Abstract
Shelterin/telosome is a multi-protein complex at mammalian telomeres, anchored to the double-stranded region by the telomeric-repeat binding factors-1 and -2. In vitro modification of these proteins by poly(ADP-ribosyl)ation through poly(ADP-ribose) polymerases-5 (tankyrases) and -1/-2, respectively, impairs binding. Thereafter, at least telomeric-repeat binding factor-1 is degraded by the proteasome. We show that pharmacological inhibition of poly(ADP-ribose) polymerase activity in cells from two different species leads to rapid decrease in median telomere length and stabilization at a lower setting. Specific knockdown of poly(ADP-ribose) polymerase-1 by RNA interference had the same effect. The length of the single-stranded telomeric overhang as well as telomerase activity were not affected. Release of inhibition led to a fast re-gain in telomere length to control levels in cells expressing active telomerase. We conclude that poly(ADP-ribose) polymerase-1 activity and probably its interplay with telomeric-repeat binding factor-2 is an important determinant in telomere regulation. Our findings reinforce the link between poly(ADP-ribosyl)ation and aging/longevity and also impact on the use of poly(ADP-ribose) polymerase inhibitors in tumor therapy.
INTRODUCTION
Telomeres are structures at the end of chromosomes, which comprise a highly repetitive DNA sequence (T2AG3 in vertebrates) and a protective, specific protein complex (shelterin/telosome) with associated nontelomere-specific proteins (1,2). The telomeric G-rich strand runs from the centromere outwards and ends in a single-stranded 3′-overhang (3). Telomeres shield chromosomal ends from degradation and undesirable repair activities, at least partially, by t-loop formation, with the 3′-overhang folding back and invading the double-stranded DNA (4). Shelterin can be divided into three subcomplexes: (i) a telomere-length regulation complex, comprising telomeric repeat-binding factor 1 (TRF1) bound to the double-stranded region and associated proteins; (ii) a telomere/t-loop stabilizing complex, comprising TRF2 bound to the double strand and associated proteins; and (iii) the single-strand binding protein POT1, associated with the TRF1 subcomplex via TPP1. The protein TIN2 interconnects the two double-strand binding complexes. Binding of TRFs to telomeres is postulated to be under control of the activity of poly(ADP-ribose) polymerases (PARPs): TRF1 interacts with PARP5 (tankyrases, TNKS) (5,6) and TRF2 with PARP1 and PARP2 (7–9). Poly(ADP-ribosyl)ation is a complex posttranslational protein modification and represents an immediate response of cells to genotoxic stress, due to the dramatic activation of PARP1 and PARP2 by DNA strand breaks. PARPs use NAD+ as substrate and synthesize a branched polymer of ADP-ribose units, with stoichiometric release of nicotinamide (10). Apart from undergoing covalent modification with poly(ADP-ribose) (PAR), proteins may also bind PAR in a noncovalent yet specific manner (11). Whereas covalent modification of a target protein renders it mostly inactive, noncovalent binding to PAR can have diverse effects, leading either to stimulation or repression of activity, probably also dependent on PAR chain length and branching frequency (11–14). The main target proteins undergoing poly(ADP-ribosyl)ation are PARPs themselves, thus creating an autoregulatory feedback loop, but many other proteins are modified in vivo and/or in vitro, e.g. histones H1 and H2B, p53, XPA, HMG2, CenpA, DNA-PKcs, topoisomerase-1 and transcription factors such as YY1 (15,16). PARP1 is the founding member of a large family of PARPs (17). The dramatic PAR formation stimulated by genotoxic agents has been associated with PARP1 and PARP2, with PARP1 being the most active protein, responsible for about 90% of cellular PAR formation observed under these conditions (18). Overall, PARP activity has been implicated in nearly all aspects of genome integrity and cell survival regulation, like repair (19,20), transcription (21–25), DNA replication (26–28), differentiation (26,29,30), mitosis and mitotic organization (31–35), choice of cell death pathway (36,37), vesicle trafficking (38) and telomere-length regulation (6,39). Furthermore, we have shown that the level of PARP activity induced by DNA strand breaks in mononuclear blood cells correlates with mammalian life span (40) and that enzymatic activity of recombinant purified human PARP1 is 2-fold higher than that of the rat protein (41,42). Thus, the PAR system seems also to be involved in cellular/organismal aging, as is the case for several other repair enzymes (i.e. WRN, BLM and ATM).
Controversial results have been published regarding the influence of Parp1 knockout on telomere length in mice. Whereas one group showed no impact (43,44), others reported shortened telomeres (45,46).
Overexpression of NLS-tagged TNKS1 leads to telomere elongation (39), whereas knockdown by siRNA leads to mitotic arrest and cell death (32), apparently by interfering with spindle organization and telomere-specific cohesion cleavage (47–49). Intriguingly, inhibitors of PARP activity do not have an impact on cell survival, although they are effective against TNKS1 in vitro (50). Thus, TNKS may not be affected within a cell at inhibitor concentrations used to block PARP1 and PARP2 activity.
To clarify the role of PARPs on telomere regulation, we used cells from two mammalian species (hamster and human) and inhibited PARP activity either pharmacologically or more selectively by siRNA against PARP1 or PARP2.
MATERIALS AND METHODS
Cell culture and treatment
Cells were grown in DMEM supplemented with 100 U/ml of penicillin and 0.1 mg/ml streptomycin and 10% FCS, at 37°C, 95% humidity and 5% CO2. Cells were counted and seeded 3 h before addition of 3-aminobenzamide (3AB) or including the inhibitor in subsequent passages. 3AB was dissolved in medium without FCS and sterile filtered.
Chromosome isolation for quantitative fluorescence in situ hybridization
COM3 hamster cells
This cell system has been described before (51,52). Cells in 75 cm2 flasks were treated with 0.01 mg/ml colcemide (Life Technologies/Invitrogen GmbH, Germany) for 1 h to stall mitosis. Then, the supernatant was removed and replaced with 4 ml of chromosome isolation buffer (CIB; 0.5 mM CaCl2, 1 mM MgCl2, 25 mM Tris–HCl, 750 mM hexane-diol, pH 7.5; 1% acetic acid added before use). The supernatant was replaced with new CIB and mitotic cells were shaken off the flask by gentle rinsing. Both CIB supernatants were pooled, cells were spun down for 10 min at 200g and the resulting pellet was resuspended in methanol + acetic acid (3 + 1). Fixed cells were stored at –20°C.
Human cells (HeLaS3, IMR90)
Cells were treated with 0.01 mg/ml colcemide (Life Technologies) for 1 h to stall mitoses. Adherent cells were harvested by trypsin (Life Technologies) treatment for 5 min at room temperature and washed 1× with PBS. Fixation was essentially done as described in (53). Fixed cells were stored at –20°C.
Quantitative fluorescence in situ hybridization analysis and evaluation
Quantitative fluorescence in situ hybridization (Q-FISH) analysis was done essentially as described (54). Metaphase spreads on Superfrost slides (VWR, Germany) were hybridized to Cy3-labeled PNA-telomere probes (Dako Cytomation, Denmark), counterstained with DAPI and analyzed with a fluorescence microscope (Zeiss Axiovert) using Axiovision software (Zeiss, Germany). Telomere signal intensities were analyzed with TeloQuant (Dako Cytomation). For any single data point the median of signal intensities of telomeres from at least 500 chromosomes was determined. Each experiment was performed in triplicates. Data are expressed as percent of control.
Telomeric-repeat-amplifying-protocol assay
Telomeric-repeat-amplifying-protocol (TRAP) assay was done according to the manufacturer's instructions (Intergen, TRAPeze telomerase detection kit). X-ray films were scanned and analyzed by using ImageQuant. Calculation of total product generated (TPG) units was done according to the manufacturer's instructions (Intergen, NY, USA TRAPeze telomerase detection kit).
Telomere oligo-ligation assay
Telomere oligo-ligation assay (T-OLA) was performed essentially as described (55). Briefly, 5 µg of genomic DNA was either treated with mung-bean exonuclease or left untreated. After phenol–chloroform purification and precipitation, DNA was hybridized to 32P end-labeled oligonucleotides (T2AG3)3 or (A2TC3)3 overnight at 50°C followed by 5 h incubation at 50°C with Taq-ligase (NEB, Germany) in Taq-ligase buffer. DNA was extracted by phenol–chloroform purification and precipitated, and ligation products were resolved on 5% polyacrylamide/6 M urea gels. DNA was blotted onto HybondN+ membranes (GE HealthCare, Europe, Germany) and exposed to X-ray films (Kodak). Scans from autoradiographs were analyzed by using ImageQuant software (GE Healthcare). The percentage of intensity of each band (relative to total lane intensity) was calculated and normalized to the length of ligation products. As we routinely detected ligation to quadruplicates also in negative controls, products of less than 5× (A2TC3)3 were excluded from the analysis.
Synthesis of small interfering RNAs and transfection
Small interfering RNAs (siRNAs) were synthesized in vitro from oligonucleotide templates (Microsynth, Switzerland) using a siRNA construction kit (Ambion, TX, USA), according to the manufacturer's instructions. The 21-nt sequences in the human coding region of PARP-1 and PARP-2 cDNA were chosen as targets for siRNA. A scrambled version of siRNA PARP-1 sequence was used as a negative control. Transfection procedure and siRNA sequences are described in ref. (56), except for siRNA P1n (2310–2329): CAUCGAGGUGGCCUACAGU.
Statistical analyses
Experiments were performed independently at least three times. Statistical analysis was done by using Prism5 or Instat3 (GraphPad Software). One-way or repeated measures ANOVA was used for calculation of significance if not stated otherwise. Error bars represent means ± SEM if not stated otherwise.
RESULTS
Dose-dependent telomere shortening by 3AB treatment
Binding of TRF proteins to the telomeric double strand are, at least in the case of TRF1, under control of PARP5 (TNKS) activity (5,6), leading to TRF1 degradation (57). Telomere elongation by overexpression of an artificially NLS-tagged version of TNKS1 is a rather slow process (39). TRF2 has been shown to interact with two other PARPs involved in DNA repair, namely PARP1 (7) and PARP2 (8). But the latter one was detected only in ALT (alternative lengthening of telomeres) cells, which use homologous recombination to maintain their telomeric DNA. So far, the impact of PARP activity on TRF2 substrate binding and telomere regulation remains elusive, although a recent publication reported the presence of PARP1 at eroded telomeres (9).
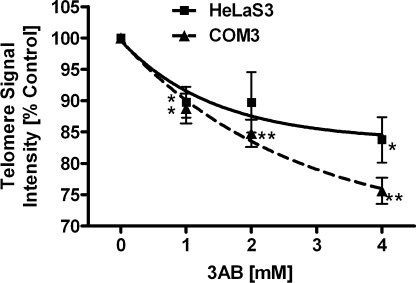
In order to test the short-term responsiveness of telomere regulation in mammalian cells to treatment with the pan-PARP inhibitor 3AB, we exposed the cells to various concentrations for 1 week and analyzed median telomere length by Q-FISH. Telomere signal intensity reduction was clearly correlated to 3AB concentration (Figure 1), leveling out at 75% of controls at the highest concentration applied (4 mM). The cell lines used in this study have a similar telomere length of ∼5 kb as measured by Southern blotting (5.5 kb HeLaS3, human) and comparison of Q-FISH signals with known standards (5 kb COM3, hamster) (data not shown). Cell numbers and cell death rates were not significantly affected, except for a minor increase in proliferation time in cells treated with highest dose of 3AB in long-term experiments (Supplementary Figure 1A and B). This indicates that the effect is not dependent on interference with cell cycle progression or cell viability, in line with the fact that mono-ADP-ribosyl transferases involved in signaling cascades are not inhibited at the 3AB concentrations used (58). Therefore, the dose–response curves observed can be attributed to gradual inhibition of overall PARP activity.
Figure 1.
Dose-dependent telomere shortening in mammalian cells exposed to increasing concentrations of 3AB. Telomere length was measured by Q-FISH and resulting values were expressed as percentage of the corresponding control. HeLaS3 and COM3 hamster cells were treated with concentrations of 3AB as indicated for 1 week. Both show a dose-dependent decrease in telomere signal intensity, with ∼85% and 75% of control level, respectively, at the highest concentration of the inhibitor. Squares, HeLaS3; triangles, COM3; *P < 0.05, **P < 0.01, compared to control.
Time course of 3AB-induced telomere shortening
Next, we studied the kinetics of telomere shortening induced by the PARP inhibitor. We wanted to address if long-term treatment with 3AB would lead to a steady decrease in telomere length and subsequent cell death or senescence, which is induced by critically short telomeres (59). Therefore, we treated cells continuously with 3AB. After the first week, treated cultures were split and one half was further inhibited, whereas the other half was released, and both were cultured for 3 additional weeks (Figure 2). In HeLaS3 and COM3 cells, we detected a fast decrease in telomeric signals by 500 bp/population doubling (PD) within 48 h. This effect cannot be explained by the mere inhibition of telomerase activity, as telomerase-negative fibroblasts show a maximal decrease of 50–200 bp/PD (60). Telomeres of long-term treated cultures stabilized at about 70 and 80% of control levels in hamster and human cells, respectively. HeLaS3 showed less pronounced telomere shortening, because the impact of 3AB treatment is dose-dependent and we used only 2 mM of 3AB as a maximal concentration for this cell line. Upon release from 3AB, telomeres were rapidly elongated to and stabilized at control levels. The speed of increase was nearly identical in hamster and in human cells with about 15% addition/PD. Interestingly, HeLa cells showed an overshooting reaction and elongated to a median of 110% of control telomeres, compared to 100% control length in COM3. Thereafter, HeLaS3 telomeres shortened again and control levels were re-established. Re-elongation was not detectable in IMR90 lung fibroblasts lacking active telomerase (Figure 3). Southern blotting of digested genomic DNA (from 204 h and 684 h postinitial treatment) and probing with a G-strand-specific oligonucleotide supported the Q-FISH data, which were obtained by using PNA probes, although differences were less pronounced and did not reach significance (Supplementary Figure 2A and B). The 3′-overhang at the time of highest divergence between treated and released cultures (204 h) was not altered, as assessed by the T-OLA method (Figure 4). Telomerase activity measured by the PCR-based TRAP method (Figure 5A and B) was not affected either, as there were no significant differences between the differently treated cultures after 4 weeks (control versus 3AB-treated versus 3AB-released). Thus, in our hands, PARP inhibitors did not downregulate telomerase activity in vitro, although opposite findings have been published by one group, using the same method (61,62).
Figure 2.
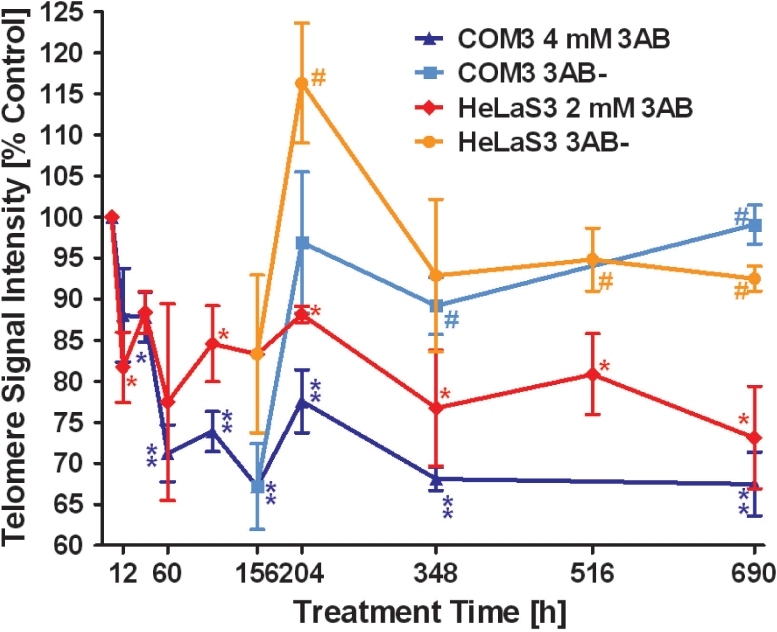
Time course of telomere shortening during 3AB treatment. Telomere length was measured and plotted as in Figure 1. HeLaS3 and COM3 cells were treated with 2 mM and 4 mM 3AB, respectively. After 156 h incubation with the inhibitor, cultures were split and one part was released, whereas the other was further incubated with 3AB. 3AB− indicates cultures released from the inhibitor. After an initial drop, telomeres are stabilizing at a new lower level. Release leads to fast increase in median length, which stabilizes at control levels. Red diamonds, HeLaS3/3AB; dark blue triangles, COM3/3AB; orange circles, HeLaS3 released; light blue squares, COM3 released. *P < 0.05, **P < 0.01, compared to control. #P < 0.05 released compared to treated cultures; unpaired two-tailed t-test.
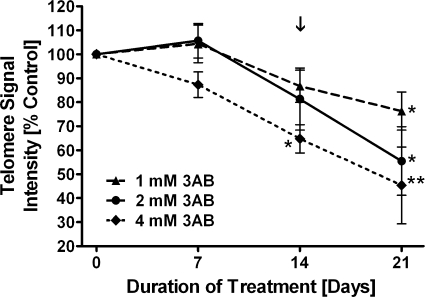
Figure 3.
Irreversible telomere shortening in IMR90 fibroblasts exposed to the PARP inhibitor 3AB. IMR90 fibroblastswere incubated with 3AB for 14 days and released thereafter. After 14 days, significant telomere shortening is seen in IMR90 cells treated with 4 mM 3AB. After 21 days, all cultures show significant telomere shortening compared to telomeres from control cells. The arrow marks the time of release from 3AB. Triangles/dashed line, 1 mM 3AB; circles/closed line, 2 mM 3AB; diamonds/dotted line, 4 mM 3AB; *P < 0.05, **P < 0.01 compared to control.
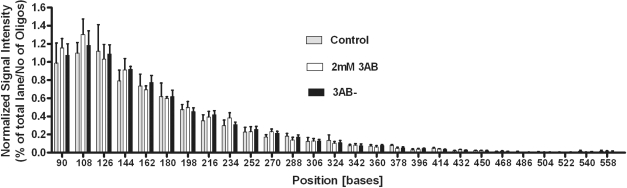
Figure 4.
3AB does not influence telomeric 3′-overhang. The length of the telomeric 3′-overhang was determined in HeLaS3 cells by using the T-OLA method at 204 h of 3AB treatment (greatest difference between treated and released cultures). Bars represent percentage of intensity of bands in one lane. Intensities were normalized to the number of ligated oligonucleotides. There is no significant difference between 3AB treated, released and control cells. Gray bars, controls; open bars, 2 mM 3AB; black bars, released.
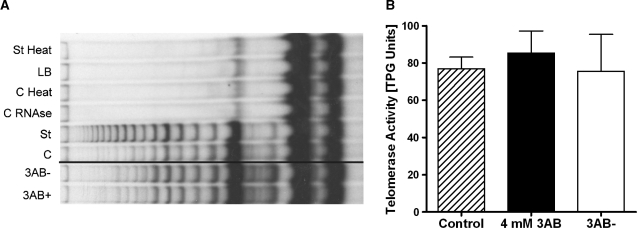
Figure 5.
3AB does not affect in vitro telomerase activity. After 4 weeks of incubation of cells, in vitro telomerase activity of COM3 cell extracts were measured by conventional PCR-based assay (TRAP). (A) Representative gel with TRAP products; 3AB+ and 3AB− lanes were from the same gel and only rearranged to the control lane. 3AB+, continuously 3AB treated; 3AB−, 3 weeks after 3AB release; C, control cells; St, standard reaction for normalization; C RNAse, control sample treated with RNaseA; C Heat, control sample treated with heat; LB, lysis buffer only; St Heat, standard reaction treated with heat. (B) Evaluation of three independent experiments. The activity is expressed as total product generated (TPG) units. No significant differences were observed. Hatched bar, control; black bar, 4 mM 3AB; open bar, released.
PARP1 is responsible for telomere shortening induced by 3AB
Knockdown of TNKS1 by siRNA approach led to mitotic failure (32,47,49) and it has been shown in vitro that TNKS1 is equally sensitive to inhibition as PARP1 (50). But as 3AB has been routinely used for continuous treatment of cells without any impact on cell viability, the situation in a cellular context may be different (see Discussion section). As PARP1 as well as PARP2 localize to telomeres by interaction with TRF2 (7,8), we wanted to determine which of these two PARPs is responsible for the observed 3AB effect on telomeres.
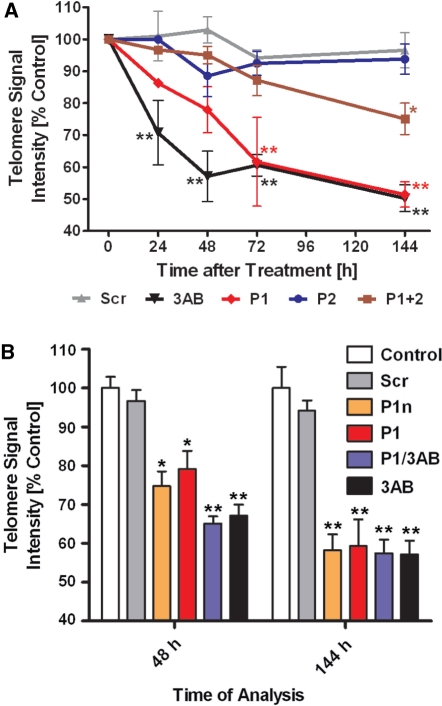
Therefore, we transfected HeLaS3 cells with siRNAs specifically targeting either PARP1 (P1) or PARP2 (P2). We also cotransfected both siRNAs in a 1:1 ratio, keeping the total siRNA concentration constant. The efficacy of siRNA-induced knockdown is shown in Supplementary Figure 3A for PARP1 and Supplementary Figure 3B for PARP2. On average, PARP1 protein was knocked down slightly more effectively than PARP2, with residual levels of 17% versus 11% at 24 h posttransfection; 9% versus 29% at 72 h posttransfection; and 37% versus 75% at 144 h posttransfection. In double transfectants, PARP1 knockdown was only half as pronounced as in single transfections, whereas PARP2 showed essentially the same knockdown levels (11%, 13% and 67%). Therefore, PARP1 siRNA was needed in the highest concentration for an effective knockdown. We monitored telomere length by Q-FISH and normalized the signal intensities to untreated controls (Figure 6A). As expected, transfection of a scrambled version of P1 (Scr) did not show any significant effect, whereas 3 mM 3AB induced a fast drop in PNA-dependent fluorescence within the first 48 h. Later, telomeres stabilized at a new length as in previous experiments. P1 siRNA also induced telomere shortening with delayed kinetics, probably because PARP1 protein had to be degraded to abrogate activity. Starting from 72 h posttransfection, there was no significant difference between the 3AB treated and with P1 siRNA transfected cells with regard to telomere length. Transfection of siRNA P2 showed no impact on PNA fluorescence compared to Scr siRNA or untreated controls. Double tranfectants (P1 + 2) showed an intermediate decrease in telomere length at later time points, further strengthening the hypothesis that only inhibition of PARP1 activity is responsible for telomere shortening. To demonstrate that PARP1 is indeed the only effector of 3AB treatment and to exclude off-target effects of the siRNA, we treated P1 siRNA transfected cells simultaneously with 3AB and we tested a second PARP1 siRNA (P1n) at two time points where 3AB mediated telomere shortening was different (48h) and identical (144h) to P1 siRNA (Figure 6B). Silencing efficiency mediated by P1n was similar to P1 (data not shown). Both siRNAs showed near-identical impact on telomere shortening and double treatment with P1 and 3AB was indistinguishable from 3AB administration alone. With these experiments we demonstrate that PARP1 and not PARP2 is responsible for the observed negative effect of 3AB on telomere length.
Figure 6.
Knockdown of PARP1 shortens telomeres similar to 3AB. (A) HeLaS3 cells were transfected with different siRNAs or incubated with 3AB as telomere shortening control. Values were normalized to untreated controls. 3AB treated cells show the same effect as siRNA against PARP1, whereas Scr and PARP2 siRNA have no effect. Black upside down triangles, 3AB treated; red diamonds, PARP1 siRNA (P1); brown squares, PARP1 + PARP2 siRNAs (P1 + 2); blue dots, PARP2 siRNA (P2); gray triangles, scrambled siRNA (Scr); *P < 0.05, **P < 0.01 compared to control. (B) HeLaS3 cells were treated as in (A), but with a second unrelated PARP1 siRNA (P1n) and double treated with P1 siRNA and 3 mM 3AB (P1/3AB). Bar-code, empty, control; light gray, Scr; orange, P1n; red, P1; blue, P1/3AB; black, 3AB; *P < 0.01, **P < 0.001, compared to control.
DISCUSSION
We found in two mammalian cell systems (hamster and human) that pharmacological inhibition of PARP1 led to a fast, dose-dependent decrease of telomere length (Figures 1 and 2). After release from the inhibitor 3AB, telomeres elongated back to control levels in telomerase-expressing cells (Figure 2), whereas they stayed short in telomerase-negative cells (Figure 3). The inhibitor neither influenced the telomeric 3′-overhang length (Figure 4) nor telomerase activity in a standard TRAP assay (Figure 5), in line with published data (45). In order to discriminate between the two different PARPs (PARP1 and PARP2) proposed to interact with TRF2 and sensitive to the used inhibitor concentration, we analyzed telomere length after knockdown of each by siRNA (Figure 6). Surprisingly, only cells transfected with PARP1 siRNA showed telomere shortening. Two independent PARP1 siRNAs produced the same effect. Furthermore, PARP1 knockdown reproduced perfectly the results with 3AB, and the effect of combination of both was identical to inhibitor treatment alone. These results strongly suggest that PARP1 is the only effector protein responsive to a standard PARP inhibitor that mediates telomere regulation, although we cannot exclude that other PARP family members—with the exception of PARP2—may contribute to a minor extent to this effect.
In 2004, Dynek and colleagues (32) published that knockdown of TNKS1 leads to mitotic failure. Several recent publications link TNKS1 with spindle formation/regulation and mitosis progression (47–49). In vitro experiments revealed that TNKS1 and PARP1 are equally sensitive to inhibition by different drugs (50). But 3AB has been widely used without any detectable influence on cell cycle progression or cell viability (including this work). Thus, most likely, TNKS1 is not inhibited in cells by the drug concentrations used. Supporting this notion, cellular PARP1 activity has been reported to be fully inhibited at a concentration of 1–2 µM of the PARP inhibitor PJ34 (63), whereas TNKS1 was only completely blocked at concentrations greater 40 µM (64). Additionally, TNKS-mediated telomere elongation is slow (significant only after several PD) and only achieved by overexpression of an NLS-tagged version, as otherwise the protein is not found in the nucleus after transfection (39,65). Therefore, we reasoned that the effect of 3AB in our experimental setup is probably not dependent on TNKS1, but on other PARPs localized at telomeres. Good candidates were PARP1 and PARP2, which have been shown to interact only with TRF2 within the shelterin complex (7–9).
We provide evidence that treatment of intact mammalian cells with the well-established PARP inhibitor 3AB leads to a reversible loss of telomere length, shown by Q-FISH (Figures 1–3) as well as by Southern blotting (Supplementary Figure 2). Southern blotting is a less sensitive method compared to Q-FISH, as subtelomeric regions, which can be of substantial size [3 kb, (66)] are also includes in length determination.
The decline in telomere length was fast (about 30% within 60 h or 2.5 PD) and led to a new setting without any obvious loss of cell viability. Both findings are perfectly in line with data published by d'Adda di Fagagna and colleagues (45), who showed in embryonic fibroblasts from Parp1 knockout mice also 30% loss of telomere length and a new stable setting. We demonstrated that in hamster cells in vitro telomerase activity is not affected (Figure 5) and that the rate of telomere length decrease is faster than expected from blocking telomerase alone. Therefore, telomere loss must be due to incomplete replication or an active degradation process. Two nucleases have been shown to localize to telomeres, i.e. the nucleotide excision repair complex nuclease ERCC1/XPF (67) as well as Apollo (68), a recently discovered ‘sibling’ of the Artemis nuclease involved in nonhomologous end-joining. Whether any of these are responsible for the observed telomere shortening awaits further investigation. 3AB treatement of telomerase-negative human fibroblasts led to accelerated telomere shortening compared to controls (Figure 3). This also excludes telomerase as main effector of PARP inhibition, although two recent publications described opposite findings using the same method (61,62). Release from 3AB led to regain in telomeric sequences with a similar rate as loss (1 kb within 48 h) in telomerase expressing cells only. The sustained loss of telomere length in IMR90 cells after release may well be due to the fact that these fibroblasts were close to senescence. Inhibiting PARP at this critical point could destabilize the telomeric structure and increase the susceptibility to degradation processes, which may be afterwards insensitive to re-established PARP activity.
In the literature, conflicting results have been reported regarding the influence of PARP1 on telomere length regulation, based on two different knockout mouse strains. One group showed no impact on telomeres (43,44), whereas the other reported shorter telomeres (45,46).
Treatment with pharmacological inhibitors has often the disadvantage of interference with a whole enzyme family, where specificity is needed for analysis of divergent activities. Therefore, a RNA interference approach is the best method to dissect in a short-term setting the influence of proteins on specific regulatory processes.
To clarify the role of PARP1 and PARP2 in telomere regulation, we used siRNA for both proteins. We showed that only PARP1 and not PARP2 is involved in telomere length regulation (Figure 6). Thus, regarding PARP1, we confirmed data from one of the knockout mouse models (45), challenged by Samper et al. (43). The observed interaction of PARP2 with TRF2 in U2OS cells (8), which use the ALT-recombination pathway to maintain telomeres, may be confined to this special case.
From our data, we propose the following model: PARP1 interacts with TRF2 at the telomere, either (A) constantly throughout the cell cycle or (B) only during a specific phase, e.g. S phase, when telomere replication takes place and the t-loop structure has to be unfolded. In the first case, PARP1 must be activated at a certain time to fulfill its action on TRF2, in the latter re-localization would bring PARP1 to its site of action. It has been shown that TRFs slow down replication fork progression on a telomeric template in vitro (69). Thus, these proteins have to be dislodged in order to facilitate proper telomere replication and maybe also for maintenance through telomerase by opening of the t-loop. In model A, slowing down replication fork progression may lead to unusual DNA structures or fork collapse, which depend on PARP1 for re-activation (70). TRF2 is subsequently modified and replication can proceed. Many other proteins and complexes involved in repair of stalled replication forks are also implicated in telomere regulation, like the 911 complex (71), BLM and WRN (72), which partially also interact directly with PARP1 (73–75). Alternatively, it has been shown that PARP1 is a member of the DNA replication complex and modifies several proteins therein (28). Thus, it is tempting to speculate that the replication fork carries its own ‘key’ for opening the t-loop, i.e. PARP1. The replication complex brings PARP1 to the telomere and the basal activity of PARP1 leads to TRF2 modification, t-loop opening and replication fork passage. In this speculative model, no DNA breaks or recombinogenic structural intermediates are needed for activation of PARP1 towards TRF2, but experimental proof awaits further investigations. Also, a recent publication argues against a constant interaction of PARP1 and TRF2 (9). If PARP1 is inhibited, telomeres shorten to the point where the t-loop formation is still possible and functions as a protective cap, but this structure would be instable (in the case of HeLaS3 and COM3 cells this would be 70% of initial telomere length). This now enables the replication fork to displace the loosely bound single-strand overhang just by passing by without detaching TRF2. The ALT pathway is unlikely to be involved in keeping telomeres stable at the new length setting, as the hallmark of this mechanism, a broad range of different telomere lengths within a cell, could not be detected.
We have previously shown that PARP activity in permeabilized PBMC stimulated by strand breaks correlates positively with mammalian life span, but during organismal aging, PARP activity declines (40). Many publications show that PARP1 (and also PARP2) activity is not only confined to cellular response to genotoxic stress. In mammalian cells, PARP1 (and PAR) has also been shown to be present at centromeres (76), centrioles (33,34) and telomeres (7). So far, inhibition of PARP activity in unstressed cells showed effects only in centriole regulation. Our present data show that PARP1 activity also positively influences telomere maintenance. Thus, during the aging process, PARP1 activity may decline below a threshold, where it is no longer able to maintain proper telomere length. As PARP inhibition also leads to accelerated telomere shortening in fibroblasts, tissue regeneration from somatic cells may be impaired as it reduces their proliferation capacity. Subsequently, telomere shortening during compensatory enhanced tissue-renewal would deplete replicating stem cells or their low-level telomerase expressing progeny by senescence/apoptosis induction. These two scenarios could lead finally to organ dysfunction and accelerated aging of the organism. It has already been shown that tissues of aged individuals have a higher number of senescent cells (77–79).
Our findings may also impact on a proposed combination therapy with genotoxic agents and PARP1 inhibitors for tumor treatment. This approach has been shown to increase the efficacy of chemotherapeutics (80–83), but with the new data presented here one should be cautious about long-term effects. Patients suffer sometimes from secondary tumors induced by cytotoxic cancer treatment. As the agents used also damage DNA in somatic, nontelomerase expressing tissues, additional accelerated telomere shortening by PARP1 inhibition may either induce an earlier onset of senescence in these cells with the above mentioned impact on organ aging or even trigger enhanced genomic instability and support thus formation of neoplasias.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EU (QLG1-CT-1999-01341 and LSHC-CT-2004-502943 to P.B.); the Swiss National Foundation for Scientific Research (to F.A.) partially. Funding for open access charge: University of Konstanz.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 2.Liu D, O’Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 3.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 5.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 6.Cook BD, Dynek JN, Chang W, Shostak G, Smith S. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell Biol. 2002;22:332–342. doi: 10.1128/MCB.22.1.332-342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J. Biol. Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 8.Dantzer F, Giraud-Panis MJ, Jaco I, Ame JC, Schultz I, Blasco M, Koering CE, Gilson E, Menissier-de Murcia J, de Murcia G, et al. Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol. Cell Biol. 2004;24:1595–1607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez M, Wu J, Schreiber V, Dunlap J, Dantzer F, Wang Y, Liu Y. PARP1 Is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol. Biol. Cell. 2006;17:1686–1696. doi: 10.1091/mbc.E05-07-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer RG, Meyer-Ficca ML, Jacobson EL, Jacobson MK. In: Poly(ADP-Ribosyl)ation. Bürkle A, editor. Landes Bioscience: Georgetown; 2006. pp. 1–12. [Google Scholar]
- 11.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 12.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Althaus FR, Kleczkowska HE, Malanga M, Muntener CR, Pleschke JM, Ebner M, Auer B. Poly ADP-ribosylation: a DNA break signal mechanism. Mol. Cell Biochem. 1999;193:5–11. [PubMed] [Google Scholar]
- 14.Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 16.Faraone-Mennella MR. Chromatin architecture and functions: the role(s) of poly(ADP-RIBOSE) polymerase and poly(ADPribosyl)ation of nuclear proteins. Biochem. Cell Biol. 2005;83:396–404. doi: 10.1139/o05-042. [DOI] [PubMed] [Google Scholar]
- 17.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. BioEssays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 18.Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, Jacobson EL. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 19.Vidakovic M, Poznanovic G, Bode J. DNA break repair: refined rules of an already complicated game. Biochem. Cell Biol. 2005;83:365–373. doi: 10.1139/o05-044. [DOI] [PubMed] [Google Scholar]
- 20.Petermann E, Keil C, Oei SL. Importance of poly(ADP-ribose) polymerases in the regulation of DNA-dependent processes. Cell Mol. Life Sci. 2005;62:731–738. doi: 10.1007/s00018-004-4504-2. [DOI] [PubMed] [Google Scholar]
- 21.Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. Biol. Chem. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 22.Simbulan-Rosenthal CM, Rosenthal DS, Luo R, Smulson ME. Poly(ADP-ribose) polymerase upregulates E2F-1 promoter activity and DNA pol alpha expression during early S phase. Oncogene. 1999;18:5015–5023. doi: 10.1038/sj.onc.1202900. [DOI] [PubMed] [Google Scholar]
- 23.Akiyama T, Takasawa S, Nata K, Kobayashi S, Abe M, Shervani NJ, Ikeda T, Nakagawa K, Unno M, Matsuno S, et al. Activation of Reg gene, a gene for insulin-producing beta – cell regeneration: poly(ADP-ribose) polymerase binds Reg promoter and regulates the transcription by autopoly(ADP-ribosyl)ation. Proc. Natl Acad. Sci. USA. 2001;98:48–53. doi: 10.1073/pnas.240458597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendoza-Alvarez H, Alvarez-Gonzalez R. Regulation of p53 sequence-specific DNA-binding by covalent poly(ADP-ribosyl)ation. J. Biol. Chem. 2001;276:36425–36430. doi: 10.1074/jbc.M105215200. [DOI] [PubMed] [Google Scholar]
- 25.Ziegler M, Oei SL. A cellular survival switch: poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. BioEssays. 2001;23:543–548. doi: 10.1002/bies.1074. [DOI] [PubMed] [Google Scholar]
- 26.Smulson ME. Poly(ADP-ribose) polymerase gene on chromosome 1q: early role in differentiation linked replication; gene on human chromosome 13q: marker of carcinogenesis. Mol. Cell Biochem. 1994;138:77–84. doi: 10.1007/BF00928446. [DOI] [PubMed] [Google Scholar]
- 27.Simbulan-Rosenthal CM, Rosenthal DS, Hilz H, Hickey R, Malkas L, Applegren N, Wu Y, Bers G, Smulson ME. The expression of poly(ADP-ribose) polymerase during differentiation-linked DNA replication reveals that it is a component of the multiprotein DNA replication complex. Biochemistry. 1996;35:11622–11633. doi: 10.1021/bi953010z. [DOI] [PubMed] [Google Scholar]
- 28.Simbulan-Rosenthal CM, Rosenthal DS, Boulares AH, Hickey RJ, Malkas LH, Coll JM, Smulson ME. Regulation of the expression or recruitment of components of the DNA synthesome by poly(ADP-ribose) polymerase. Biochemistry. 1998;37:9363–9370. doi: 10.1021/bi9731089. [DOI] [PubMed] [Google Scholar]
- 29.Ohashi Y, Ueda K, Hayaishi O, Ikai K, Niwa O. Induction of murine teratocarcinoma cell differentiation by suppression of poly(ADP-ribose) synthesis. Proc. Natl Acad. Sci. USA. 1984;81:7132–7136. doi: 10.1073/pnas.81.22.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhatia M, Kirkland JB, Meckling-Gill KA. Overexpression of poly(ADP-ribose) polymerase promotes cell cycle arrest and inhibits neutrophilic differentiation of NB4 acute promyelocytic leukemia cells. Cell Growth Differ. 1996;7:91–100. [PubMed] [Google Scholar]
- 31.Smith S, de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 1999;112(Pt 21):3649–3656. doi: 10.1242/jcs.112.21.3649. [DOI] [PubMed] [Google Scholar]
- 32.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 33.Kanai M, Uchida M, Hanai S, Uematsu N, Uchida K, Miwa M. Poly(ADP-ribose) polymerase localizes to the centrosomes and chromosomes. Biochem. Biophys. Res. Commun. 2000;278:385–389. doi: 10.1006/bbrc.2000.3801. [DOI] [PubMed] [Google Scholar]
- 34.Kanai M, Tong WM, Sugihara E, Wang ZQ, Fukasawa K, Miwa M. Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-Ribosyl)ation in regulation of centrosome function. Mol. Cell Biol. 2003;23:2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 36.Koh DW, Dawson TM, Dawson VL. Mediation of cell death by poly(ADP-ribose) polymerase-1. Pharmacol. Res. 2005;52:5–14. doi: 10.1016/j.phrs.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 37.Bürkle A. Poly(APD-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett. 2001;163:1–5. doi: 10.1016/s0304-3835(00)00694-7. [DOI] [PubMed] [Google Scholar]
- 38.Chi NW, Lodish HF. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 39.Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- 40.Grube K, Bürkle A. Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc. Natl Acad. Sci. USA. 1992;89:11759–11763. doi: 10.1073/pnas.89.24.11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beneke S, Alvarez-Gonzalez R, Bürkle A. Comparative characterisation of poly(ADP-ribose) polymerase-1 from two mammalian species with different life span. Exp. Gerontol. 2000;35:989–1002. doi: 10.1016/s0531-5565(00)00134-0. [DOI] [PubMed] [Google Scholar]
- 42.Burkle A, Diefenbach J, Brabeck C, Beneke S. Ageing and PARP. Pharmacol. Res. 2005;52:93–99. doi: 10.1016/j.phrs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Samper E, Goytisolo FA, Menissier-de Murcia J, Gonzalez-Suarez E, Cigudosa JC, de Murcia G, Blasco MA. Normal telomere length and chromosomal end capping in poly(ADP-ribose) polymerase-deficient mice and primary cells despite increased chromosomal instability. J. Cell Biol. 2001;154:49–60. doi: 10.1083/jcb.200103049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Espejel S, Klatt P, Menissier-de Murcia J, Martin-Caballero J, Flores JM, Taccioli G, de Murcia G, Blasco MA. Impact of telomerase ablation on organismal viability, aging, and tumorigenesis in mice lacking the DNA repair proteins PARP-1, Ku86, or DNA-PKcs. J. Cell Biol. 2004;167:627–638. doi: 10.1083/jcb.200407178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.d’Adda di Fagagna F, Hande MP, Tong WM, Lansdorp PM, Wang ZQ, Jackson SP. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat. Genet. 1999;23:76–80. doi: 10.1038/12680. [DOI] [PubMed] [Google Scholar]
- 46.Tong WM, Hande MP, Lansdorp PM, Wang ZQ. DNA strand break-sensing molecule poly(ADP-Ribose) polymerase cooperates with p53 in telomere function, chromosome stability, and tumor suppression. Mol. Cell Biol. 2001;21:4046–4054. doi: 10.1128/MCB.21.12.4046-4054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang W, Dynek JN, Smith S. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem. J. 2005;391:177–184. doi: 10.1042/BJ20050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 49.Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. EMBO J. 2007;26:4867–4878. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rippmann JF, Damm K, Schnapp A. Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J. Mol. Biol. 2002;323:217–224. doi: 10.1016/s0022-2836(02)00946-4. [DOI] [PubMed] [Google Scholar]
- 51.Küpper JH, de Murcia G, Bürkle A. Inhibition of poly(ADP-ribosyl)ation by overexpressing the poly(ADP-ribose) polymerase DNA-binding domain in mammalian cells. J. Biol. Chem. 1990;265:18721–18724. [PubMed] [Google Scholar]
- 52.Küpper JH, Müller M, Jacobson MK, Tatsumi-Miyajima J, Coyle DL, Jacobson EL, Bürkle A. Trans-dominant inhibition of poly(ADP-ribosyl)ation sensitizes cells against gamma-irradiation and N-methyl-N'-nitro-N-nitrosoguanidine but does not limit DNA replication of a polyomavirus replicon. Mol. Cell Biol. 1995;15:3154–3163. doi: 10.1128/mcb.15.6.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mai S, Wiener F. In: FISH: A Practical Approach. Beatty B, Mai S, Squire J, editors. Oxford University Press: Oxford; 2002. pp. 55–76. [Google Scholar]
- 54.Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 55.Cimino-Reale G, Pascale E, Battiloro E, Starace G, Verna R, D’Ambrosio E. The length of telomeric G-rich strand 3′-overhang measured by oligonucleotide ligation assay. Nucleic Acids Res. 2001;29:E35. doi: 10.1093/nar/29.7.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohausz O, Blenn C, Malanga M, Althaus FR. The roles of poly(ADP-ribose)-metabolizing enzymes in alkylation-induced cell death. Cell Mol. Life Sci. 2008;65:644–655. doi: 10.1007/s00018-008-7516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang W, Dynek JN, Smith S. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 2003;17:1328–1333. doi: 10.1101/gad.1077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rankin PW, Jacobson EL, Benjamin RC, Moss J, Jacobson MK. Quantitative studies of inhibitors of ADP-ribosylation in vitro and in vivo. J. Biol. Chem. 1989;264:4312–4317. [PubMed] [Google Scholar]
- 59.d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 60.von Zglinicki T, Pilger R, Sitte N. Accumulation of single-strand breaks is the major cause of telomere shortening in human fibroblasts. Free Radic. Biol. Med. 2000;28:64–74. doi: 10.1016/s0891-5849(99)00207-5. [DOI] [PubMed] [Google Scholar]
- 61.Ghosh U, Bhattacharyya NP. Benzamide and 4-amino 1,8 naphthalimide treatment inhibit telomerase activity by down-regulating the expression of telomerase associated protein and inhibiting the poly(ADP-ribosyl)ation of telomerase reverse transcriptase in cultured cells. Febs J. 2005;272:4237–4248. doi: 10.1111/j.1742-4658.2005.04837.x. [DOI] [PubMed] [Google Scholar]
- 62.Ghosh U, Das N, Bhattacharyya NP. Inhibition of telomerase activity by reduction of poly(ADP-ribosyl)ation of TERT and TEP1/TP1 expression in HeLa cells with knocked down poly(ADP-ribose) polymerase-1 (PARP-1) gene. Mutat. Res. 2007;615:66–74. doi: 10.1016/j.mrfmmm.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 63.Jagtap P, Soriano FG, Virag L, Liaudet L, Mabley J, Szabo E, Hasko G, Marton A, Lorigados CB, Gallyas F., Jr., et al. Novel phenanthridinone inhibitors of poly (adenosine 5′-diphosphate-ribose) synthetase: potent cytoprotective and antishock agents. Crit. Care Med. 2002;30:1071–1082. doi: 10.1097/00003246-200205000-00019. [DOI] [PubMed] [Google Scholar]
- 64.Yeh TY, Meyer TN, Schwesinger C, Tsun ZY, Lee RM, Chi NW. Tankyrase recruitment to the lateral membrane in polarized epithelial cells: regulation by cell-cell contact and protein poly(ADP-ribosyl)ation. Biochem. J. 2006;399:415–425. doi: 10.1042/BJ20060713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seimiya H, Muramatsu Y, Ohishi T, Tsuruo T. Tankyrase 1 as a target for telomere-directed molecular cancer therapeutics. Cancer Cell. 2005;7:25–37. doi: 10.1016/j.ccr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 66.Hultdin M, Gronlund E, Norrback K, Eriksson-Lindstrom E, Just T, Roos G. Telomere analysis by fluorescence in situ hybridization and flow cytometry. Nucleic Acids Res. 1998;26:3651–3656. doi: 10.1093/nar/26.16.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 68.van Overbeek M, de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr. Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 69.Ohki R, Ishikawa F. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004;32:1627–1637. doi: 10.1093/nar/gkh309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004;23:3872–3882. doi: 10.1038/sj.onc.1207491. [DOI] [PubMed] [Google Scholar]
- 71.Francia S, Weiss RS, Hande MP, Freire R, d’Adda di Fagagna F. Telomere and telomerase modulation by the mammalian Rad9/Rad1/Hus1 DNA-damage-checkpoint complex. Curr. Biol. 2006;16:1551–1558. doi: 10.1016/j.cub.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 72.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 73.Lebel M, Lavoie J, Gaudreault I, Bronsard M, Drouin R. Genetic cooperation between the Werner syndrome protein and poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice. Am. J. Pathol. 2003;162:1559–1569. doi: 10.1016/S0002-9440(10)64290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.von Kobbe C, Harrigan JA, May A, Opresko PL, Dawut L, Cheng WH, Bohr VA. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol. Cell Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.von Kobbe C, Harrigan JA, Schreiber V, Stiegler P, Piotrowski J, Dawut L, Bohr VA. Poly(ADP-ribose) polymerase 1 regulates both the exonuclease and helicase activities of the Werner syndrome protein. Nucleic Acids Res. 2004;32:4003–4014. doi: 10.1093/nar/gkh721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saxena A, Saffery R, Wong LH, Kalitsis P, Choo KH. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J. Biol. Chem. 2002;277:26921–26926. doi: 10.1074/jbc.M200620200. [DOI] [PubMed] [Google Scholar]
- 77.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Melk A, Kittikowit W, Sandhu I, Halloran KM, Grimm P, Schmidt BM, Halloran PF. Cell senescence in rat kidneys in vivo increases with growth and age despite lack of telomere shortening. Kidney Int. 2003;63:2134–2143. doi: 10.1046/j.1523-1755.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- 79.Wong KK, Maser RS, Bachoo RM, Menon J, Carrasco DR, Gu Y, Alt FW, DePinho RA. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature. 2003;421:643–648. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- 80.Delaney CA, Wang LZ, Kyle S, White AW, Calvert AH, Curtin NJ, Durkacz BW, Hostomsky Z, Newell DR. Potentiation of temozolomide and topotecan growth inhibition and cytotoxicity by novel poly(adenosine diphosphoribose) polymerase inhibitors in a panel of human tumor cell lines. Clin. Cancer Res. 2000;6:2860–2867. [PubMed] [Google Scholar]
- 81.Bowman KJ, White A, Golding BT, Griffin RJ, Curtin NJ. Potentiation of anti-cancer agent cytotoxicity by the potent poly(ADP-ribose) polymerase inhibitors NU1025 and NU1064. Br. J. Cancer. 1998;78:1269–1277. doi: 10.1038/bjc.1998.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, Durkacz BW, Hostomsky Z, Kumpf RA, Kyle S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J. Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 83.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.