Abstract
Using a suppressive subtractive hybridization system, we identified CSIG (cellular senescence-inhibited gene protein; RSL1D1) that was abundant in young human diploid fibroblast cells but declined upon replicative senescence. Overexpression or knockdown of CSIG did not influence p21Cip1 and p16INK4a expressions. Instead, CSIG negatively regulated PTEN and p27Kip1 expressions, in turn promoting cell proliferation. In PTEN-silenced HEK 293 cells and PTEN-deficient human glioblastoma U87MG cells, the effect of CSIG on p27Kip1 expression and cell division was abolished, suggesting that PTEN was required for the role of CSIG on p27Kip1 regulation and cell cycle progression. Investigation into the underlying mechanism revealed that the regulation of PTEN by CSIG was achieved through a translational suppression mechanism. Further study showed that CSIG interacted with PTEN mRNA in the 5′ untranslated region (UTR) and that knockdown of CSIG led to increased luciferase activity of a PTEN 5′ UTR-luciferase reporter. Moreover, overexpression of CSIG significantly delayed the progression of replicative senescence, while knockdown of CSIG expression accelerated replicative senescence. Knockdown of PTEN diminished the effect of CSIG on cellular senescence. Our findings indicate that CSIG acts as a novel regulatory component of replicative senescence, which requires PTEN as a mediator and involves in a translational regulatory mechanism.
Replicative senescence is defined as a state of proliferative arrest accompanying the replicative exhaustion of cultured human fibroblast cells (16). The stable state of proliferation arrest limits the proliferation of damaged cells and may act as a natural barrier to cancer progression. Some genes linked to senescence in vitro also influence organismal life span in vivo, raising the possibility that replicative senescence underlies organismal aging (5, 8, 22, 31). After a finite number of population doublings, human diploid fibroblasts exhibit senescent features (6) such as growth inhibition, cell cycle arrest (16), enlarged and flat morphology (3), elevated senescence-associated β-galactosidase (SA-β-gal) activity (10), and accumulation of senescence-associated heterochromatin foci (26, 30). Although viable and metabolically active, senescent cells remain permanently insensitive to mitogenic signals (7).
Over the years, the molecular mechanisms that regulate the expression of genes whose levels are altered in senescent cells relative to those in young cells have been extensively studied. It is well accepted that decreased levels of cell cycle regulatory proteins such as cyclin A, CAK, Cdc2, E2F-1, E2F-2, and PCNA, as well as increased abundance of cyclin-dependent kinase (CDK) inhibitors, including p16INK4a and p21Cip1, contribute to the growth inhibition of senescent cells (36, 38-39). In addition, the activities of Rb and p53 tumor suppressors are constitutively elevated in senescent cells and promote cell senescence (13, 17, 26). In recent years, however, emerging evidence revealed that the tumor suppressor phosphatase and tensin homolog PTEN and its downstream effector p27Kip1 are also critical for replicative senescence, although the upstream regulatory events remain largely unknown (1, 4, 34). PTEN, a lipid phosphatase, is the central negative regulator of the phosphatidylinositol-3-kinase (PI3K) signal transduction cascade. PI3K catalyzes the conversion of phosphatidylinositol 4,5-phosphate (PIP2) to phosphatidylinositol 3,4,5-phosphate (PIP3) and activates AKT kinase and other downstream effectors (12). PTEN dephosphorylates PIP3 at the plasma membrane, thereby inhibiting PI3K-mediated signals for cell growth, proliferation, and survival (32). Despite its regulatory function on the PI3K/AKT pathway, recent studies strongly suggest that a phosphatase-independent mechanism is important for the biological functions of PTEN in the nucleus (2). By inhibiting ubiquitin-dependent proteasome turnover, PTEN elevates the p27Kip1 level and induces G1 arrest (23). In addition, PTEN was also found to interact with and regulate p53 expression (14, 18). Therefore, PTEN may act as a common regulator of both p53 and p27Kip1, which in turn, lead to cell cycle arrest and induce cell senescence.
Using a suppressive subtractive hybridization (15), we have identified and cloned a cellular senescence-inhibited gene (encoding CSIG; GenBank accession no. AY154473) (http://www.ncbi.nim.nih.gov). CSIG is abundantly expressed in young fibroblasts, but its expression declines during cellular senescence. CSIG is a ribosomal L1 domain-containing protein and therefore was also named as RSL1D1 in the Human Genome Organization (HUGO) Nomenclature Committee database. A BLAST search of the human genome database revealed that the human CSIG gene localizes on chromosome 16p13, spans 5,137 bp, and is composed of nine exons which encode 490 amino acid residues. According to informatics analysis (available at http://www.expasy.org), CSIG is evolutionarily conserved, and the human CSIG protein contains part of the ribosomal L1p/L10e consensus sequence (residues 30 to 260) in the N terminus and a long lysine-rich domain (residues 280 to 485) in the C terminus, suggesting that it may participate in ribosome biosynthesis or act as a transcriptional cofactor. As a differentially expressed gene involved in replicative senescence, the CSIG gene level is higher in young fibroblasts, while it is lower in senescent cells (15). However, whether CSIG is an active regulator of replicative senescence remains to be addressed. In this study, we have investigated the role of CSIG in senescence and its underlying mechanisms using the human 2BS fibroblast model of replicative senescence. We demonstrate that CSIG potently regulates p27Kip1 levels and the progression of cell senescence by regulating the translational level of PTEN.
MATERIALS AND METHODS
Cell culture, FACS analysis, cell proliferation, and SA-β-gal activity.
Early-passage (young; ∼28 population doublings [pdl]), middle-passage (middle-aged; ∼45 pdl), and late-passage (senescent; ∼58 pdl) human diploid 2BS fibroblasts (National Institute of Biological Products, Beijing, China), human glioblastoma U87MG cells (Chinese Academy of Medical Science, Beijing, China), human embryonic kidney HEK 293 cells, and HeLa cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin, at 37°C in 5% CO2. Fluorescence-activated cell sorting (FACS) analysis, cell proliferation assessment, and SA-β-gal staining were performed as described previously (11, 38).
Construction of pIRES-CSIG and pSilencer-CSIG and transfection.
For the construction of the vector expressing CSIG, the full-length CSIG cDNA was amplified by PCR and inserted into the EcoRI site in the pIRES neo2 vector (Clontech). For the construction of pSilencer-CSIG, oligonucleotides corresponding to small interfering RNA (siRNA) targeting the CSIG coding region (AGAAGGAACAGACGCCAGA) and a control (Ctrl) siRNA (AAGTGTAGTAGATCACCAGGC) were inserted into the BamHI and HindIII sites in the pSilencer 2.1-U6 neo vector (Ambion) to generate vectors expressing CSIG and Ctrl siRNAs by following the manufacturer's instructions.
To establish lines stably expressing CSIG or CSIG siRNA, early-passage (∼28 pdl) 2BS cells were transfected with either the pIRES-CSIG and pSilencer-CSIG constructs or the respective Ctrl vectors by the Lipofectamine 2000 reagent (Invitrogen) by following the manufacturer's instructions, selected by the G418 reagent (300 μg/ml; Invitrogen) for 2 to 4 weeks, and maintained in medium supplemented with 50 μg/ml G418.
To transiently silence CSIG, siRNA targeting CSIG (CSIG siRNA; AGAAGGAACAGACGCCAGA) and PTEN (PTEN siRNA; GUUAGCAGAAACAAAAGGAG) as well as a Ctrl siRNA (AAGTGTAGTAGATCACCAGGC) were transfected at concentrations indicated by the Oligofectamine reagent (Invitrogen) by following the manufacturer's recommendations. Cells were collected 48 to 72 h after transfection for further analysis.
Construction of pGEX-GST-CSIG, protein purification, and development of CSIG polyclonal antibody.
For the construction of the pGEX-GST-CSIG vector expressing a glutathione S-transferase (GST)-CSIG fusion protein, the CSIG cDNA sequence corresponding to amino acid residues 320 to 365 was amplified by PCR using primers 5′-CGGGATCCGATGTGGCACCTGAAAGTG-3′ and 5′-CCGCTCGAGTTAGATTTCGTCTTCGGATTCATTT-3′ and cloned into the pGEX-4T-2 vector (Amersham-Pharmacia Biotech) at BamHI and XhoI sites. The pGEX-GST-CSIG vector was transformed into Escherichia coli BL21. Expression of GST-CSIG was induced by isopropyl-1-thio-β-d-galactopyranoside (IPTG) and purified with glutathione-conjugated Sepharose beads (Pharmacia) by following the manufacturer's instructions.
To develop polyclonal antisera recognizing CSIG, purified GST-CSIG recombinant protein was emulsified in complete Freund's adjuvant as soluble antigen and injected into two rabbits. The animals were reinjected at 3-week intervals with the same dose of antigen in incomplete Freund's adjuvant, and the sera were collected 2 weeks after the second reinjection.
Immunofluorescence.
To determine the subcellular localization of endogenous CSIG, 2BS and HeLa cells were fixed with cold methanol for 5 min at room temperature, washed with phosphate-buffered saline (PBS) twice for 5 min, permeabilized with 0.5% Triton X in 100 in PBS for 15 min, and blocked with 0.5% bovine serum albumin in PBS for 1 h. Immunofluorescence assay was carried out by incubating cells with antisera recognizing CSIG at room temperature for 2 h and then rinsing and incubating them with fluorescein isothiocyanate-conjugated secondary antibody for 1 h. The nucleoli were immunostained with antinucleolin antibody (sc-8031; Santa Cruz) and subsequent tetramethyl rhodamine isothiocyanate-conjugated secondary antibody. Signals were visualized by fluorescence confocal microscopy. DAPI (4′,6-diamidino-2-phenylindole) was used to visualize nuclei.
Northern and Western blot analysis.
For Northern blot analysis, total RNA was isolated using an RNeasy mini kit (Qiagen) by following the manufacturer's protocol. Northern blot analysis was performed as previously described (15). The tissue expression pattern was assessed by using a premade human 12-lane multiple tissue Northern blot (MTN; Clontech) which contains approximately 1 μg of poly (A)+ RNA per lane from 12 different human tissue samples.
For Western blot analysis, lysates were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes. Monoclonal antibodies recognizing PTEN, p27Kip1, p16INK4a, p21Cip1, and β-actin were from Santa Cruz Biotechnologies (Santa Cruz, CA). After secondary antibody incubation, signals were detected by the SuperSignal West Pico chemiluminescent substrate (Pierce) by following the manufacturer's instructions and quantitated by densitometric analysis with the ImageMaster VDS software.
Preparation of polysomal fractions.
A total of 50 million cells were incubated for 15 min with 100 mg/ml cycloheximide, and total lysates (500 μl) were layered onto a cushion of 30% sucrose in ice-cold buffer containing 20 mM HEPES (pH 7.4), 50 mM potassium acetate, 5 mM magnesium acetate, 1 mM dithiothreitol, 1 unit of RNasin per μl, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml, and 0.5 mM phenylmethylsulfonyl fluoride. After centrifugation (Beckman SW40; 100,000 × g for 2 h, 4°C), the supernatant was collected as a nonpolysomal fraction; the pellet was resuspended in ice-cold buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2) containing 0.3 M NaCl, incubated on ice for 1 h, and centrifuged at 10,000 × g for 15 min at 4°C; and the resulting supernatant (polysomal extract) was collected for Western blot analysis.
Analysis of nascent protein.
A total of 1 × 106 cells were incubated with 1 mCi (1 Ci = 37 GBq) l-[35S]methionine and l-[35S]cysteine (Easy Tag Express; NEN/Perkin-Elmer) per 60-mm plate for 20 min, whereupon cells were lysed by using TSD lysis buffer (50 mM Tris [pH 7.5], 1% SDS, 5 mM dithiothreitol), and lysates were immunoprecipitated by using either monoclonal anti-PTEN antibody (Santa Cruz Biotechnology), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (BiOS Biotechnology), or immunoglobulin G (IgG) for 1 h at 4°C. After extensive washes in TNN buffer (50 mM Tris [pH 7.5], 250 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40), immunoprecipitated material was resolved by 12% SDS-PAGE, transferred onto polyvinylidene difluoride membranes, and visualized by using a PhosphorImager (Molecular Dynamics).
RNA-protein binding assays.
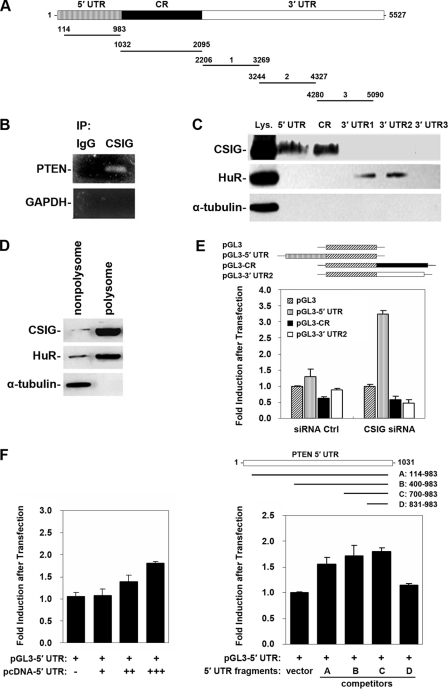
cDNA was used as a template for PCR amplification of different PTEN untranslated regions (UTR) and coding regions (CR). All forward primers contained the T7 promoter sequence 5′-CCAAGCTTCTAATACGACTCACTATAGGGAGA-3′ (T7). To prepare the PTEN 5′ UTR (positions 114 to 983), primers (T7)GGGCGGTGATGTGGCGGGACTCTT and CTGCTGGTGGCGGGGCTTCTTCTG were used. To prepare the PTEN CR (positions 1032 to 1095), primers (T7)GGAGATATCAAGAGGATGGATTCG and ATGCTGATCTTCATCAAAAG were used. To prepare the 3′ UTR-1 (positions 2206 to 3269), 3′ UTR-2 (positions 3244 to 4327), and 3′ UTR-3 (positions 4280 to 5090) fragments, primers (T7)TTGATGAAGATCAGCATA and CATCCACAGCAGGTATTA, (T7)TTCAATCATAATACCTGCTG and TATGAAAGACAAGGTGGC, and (T7)ATTCTAACACCTCACCAT and AATCAGTTTTAAGTGGAGT were used, respectively. For biotin pulldown assays, PCR-amplified DNA was used as a template to transcribe biotinylated RNA by using T7 RNA polymerase in the presence of biotin-UTP and was purified as described previously (38). Six micrograms of biotinylated transcripts were incubated with 120 μg of total cell lysates for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway), and pulldown material was analyzed by Western blotting.
To assess the association of endogenous CSIG with endogenous PTEN mRNA, immunoprecipitation (IP) of endogenous CSIG-mRNA complexes was performed as described previously (38). HEK 293 cells were collected, and total cell lysates (100 μg) were used for IP for 4 h at room temperature in the presence of excess (3 μg) IP antibody (either rabbit polyclonal anti-CSIG antibody or IgG). RNA in IP material was used in reverse transcription (RT)-PCRs to detect the PTEN mRNA by using primers 5′-CCGTTCCACCCTTTTGACC-3′ and 5′-TCGGGGGAGCACTATGAA-3′ and the following amplification conditions: 30 s at 94°C, 30 s at 56°C, and 30 s at 72°C, for 30 cycles. PCR products were visualized by ethidium bromide staining of 1% agarose gels.
Reporter gene and competition assays.
The pGL3-5′ UTR was constructed by inserting the PTEN 5′ UTR (positions 114 to 983) into the NcoI site of the pGL3-promoter (Promega). Plasmids pGL3-CR and pGL3-3′ UTR-1/-2/-3 were constructed by inserting the PTEN CR (positions 1032 to 2242), 3′ UTR-1 (positions 2206 to 3269), 3′ UTR-2 (positions 3244 to 4327), and 3′ UTR-3 (positions 4280 to 5090) into the XbaI site of the pGL3-promoter vector. Transient transfection of 293 cultures with either pGL3, pGL3-5′ UTR, pGL3-CR, or pGL3-3′ UTR-2 was carried out by using the Lipofectamine 2000 reagent (Invitrogen). Cotransfection of pRL-CMV served as an internal control. For competition assays, PTEN 5′ UTR fragments A (nucleotide positions 114 to 983), B (nucleotide positions 400 to 983), C (nucleotide positions 700 to 983), and D (nucleotide positions 831 to 983) were inserted into the BamHI and XbaI sites of pcDNA3.1 vectors. HEK 293 cells used for pGL3-5′ UTR reporter gene assays were cotransfected with pcDNA vectors expressing PTEN 5′ UTR fragment A, B, C, or D individually, and cell lysates were prepared and subjected to luciferase assays. Firefly and Renilla luciferase activities were measured with a double luciferase assay system (Promega, Madison, WI) by following the manufacturer's instructions. All firefly luciferase measurements were normalized to Renilla luciferase measurements from the same sample.
RESULTS
Expression of CSIG during replicative senescence and its subcellular localization.
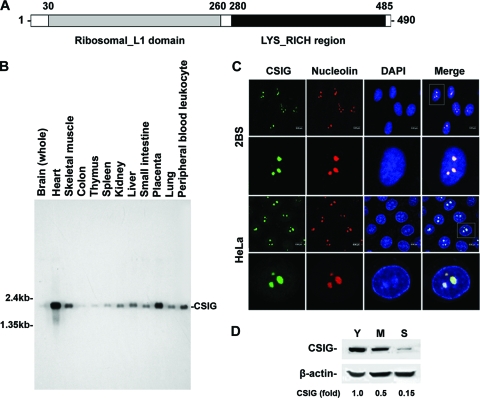
According to bioinformatic analysis, the CSIG protein contains a ribosomal L1 domain at the N terminus and a lysine-rich region in the C terminus and is predicted to localize in the nucleus or nucleolus (Fig. 1A). We began our studies with the analysis of the distribution of CSIG in various human tissues by Northern blot analysis. As shown in Fig. 1B, CSIG was expressed at high intensities in the heart, skeletal muscle, and placenta. To confirm the subcellular localization of CSIG, the intracellular localization of human CSIG in both 2BS and HeLa cells was tested by immunofluorescence assays; the signal of CSIG was compared with the staining of nucleolin (a marker for nucleolus localization) and DAPI. The results shown in Fig. 1C suggest that human CSIG is predominantly localized in the nucleolus.
FIG. 1.
The tissue expression patterns, senescence-associated expression and subcellular localization of CSIG. (A) Schematic representation of human CSIG. (B) Multiple-tissue expression of CSIG. Results of Northern blot analysis using human multiple-tissue blot (Clontech). (C) Representative confocal images for indirect immunofluorescence of CSIG (green) in 2BS (top two rows) and HeLa (bottom two rows) cells. Cells were immunolabeled for endogenous CSIG with antisera against CSIG. Nucleoli (red) were immunolabeled with antinucleolin antibody, and nuclei (blue) were counterstained with DAPI. Right panels show merged confocal images. (D) Age-dependent decrease of CSIG. Results of Western blot analysis of expression of CSIG in young (Y; ∼28 pdl), middle-aged (M; ∼45 pdl), and senescent (S; ∼58 pdl) 2BS cells. β-Actin was served as a loading Ctrl. Relative abundances of CSIG were assessed by densitometry and are expressed as increases relative to the CSIG level in young 2BS cells.
Given our previous findings by suppressive subtractive hybridization that CSIG expression was significantly reduced during replicative senescence (15), we examined the expression of CSIG in early-passage (young; ∼28 pdl), middle-passage (middle-aged; ∼45 pdl), and late-passage (senescent; ∼58 pdl) human diploid 2BS fibroblasts by Western blot analysis. As shown in Fig. 1D, a CSIG protein band of about 55 kDa was observed. In agreement with our previous observations (15), the expression of CSIG was high in young cells but decreased significantly during replicative senescence (∼2.5-fold decrease in middle-aged cells, ∼6.6-fold decrease in senescent cells), this age-dependent reduction suggesting that CSIG may be implicated in the process of cellular senescence.
CSIG suppresses PTEN and p27Kip1 in replicative senescence.
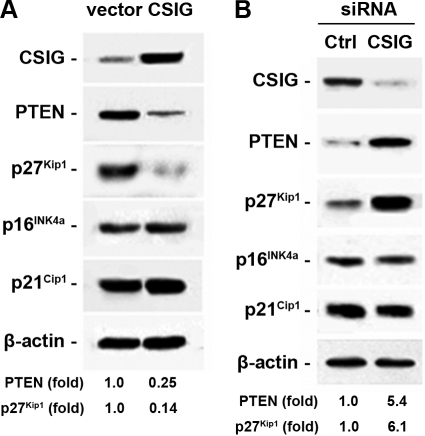
Given that CSIG was abundant in young human fibroblasts and reduced in senescent cells, we next tested the expression of several key mediators of replicative senescence (4, 11), including PTEN and the CDK inhibitors p16INK4a, p21Cip1, and p27Kip1, in 2BS lines with either elevated (Fig. 2A) or reduced (Fig. 2B) levels of CSIG. Western blot analyses showed that the expression levels of p16INK4a and p21Cip1 were not affected by CSIG abundance. However, p27Kip1 and PTEN protein levels were greatly decreased (∼4-fold for PTEN and ∼7-fold for p27Kip1) in cells with CSIG overexpression (Fig. 2A). In contrast, their levels were markedly increased (∼5.4-fold for PTEN and ∼6.1-fold for p27Kip1) after CSIG knockdown. These results suggest that CSIG functions, at least in part, by suppressing PTEN and p27Kip1 expression.
FIG. 2.
Influence of CSIG levels on PTEN expression. 2BS cells were stably transfected with pIRES-CSIG (A) or pSilencer-CSIG (B) or the respective empty vectors. Whole-cell lysates were prepared and subjected to Western blot analysis to determine PTEN, p27Kip1, p16INK4a, and p21Cip1 levels, and β-actin was served as a loading Ctrl. Relative abundances of PTEN and p27Kip1 were measured by densitometry and are expressed as increases relative to their respective levels in empty-vector-transfected cells.
PTEN is required for the effect of CSIG on p27Kip1 expression and cell cycle progression.
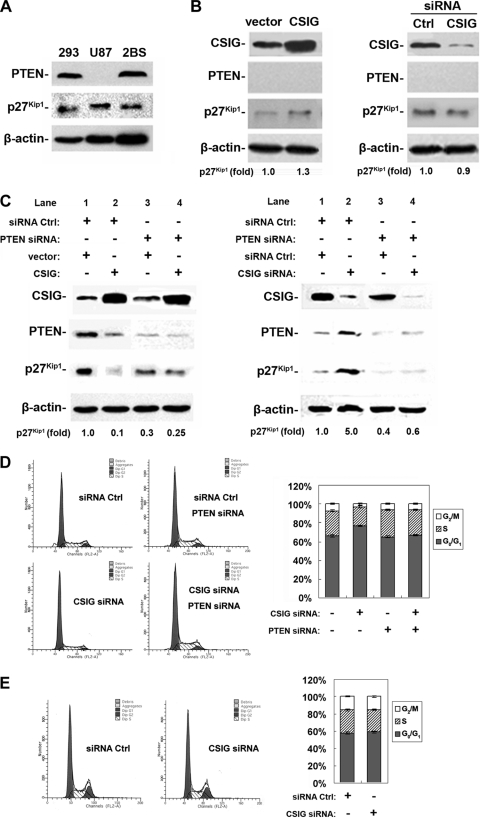
It has been reported that PTEN regulates p27Kip1 expression by inhibiting its proteasome degradation (23). Therefore, we set out to investigate whether CSIG directly regulates p27Kip1 expression levels or its effect is mediated by PTEN. To this end, human glioblastoma U87MG cells (U87) were utilized. U87 cells do not express detectable levels of PTEN but express levels of p27Kip1 similar to those of HEK 293 and 2BS cells (Fig. 3A). CSIG was overexpressed in U87 cells by transient transfection with pIRES-CSIG, or knocked down by transfection of siRNA targeting CSIG, as described in Materials and Methods. Parallel transfections using an enhanced green fluorescent protein-expressing vector revealed that over 90% of cells were transfected under our experimental conditions (data not shown). As shown in Fig. 3B, transfection of pIRES-CSIG in U87 cells resulted in an ∼7-fold increase in CSIG expression relative to the CSIG levels in empty-vector-transfected cells (Fig. 3B, left panel). Conversely, transfection of CSIG siRNA led to an ∼85% (∼6.7-fold) reduction in CSIG levels (Fig. 3B, right panel), compared with transfection of Ctrl siRNA. However, neither overexpression (Fig. 3B, left panel) nor silencing (Fig. 3B, right panel) of CSIG in U87 cells could remarkably alter the expression of p27Kip1, suggesting an essential role of PTEN in CSIG-dependent expression of p27Kip1. To further test the function of PTEN as a key mediator, we knocked down PTEN by transient transfection with its siRNA in HEK 293 cells, which express a level of PTEN similar to that expressed in 2BS cells (Fig. 3A). Knockdown of PTEN in HEK 293 cells greatly diminished the effect of either overexpression (Fig. 3C, left panel, lanes 3 and 4) or knockdown (Fig. 3C, right panel, lanes 3 and 4) of CSIG on p27Kip1 expression. As a Ctrl, both PTEN and p27Kip1 levels are affected by CSIG in HEK 293 cells transfected with Ctrl siRNA, consistent with our observations in 2BS cells (Fig. 2A and B). As shown in Fig. 3C, overexpression of CSIG (Fig. 3C, left panel, lanes 1 and 2) in HEK 293 cells inhibited PTEN by ∼3-fold and inhibited p27Kip1 level by about 10-fold relative to Ctrls (empty-vector transfection), while silencing CSIG (Fig. 3C, right panel, lanes 1 and 2) by an siRNA approach led to an ∼3-fold increase of PTEN and an ∼5-fold increase of p27Kip1. As expected, silencing of CSIG in HEK 293 cells led to increased G0/G1 and reduced S compartments (Fig. 3D). However, in U87 or HEK 293 cells transfected with PTEN siRNA, silencing of CSIG had no significant effect on cell cycle distribution (Fig. 3D and E). These findings support the notion that CSIG directly regulates PTEN expression, which in turn regulates p27Kip1 and cell cycle progression, and that this regulation likely occurs broadly, not limited to human fibroblasts.
FIG. 3.
PTEN is required for CSIG to regulate p27Kip1 expression. (A) Results of Western blot analysis of endogenous PTEN and p27Kip1 expression in HEK 293, U87, and 2BS cells. (B) U87 cells were transfected with pIRES-CSIG (CSIG) versus empty vector (vector) (left panel) or with 20 nM siRNA targeting CSIG (siRNA CSIG) versus Ctrl siRNA (siRNA Ctrl) (right panel) for 48 h. Expressions of CSIG, PTEN, and p27Kip1 were assessed by Western blot analysis; β-actin served as a loading Ctrl. Relative p27Kip1abundances are expressed as increases relative to its level in control cells. (C) HEK 293 cells were transiently transfected with siRNA (20 nM) targeting PTEN (PTEN siRNA) (lanes 3 and 4) or siRNA Ctrl (lanes 1 and 2). Twenty-four hours later, cells were further transfected with pIRES-CSIG (CSIG) (left panel, lanes 2 and 4) versus an empty vector (vector; left panel, lanes 1 and 3) or with siRNA (20 nM) targeting CSIG (CSIG siRNA) (right panel, lanes 2 and 4) versus siRNA Ctrl (right panel, lanes 1 and 3). Cells were then cultured for an additional 48 h. Expression of CSIG, PTEN, and p27Kip1 as well as the relative abundances of p27Kip1 were evaluated as described for panel B. (D and E) FACS analysis of cells used in the right panel of panel C (D) and in the right panel of panel B (E).
CSIG regulates PTEN in translational levels.
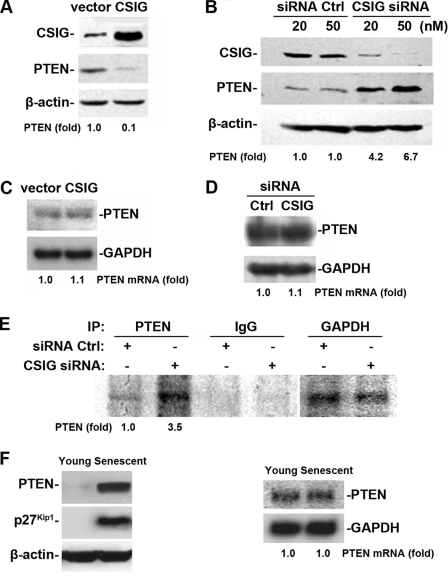
Next, we extended our study to the mechanism for how CSIG regulates PTEN expression. We first tested if the regulation is at the transcriptional level. HEK 293 cells were transiently transfected either with a pIRES-CSIG vector expressing CSIG to elevate CSIG level or with siRNA targeting CSIG (20 and 50 nM) to silence CSIG expression, as described in Fig. 3C. Two days after the transfection, mRNA levels of PTEN in transfected HEK 293 cells were tested. To our surprise, neither overexpression nor silencing of CSIG significantly affected the mRNA levels of PTEN (Fig. 4C and D) although the PTEN protein level was decreased by 10-fold in CSIG overexpression (Fig. 4A) and increased in CSIG silenced cells (∼4-fold for 20 nM siRNA and ∼10-fold for 50 nM siRNA) (Fig. 4B) by Western blot analysis. These results suggest that CSIG regulated PTEN at the posttranscriptional level. We further investigated whether CSIG affects the translation of PTEN. HEK 293 cells were first transiently transfected with CSIG siRNA (50 nM) or Ctrl siRNA for 48 h followed by incubation with l-[35S]methionine and l-[35S]cysteine for 20 min. The brief incubation period was chosen to minimize the contribution of PTEN degradation in our analysis. Cell lysates were then prepared and the nascent PTEN protein was analyzed by IP. As shown in Fig. 4E, the nascent PTEN protein level in CSIG-silenced cells was ∼3.5-fold higher than in control cells. As a control, silencing of CSIG did not influence the nascent protein levels of GAPDH. Therefore, CSIG may regulate PTEN at the translational level. In agreement with this view, although both PTEN and p27Kip1 protein levels in senescent 2BS cells (∼58 pdl) were much higher (both exhibited a >20-fold increase) than those in young cells (∼28 pdl) (Fig. 4F, left panel), PTEN mRNA levels detected by Northern blot analysis stayed unchanged in the process of cellular senescence (Fig. 4F, right panel). Taken together, our observations suggest that the passage-dependent elevation of PTEN levels in senescent cells is basically a result of translational regulation, and the reduction of CSIG during cell aging contributes to the age-dependent elevation of PTEN translation.
FIG. 4.
CSIG expression regulates PTEN in the translational level without influencing PTEN mRNA levels. (A and B) Ectopic intervention of CSIG expression was achieved by transiently transfecting HEK 293 cells either with pIRES-CSIG along with an empty vector (A) or with siRNA targeting CSIG (20 nM [lane 3] and 50 nM [lane 4]) along with a Ctrl siRNA (20 nM [lane 1] and 50 nM [lane 2]) for 48 h. (B) CSIG and PTEN expression were detected by Western blot analysis; β-actin was served as a loading Ctrl. Relative PTEN abundances were estimated by densitometry and are expressed as increases relative to PTEN levels in control cells. (C and D) Results of Northern blot analysis of PTEN mRNA levels after CSIG overexpression (C) or CSIG knockdown (D); GADPH served as a loading Ctrl. The PTEN mRNA abundance is expressed as the increase relative to PTEN mRNA levels in Ctrl cells. (E) Analysis of PTEN translation in CSIG-silenced HEK 293 cells. Newly translated PTEN was measured by incubating cells with l-[35S]methionine and l-[35S]cysteine for 20 min, followed by IP using either anti-PTEN antibody, anti-GAPDH antibody, or IgG; resolving immunoprecipitated samples by SDS-PAGE; and transferring for visualization of signals by using a PhosphorImager. (F) Results of Western blot analysis of PTEN and p27Kip1 (left panel), and results of Northern blot analysis (panel right) of PTEN in young (∼28 pdl) and senescent (∼58 pdl) 2BS cells. β-Actin and GADPH served as loading Ctrls for Western or Northern blot analysis, respectively.
CSIG associates with the 5′ UTR of the PTEN mRNA and modulates its translation.
Because CSIG localized in the nucleolus (Fig. 1C) and regulates specifically PTEN translation (Fig. 4), we further asked if CSIG could interact with PTEN mRNA or PTEN translation complex. First, IP assays against CSIG polyclonal antibody under conditions that preserved endogenous protein-RNA complexes were performed, followed by RT-PCR amplification of the IP materials using sequence-specific primers. As shown, the PTEN mRNA was predominantly bound to CSIG, as a significant amount of PTEN PCR product was amplified from anti-CSIG IP reactions (Fig. 5B). By contrast, IP assays using IgG showed undetectable signals, a Ctrl PCR product corresponding to the housekeeping GADPH mRNA serving to monitor background mRNA binding to IP material showed only weak background mRNA binding, and the analysis of GAPDH served to verify that the interaction between CSIG and PTEN mRNA was specific and the sample input was even. Further demonstration of the existence of the CSIG-PTEN mRNA complex was obtained through pulldown assays using biotinylated transcripts of PTEN 5′ UTR, CR, and 3′ UTR (3′ UTR-1, 3′ UTR-2, and 3′ UTR-3). As shown in Fig. 5C, by Western blotting of CSIG in the pulldown materials, PTEN 5′ UTR and CR (but not 3′ UTR) were targets of CSIG. As a Ctrl, RNA binding protein HuR was found to interact with PTEN 3′ UTR-1 and 3′ UTR-2, but not with 5′ UTR and CR. Because of the role of CSIG on PTEN translation, we isolated the polysomal fraction from HEK 293 cell lysates and studied the presence of CSIG in it. As shown in Fig. 5D, similar to HuR, which was proved to be a polysomal component (24), CSIG was also present in the polysomal fraction. Together, these results showed that CSIG was capable of interacting with PTEN mRNA and that they colocalized within the polysomal compartment of the cell.
FIG. 5.
Binding of CSIG to PTEN mRNA and influence of PTEN 5′ UTR on the expression of a chimeric luciferase reporter construct after CSIG silencing. (A) Schematic presentation of the full-length PTEN cDNA and various transcripts derived from the 5′ UTR, CR, and 3′ UTR used in this study. (B) Whole-cell lysates (100 μg) were prepared from HEK 293 cells, and endogenous target transcripts were detected by RT-PCR assay of the corresponding IP materials; PCR products corresponding to PTEN mRNA were visualized on agarose gel. The PCR product of GAPDH served as a negative Ctrl. (C) Results of a pulldown assay using biotinylated fragments to detect bound CSIG by Western blotting. A 10-μg portion of whole-cell lysates (Lys.) and binding of HuR (positive Ctrl) and α-tubulin (negative Ctrl) to PTEN mRNA were included. (D) A polysomal fraction from HEK 293 cell lysates was prepared and subjected to Western blot analysis to evaluate the presence of CSIG. (E) pGL3, pGL3-5′ UTR, pGL3-CR, and pGL3-3′ UTR-2 plasmid (0.1 μg/ml) were transiently transfected into HEK 293 cells along with the pRL-CMV reporter (5 ng/ml) as a Ctrl. Twenty-four hours later, transfected cells were cotransfected either with CSIG siRNA to silence CSIG or with a Ctrl siRNA for 48 h; firefly luciferase activities were determined and normalized against Renilla luciferase activity. Values represent means ± standard errors of the means (SEM) of the results for five independent experiments. (F) Left panel: pGL3-5′ UTR (0.1 μg/ml), pRL-CMV (5 ng/ml) plasmids, and a different dose of pcDNA-5′ UTR [0 μg/ml (−), 2 μg/ml (+), 4 μg/ml (++), or 6 μg/ml (+++)] were transiently cotransfected into HEK 293 cells for 48 h. Right panel: pGL3-5′ UTR (0.1 μg/ml), pRL-CMV (5 ng/ml) plasmids, and pcDNA vectors expressing PTEN 5′ UTR fragments A, B, C, or D (6 μg/ml) were transiently cotransfected into HEK 293 cells for 48 h; firefly luciferase activities then were determined and normalized against Renilla luciferase activity. Values represent means ± SEMs of the results for five independent experiments.
To address if PTEN 5′ UTR or CR contains the CSIG response elements, luciferase reporter constructs expressing chimeric mRNAs, including PTEN 5′ UTR-luciferase (pGL3-5′ UTR), luciferase-PTEN CR (pGL3-CR), or luciferase-PTEN 3′ UTR-2 (pGL3-3′ UTR-2), were transiently transfected into either CSIG-silenced or Ctrl HEK 293 cells, followed by reporter gene assays. As shown in Fig. 5E, after CSIG silencing, luciferase activities of pGL3 (empty vector), pGL3-CR, and pGL-3′ UTR-2 did not show a big difference compared with that of Ctrl cells, although protein-RNA binding assays showed that CSIG also bound to PTEN coding region (Fig. 5C). By contrast, pGL3-5′ UTR, expressing a chimeric mRNA encoding PTEN 5′ UTR and luciferase, exhibited an ∼2.5-fold increase in luciferase activity in CSIG-silenced cells compared with what was seen in Ctrl cells (Fig. 5E). To confirm the specificity of the PTEN 5′ UTR reporter activity and to identify CSIG response elements in the 5′ UTR of PTEN, pcDNA3.1 vectors expressing series of PTEN 5′ UTR fragments (fragments A [nucleotide positions 114 to 983], B [nucleotide positions 400 to 983], C [nucleotide positions 700 to 983], or D [nucleotide positions 831 to 983]) were cotransfected with pGL3-5′ UTR and employed for competition assays. As shown in Fig. 5F (left panel), as a result of competitive interaction with endogenous CSIG, expression of the PTEN 5′ UTR fragment A increased the luciferase activity of pGL3-5′ UTR in a dose-dependent (0, 2, 4, or 6 μg/ml) manner. Further study revealed that expressions of fragments A, B, and C, but not D, elevated the luciferase activity of pGL3-5′ UTR to a similar extent (∼1.65-fold, ∼1.7-fold, and ∼1.85-fold, respectively) compared with that observed in empty-vector-cotransfected cells (Fig. 5F, right panel). These results suggest that the luciferase activity of pGL3-5′ UTR is specific and that the CSIG response element may localize between nucleotide positions 700 and 830 in the PTEN 5′ UTR.
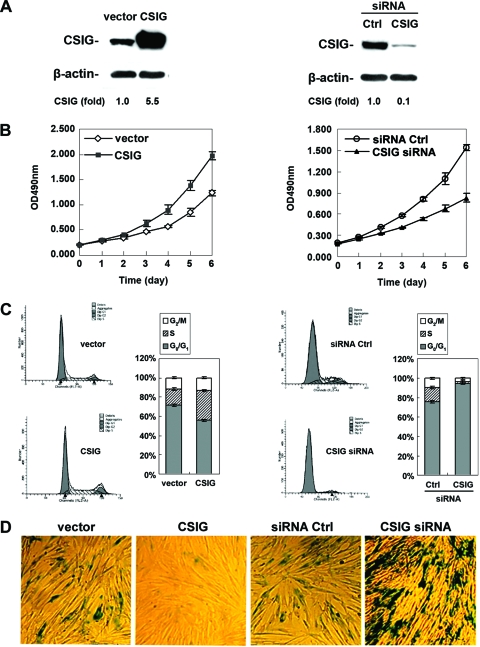
CSIG is a regulator of cell proliferation and replicative senescence.
Since CSIG repressed the PTEN/p27Kip1 pathway, we further investigated if the regulation of PTEN/p27Kip1 by CSIG had an impact on cellular senescence. To this end, CSIG expression in 2BS cells was either elevated by stable transfection with pIRES-CSIG or reduced by stable transfection with pSilencer-CSIG plasmids (see Materials and Methods). Expression of CSIG was monitored by Western blot analysis. As depicted in Fig. 6A, CSIG expression was about 5.5-fold higher in pIRES-CSIG-transfected cells (Fig. 6A, left panel), but about 90% (10-fold) (Fig. 6A, right panel) lower in pSilencer-CSIG-transfected cells, than that seen in the Ctrl cells (transfected with empty vectors). These alterations were specific for CSIG since β-actin levels remained unchanged. Importantly, these interventions to either elevate or reduce CSIG expression levels influenced cell growth and the process of cellular senescence. As shown in Fig. 6, cells overexpressing CSIG exhibited markedly elevated proliferation rates, showed increased S and reduced G1 compartments, and displayed a feature of young cells (lower SA-β-gal activity) compared with the empty-vector-transfected cells (Fig. 6B, C, and D, left panels). By contrast, cells expressing reduced CSIG levels displayed decreased proliferation rates, smaller S and G2/M compartments, and enhanced senescent phenotype (high SA-β-gal activity) (Fig. 6B, C, and D, right panels). These findings suggest that CSIG potentially regulates cell proliferation and replicative senescence in 2BS cells by suppressing PTEN translation.
FIG. 6.
Influences of CSIG levels on replicative senescence. (A) Early passage 2BS (∼28 pdl) cells were transfected with either pIRES-CSIG (CSIG) versus empty vector (vector) (left panel) or pSilencer-CSIG (CSIG siRNA) versus vector expressing Ctrl siRNA (siRNA Ctrl) (right panel) and then selected by G418 for 3 weeks. Expression of CSIG was monitored by Western blot analysis; β-actin served as a loading Ctrl. Relative CSIG abundances were assessed by densitometry and are expressed as increases relative to CSIG levels in cells transfected with the empty vector. (B) Effect of overexpression (left panel) or silencing (right panel) of CSIG on cell growth. Transfected cells were seeded 2 × 103 per well in 96-well plates, and cell number was assessed by MTT (methyl thiazolyldiphenyl-tetrazolium bromide) method at the times indicated. Values represent the means ± SEMs of the results for three independent experiments. (C) Transfected cells were subjected to FACS analysis to evaluate the effect of ectopic intervention to either overexpress (left panel) or knock down (right panel) CSIG on cell cycle distribution. Bars represent the means ± SEMs of the results for three independent experiments. (D) Influence of CSIG levels on SA-β-gal activity. Transfected 2BS cells that either elevate (left panel) or reduce (right panel) CSIG levels were stained to assess SA-β-gal activity. Data are representative of the results for three independent experiments.
PTEN is required for CSIG's role in regulating replicative senescence.
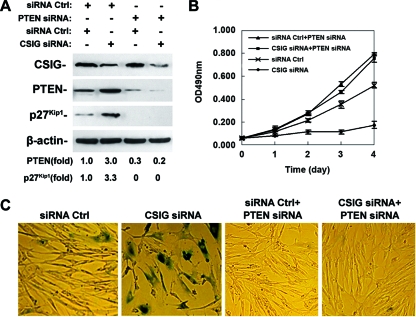
We next addressed whether PTEN is required for CSIG to regulate cellular senescence. For this purpose, 2BS cells were stably transfected with vectors expressing Ctrl siRNA or CSIG siRNA and cotransfected with vectors expressing either Ctrl siRNA plus PTEN siRNA or CSIG siRNA plus PTEN siRNA. Expression of CSIG, PTEN, and p27Kip1 was monitored by Western blot analysis. As shown in Fig. 7A, consistent with our results from 2BS and HEK 293 cells (Fig. 2B and 3C), transfection with vectors expressing PTEN siRNA led to an ∼70 to 80% reduction of PTEN and a near loss of p27Kip1 expression; by contrast, knockdown of CSIG increased p27Kip1 by ∼3.3-fold in Ctrl cells but failed to increase p27Kip1expression in PTEN- silenced cells. Similar to our results in Fig. 6B and D, silencing of CSIG resulted in reduced proliferation rates and increased SA-β-gal activity compared with the Ctrl siRNA-expressing cells (Fig. 7B and C). However, 2BS cells cotransfected either with Ctrl siRNA plus PTEN siRNA or with CSIG siRNA plus PTEN siRNA displayed higher cell proliferation rates than those observed in Ctrl siRNA-expressing cells (Fig. 7B). More importantly, knockdown of CSIG failed to elevate SA-β-gal activity in PTEN-silenced 2BS cells (Fig. 7C). Therefore, PTEN is not only required for CSIG to regulate p27Kip1 and cell proliferations but is essential for CSIG's role in regulating replicative senescence.
FIG. 7.
PTEN is required for CSIG to regulate cell proliferation and senescence. 2BS cells were stably transfected with vectors expressing Ctrl siRNA (siRNA Ctrl) or CSIG siRNA and were cotransfected with vectors expressing Ctrl siRNA plus PTEN siRNA (siRNA Ctrl+PTEN siRNA) or CSIG siRNA plus PTEN siRNA (CSIG siRNA+PTEN siRNA). (A) Expression of CSIG, PTEN, and p27Kip1 was monitored by Western blot analysis; β-actin served as a loading Ctrl. (B) Transfected cells were seeded 2 × 103 per well in 96-well plates, and cell numbers were assessed at the times indicated by the MTT method. Values represent the means ± SEMs of the results for three independent experiments. (C) Transfected cells were stained to assess SA-β-gal activity. Data were representative of the results for three independent experiments.
DISCUSSION
The present study stemmed from our previous findings using suppressive subtractive hybridization, whereby the CSIG transcript was dramatically higher in young 2BS cells than in senescent cells. Here, we set out to test whether CSIG is important for the progression to senescence. We originally demonstrated an age-dependent reduction of CSIG expression in 2BS fibroblasts, a model cell line that is routinely used for the study of replicative senescence (11, 20, 33, 36, 39-40) (Fig. 1D). We further demonstrate that overexpression of CSIG in 2BS cells promotes cell growth, increases the S phase compartment, and delays the progression to senescence. In contrast, silencing of CSIG led to slow cell growth, decreased the S and G2/M compartments, and accelerated cellular senescence (Fig. 6). Together, this evidence indicates that CSIG is capable of modulating the process of replicative senescence. While elevation of both p16INK4a and p21Cip1 are key hallmarks of entry into cell senescence, neither of them appear to mediate the effect of CSIG on senescence, as ectopic intervention to either elevate or reduce CSIG levels failed to influence their levels. Instead, elevating CSIG expression negatively influences PTEN and p27Kip1 (Fig. 2A and B), suggesting that these proteins are downstream effectors of CSIG's influence on cell proliferation and senescence. It is likely that CSIG is also involved in the regulation of in vivo aging and that additional targets of CSIG may also participate in the implementation of cell proliferation and senescence. In this regard, studies to assess the CSIG levels in young and aged mouse tissues, to develop a knockout system, and to screen more targets of CSIG during cellular and tissue aging are under way in our laboratory. The results of these studies will hopefully provide a more complete understanding of the role of CSIG and its mechanisms of action.
The CDK inhibitor p27Kip1 is important for G0/G1 arrest during cell aging (1, 4). Degradation of p27Kip1 is a critical event for the G1/S transition and occurs through ubiquitination by SCFSkp2 and subsequent degradation by the 26S proteasome (27). Regulation of p27Kip1 by PTEN has been reported broadly. For example, ectopic expression of PTEN in human glioblastoma cells led to accumulation of p27Kip1 (19, 29), while PTEN deficiency in mouse embryonic stem cells led to decreased p27Kip1 levels (32). Emerging evidence suggests that PTEN inhibits the ubiquitin-dependent degradation of p27Kip1, elevates p27Kip1 levels, which in turn, negatively regulates the G1/S cell cycle transition (9, 21, 23, 32). The PTEN/p27Kip1 pathway is important for controlling the replicative senescence and life span of normal human fibroblasts (1, 4, 28). Although both PTEN and p27Kip1 levels are influenced by CSIG, our findings clearly indicate that PTEN, but not p27Kip1, is the direct target of CSIG. In other words, PTEN is required for the regulation of p27Kip1 by CSIG. We arrive at this conclusion from the following two observations: (i) ectopic intervention to either elevate or reduce CSIG expression failed to significantly change p27Kip1 expression in PTEN-deficient U87 cells (Fig. 3B), and (ii) silencing of PTEN in HEK 293 cells by an siRNA approach also dramatically diminished the effect of ectopic intervention to either elevate or reduce CSIG levels on p27Kip1 expression (Fig. 3C). Not surprisingly, the PTEN/p27Kip1 pathway acts as a major mediator for CSIG-regulated cell proliferation and senescence, as knockdown of CSIG failed to affect cell cycle progression and proliferation (Fig. 3D and E and 7B) as well as the senescent phenotype (Fig. 7C) in PTEN-deficient or -silenced cells.
According to the model that emerges from this study, we propose that high CSIG expression in young cells contributes to the low levels of PTEN and keeps cells from aging, as characterized by high proliferation state and low SA-β-gal activity. In accordance with the age-dependent decrease of CSIG, expression levels of PTEN increased with replicative senescence (Fig. 4F, left panel), which in turn leads to increased levels of p27Kip1 and its downstream effectors. Interestingly, the robust increase of PTEN in senescent cells was not accompanied by a concomitant elevation in PTEN mRNA abundance (Fig. 4F, right panel), suggesting that a posttranscriptional mechanism is involved in the regulation of PTEN by CSIG. Accordingly, either overexpression or knockdown of CSIG failed to affect PTEN mRNA levels (Fig. 4C and D). Therefore, CSIG-regulated expression of PTEN must be achieved by either translational or posttranslational events. Indeed, newly synthesized PTEN is much more abundant after silencing of CSIG (Fig. 4E), indicating that enhanced PTEN translation is a critical mechanism leading to PTEN accumulation. Although ubiquitin-dependent proteasome degradation has been described as an important mechanism for the regulation of PTEN (35, 37), CSIG is unlikely to regulate PTEN through this mechanism because silencing of CSIG expression by RNA interference in 2BS cells did not affect the PTEN proteasome turnover rate (our unpublished data). It is likely that CSIG downregulates PTEN by inhibiting its translation during replicative senescence. Consistently, our results showed that CSIG predominantly localized in the nucleolus (Fig. 1C), the major site for synthesizing and assembling ribosomal subunits. This result was recently confirmed by another group showing that CSIG/RSL1D1 colocalizes with nucleostemin in the nucleolus (25).
The structural feature of CSIG with the N-terminal ribosomal L1p/L10e consensus sequence (residues 30 to 260) and the predominant nucleolus localization suggest that CSIG acts as a ribosome-associated protein. This is supported by the evidence that CSIG is present in the cell's polysomal fraction (Fig. 5D) and formed a complex with PTEN 5′ UTR (Fig. 5B and C). The PTEN 5′ UTR confers higher luciferase activity to PTEN 5′ UTR-luciferase chimeric mRNA. More importantly, the PTEN 5′ UTR-related luciferase activity increased significantly after CSIG silencing (Fig. 5E). Therefore, interaction of CSIG with PTEN 5′ UTR may be critical for the role of CSIG in PTEN translational regulation. The CSIG response element in PTEN 5′ UTR appears to localize between positions 700 and 830 because the luciferase activity of PGL3-5′UTR could be competed by coexpression of PTEN 5′ UTR fragments A, B, and C, but not D (Fig. 5F, right panel). However, in vitro RNA-protein binding assays using in vitro-expressed GST-CSIG recombinant protein and 32P-labeled PTEN 5′ UTR did not show direct binding between CSIG and PTEN 5′ UTR (our unpublished results), suggesting that CSIG may not act as an RNA binding protein. It remains possible that posttranslational modifications are absent in the purified, bacterially expressed CSIG protein in order for CSIG to bind a target transcript; however, we favor the view that CSIG may instead interact either with bona fide RNA binding proteins implicated in regulating translation or with components of PTEN translation complex. It will be of great interest to investigate whether CSIG is a common regulator of translation and how CSIG selectively exerts its effect on certain targets, such as PTEN, during cellular senescence. In our ongoing studies, we are screening targets of CSIG other than PTEN by using cDNA/protein arrays and investigating whether CSIG acts as a ribosomal component or as a cofactor of translation. The results of this work will likely enhance our understanding of CSIG function during the process of replicative senescence.
Acknowledgments
This work was supported by grant 2007CB507400 from the Major State Basic Research Development Program of China and grant 30671064 from the National Science Foundation of China.
We are grateful to Dalong Ma for technical help, Yongfeng Shang for providing the pSilencer vector, and Chunyan Zhou for the pGL3 promoter vector.
Footnotes
Published ahead of print on 4 August 2008.
REFERENCES
- 1.Alexander, K., and P. W. Hinds. 2001. Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol. Cell. Biol. 213616-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S. J. 2007. PTEN enters the nuclear age. Cell 12825-28. [DOI] [PubMed] [Google Scholar]
- 3.Bayreuther, K., H. P. Rodemann, R. Hommel, K. Dittmann, M. Albiez, and P. I. Francz. 1988. Human skin fibroblasts in vitro differentiate along a terminal cell lineage. Proc. Natl. Acad. Sci. USA 855112-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bringold, F., and M. Serrano. 2000. Tumor suppressors and oncogenes in cellular senescence. Exp. Gerontol. 35317-329. [DOI] [PubMed] [Google Scholar]
- 5.Campisi, J. 2001. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 11S27-S31. [DOI] [PubMed] [Google Scholar]
- 6.Campisi, J. 2001. From cells to organisms: can we learn about aging from cells in culture? Exp. Gerontol. 36607-618. [DOI] [PubMed] [Google Scholar]
- 7.Campisi, J. 1997. The biology of replicative senescence. Eur. J. Cancer 33703-709. [DOI] [PubMed] [Google Scholar]
- 8.Campisi, J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120513-522. [DOI] [PubMed] [Google Scholar]
- 9.Chung, J. H., and C. Eng. 2005. Nuclear-cytoplasmic partitioning of phosphatase and tensin homologue deleted on chromosome 10 (PTEN) differentially regulates the cell cycle and apoptosis. Cancer Res. 658096-8100. [DOI] [PubMed] [Google Scholar]
- 10.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, M. Peacocke, and J. Campisi. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 929363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan, J., Z. Zhang, and T. Tong. 2001. Senescence delay of human diploid fibroblast induced by anti-sense p16INK4a expression. J. Biol. Chem. 27648325-48331. [DOI] [PubMed] [Google Scholar]
- 12.Engelman, J. A., J. Luo, and L. C. Cantley. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7606-619. [DOI] [PubMed] [Google Scholar]
- 13.Ferbeyre, G., E. de Stanchina, A. W. Lin, E. Querido, M. E. McCurrach, G. J. Hannon, and S. W. Lowe. 2002. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol. Cell. Biol. 223497-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freeman, D. J., A. G. Li, G. Wei, H. H. Li, N. Kertesz, R. Lesche, A. D. Whale, H. Martinez-Diaz, N. Rozengurt, R. D. Cardiff, X. Liu, and H. Wu. 2003. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell 3117-130. [DOI] [PubMed] [Google Scholar]
- 15.Guo, S. Z., Z. Y. Zhang, and T. J. Tong. 2004. Cloning and characterization of cellular senescence-associated genes in human fibroblasts by suppression subtractive hybridization. Exp. Cell Res. 298465-472. [DOI] [PubMed] [Google Scholar]
- 16.Hayflick, L., and P. S. Moorhead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25585-621. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S. W., L. Fang, M. Igarashi, T. Ouchi, K. P. Lu, and S. A. Aaronson. 2000. Sustained activation of Ras/Raf/mitogen-activated protein kinase cascade by the tumor suppressor p53. Proc. Natl. Acad. Sci. USA 978302-8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, A. G., L. G. Piluso, X. Cai, G. Wei, W. R. Sellers, and X. Liu. 2006. Mechanistic insights into maintenance of high p53 acetylation by PTEN. Mol. Cell 23575-587. [DOI] [PubMed] [Google Scholar]
- 19.Li, D. M., and H. Sun. 1998. PTEN/MMAC1/TEP1 suppresses the tumorigenicity and induces G(1) cell cycle arrest in human glioblastoma cells. Proc. Natl. Acad. Sci. USA 9515406-15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, J., Z. Zhang, and T. Tong. 1995. The proliferative response and anti-oncogene expression in old 2BS cells after growth factor stimulation. Mech. Ageing Dev. 8025-34. [DOI] [PubMed] [Google Scholar]
- 21.Liu, J. L., X. Sheng, Z. K. Hortobagyi, Z. Mao, G. E. Gallick, and W. K. Yung. 2005. Nuclear PTEN-mediated growth suppression is independent of Akt down-regulation. Mol. Cell. Biol. 256211-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ly, D. H., D. J. Lockhart, R. A. Lerner, and P. G. Schultz. 2000. Mitotic misregulation and human aging. Science 2872486-2492. [DOI] [PubMed] [Google Scholar]
- 23.Mamillapalli, R., N. Gavrilova, V. T. Mihaylova, L. M. Tsvetkov, H. Wu, H. Zhang, and H. Sun. 2001. PTEN regulates the ubiquitin-dependent degradation of the CDK inhibitor p27KIP1 through the ubiquitin E3 ligase SCFSKP2. Curr. Biol. 11263-267. [DOI] [PubMed] [Google Scholar]
- 24.Mazan-Mamczarz, K., S. Galban, I. Lopez-de-Silanes, J. Martindale, U. Atasoy, J. Keene, and M. Gorospe. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 1008354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng, L., H. Yasumoto, and R. Y. Tsai. 2006. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J. Cell Sci. 1195124-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narita, M., S. Nunez, E. Heard, M. Narita, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113703-716. [DOI] [PubMed] [Google Scholar]
- 27.Pagano, M., S. W. Tam, A. M. Theodoras, P. Beer-Romero, S. G. Del, V. Chau, P. R. Yew, G. F. Draetta, and M. Rolfe. 1995. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269682-685. [DOI] [PubMed] [Google Scholar]
- 28.Rohme, D. 1981. Evidence for a relationship between longevity of mammalian species and life spans of normal fibroblasts in vitro and erythrocytes in vivo. Proc. Natl. Acad. Sci. USA 785009-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sano, T., H. Lin, X. S. Chen, L. A. Langford, D. Koul, M. L. Bondy, K. R. Hess, J. N. Myers, Y. K. Hong, W. K. Yung, and P. A. Steck. 1999. Differential expression of MMAC/PTEN in glioblastoma multiforme: relationship to localization and prognosis. Cancer Res. 591820-1824. [PubMed] [Google Scholar]
- 30.Schulz, L., and J. Tyler. 2005. Heterochromatin focuses on senescence. Mol. Cell 17168-170. [DOI] [PubMed] [Google Scholar]
- 31.Smith, J. R., and O. M. Pereira-Smith. 1996. Replicative senescence: implications for in vivo aging and tumor suppression. Science 27363-67. [DOI] [PubMed] [Google Scholar]
- 32.Sun, H., R. Lesche, D. M. Li, J. Liliental, H. Zhang, J. Gao, N. Gavrilova, B. Mueller, X. Liu, and H. Wu. 1999. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5-trisphosphate and Akt/protein kinase B signaling pathway. Proc. Natl. Acad. Sci. USA 966199-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang, Z., Z. Zhang, Y. Zheng, M. J. Corbley, and T. Tong. 1994. Cell aging of human diploid fibroblasts is associated with changes in responsiveness to epidermal growth factor and changes in Her-2 expression. Mech. Ageing Dev. 7357-67. [DOI] [PubMed] [Google Scholar]
- 34.Tresini, M., M. Mawal-Dewan, V. J. Cristofalo, and C. Sell. 1998. A phosphatidylinositol 3-kinase inhibitor induces a senescent-like growth arrest in human diploid fibroblasts. Cancer Res. 581-4. [PubMed] [Google Scholar]
- 35.Trotman, L. C., X. Wang, A. Alimonti, Z. Chen, J. Teruya-Feldstein, H. Yang, N. P. Pavletich, B. S. Carver, C. Cordon-Cardo, H. Erdjument-Bromage, P. Tempst, S. G. Chi, H. J. Kim, T. Misteli, X. Jiang, and P. P. Pandolfi. 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128141-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, W., J. Wu, Z. Zhang, and T. Tong. 2001. Characterization of regulatory elements on the promoter region of p16INK4a that contribute to overexpression of p16 in senescent fibroblasts. J. Biol. Chem. 27648655-48661. [DOI] [PubMed] [Google Scholar]
- 37.Wang, X., L. C. Trotman, T. Koppie, A. Alimonti, Z. Chen, Z. Gao, J. Wang, H. Erdjument-Bromage, P. Tempst, C. Cordon-Cardo, P. P. Pandolfi, and X. Jiang. 2007. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, W., J. L. Martindale, X. Yang, F. J. Chrest, and M. Gorospe. 2005. Increased stability of the p16 mRNA with replicative senescence. EMBO Rep. 6158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, Q. H., L. W. Ma, W. G. Zhu, Z. Y. Zhang, and T. J. Tong. 2006. p21(waf1/cip1) plays a critical role in modulating senescence through changes of DNA methylation. J. Cell. Biochem. 981230-1248. [DOI] [PubMed] [Google Scholar]
- 40.Zheng, W., H. Wang, L. Xue, Z. Zhang, and T. Tong. 2004. Regulation of cellular senescence and p16INK4a expression by Id1 and E47 proteins in human diploid fibroblast. J. Biol. Chem. 27931524-31532. [DOI] [PubMed] [Google Scholar]