) complex involved in mitotic control (42). As a preliminary investigation into their conjoint nuclear function, we tested whether the RHA-topoisomerase IIα interaction can be recapitulated in vitro using purified proteins. This study led us to an unexpected discovery that an RNA-binding activity is intrinsically associated with topoisomerase IIα.
MATERIALS AND METHODS
Protein, DNA and RNA
Preparation of recombinant RHA and MLE used in the present study is described previously (26,29,30). Topoisomerase IIα was either prepared as described previously (26) or obtained from Vaxron (USA). Supercoiled pBluescript II KS− (pBSKS−) used in relaxation assay was prepared using the Qiagen plasmid kit and Tetrahymena kinetoplast DNA was obtained from Vaxron. All synthetic homopolymer RNAs were purchased from GE Healthcare. Synthetic RNAs used in the electrophoresis mobility shift assay (EMSA), including the 98 mer ssRNA, a partial dsRNA, the 91 mer ssRNA (Figure 3C) and 38 mer ssRNA were all prepared as described previously (27,28). Unless otherwise indicated, the specific radioactivity of all RNA substrates was adjusted to 100 000 cpm/50 fmol of RNA. To prepare the 39 mer ssRNA, 38 mer ssRNA was first transcribed at a specific radioactivity of 250 c.p.m./50 fmol. After gel purification, 20 pmol of 38 mer ssRNA was end-labeled at 37°C for 1 h in a reaction mixture (100 μl) containing T4 RNA ligase (NEB, 20 U) and α-32P-Cp (200 μCi). Specific radioactivity was ≈300 000 cpm/50 fmol, as determined by standard TCA precipitation. End-labeled 39 mer ssRNA was again gel-purified and resuspended at 100 fmol/μl as described previously (27,28).
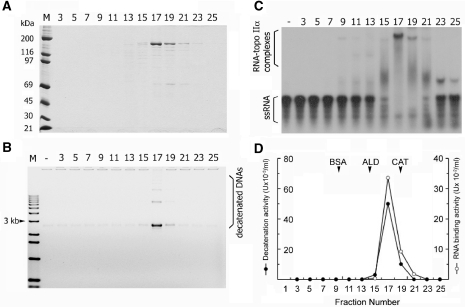
Figure 3.
Sedimentation analysis of biochemical activities associated with topoisomerase IIα. Sedimentation analysis was performed using topoisomerase IIα (60 μg) and a linear glycerol gradient (4.8 ml, 20–40%). After centrifugation at 45 000 r.p.m. for 27 h at 4°C in a Beckman 55TiSw rotor, 27 fractions were collected. All numbers represent the fraction numbers from the glycerol gradient. (A) Aliquots (10 μl) were subjected to 7.5% SDS–PAGE, and proteins were visualized by Coomassie staining. (B) Glycerol fractions were diluted 1:100, and 1-μl aliquots were tested for decatenation activity of kinetoplast DNA (0.5 μg). After a 30-min incubation at 37°C, reaction mixtures were resolved on 1% agarose gel, and monomer circles were quantified using a Gel Doc system (Bio-Rad) and presented in (D). (C) Aliquots (0.5 μl) of glycerol fractions were examined for RNA-binding activity using a 91 mer ssRNA (1.7 nM), which is derived from the 3′-UTR of human TNF-α mRNA, as described in Materials and methods section. RNA–topoisomerase IIα complexes were resolved on a native gel, detected by autoradiography and quantified as described in Figure 1.
A flushed-ended dsRNA (60 bp) was prepared as follows. In brief, two complementary ssRNAs were transcribed using pBSKS+ linearized with KpnI or BamHI. In vitro transcription reactions were conducted at 37°C for 90 min in reactions (100 μl) containing 40 mM Tris–HCl, pH 7.8, 10 mM DTT, 0.5 mM GTP/ATP/CTP, 50 μM UTP, 180 μCi α-32P-UTP, 0.5 U/ml RNasin (Promega), 2 mM spermidine, 14 mM MgCl2 and either T3 or T7 RNA polymerase (Promega). Following treatment with DNase I (20 μg/ml) and 1 mM CaCl2 at 37°C for 30 min, RNAs were isolated through phenol/chloroform extraction and ethanol precipitation, and subsequently they were resuspended in DEPC-treated water (3.3 μl) and combined. The RNA solution was heated to 100°C and mixed with 16.7 μl of a hybridization buffer (0.24 M PIPES, pH 6.4, 2.4 M NaCl and 6 mM EDTA) and formamide (80 μl). After incubation at 50°C for 1 h, RNAs were repurified and digested with RNase A (1 μg/ml) and RNase T1 (10 U) at 20°C for 1 h. The dsRNA was then gel-purified as described previously (28). Unless otherwise indicated, the specific radioactivity of all RNA substrates was adjusted to 100 000 c.p.m./50 fmol of RNA.
Electrophoresis mobility shift assay (EMSA)
A standard EMSA reaction (20 μl) contained 20 mM HEPES-NaOH, pH 7.4, 2 mM DTT, 3 mM MgCl2, 0.05% NP-40, substrate RNA (1.25–2.5 nM), and indicated amounts of topoisomerase IIα, MLE or RHA. After incubation for 30 min at 37°C, reaction mixtures were transferred on ice and mixed with 5 μl of 5 × dye-mix consisting of 0.1 M Tris–HCl, pH 7.4, 0.1% bromophenol blue, 0.1% xylene cyanol, 0.1% NP-40 and 50% glycerol. RNA–protein complexes were resolved on a composite gel containing 5% (60:1) polyacrylamide and 5% glycerol in 0.5 × TBE. RNA–protein complexes were visualized by autoradiography and quantified using a PhosphorImager (Molecular Dynamics).
Decatenation and relaxation assays
Standard reaction mixture (10 μl), used to measure both decatenation and relaxation activities of topoisomerase IIα, consisted of 50 mM Tris–HCl, pH 8.0, 0.1 M NaCl, 10 mM MgCl2, 0.5 mM DTT, 0.5 mM ATP, 0.2 mg/ml BSA with indicated amounts of topoisomerase IIα and either Tetrahymena kinetoplast DNA (0.5 μg) or pBSKS− (0.25 μg) as substrate. Unless otherwise indicated, decatenation and relaxation reactions were carried out for 15 min and 30 min at 37°C, respectively. Subsequently they were mixed with 1 μl each of SDS (6%) and proteinase K (3 mg/ml). Following a 30-min incubation at 37°C, reaction mixtures were provided with 3 μl of 5 × dye-mix described above and loaded on a 1% agarose gel in 1 × Tris–acetate/EDTA (TAE) buffer, pH 8.0. DNAs were visualized by ethidium bromide (0.5 μg/ml) staining for 20–45 min and quantified using a Gel Documentation System (Bio-Rad).
Sedimentation analysis
An aliquot (60 μg) of topoisomerase IIα was loaded on a linear glycerol gradient (4.8 ml, 20–40%) in a buffer containing 20 mM HEPES-NaOH, pH 7.4, 0.2 M KCl, 0.2 mM EDTA, 1 mM DTT and 0.05% NP-40. After centrifugation at 45 000 r.p.m. for 27 h at 4°C in a Beckman 55TiSw rotor, 27 fractions were collected and tested for the presence of topoisomerase IIα, decatenation activity and RNA-binding activity. First, distribution of topoisomearse IIα was determined by analyzing aliquots (10 μl) of glycerol fractions on a 7.5% SDS–PAGE, followed by Coomassie staining. Decatenation activity of each glycerol fraction was examined with aliquots (1 μl) of 1:100 diluted fraction in the standard decatenation assay described above. One unit of decatenation activity equals complete decatenation of 1 μg of kinetoplast DNA in 1 h. Aliquots (0.5 μl) of glycerol fractions were assayed for binding activity to a 91 mer ssRNA (1.7 nM) in a standard EMSA as described above. RNA–topoisomerase IIα complexes, resolved on a native gel, were quantified using a PhosphorImager. One unit of RNA-binding activity is defined as forming complexes with 10 fmol of ssRNA in 30 min at 37°C.
RESULTS
Topoisomerase IIα directly interacts with RNA
We previously reported that the in vivo interaction of topoisomerase IIα with RHA is sensitive to RNase A treatment (26). Several possibilities can be considered for the molecular nature of their interaction. For example, since RHA possesses a demonstrated RNA-binding activity (28–30) and the RHA–topoisomerase IIα interaction is cell cycle-specific (25), there might exist RNAs that associate with RHA, spatiotemporally enabling it to interact with topoisomerase IIα. Alternatively, there might be an as-yet-unidentified factor that bridges the RHA–topoisomerase IIα interaction in an RNA-dependent manner. To test these possibilities, we explored whether RHA–topoisomerase IIα interaction can be recapitulated utilizing in vitro transcribed synthetic RNAs.
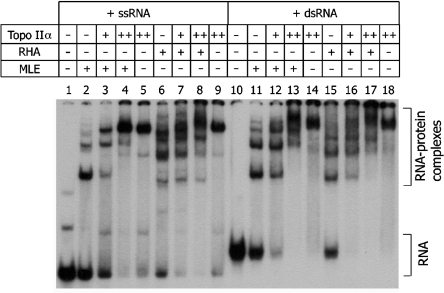
As described in our previous report, RHA and topoisomerase IIα were isolated from HeLa cells (26), whereas MLE, the Drosophila counterpart of RHA, was prepared as a recombinant protein using baculovirus (30). In an EMSA, both RHA and MLE (6.9 nM) stably interacted with either a 98 mer ssRNA (1.25 nM) or a partial dsRNA (2.5 nM) (Figure 1, lanes 2, 6, 11 and 15). Unexpectedly, increasing concentrations of topoisomerase IIα (1.47–5.88 nM) appeared to compete out RHA and MLE for the interaction with ssRNA (lanes 3–4 and 7–8) or dsRNA (lanes 12–13 and 16–17). More intriguingly, topoisomerase IIα alone formed stable complexes with both ssRNA (lanes 5 and 9) and dsRNA (lanes 14 and 18). Although it is yet to be determined whether there exists any physical contact between topoisomerase IIα and RHA or MLE within such complexes, a direct binding of topoisomerase IIα to RNA suggests that a more complex interaction between topoisomerase IIα and RHA could exist than was initially speculated.
Figure 1.
Detection of an RNA-binding activity associated with topoisomerase IIα. EMSA was performed with 98 mer ssRNA (1.25 nM, lanes 1–9) or a partial dsRNA (2.5 nM, lanes 10–18), and MLE (10 ng, lanes 2–4 and 11–13) or RHA (10 ng, lanes 6–8 and 15–17). Topoisomerase IIα (5 or 20 ng) was also included in the indicated reactions (lanes 3–5, 7–9, 12–14 and 16–18). RNA–protein complexes were resolved on a composite gel, as described in Materials and Methods section, and visualized by autoradiography and quantified by PhosphorImager analysis (Molecular Dynamics).
Characterization of RNA-binding activity of topoisomerase IIα
That topoisomerase IIα can react with RNA is not unprecedented. Wang et al. (43) reported that human topoisomerase IIα and IIβ catalyze the cleavage of ribonucleotide-containing substrate more efficiently than DNA substrate. Earlier work also described that Escherichia coli topoisomerase III, a type IA topoisomerase, catalyzes an RNA strand passage reaction, forming covalently linked RNA–protein intermediates (44). In addition, type IB topoisomerases, human and vaccinia virus topoisomerase I, catalyze site-specific endoribonucleolytic cleavage of synthetic linear substrate, DNApRNA, harboring ribonucleotide sequence in the pentapyrimidine recognition site, 5′-CCCTU or 5′-CCCUU (45). In further support of the topoisomerase–RNA interaction, Fisher and coworkers (46) showed that the majority of Drosophila topoisomerase II is associated with DNA in interphase but in mitosis the vast majority of soluble topoisomerase II associates with RNA.
Although topoisomerases all share an ability to react with RNA, be it as part of linear or circular DNA–RNA hybrid, their interaction with RNA, in particular with cellular RNA, has never been studied. Therefore, we attempted to further characterize the RNA-binding activity of topoisomerase IIα utilizing linear RNAs in different conformations such as ssRNA (Figure 2A, lanes 1–4), partial duplex RNA (dsRNA) (Figure 2A, lanes 5–8) and flush-ended dsRNA (Figure 2B). Both ssRNA and dsRNA (2.5 nM) were efficient in interacting with topoisomerase IIα (7.35–29.4 nM) (Figure 2A). Interaction of topoisomerase IIα (7.35–29.4 nM) with a 60-bp flush-ended RNA (2.5 nM) was less efficient than that with RNAs containing single-stranded regions (Figure 2B). Despite apparent engagement of all ssRNA or dsRNA in the formation of complex with topoisomerase IIα (7.35 nM) (Figure 2A, lanes 2 and 6), higher concentrations of topoisomerase IIα (14.7–29.4 nM) resulted in further retardation of RNA–protein complexes (compare lanes 2 and 6 to lanes 3–4 and 7–8, respectively). This result suggests that a 98-mer ssRNA is capable of associating with more than one topoisomerase IIα molecule. At the highest concentration (29.4 nM) of topoisomerase IIα, all RNA substrates failed to enter the gel (Figure 2A, lanes 4 and 8; Figure 2B, lane 4). We also determined the influence of ATP or increasing salt concentration on the topoisomerase IIα–RNA interaction. Neither the presence of ATP (2 mM) (Figure 2B, lanes 5–8) nor increasing concentration of NaCl up to 100 mM (Figure 2C) significantly affected the topoisomerase IIα–RNA interaction. The topoisomerase IIα–RNA interaction persisted even in the presence of higher NaCl concentration up to 300 mM, though with reduced efficiency (Figure 2C). These results indicate that the topoisomerase IIα–RNA interaction can occur under normal physiological conditions even in the absence of ATP.
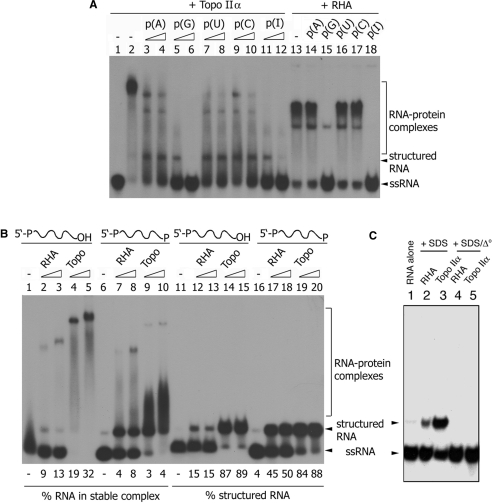
Figure 2.
Characterization of the RNA-binding activity associated with topoisomerase IIα. (A) Either ssRNA or a partial dsRNA (2.5 nM) was incubated with an increasing amount of topoisomerase IIα (7.35, 14.7 or 29.4 nM) in a reaction mixture (10 μl) consisting of 20 mM HEPES-NaOH, pH 7.4, 2 mM DTT, 0.05% NP-40 and 7 mM MgCl2. (B) ATP (2 mM) or (C) increasing concentration of salt (0–300 mM) was included in a standard reaction mixture (see Materials and Methods section), containing topoisomerase IIα (7.35 nM) and 60-bp flush-ended dsRNA. (D) Supershift assay was performed by sequentially incubating topoisomerase IIα (7.35 nM) (lanes 2–4 and 6–7) with 60-bp flush-ended dsRNA and monoclonal antibody (0.125 or 0.25 μg) specific for the C-terminal domain of topoisomerase IIα (aa 1245–1361) (lanes 3–5). All RNA–topoisomerase IIα complexes were detected as described in Figure 1.
RNA-binding activity is intrinsically associated with topoisomerase IIα
Since a direct and stable association with RNA has not been reported for any topoisomerases prior to our work, we asked whether such RNA-binding activity is intrinsically associated with topoisomerase IIα or with an as-yet-unidentified factor that is co-purified with topoisomerase IIα. First, we performed an antibody-driven super-shift assay. Topoisomerase IIα (7.35 nM) was sequentially incubated with a 60-bp flush-ended RNA and monoclonal antibody (0.125 or 0.25 μg) specific for the C-terminal domain of topoisomerase IIα (aa 1245–1361) or for hemaglutinin (HA) epitope, KAFSNCYPYDVPDYASLRS. As shown in Figure 2D, topoisomerase IIα–RNA complex was shifted only by monoclonal antibody specific for topoisomerase IIα (compare lanes 3–4 and 6–7).
Second, topoisomerase IIα was subjected to ultracentrifugal separation through glycerol gradient (20–40%), and subsequently, distribution of decatenation activity, a signature and intrinsic function of topoisomerase IIα, was determined in comparison with that of RNA-binding activity. When aliquots of gradient fractions were analyzed by SDS–PAGE, topoisomerase IIα was enriched in fractions 17–19 with an estimated sedimentation coefficient of 10.83 S (Figure 3A). Together with a Stokes radius of 7.2 nm, obtained by Superose 6HR chromatography (data not shown), a native molecular weight of topoisomerase IIα was calculated to be ≈322 kDa with a friction coefficient of 3.4 according to the method of Siegel and Monty (47). Since the predicted molecular weight of topoisomerase IIα is 174.4 kDa (GenBank accession no. NP_001058), our result indicates that human topoisomerase IIα enriched in gradient fractions 17–19, is a rod-shape homodimer, similar to topoisomerase II from other organisms, including Drosophila (48) and bovine (49). Distribution of decatenation activity (Figure 3B), measured with catenated Tetrahymena kinetoplast DNA, and RNA-binding activity (Figure 3C) exhibited a sedimentation pattern identical to each other (Figure 3D) and to that of topoisomerase IIα (Figure 3A). Since there was no other protein that co-sedimented with topoisomerase IIα or co-migrated with RNA-binding activity on glycerol gradient, we conclude that RNA-binding activity is associated with a homodimeric topoisomerase IIα.
Topoisomerase IIα and RHA exhibit differential RNA-binding specificity
As discussed above, stable interaction between topoisomerase IIα and RHA is dependent upon the presence of RNA (26). Although it remains unknown whether specific RNAs bridge their interaction, it would be informative to compare RHA and topoisomerase IIα for their RNA-binding property, in particular, from the aspect of RNA sequence specificity. As shown in Figure 4A, topoisomerase IIα (7.35 nM) was incubated with a 98 mer ssRNA (2.5 nM) in the presence of increasing concentration (6 or 30 nM) of the indicated synthetic homopolymer RNA. Parallel reactions were provided with RHA (6.9 nM) in place of topoisomerase IIα and a fixed concentration (30 nM) of the indicated competitor RNA. Among all competitor RNAs tested, only poly(G) and poly(I) were inhibitory to the RHA–RNA interaction (lanes 13–18). Similarly, topoisomerase IIα–RNA interaction was severely impaired by poly(G) and poly(I). However, all competitor RNAs seemed to affect the topoisomerase IIα–RNA interaction to varying degrees. This result indicates that the RHA–RNA interaction is more sequence-specific than the topoisomerase IIα–RNA interaction.
Figure 4.
Comparative analysis of RNA-binding activities associated with topoisomerase IIα and RHA. (A) Topoisomerase IIα (7.35 nM) or RHA (13.8 nM) was incubated in a standard reaction mixture (10 μl) containing 98 mer ssRNA (2.5 nM). Increasing concentration (6–30 nM) of indicated homopolymer RNA was included as competitor in reactions containing topoisomerase IIα (lanes 3–12), whereas a fixed concentration (30 nM) of competitor RNA was added to reactions with RHA (lanes 14–18). After a 30-min incubation at 37°C, RNA–protein complexes were analyzed as described in Figure 1. (B) Increasing concentrations of topoisomerase IIα (3.7–7.35 nM) or RHA (13.8–27.6 nM) were incubated in a standard reaction mixture (10 μl) containing either 38 mer ssRNA with a hydroxyl group at the 3′-end (lanes 1–5 and 11–15) or 39 mer ssRNA with a phosphate group at the 3′-end (lanes 6–10 and 16–20). After a 30-min incubation at 37°C, reactions were mixed with either 5 × dye mix alone (lanes 1–10) or in combination with SDS to a final concentration of 0.5%. Due to the difference in the specific radioactivity between 38 mer and 39 mer ssRNA, as described in Materials and Methods section, the entire mixture was loaded on a gel for reactions containing 38 mer ssRNA (lanes 1–5 and 11–15), whereas 5-μl aliquots were analyzed for reactions provided with 39 mer ssRNA (lanes 6–10 and 16–20). Subsequently, RNA–protein complexes were examined as described in Figure 1. (C) RHA (13.8 nM) (lanes 2 and 4) or topoisomerase IIα (3.7 nM) (lanes 3 and 5) was reacted with 38 mer ssRNA as described in (B). After a 30-min incubation at 37°C, all reactions were mixed with SDS (0.5%) and proteinase K (0.25 mg/ml) and incubated for an additional 30 min. Subsequently, 5 × dye mix was added, and the mixture was either directly loaded on a gel (lanes 1–3) or preheated at 95°C for 5 min (lanes 4–5).
Recently, Dong et al. (50) reported that La protein possesses the ability to discriminate between RNAs that bear 3′-OH versus 3′-PO4. They proposed that this discrimination is potentially important for La proteins to recognize newly synthesized RNAs and to distinguish them from RNase-degraded RNA that contains 3′-PO4. We speculated that like La protein, topoisomerase IIα–RNA interaction might be influenced by the identity of the 3′-end of RNA. To test this possibility, a 39 mer ssRNA containing 3′-PO4 was generated by ligating 38 mer ssRNA to cytidine 3′,5′-bisphosphate using T4 RNA ligase. The resulting 39 mer RNA was compared to its precursor 38 mer RNA for the ability to associate with RHA and topoisomerase IIα.
Both 38 and 39 mer RNA were poorly engaged in the formation of stable complex with RHA (13.8–27.6 nM) without noticeable difference (Figure 4B, compare lanes 2–3 and 7–8). Overall efficiency of the formation of stable complexes with RHA was much lower for both 38 and 39 mer RNAs than for 98 mer RNA (Figure 4A, lane 13), suggesting that the length of RNA is a factor that influences efficient association with RHA. In contrast, 38 mer and 39 mer RNA exhibited a noticeable difference in the formation of stable complex with topoisomerase IIα (7.35–14.7 nM). Compared to RHA, the ability of topoisomerase IIα to bind RNA was relatively unaffected by RNA length (compare Figure 4A, lane 2 and Figure 4B, lanes 4–5). Although the vast majority of 38 mer RNA appeared to interact with topoisomerase IIα only a small fraction (19 or 32%) of 38 mer RNA was engaged in the formation of ‘stable’ complex with topoisomerase IIα (Figure 4B, lanes 4–5). In addition, quantifiably more stable topoisomerase IIα–38 mer RNA complex was formed than was topoisomerase IIα–39 mer RNA complex (Figure 4B, compare lanes 4–5 and 9–10). Coincidently, the formation of unstable complexes, giving rise to smeared RNA signals, was increased when topoisomerase IIα was bound to 39 mer RNA (Figure 4B, lanes 9–10). These results suggest that the stable association of topoisomerase IIα with RNA is more influenced by the 3′-OH group than RNA length.
DiGate and Marians (44) reported that E. coli topoisomerase III catalyzes site-specific cleavage of ssRNA as short as 30-nt long, forming a covalent RNA–topoisomerase III intermediate. Sekiguchi and Shuman (45) made a similar observation with Vaccinia virus topoisomerase I. To examine whether the topoisomerase IIα–RNA interaction involves the formation of covalent linkage similar to type I topoisomerases, topoisomerase IIα- and RHA–RNA complexes were treated with SDS and analyzed for RNA (Figure 4B, lanes 11–20). Treatment with SDS (0.5%) and proteinase K (0.25 mg/ml) did not alter the migration of substrate RNAs (compare lanes 1, 6, 11 and 16). However, interaction with topoisomerase IIα generated a slowly migrating form (lanes 14–15 and 19–20). RHA yielded a similar result but more efficiently with 39 mer RNA than with 38 mer RNA (compare lanes 12–13 and 17–18). The slowly migrating RNA is likely to be a structured form of RNA since heat treatment (95°C, 5 min) resulted in their conversion to a faster migrating form of RNA (Figure 4C). Although the identity of structured RNA remains to be determined, the above result excludes the possibility of covalent modification of ssRNA substrates by topoisomerase IIα.
Implication of topoisomerase IIα in RNA metabolism
What would be the physiological significance of topoisomerase IIα binding to RNA? There might exist an as-yet-unknown RNA that modulates the ability of topoisomerase IIα to interact with other factors such as RHA or to regulate DNA topology. Alternatively, RNA metabolism might be influenced by topoisomerase IIα. As an initial attempt toward testing these possibilities, we first explored whether RNA influences the ability of topoisomerase IIα to regulate DNA topology. For this purpose, we measured the decatenation activity of topoisomerase IIα (0.6 nM) in reactions containing Tetrahymena kinetoplast DNA (0.5 μg) and varying concentrations (0.8–57 nM) of synthetic homopolymer RNAs. As shown in Figure 5, RNA brought about diverse effects on topoisomerase IIα. In brief, poly(A) and poly(C) were noninfluential, whereas poly(G), poly(I) and poly(U) were inhibitory (Figure 5A). IC50 was estimated to be 0.5–1.5 nM, which was obtained from four independent experiments provided with the indicated RNA (0.6–8 nM) (Figure 5B). We asked whether this diverse effect is governed by base sequence alone or in combination with the sugar moiety (i.e. ribose versus deoxyribose). In the former case, both RNA and DNA homopolymer of the same sequence would elicit similar effects on the decatenation activity of topoisomerase IIα. In order to test this possibility, a kinetic analysis was performed in reactions for the decatenation activity in the presence of poly(A) or poly(dA). As shown in Figure 5C, in contrast to poly(A), which even appeared to be slightly stimulatory, poly(dA) inhibited decatenation activity of topoisomerase IIα (Figure 5C). This result indicates that the effects observed in Figure 5A and B have to do with both the base sequence and identity of sugar moiety.
Figure 5.
Influence of homopolymer RNA on the ability of topoisomerase IIα function to regulate DNA topology. (A) Decatenation reaction was carried out in a standard reaction mixture (10 μl) containing kinetoplast DNA (0.5 μg) and the indicated RNA homopolymer. Lane 1; control reaction without topoisomerase IIα. Lanes 2–17; with topoisomerase IIα (0.6 nM). Lanes 3–5; 0.8, 3.1 and 7.70 nM poly(A). Lanes 6–8; 5.7, 23 and 57 nM poly(G). Lanes 9–11; 1.1, 4.5 and 11 nM poly(C). Lanes 12–13; 1.5, 6.2 and 15 nM poly(U). Lanes 15–17; 0.6, 2.6 and 6.4 nM poly(I). (B) Decatenation reaction was performed at 37°C for 20 min in a standard reaction mixture containing topoisomerase IIα (0.6 nM) and varying concentrations (0–8 nM) of homopolymer RNA. Following treatment with proteinase K (0.25 mg/ml) and SDS (0.5%), reaction mixtures were resolved on a 1% agarose gel, and both kinetoplast DNA and decatenated monomer were visualized by ethidium bromide staining and quantified using a Gel-Documentation System (Bio-Rad), as described in Materials and Methods section. The 100% decatenation activity is an arbitrary unit that equals the amount of monomer circle produced by topoisomerase IIα in the absence of synthetic homopolymer RNA (lane 2). Decatenation activity was measured in four independent experiments, and their average values are presented with error bars. (C) Decatenation reaction, provided with topoisomerase IIα (0.6 nM) and indicated homopolymer (0.1 μg) in a standard reaction mixture (10 μl), was incubated at 37°C for increasing periods of time (2, 5, 10, 15, 22 and 30 min). After resolving on a 1% agarose gel, decatenated DNAs were visualized by ethidium bromide staining and quantified as described in (B). The 100% of DNA equals to the summed density of all DNA bands detected in lane 7, of which monomer circle accounts for 94% of the total DNA. Percent-decatenated DNA represents the relative density of monomer circle obtained from each reaction. (D) Relaxation reaction was carried out at 37°C for 30 min in a standard reaction mixture (10 μl) containing topoisomerase IIα (11.8 nM), 0.25 μg of supercolied pBSKS− DNA (0.25 μg), and varying concentrations (0.6, 2.4 and 6.0 nM) of the indicated RNA homopolymer. Supercoiled DNA and its topoisomers were analyzed as described in (B).
The effects of RNA were also examined for the relaxation activity of topoisomerase IIα. For this purpose, in vitro relaxation of supercoiled plasmid DNA (pBSKS−) was measured in reactions provided with three different concentrations (0.6, 2.4 and 6.0 nM) of synthetic RNA. As shown in Figure 5D, poly(G) and poly(I) but not poly(A) and poly(C) were inhibitory. In contrast to its effect on the decatenation activity of topoisomerase IIα, poly(U) did not significantly affect the relaxation activity of topoisomerase IIα. This result raises an intriguing possibility that the decatenation and relaxation activity of topoisomerase IIα could be subject to differential regulation by an RNA molecule, depending on the base sequence. Together with the result in Figure 4A, the above result may also indicate that the stability and specificity of the RNA-binding activity of topoisomerase IIα is RNA sequence-specific. In this regard, a report by Ideue et al. (24) is particularly noteworthy. In an attempt to identify novel factors involved in mRNA export, they screened a yeast ts mutant bank using in situ hybridization (FISH) with an oligo(dT) probe. The authors described one particular mutant ptr11 and two of its alleles that displayed both the ‘cut’ phenotype and nuclear accumulation of poly(A)+ RNA. Cloning of ptr11 revealed that the ORF is top2+ coding for topoisomerase II. The finding that nuclear accumulation of poly(A)+ RNA is observed only in the anucleolate nuclei led the authors to hypothesize that the nucleolus is required for mRNA export in yeast. Although the underlying mechanisms remain unknown, this observation strongly suggests a role played by topoisomerase II in poly(A)+ RNA export. Based on the results shown in Figure 5, we predict that topoisomerase II sequentially interacts with the (G/U)-rich region and poly(A)+ tail of the transcript to regulate the template DNA topology in coordination with transcription termination and poly(A)+ RNA export (see Discussion section).
DISCUSSION
In this report, we identify a novel RNA-binding activity of topoisomerase IIα. Our observation that RNA association with topoisomerase IIα modulated the ability of topoisomerase IIα to regulate DNA topology might suggest the RNA can regulate topoisomerase IIα function or that topoisomerase IIα has a role in RNA metabolism through its direct association with RNA in a sequence-specific manner.
At present, the molecular basis by which poly(I), poly(G) and poly(U) inhibit topoisomerase IIα remains unknown. Since poly(G) and poly(I), the two highly inhibitory RNAs, potentially form quadruple structure (51), they may compete with DNA substrate for topoisomerase IIα. Consistent with this possibility, inhibitory poly(U) forms antiparallel A-type double helices, whereas noninfluential poly(A) and poly(C) form only a single helix (51). This competition model presupposes the presence of a binding site in topoisomerase IIα commonly occupied by structured RNA and DNA. However, it cannot explain why poly(dA), which forms a single-stranded B-type helix (52), was also inhibitory to topoisomerase IIα. Alternatively, it is possible that the catalytic activity of topoisomerase IIα may be regulated by RNA bound on an as yet unknown domain. If this is the case, are there any features common to the inhibitory or non-nhibitory RNAs described here? In this regard, it should be noted that an amino group (–NH2) is commonly present in bases of noninhibitory RNA, whereas a keto (C = O) group is present in bases of inhibitory RNA. Current models on DNA transaction catalyzed by topoisomerase II envision the formation of scissile breaks in the phosphate backbone of DNA by nucleophilic attack of an oxygen atom in a conserved tyrosine residue. In bases of inhibitory RNA such as poly(G), a change in hydrogen atom position can lead to transformation of a keto (C = O) group to an enol (C–OH) group. Although the enol configuration is extremely rare in solution, interaction with topoisomerase IIα might increase its population sufficiently to interfere with the nucleophilic attack of the tyrosine oxygen atom.
Recent studies have provided several lines of evidence indicating a mechanistic coupling of transcription to DNA supercoiling. In 1987, Liu and Wang (11) proposed the twin supercoil model to explain a higher level of negative or positive supercoils in intracellular plasmid DNA when topoisomerase I or gyrase is inhibited. This model predicts that the transcription process simultaneously generates positive and negative supercoils in regions of DNA behind and in front of the transcription elongation complex, respectively. In prokaryotes, topoisomerase I preferentially removes negative supercoils, while DNA gyrase removes positive supercoils, maintaining the proper level of supercoils in genomic DNA. Despite technical difficulties due to the activity of gyrase to convert positive supercoil to negative supercoil, there is substantial evidence supporting the twin transcription-loop model. For example, employing a defined in vitro transcription system utilizing sequence-specific DNA-binding proteins and DNA gyrase, Leng et al. (12) found that DNA supercoiling is induced by transcription in two pathways: the first is dependent on the formation of the R-loop and the second depends on a substantially long RNA transcript (>3 kb) and the presence of DNA-binding protein.
Accumulating evidence indicates critical roles for topoisomerases in eukaryotic transcription. In 1987, Brill et al. (53) reported severe impairment of ribosomal RNA and poly(A)+ RNA synthesis in the Saccharomyces cerevisiae top1top2 double mutant. Recently, Huertas and Aguilera (54) performed a thorough phenotypic analysis of S. cerevisiae mutants of THO/TREX, a conserved protein complex functioning at the interface of transcription and RNA export. They found that impaired transcription elongation leads to the formation of DNA: RNA hybrids, promoting hyper-recombination. Similar to its suppressive effect on the formation of negative supercoils induced by the R-loop in prokaryotes, overexpression of RNase H abolished transcription-associated recombination in S. cerevisiae. In 2005, Li and Manley (55) reported a similar observation in higher eukaryotic cells. In brief, depletion of the splicing factor ASF/SF2 resulted in genomic instability, causing accumulation of high-molecular-weight DNA fragments. Detailed sequence analysis, employing bisulfite-modification of genomic DNA followed by PCR sequencing, showed that template DNA was protected from bisulfite modification, most probably due to base pairing between RNA and the DNA template. They also reported that the formation of R-loop associated with impaired RNA maturation was suppressed by overexpression of RNase H.
The above observations underline that the steady-state level of superhelicity of genomic DNA is maintained through an intimate functional coordination of topoisomerases and the transcription machinery, and any perturbation leads to the formation of the R-loop, causing genomic instability. That R-loop formation was primarily observed distal from the promoter but close to the 3′-end of the gene (54), indicates a possible function for topoisomerase at transcription termination. Since transcription termination, mRNA 3′-end cleavage, polyadenylation and mRNA export occur in a coupled reaction (56–58), could topoisomerases exert their influence on this concerted process? This scenario is supported by a recent report by Ideue et al. (24). Their efforts toward uncovering novel factors involved in mRNA export led to the identification of one particular mutant, ptr11-1, and its two other alleles that displayed both ‘cut’ phenotype and the nuclear accumulation of poly(A)+ RNA. The ptr11 gene was identified as top2+, coding for topoisomerase II.
Although it is unknown how topoisomerase II influences transcription termination and mRNA export, the reactivity of topoisomerase IIα with RNA may provide a clue. Upon arrival of the transcription elongation complex at the termination site, the growing chain of mRNA undergoes drastic changes; mRNA is polyadenylated after removal of the G/U-rich downstream element (DSE) in the 3′-untranslated region (3′-UTR). Intriguingly, this event can be viewed as sequential presentation of the G/U-rich DSE and poly(A) tail to topoisomerase II, through which, perhaps, the function of topoisomerase II to regulate superhelicity of the template DNA becomes coordinated with transcription termination, polyadenylation and mRNA export.
In addition to transcription, topoisomerase IIα has been implicated in the formation of heterochromatin (19,59). Intriguingly, there is increasing evidence that small noncoding RNAs are involved in heterochromatin formation and maintenance (60,61). In addition, RHA interacts with both topoisomerase IIα and the RISC complex containing siRNA (41). Taken together, it would be worthwhile testing whether topoisomerase IIα is required for the formation or maintenance of heterochromatin involving the function of the RISC complex. In addition, it can also be tested whether topoisomerase IIα directly interacts with noncoding RNAs such as siRNA on heterochromatin. This future study will be greatly facilitated by the identification of domains or amino acids critical for the topoisomerase IIα–RNA interaction.
FUNDING
American Heart Association (AHA0535548T to Y.P.); the National Science Foundation (MCB-0818464 to Y.P.); and a National Institutes of Health Grant (AI34552 to M.B.M.). Funding for open access charge: AHA0535548T.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr Charles Reichman for his comments and critical reading of the manuscript.
REFERENCES
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:69–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 3.Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK. Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 1986;188:613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- 4.Caron PR, Watt P, Wang JC. The C-terminal domain of Saccharomyces cerevisiae DNA topoisomerase II. Mol. Cell. Biol. 1994;14:3197–3207. doi: 10.1128/mcb.14.5.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uemura T, Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uemura T, Morikawa K, Yanagida M. The nucleotide sequence of the fission yeast DNA topoisomerase II gene: structural and functional relationships to other DNA topoisomerases. EMBO J. 1986;5:2355–2361. doi: 10.1002/j.1460-2075.1986.tb04504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M, Beck WT. Teniposide-resistant CEM cells, which express mutant DNA topoisomerase II alpha, when treated with non-complex-stabilizing inhibitors of the enzyme, display no cross-resistance and reveal aberrant functions of the mutant enzyme. Cancer Res. 1993;53:5946–5953. [PubMed] [Google Scholar]
- 8.Ishida R, Takashima R, Koujin T, Shibata M, Nozaki N, Seto M, Mori H, Haraguchi T, Hiraoka Y. Mitotic specific phosphorylation of serine-1212 in human DNA topoisomerase IIa. Cell Struct. Funct. 2001;26:215–226. doi: 10.1247/csf.26.215. [DOI] [PubMed] [Google Scholar]
- 9.Shamu CE, Murray AW. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J. Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 11.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leng F, Amado L, McMacken R. Coupling DNA supercoiling to transcription in defined protein systems. J. Biol. Chem. 2004;279:47564–47571. doi: 10.1074/jbc.M403798200. [DOI] [PubMed] [Google Scholar]
- 13.Masse E, Drolet M. Relaxation of transcription-induced negative supercoiling is an essential function of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1999;274:16654–16658. doi: 10.1074/jbc.274.23.16654. [DOI] [PubMed] [Google Scholar]
- 14.Mondal N, Zhang Y, Jonsson Z, Dhar SK, Kannapiran M, Parvin JD. Elongation by RNA polymerase II on chromatin templates requires topoisomerase activity. Nucleic Acids Res. 2003;31:5016–5024. doi: 10.1093/nar/gkg705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hraiky C, Raymond M.-A, Drolet M. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J. Biol. Chem. 2000;275:11257–11263. doi: 10.1074/jbc.275.15.11257. [DOI] [PubMed] [Google Scholar]
- 16.Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi M, Aita N, Hayashi S, Okada K, Ohta T, Hirose S. DNA supercoiling factor localizes to puffs on polytene chromosomes in Drosophila melanogaster. Mol. Cell. Biol. 1998;18:6747–6744. doi: 10.1128/mcb.18.11.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. hromatin-remodeling factor CHRAC contains the ATPase ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 19.Tsai SC, Valkov N, Yang WM, Gump J, Sullivan D, Seto E. Histone deacetylase interacts directly with topoisomerase II. Nat. Genet. 2000;26:349–353. doi: 10.1038/81671. [DOI] [PubMed] [Google Scholar]
- 20.Kal AJ, Mahmoudi T, Zak NB, Verrijzer CP. The Drosophila brahma complex is an essential cofactor for the trithorax group protein zeste. Genes Dev. 2000;14:1058–1071. [PMC free article] [PubMed] [Google Scholar]
- 21.Tavormina PA, Come M.-G, Hudson JR, Mo Y.-Y, Beck WT, Gorbsky GJ. Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J. Cell. Biol. 2002;158:23–29. doi: 10.1083/jcb.200202053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dundr M, Misteli T, Olson MO. The dynamics of postmitotic reassembly of the nucleolus. J. Cell. Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuwaki M, Iwamatsu A, Tsujimoto M, Nagata K. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 2001;311:41–55. doi: 10.1006/jmbi.2001.4812. [DOI] [PubMed] [Google Scholar]
- 24.Ideue T, Azad AK, Yoshida J, Matsusaka T, Yanagida M, Ohshima Y, Tani T. The nucleolus is involved in mRNA export from the nucleus in fission yeast. J. Cell. Sci. 2004;117:2887–2895. doi: 10.1242/jcs.01155. [DOI] [PubMed] [Google Scholar]
- 25.Lee CG, Hague LK, Li H, Donnelly R. Identification of toposome, a novel multisubunit complex containing topoisomerase IIalpha. Cell Cycle. 2004;3:638–647. [PubMed] [Google Scholar]
- 26.Zhou K, Choe K.-T, Zaidi Z, Wang Q, Mathews MB, Lee C-G. RNA helicase A interacts with dsDNA and topoisomerase II{alpha} Nucleic Acids Res. 2003;31:2253–2260. doi: 10.1093/nar/gkg328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fritz DT, Ford LP, Wilusz J. An in vitro assay to study regulated mRNA stability. Sci. STKE. 2000;2000 doi: 10.1126/stke.2000.61.pl1. PL1. [DOI] [PubMed] [Google Scholar]
- 28.Lee C, Hurwitz J. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J. Biol. Chem. 1992;267:4398–4407. [PubMed] [Google Scholar]
- 29.Lee C, Hurwitz J. Human RNA helicase A is homologous to the maleless protein of Drosophila. J. Biol. Chem. 1993;268:16822–16830. [PubMed] [Google Scholar]
- 30.Lee C-G, Chang KA, Kuroda MI, Hurwitz J. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 1997;16:2671–2681. doi: 10.1093/emboj/16.10.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Grosse F. Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry. 1994;33:3906–3912. doi: 10.1021/bi00179a016. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S, Maacke H, Grosse F. Molecular cloning of the gene encoding nuclear DNA helicase II. A bovine homologue of human RNA helicase A and Drosophila Mle protein. J. Biol. Chem. 1995;270:16422–16427. doi: 10.1074/jbc.270.27.16422. [DOI] [PubMed] [Google Scholar]
- 33.Lee CG, Soares V, Newberger C, Manova K, Lacy E, Hurwitz J. RNA helicase A is essential for normal gastrulation in mice. PNAS USA. 1998;95:13709–13713. doi: 10.1073/pnas.95.23.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 35.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 36.Akimitsu N, Adachi N, Hirai H, Hossain MS, Hamamoto H, Kobayashi M, Aratani Y, Koyama H, Sekimizu K. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIalpha. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 37.Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 38.Yang JP, Tang H, Reddy TR, Wong-Staal F. Mapping the functional domains of HAP95, a protein that binds RNA helicase A and activates the constitutive transport element of type D retroviruses. J. Biol. Chem. 2001;276:30694–30700. doi: 10.1074/jbc.M102809200. [DOI] [PubMed] [Google Scholar]
- 39.Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol. 2001;152:75–86. doi: 10.1083/jcb.152.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Argasinska J, Zhou K, Donnelly RJ, Hay RT, Lee C-G. A functional interaction between RHA and Ubc9, an E2-like enzyme specific for Sumo-1. J. Mol. Biol. 2004;341:15–25. doi: 10.1016/j.jmb.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 41.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 42.Guan D, Altan-Bonnet N, Parrott AM, Arrigo CJ, Li Q, Khaleduzzaman M, Li H, Lee CG, Pe’ery T, Mathews MB. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol. Cell. Biol. 2008;28:4629–4641. doi: 10.1128/MCB.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Knudsen BR, Bjergbaek L, Westergaard O, Andersen AH. Stimulated activity of human topoisomerases IIalpha and IIbeta on RNA-containing substrates. J. Biol. Chem. 1999;274:22839–22846. doi: 10.1074/jbc.274.32.22839. [DOI] [PubMed] [Google Scholar]
- 44.DiGate RJ, Marians KJ. Escherichia coli topoisomerase III-catalyzed cleavage of RNA. J. Biol. Chem. 1992;267:20532–20535. [PubMed] [Google Scholar]
- 45.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 46.Rzepecki R, Fisher P. During both interphase and mitosis, DNA topoisomerase II interacts with DNA as well as RNA through the protein's C-terminal domain. J. Cell Sci. 2000;113:1635–1647. doi: 10.1242/jcs.113.9.1635. [DOI] [PubMed] [Google Scholar]
- 47.Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 48.Shelton ER, Osheroff N, Brutlag DL. DNA topoisomerase II from Drosophila melanogaster. Purification and physical characterization. J. Biol. Chem. 1983;258:9530–9535. [PubMed] [Google Scholar]
- 49.Schomburg U, Grosse F. Purification and characterization of DNA topoisomerase II from calf thymus associated with polypeptides of 175 and 150 kDa. Eur. J. Biochem. 1986;160:451–457. doi: 10.1111/j.1432-1033.1986.tb10061.x. [DOI] [PubMed] [Google Scholar]
- 50.Dong G, Chakshusmathi G, Wolin SL, Reinisch KM. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 2004;23:1000–1007. doi: 10.1038/sj.emboj.7600115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanger W. Principles of Nucleic Acid Structure. 1984. Springer, New York INc, New York. [Google Scholar]
- 52.Isaksson J, Acharya S, Barman J, Cheruku P, Chattopadhyaya J. Single-stranded adenine-rich DNA and RNA retain structural characteristics of their respective double-stranded conformations and show directional differences in stacking pattern. Biochemistry. 2004;43:15996–16010. doi: 10.1021/bi048221v. [DOI] [PubMed] [Google Scholar]
- 53.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 54.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 55.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 56.Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell. Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.West S, Gromak N, Proudfoot NJ. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 59.Lupo R, Breiling A, Bianchi ME, Orlando V. Drosophila chromosome condensation proteins topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol. Cell. 2001;7:127–136. doi: 10.1016/s1097-2765(01)00161-7. [DOI] [PubMed] [Google Scholar]
- 60.Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- 61.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]