Abstract
Pax6 regulates eye development in many animals. In addition, Pax6 activates atonal transcription factors in both invertebrate and vertebrate eyes. Here we investigate the roles of Pax6 and atonal during embryonic development of Limulus polyphemus rudimentary lateral, medial and ventral eyes, and the initiation of lateral ommatidial eye and medial ocelli formation. Limulus eye development is of particular interest because these animals hold a unique position in arthropod phylogeny and possess multiple eye types. Furthermore, the molecular underpinnings of eye development have yet to be investigated in chelicerates. We characterized a Limulus Pax6 gene, with multiple splice products and predicted protein isoforms, and one atonal homologue. Unexpectedly, neither gene is expressed in the developing eye types examined, although both genes are present in the lateral sense organ, a structure of unknown function.
Keywords: Limulus, Pax6, Atonal, chelicerate, eye development
INTRODUCTION
Eye morphology, tissue and cell type composition, and mechanisms of photoreception and phototransduction are features that define the diversity of eye types found in different organisms. Eye development also provides valuable data for phylogenetic analyses, particularly those concerning bilaterian relationships (reviewed in Arendt and Wittbrodt, 2001; Arendt, 2003). In recent years, evolutionary comparisons of developmental gene hierarchies have contributed to theories of eye evolution, to the extent that several long-standing views were revised (Halder et al., 1995b; Gehring, 1996; Callaerts et al., 1997; Pichaud et al., 2001). One transcription factor with a highly conserved and essential role in eye development is Pax6. Mutational analyses have demonstrated that Pax6 is critical for initial eye specification and differentiation (Hill et al., 1991; Ton, 1991; Glaser et al., 1992; Quiring et al., 1994). Conversely, misexpression of Pax6 revealed its sufficiency, since it respecifies ectodermal tissues into partial or complete ectopic eyes (Halder et al., 1995a; Tomarev et al., 1997; Chow et al., 1999). However, Pax6 is not universally expressed during metazoan visual system development. For example, particular jellyfish, polychaete worm, myriapod and amphioxus eyes or visual organs lack Pax6 expression (Glardon et al., 1998; Arendt et al., 2002; Piatigorsky and Kozmik, 2004; Prpic, 2005). Instead, the expression of Pax-B and So within embryonic jellyfish or adult Platyneris eyes respectively, point to alternative mechanisms of eye development in some species (Arendt et al., 2002; Piatigorsky and Kozmik, 2004).
Nonetheless, multiple features of the Pax6 gene are well conserved among bilaterians. First, the amino acid sequence of the encoded protein retains high identity, particularly within the paired- and homeodomains that bind DNA (reviewed in Callaerts et al., 1997). Moreover, the exon/intron structure of numerous Pax6 genes are strongly conserved, as are distinct cis-regulatory elements and trans-acting factors that bind them (Callaerts et al., 1997; Kammandel et al., 1999; Xu et al., 1999). Some invertebrate species have duplicate Pax6 genes (Czerny and Busslinger, 1995; Jun et al., 1998; Czerny et al., 1999; Prpic, 2005), whereas most chordates have a single Pax6 gene (reviewed in Callaerts et al., 1997; van Heyningen and Williamson, 2002). Instead of gene duplication and functional divergence, vertebrate Pax6 functions are divided among distinct splice products and/or protein isoforms (Carriere et al., 1993; Epstein et al., 1994; Glardon et al., 1998; Mishra et al., 2002; Lakowski et al., 2007).
Concomitant with Pax6 gene conservation is its maintained regulation of orthologous downstream genes (reviewed in Wawersik and Maas, 2000; Hanson, 2001; Friedrich, 2006). For instance, eya-Eya, so-Six and atonal-Ath5 gene families all act immediately downstream of Pax6 in both fruit fly and mouse eye development. These downstream genes also perform conserved roles in specific aspects of eye development. For example, the atonal-Ath5 family encodes basic helix-loop-helix (bHLH) factors that specify the first retinal neuron class during fruit fly, zebrafish, chick, frog and mouse eye development (reviewed in Vetter and Brown, 2001; Mu and Klein, 2004).
The visual system of the chelicerate Limulus polyphemus (Lp) has been studied for decades, especially with respect to its circadian biology, neuroanatomy and physiology (H.K. Hartline, 1956; Barlow, 1969; Snodderly and Barlow, 1970; French, 1980; Sekiguchi et al., 1982; Barlow et al., 2001). More recently, molecular approaches have been applied to studies of Limulus eye development (Harzsch et al., 2006). During embryogenesis three types of rudimentary eyes (lateral, median and ventral) develop and innervate the brain (see Figure 1 of Harzsch et al., 2006 for detailed diagrams). All are closely associated with pigmented cells termed guanophores, and these eyes seem to provide phototic input during embryonic and early larval life (French, 1980; Harzsch et al., 2006). Subsequently, the lateral compound eye and median ocellar eyes develop during larval stages and provide major visual inputs in adult animals. The lateral compound eyes, median ocelli, lateral and ventral rudimentary eyes express visible light opsins (Smith et al., 1993; Battelle et al., 2001; Dalal et al., 2003), and median ocelli also display UV-sensitivity (Nolte and Brown, 1969). The opsins in the median rudimentary eyes are not yet known. Two Limulus opsin genes encoding visible light proteins have been characterized (reviewed in Batelle, 2006), and the photoreceptor cells within the various eyes classified as rhabdomeric (Arendt and Wittbrodt, 2001). In an evolutionary context, Limulus is interesting because it has both multiple eyes and eye types, which raises the possibility for heterogeneity in the development of different eyes and types of visual organs. In addition, regulatory genes involved in eye development have been described for all major arthropod lineages, except chelicerates.
Here we report isolation and embryonic expression of Lp Pax6 and atonal genes. Paradoxically, we found that neither gene is expressed during eye development in the Limulus embryo or young larvae. Instead, we found Pax6 expression in the developing brain, ventral nerve cord, and lateral sense organ. In addition a Lp atonal family member was identified, but is only expressed in the developing lateral sense organ, a sensory structure whose function remains unresolved. These findings suggest that particular aspects of Limulus embryogenesis are divergent from other arthropods.
RESULTS
Molecular cloning of Limulus Pax6
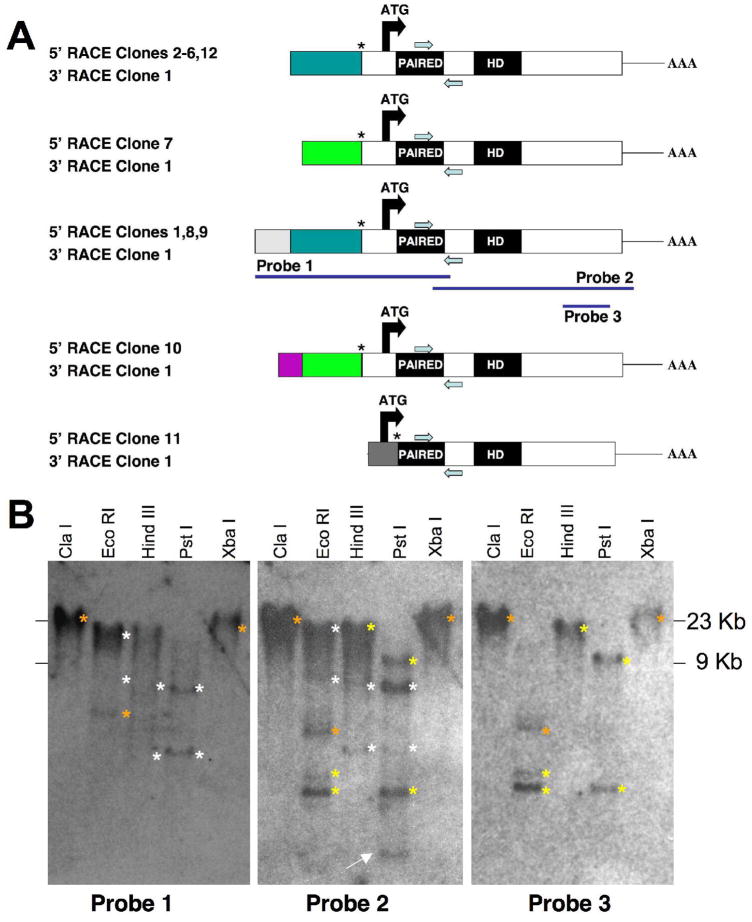
A Limulus polyphemus (Lp) Pax6 800 bp fragment, encoding partial paired- and homeodomains, was isolated by low-stringency RT-PCR using the strategy, nested primers and PCR conditions previously described by Arendt et al. (2002). Multiple subclones of this fragment had identical nucleotide sequences. 5′ and 3′ RACE were then performed on mixed stage embryo cDNA. We isolated distinct 5′ RACE products (Figures 1 and 2), but only one 3′ RACE product of 1.2 Kb in length. DNA sequence analysis indicated that this product contains a partial Pax6 paired domain, homeodomain, stop codon and 3′ UTR (Figures 1A, 2A). Twelve different 5′ RACE subclones were DNA sequenced, their nucleotide sequences compared and then divided into five classes (Figure 1A). Four of five classes differ only in their 5′ UTR, with all subclones converging at the same predicted splice acceptor site, 91 nucleotides upstream of the ATG start codon (asterisk in top four diagrams of Figure 1A). The 5′ RACE products of these four classes have identical nucleotide sequences, from the putative splice site to their 3′ end within the coding region (Figure 1A). The fifth 5′ RACE product has a shorter 5′ UTR, predicted alternative ATG start codon and four unique amino acids, which are in frame with the paired domain (boldfaced amino acids in Clone 11, Figure 2A). This RACE product is predicted to use a separate downstream splice acceptor site (asterisk in Clone 11 in Figure 1A). This 5′ RACE product has identical nucleotide sequence to that of the other four classes, from the first codon of the paired domain to its 3′ end.
Figure 1. Lp Pax6 encodes multiple isoforms.
A) Diagram of five different types Lp Pax6 5′ RACE products with a single 3′ RACE product. Color-coding indicates different 5′ UTR sequences. Use of the same color among different classes of clones indicates stretches of exact nucleotide identity. The positions of gene-specific RACE primers are indicated by blue arrows, below (5′ RACE) or above (3′ RACE) each diagram. 11 of 12 5′ RACE products have identical sequences beginning at a putative splice acceptor site (asterisk) upstream of the start ATG codon. One 5′ RACE subclone (#11) has an alternative in frame ATG start, plus 4 unique amino acids immediately upstream of the paired domain (bold-faced amino acids in Figure 2A). The longest 5′ RACE product (probe 1) was used for Southern blot analyses. B) Autoradiographs of replicate Limulus genomic Southern blots hybridized with the cDNA probes shown in A. Enzymes used to digest Limulus genomic DNA are indicated above each lane. White asterisks to the right of bands indicate those in common for Probes 1 and 2, yellow asterisks mark bands in common between Probes 2 and 3, and orange asterisks indicate bands common to all three probes. Several of the bands marked by white asterisks gave a weaker signal on the Southern blot autoradiogram. The white arrow in the middle blot points to the only band that uniquely hybridizes with Probe 2.
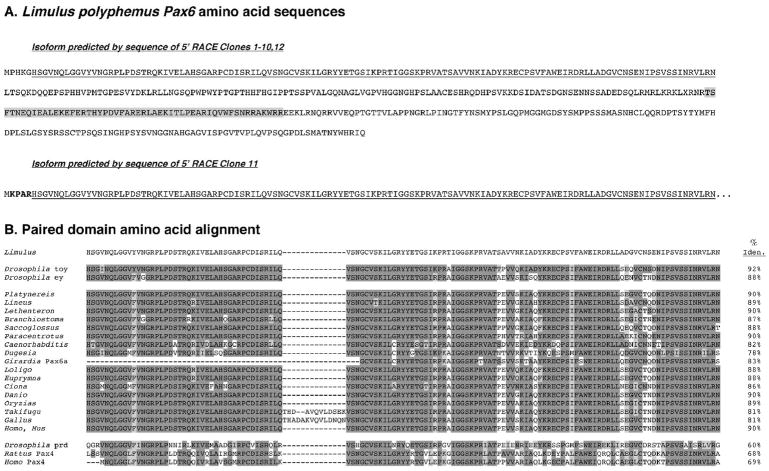
Figure 2. Amino acid sequence of Lp Pax6.
(A) Deduced amino acid sequence of two predicted protein isoforms of Limulus Pax6 that only differ by four amino acids (bold-faced in clone 11). The paired domain is underlined and the homeodomain shaded. (B) Amino acid sequence comparison of the paired domain for Lp Pax6 with various invertebrate and vertebrate Pax6 proteins in the NIH/NCBI database. Dark shading indicates identical amino acids, light shading similar amino acids to Lp Pax6. Percent identities (uncorrected pairwise sequence divergences) are listed at the right. The Lp Pax6 paired domain shares the highest percent identity with that of Drosophila toy.
To characterize the 5′ end of Lp Pax6 further, we sequentially probed a genomic Southern blot at high stringency, with three cDNA probes (Figures 1A–B). When the hybridization patterns of Probes 1 and 2 were compared, a subset of bands are shared between these probes (white asterisks), while some bands were hybridized strictly by Probe 2 (yellow asterisks or a white arrow). To understand whether Probe 2-specific bands represent restriction fragments containing 3′ nucleotides beyond the end of probe 1, we next determined the genomic Southern blot pattern of probe 3 (Figure 1A probe diagram). Comparison of Probe 2 and 3 hybridization patterns (Figure B) indicates that all bands, except one (white arrow in middle blot), contain Lp Pax6 3′-specific sequences. Bands denoted with orange asterisks are common to all three probes. To explore the possibility that the Lp genome contains more than one Pax6 gene, we probed a genomic Southern blot with Probe 2 at low stringency, and observed the identical hybridization pattern found at high stringency in Figure 1B (data not shown). We conclude that Lp embryos possess at least one Pax6 gene, with multiple 5′ splice products, which are predicted to produce two distinct protein isoforms (Figures 1A, 2A).
Evolutionary comparison of Pax6 paired domains
Overall Lp Pax6 shares significant amino acid identity in the paired domain with all Pax6 proteins aligned (Figure 2B). The Lp Pax6 paired domain has highest amino acid identity (92%) with Drosophila toy, and lowest (78%) with the flatworm Dugesia (Figure 2B). Both parsimony and Bayesian analyses provide strong support that among the Pax6 paired domains analyzed, Lp Pax6 is closely related to Drosophila toy. Moreover, within the paired domains of toy and ey, there is a critical difference of one amino acid, which has been previously demonstrated as critical for DNA binding affinity (Czerny et al., 1999). In both Lp Pax6 and Drosophila toy this residue is an asparagine, which is typical of most non-arthropod Pax6 genes. However, Drosophila ey and amphioxus Pax6 encode a glycine at this position. This further suggests that Lp Pax6 is more closely related to toy than it is to ey. The paired domains of these two genes may also be closely related to Pax6.2 of the myriapod Glomeris. However, this interpretation is tentative since the complete amino acid sequences of the Glomeris Pax6 paired domains may demonstrate that Pax6.2 is not closely related to Lp Pax6 and toy (Figure 3).
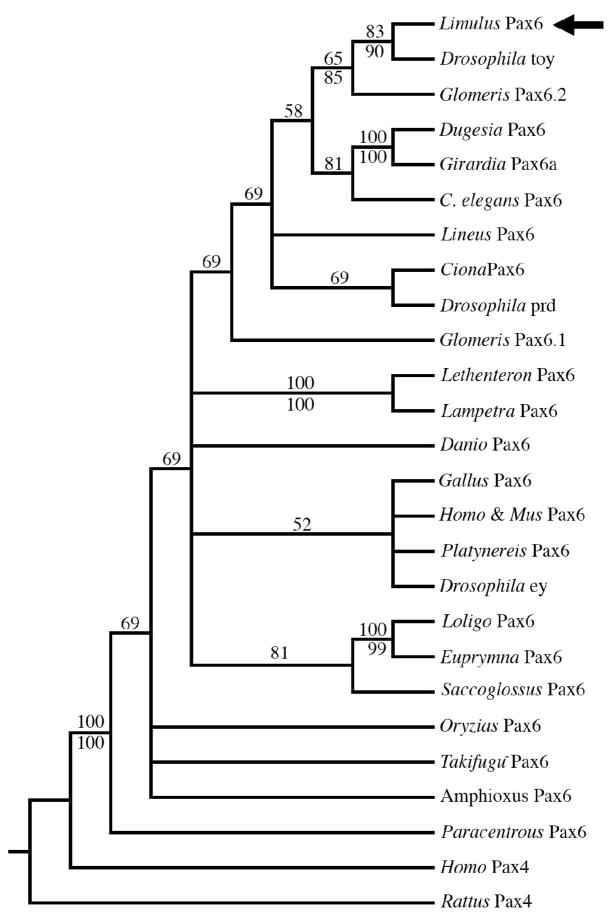
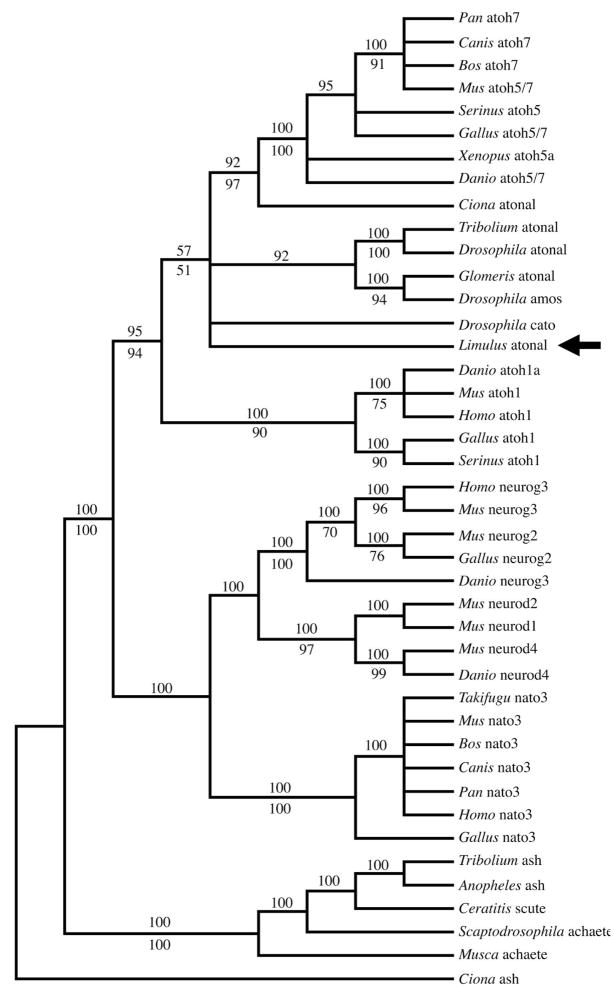
Figure 3. Phylogenetic analyses of Lp Pax6.
Majority rule consensus tree of the 114805 most parsimonious trees resulting from a heuristic search using maximum parsimony; numbers above branches are the percentage of trees in which the topology appears. Numbers below branches are the posterior probabilities for that topology found through a Bayesian phylogenetic analysis using a mixed amino acid model. Pax6 paired domains form a clade that is strongly supported by both analyses. Among the Pax6 paired domains, there is strong support that the Lp Pax6 paired domain (black arrow) is most similar to the Drosophila toy gene. There is also strong support from both analyses for clades comprising the paired domains of planarian flatworms (Dugesia and Girardia), lampreys (Lethenteron and Lampetra) and mollusks (Loligo and Euprymna).
Although our data imply a monophyly of Pax6-eye genes among ecdyzosoans, the relationship of Drosophila paired, to that of Ciona Pax6 and all Pax6 sequences analyzed, is puzzling (Figure 3). The positions of Drosophila paired and ey are not well supported by our analyses, since the bootstrap values were low. Because bootstrap values >70% and Bayesian posterior probabilities >0.95 are needed to show strong support in phylogenetic analyses (Hillis and Bull, 1993; Wilcox et al., 2002), an expansion of the Pax6 paired domain dataset and its outgroups might resolve these discrepancies. Interestingly, there is strong support from both phylogenetic analyses for the close relationship among hemichordate and mollusk Pax6-eye paired domains (Figure 3).
Cloning and evolutionary comparison of an Lp atonal homologue
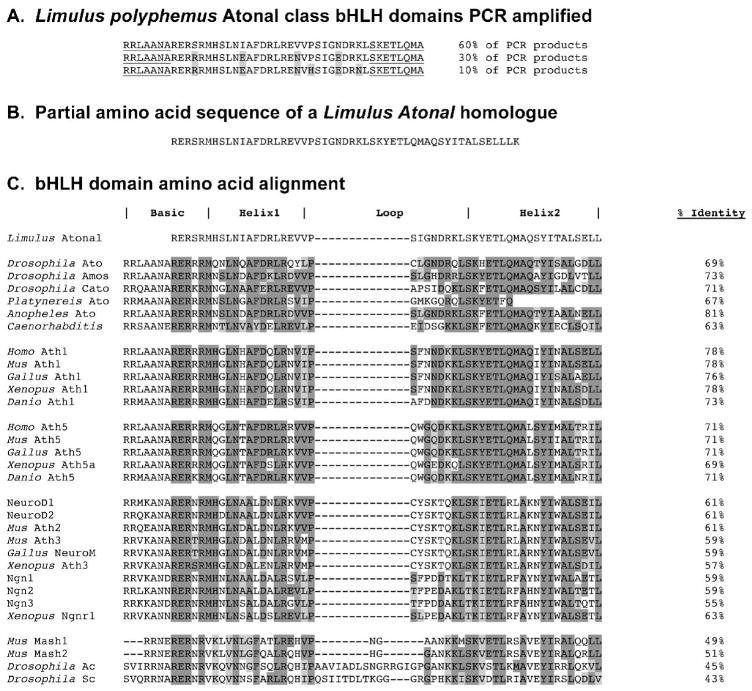
To isolate Lp Atonal bHLH homologues, degenerate bHLH domain primers and standard PCR conditions were used with Lp genomic DNA (Brown et al., 1998), since atonal gene orthologues contain no introns. Ten subclones containing the predicted 135 bp product were DNA sequenced and the nucleotide sequences compared. We found three highly related, but distinct, bHLH domains encoded (Figure 4A). The nested RT-PCR strategy of Arendt et al. (2002) was tried several times on mixed embryo cDNA, but no PCR products recovered contained bHLH domains. To obtain additional Lp atonal coding sequences, gene specific primers were designed from the bHLH domain isolated in 6 of 10 subclones (Figure 4A) and 5′ and 3′ RACE performed with mixed stage embryo cDNA. For 3′ RACE one 450 bp product was obtained that is predicted to encode the 3′ end of a bHLH domain, plus two additional amino acids (Figure 4B), immediately followed by a stop codon and 3′ UTR. We were unable to isolate any 5′ RACE products, despite repeated attempts with several different gene-specific primers and a variety of PCR conditions. The partial coding region obtained is highly similar to Drosophila Atonal (69%) Cato (71%) and Amos (73%), but shares the highest amino acid identity with Anopheles Atonal (81%)(Figure 4C). Interestingly, this Lp Atonal partial cDNA encodes a polypeptide with higher amino acid identity to mouse Math1 (78%) than to Math5 (71%). This implies that Limulus atonal is more related to Math1.
Figure 4. Molecular characterization of Lp atonal.
A) Amino acid sequences of three types of atonal-related bHLH domains PCR amplified from Limulus genomic DNA. Degenerate PCR primers were designed for the underlined amino acids, based on Drosophila atonal and Xenopus Ath5 bHLH domains (Brown et al., 1998). B) Partial amino acid sequence of a Limulus atonal homologue partially cloned. C) Comparison of bHLH domain amino acid sequence for Lp atonal with those of various invertebrate and vertebrate Atonal/Ath/ATOH proteins in the NIH/NCBI database. Dark shading indicates identical amino acids, light shading similar amino acids to Lp Atonal. Percent identities (uncorrected pairwise sequence divergences) are listed in the right column. The Limulus bHLH domain shares the highest percent identity with the Anopheles (mosquito) atonal gene.
Both parsimony and Bayesian analyses provide strong support that among bHLH domains, Lp atonal is most closely related to atonal, Ath1, and Ath5 (Figure 5). While there is strong support for the monophyly of Ath1 genes, there is only very weak support for the inclusion of Lp atonal in a lineage comprising both fruit fly atonal and vertebrate Ath5 genes. Interestingly, there is strong support for a lineage each of atonal, Ath1, and Ath5 (the latter is found only in deuterostomes), as well as a lineage for arthropod amos and atonal, excluding the Limulus gene described here. Lp Atonal does not share any of the autapomorphic amino acids found in the loop region of Drosopila Cato, which suggests that these proteins are not closely related. Likewise, the first amino acid in Lp Atonal Helix1 is a histidine, whereas in Drosophila Cato and the arthropod Amos-Atonal lineage this residue is an asparagine (although in Drosophila Atonal it is a glutamine). Therefore, the phylogenetic affinities of this Lp Atonal bHLH domain remain obscure.
Figure 5. Phylogenetic analyses of Lp Atonal.
Majority rule consensus tree of the 37 most parsimonious trees found in a heuristic search using maximum parsimony; numbers above branches are the percentage of the trees in which the topology appears. Numbers below branches are the posterior probabilities for that topology found through a Bayesian phylogenetic analysis, using a mixed amino acid model. Ath5/ATOH7, Ath1/ATOH1, and atonal bHLH domains form a clade that is strongly supported by both analyses. Among these bHLH domains, the relationship of the Lp atonal bHLH domain (black arrow) to those of other atonal genes is unresolved, although there is weak support that it may be more closely related to vertebrate Ath5 and atonal genes.
Lp Pax6 and atonal expression during embryogenesis
By RT-PCR, both Lp Pax6 and atonal mRNAs are expressed during early (st 0–12) mid (st 13–17) and late (st 18–20) horseshoe crab embryogenesis (Figure 6A). To determine the tissue localization of these transcripts, we also performed whole mount in situ hybridization. Unfortunately, we were unable to analyze embryos younger than st 13 due to their fragility during riboprobe hybridization at stringent temperatures. Beginning around stages 16/17, we observed Lp-Pax6 mRNA expression in the forming brain (white arrows in Figure 6B,D,F,H), ventral nerve cord (black arrows in Figure 6B,D,F,H) and lateral sense organs (Figure 7K,L)(n=25 experiments). All three expression domains were absent in age matched sense controls (Figure 6C,E,G,I and data not shown). Among the different Pax6 expression domains, the lateral sense organ exhibited the most robust expression (Figure 7K). However, the alkaline phosphatase color reaction product that appeared in evaginating appendages from stage 14 to trilobite larvae was not real Pax6 mRNA expression, since sense control embryos had the same staining (asterisks in Figure 6B-I). We also analyzed Lp atonal expression in parallel to that of Pax6. By contrast, atonal is only expressed in the forming lateral sense organ, from st 17/18 to hatching trilobite larvae (Figures 7I,J and data not shown; n ≥ 25 experiments). For both Lp Pax6 and atonal, no expression was found in any of the diverse eyes developing from stage 13 to trilobite larvae hatching (Figures 6, 7 and data not shown).
Figure 6. Embryonic expression of Limulus Pax6 and Atonal mRNA.
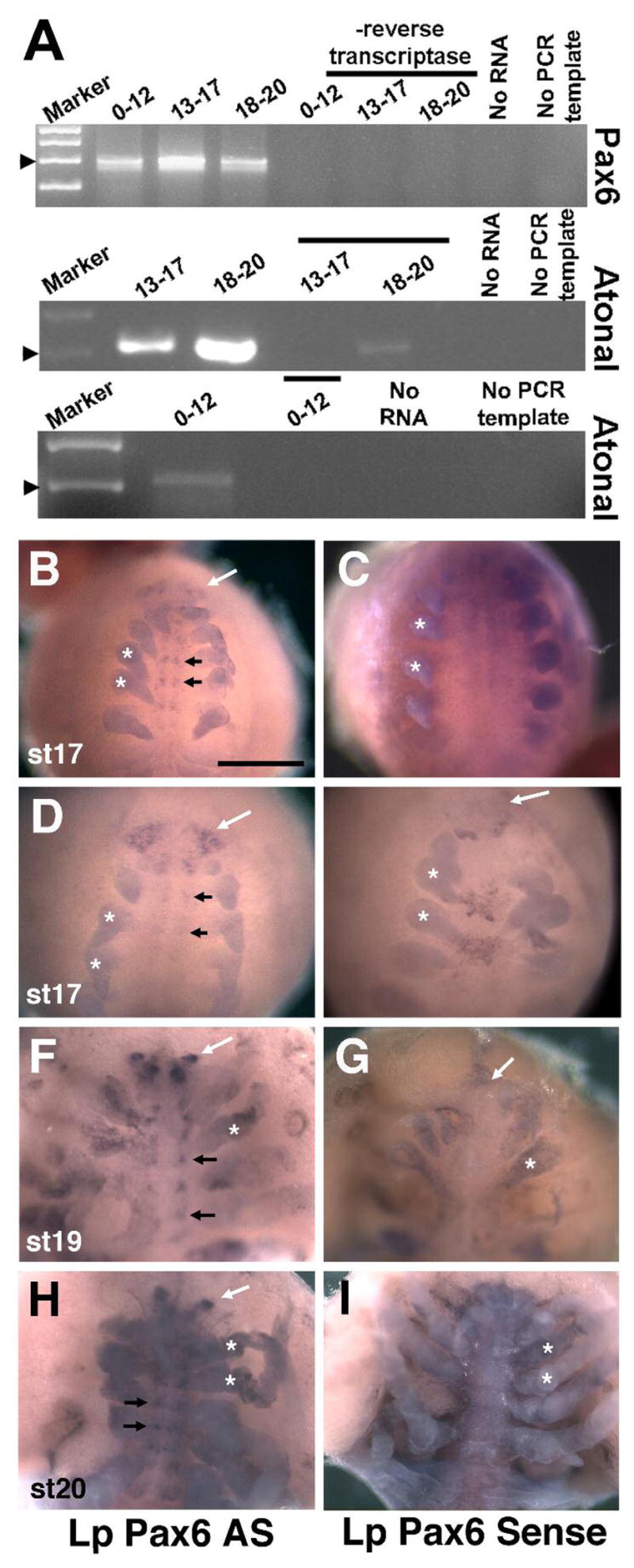
A) RT-PCR for Lp Pax6 and Lp Atonal indicating both genes are expressed throughout embryogenesis to hatching at late st 20. All PCR and RT controls exhibited little to no background amplification. B–I) In situ hybridization for Lp Pax6 antisense (AS) and sense probes, with all panels showing ventral views. B–E) St 17 embryos with Lp Pax6 mRNA expression in the forming brain (white arrows) and ventral nerve cord (black arrows), compared to sense controls. F–G) St 19 embryo with discrete brain (white arrow) and clustered ventral nerve cord (black arrows) expression domains. H,I) St 20 embryo with brain lobe (white arrows) and clustered ventral nerve cord expression (black arrows). Asterisks in all panels label forming appendages that are nonspecifically stained in both antisense and sense probed embryos. Embryo staging followed Harzsch et al, 2006. Bar in B = 500μm.
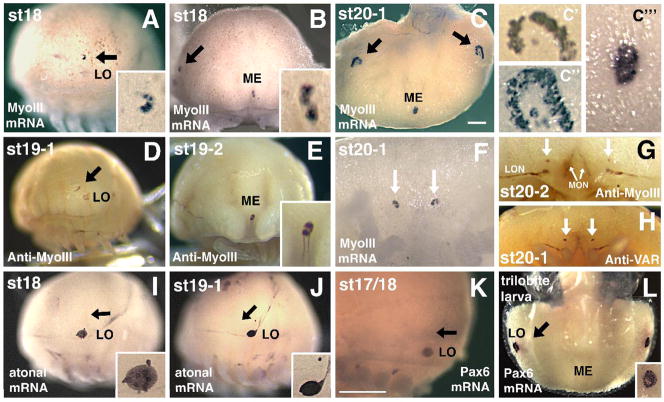
Figure 7. Limulus Pax6 and atonal are not expressed in developing eyes.
A–C,F) Myosin III mRNA expression in the rudimentary lateral eye (black arrows in A–C), early lateral ommatidial eyes (C″, C‴ show first ommatidial photoreceptors surrounded by crescent of rudimentary photoreceptors), rudimentary median eye (ME; B, C, C‴) and rudimentary ventral photoreceptors (arrows in F). The lateral sense organ (LO) is not involved in vision and does not express MyoIII mRNA (A and data not shown). D,E) Anti-Myo III antibody labels the embryonic rudimentary lateral eye (arrow in D), median eye (E) and more weakly the LO (D). F) High magnification of ventral photoreceptors (arrows) expressing MyoIII mRNA. G–H) Anti-MyoIII (G) and Anti-Visual Arrestin (VAR) staining of ventral photoreceptors (arrows) at approximately the same age as in F. These antibodies also label forming optic nerves innervating the brain from the rudimentary lateral (LON) and median (MON) eyes. I–J). Lp Atonal expression in the LO but not rudimentary lateral eye (black arrows point to eye). Insets show higher magnification of LO. K–L) Throughout ages when MyoIII mRNA and protein are expressed in rudimentary eyes or forming lateral ommatidial eyes, Pax6 mRNA is observed in the LO but is absent from these eyes (arrow points to rudimentary lateral eye). Panels A,D,I,J and K are lateral, B and E frontal, C and L are top-down dorsal, and F–H are ventral views. Scale bars in A, C = 500 μm, inset magnification is an additional 400X.
Comparison of Lp Pax6, Atonal and MyoIII expression during eye formation
Lp MyoIII protein expression has been described for the embryonic lateral, median and ventral rudimentary photoreceptors, and larval and adult compound lateral eye ommatidial and medial ocellar photoreceptors (Battelle et al., 2001; Harzsch et al., 2006). The Lp MyoIII gene encodes a photoreceptor cell-specific, unconventional myosin that is under circadian regulation (Battelle et al., 1998). To establish conditions for Limulus whole mount in situ hybridization and to follow embryonic and larval eye development, we compared MyoIII mRNA and protein expression from st 13 through newly hatched larval development (Figures 7A–H). At st 18, MyoIII mRNA expression first appears in rudimentary lateral and median photoreceptor cells (Figures 7A–B), but no expression was observed in the lateral sense organ. However, the anti-MyoIII antibody weakly labels the rudimentary lateral and median eyes, as well as the lateral sense organ beginning at st 18 (Figure 7D and Harzsch et al., 2006). Because the MyoIII antisera, but not our MyoIII riboprobe, labeled the lateral sense organ it suggests that another MyoIII-like protein is present in this sensory structure.
At st 20–1, MyoIII mRNA is expressed within the first lateral eye ommatidial cells, situated in the center of a crescent of rudimentary photoreceptors (C′ and C″ high magnifications of the lateral eyes in C). MyoIII is also maintained in the rudimentary median eye at this older age (Figure 7C‴). During st 20 a small number of ventral photoreceptor cells express MyoIII mRNA (Figure 7F) and protein (Figure 7G). MyoIII protein is localized similarly to visual arrestin (VAR, a photoreceptor-specific protein) in the rudimentary ventral eyes, lateral and median optic nerves (Figures 7G–H). In animals st 18 and older, Lp atonal and Pax6 mRNA expression was observed in the lateral sense organ that lies ventral to the lateral rudimentary eye (black arrows in Figures 7I–L). These two sense organs are readily distinguished by their dorsal-ventral positions and distinct morphologies (compare insets in Figures 7A and 7I). Because we found no evidence of Lp Pax6 or atonal expression analogous to that of MyoIII, we conclude that neither gene is expressed during the formation of the three different rudimentary eyes or early lateral compound eye and median ocellar development.
DISCUSSION
Limulus polyphemus (Lp) embryology has been studied since the late 1800s. Much more recently, investigations of chelicerate embryonic body patterning and neural development were reported by multiple groups (Telford and Thomas, 1998; Abzhanov et al., 1999; Stollewerk et al., 2001; Dearden et al., 2002; Mittmann, 2002; Dearden et al., 2003; Mittmann and Scholtz, 2003; Maxmen et al., 2005; Pioro and Stollewerk, 2006). However, only a handful of developmentally expressed Limulus genes have been characterized (Cartwright et al., 1993; Cook et al., 2001; Mittmann and Scholtz, 2001; Davis et al., 2005). Likewise, the Lp visual system has been investigated for decades (reviewed in Barlow, 2003; Batelle, 2006) but, the molecular mechanisms of eye development in Limulus and other chelicerates, is still unknown. The goal of this study was to link Lp Pax6 and atonal genes to the formation of one or more types of Limulus eyes. Much to our surprise, we found neither gene is expressed during the formation of different eyes. Instead, we observed Lp Pax6 expression in the brain and ventral nerve cord and the expression of both genes in the lateral sense organ.
There are multiple explanations for these findings that are not mutually exclusive. First, since these transcription factors are expressed at younger ages than our in situ experiments could assay, it is plausible they act in the eye primoridia(s) prior to stage 13. Second, there are other instances of bilaterian eyes that do not express Pax6, and this may also be the case for Limulus. For example, Platynereis larval eyes express Pax6, but their adult eyes form via a Pax6-independent mechanism that involves So/Six gene expression (Arendt et al., 2002). The myriapod Glomeris has two Pax6 genes, yet neither is associated with eye formation, and amphioxus Pax6 is present in its multiple eye types, but not the Organ of Hesse that contains photoreceptors (Glardon et al., 1998; Prpic, 2005). Analogously, deuterostome eye specification requires the Rx gene that acts upstream of Pax6 (Furukawa et al., 1997; Mathers et al., 1997; D’Aniello et al., 2006). Finally, multiple Pax6 and/or atonal genes may exist now or during the evolutionary history of Limulus, meaning a Pax6 or atonal paralogue may regulate eye development. Indeed, our isolation of three different atonal-class bHLH domains, suggests there are multiple Lp atonal-like genes. The Lp atonal described here is only expressed in the lateral sense organ, a structure of unknown function. The high amino acid identity among Lp Atonal, Drosophila Amos and Mus Math1 proteins, whose functions in auditory or olfactory development are known (Bermingham et al., 1999; zur Lage et al., 2003), may indicate that the lateral sense organ is involved in auditory or olfactory sensation.
Horseshoe crabs are often considered “living fossils” as they obtained their present morphological form at least by the mid Mesozoic (ca. 160 million years ago, Fisher, 1984), but possibly as early as the late Ordovican (ca. 445 million years ago, Rudkin et al., 2008). However, the last common ancestor of living taxa was more recent, probably within the last 80 million years (Avise et al., 1994). Here we describe several distinct features of the Limulus Pax6 gene, as compared to those of other arthropods. Along with the absence of Pax6 mRNA expression in multiple developing eye types, another unanticipated characteristic of Lp Pax6 is its predicted multiple splice products that would give rise to distinct isoforms. Intriguingly, distinct splice products and protein isoforms are features of vertebrate Pax6 genes. Together our findings suggest that Limulus embryos employ divergent gene networks in their developing visual systems and appendages. In the future it will be exciting to understand whether these are also characteristics of other chelicerate species. While morphological stasis may characterize horseshoe crab evolution, further research on their embryology may provide important insight into the evolution of novel gene networks.
EXPERIMENTAL PROCEDURES
Animals
The collection protocols of Harzsch et al. (2006) were followed, except that water was changed every other day with artificial seawater plus 100U/ml of penicillin (Sigma) and 100 μg/ml of streptomyocin (Sigma), to prevent microorganism growth that reduced viability. Published embryo staging criteria were used (Sekiguchi et al., 1982; Harzsch et al., 2006). Stage 13–17 embryos were dechorionated in 50% bleach for 5 minutes, rinsed three times in dH20 and fixed for one hour, rocking in a 50:50 mixture of 4% paraformaldehyde/PBS and heptane. The aqueous phase was replaced with methanol and embryos vigorously shaken in heptane-methanol for 30 seconds to rupture outer membranes. Both heptane and methanol were replaced with three changes of fresh methanol and embryos stored in methanol at −20°C. Older embryos and trilobite larvae were hand dissected from outer membranes and processed as younger embryos minus dechorionation.
Molecular Cloning
A previously published PCR strategy (Arendt et al., 2002) was used to amplify Pax6 conserved domains from mixed stage Limulus (Lp) cDNA. The remainder of the Pax6 coding region was isolated via 5′ and 3′ RACE (rapid amplification of cDNA ends). Lp atonal bHLH domains were PCR amplified from genomic DNA, with degenerate primers (Brown et al., 1998) and 40 cycles of 95°C × 30 sec, 60°C × 60 sec, 68°C × 2 min. Three highly related bHLH domains were found. Primers for RACE cloning were designed from the most frequently isolated bHLH domain. 5′ and 3′ RACE for Lp Pax6 and Lp atonal used the SMART RACE cloning kit and protocol (BD/Invitrogen) and mixed embryo cDNA templates. Pax6: 5′RACE 5′-TAGAGCAACTGGCGATGATGTCGGAGG-3′; 3′RACE 5′-CAGGAGTGTCCTTCCGTATTTGCCTGG-3′; atonal: 3′RACE 5′-CCGTGAACGAAGCCGAAGCACAGTC-3′ gene-specific primers were used. Multiple products from each RACE experiment were DNA sequenced and analyzed with MacVector (v9.0) and AssemblyLIGN (v1.1, Accelrys) computer programs. GenBank accession number for Lp-atonal is EU673469. The GenBank accession numbers for Lp-Pax6 are: EU673470 Pax6 mRNA predicted to encode the predominant isoform; EU673471 Pax6 mRNA predicted to encode alternate isoform; EU673472 Pax6 5′ RACE clone 2–6,12; Pax6 EU673473 5′ RACE clone 7; EU673474 Pax6 5′ RACE clone 1,8,9; EU673475 Pax6 5′ RACE clone 10; and EU673476 Pax6 5′RACE clone 11.
Genomic Southern blots
Lp genomic DNA was isolated from six juvenile crabs. Nylon blots (Pall Corp.) containing 5 μg of digested, transferred and immobilized genomic DNA were prehybridized in 50% formamide, 6X SSC, 5X Denhardts, 0.5% SDS, 100μg/ml herring sperm DNA (Sigma) at 42°C, and then hybridized at either 37°C or 42°C (Glaser et al., 1992). 1–2 × 107 CPM of each gel purified and 32P-labeled cDNA probe was added to 10 ml of hybridization buffer (40% formamide, 4X SSC, 0.8X Denhardt’s, 10% dextran sulphate (American Bioanalytical), 7mM Tris (pH7.4), 100 μg/ml herring sperm DNA). Blots were washed at high (3 times in 2XSSC/0.2%SDS at 55°C, twice in 0.2XSSC/0.2%SDS at 65°C) or low (2 times in 2XSSC/0.2%SDS at 50°C, twice in the same buffer at 55°C) stringency and exposed to x-ray film.
Phylogenetic Analysis
A heuristic search using maximum parsimony in PAUP (v 4.0b10, Altivec) and a Bayesian analysis using MrBayes (v 3.0b4) were performed for both a matrix of Pax6 and related paired domains and a matrix of atonal and related bHLH domains. Amino acid sequences were downloaded from GenBank in FASTA format and aligned using CLUSTALX. For paired domain analyses, the dataset was defined as 139 amino acids comprising the paired domain and homeodomain in Pax6, Drosophila eyeless (ey) and twin of eyeless (toy), with human and rat Pax4 included as outgroups. For atonal bHLH analyses, the dataset was limited to 70 amino acids that comprise the bHLH domain in atonal homologues and six achaete-scute homologues included as an outgroup. In the Bayesian analyses, the prior probability model for the amino acid rate matrix was set to mixed, allowing the analysis to sample a range of different models for molecular evolution. Bayesian analyses were conducted for one million generations, with trees sampled every 100 generations; stationarity was obtained after approximately 10,000 generations, thus the first 100 trees were discarded as burnin.
RT-PCR
Total RNA was isolated from > 100 embryos stage 0–12, and at least 50 embryos for the older age groups, These RNAs were pretreated with DNase prior to reverse transcription into cDNA. PCR parameters were 40 cycles of 95°C × 30 sec, 60°C × 60 sec, 68°C × 2 min with primers PAX6FOR 5′-TAGCACCACCTAACGGACGACTTC-3′, PAX6REV 5′-CACCTGGAGAAATGACACCTGC-3′; or ATOFOR 5′-CGTGAACGAAGCCGAATGCACAGTCT-3′, ATOREV 5′-TGTAAATGAGGCTTTCCCACATGTATT-3′.
In situ hybridization and immunohistochemistry
Lp Pax6 and atonal antisense and sense cRNA digoxygenin-labeled riboprobes contained coding plus 3′UTR nucleotide sequences. A MyoIII antisense riboprobe was synthesized from a full-length cDNA clone (Accession Number AF062069). Several modifications were made to a whole mount in situ protocol (Hargrave and Koopman, 2000). After rehydration into PBS/0.1% Triton X-100, (PBTX) embryos ≥ stage 18 were incubated overnight at room temperature in a 1:50 dilution of 5Units/ul chitinase (Sigma) in DEPC-dH20, to remove a nearly invisible chitin cuticle. Then embryos were washed in PBTX and proteinase K digested. After refixation, but prior to prehybridization, embryos and larvae were heated to 75°C for 30 minutes to quench endogenous alkaline phosphatase. Nonspecific background was lowered by preincubating a 1:500 dilution of sheep anti-digoxygenin antibody (Roche) in 50mM Tris (pH7.5), 150mM NaCl, 0.1% Triton X-100, 10% Sheep serum (Sigma), 2% BSA, 0.2% sodium azide (Sigma) plus 1mg of horseshoe crab protein powder, via rocking at 4°C for ≥4 hours. Powder was removed by brief centrifugation, and the antibody used at 1:2000. Crab powder was made by cold acetone extraction of homogenized juvenile Lp soft body parts. Anti MyoIII (1:1000) or Anti-Arrestin (VAR, 1:50) antibody labeling of Lp embryos used published protocols (Davis et al., 2001; Harzsch et al., 2006) and HRP color development.
Acknowledgments
NIH R01 EY13612 to NLB; NSF grant 0094428 to BAB
The authors thank Nipam Patel for Limulus embryo fixation, antibody and in situ staining protocols, Todd Oakley for sharing unpublished data, Tom Glaser for advice on genomic Southern blots and valuable discussion, Jon Currie for technical support and Brian Gebelein and Teresa Orenic for critical comments.
References
- Abzhanov A, Popadic A, Kaufman TC. Chelicerate Hox genes and the homology of arthropod segments. Evol Dev. 1999;1:77–89. doi: 10.1046/j.1525-142x.1999.99014.x. [DOI] [PubMed] [Google Scholar]
- Arendt D. Evolution of eyes and photoreceptor cell types. Int J Dev Biol. 2003;47:563–571. [PubMed] [Google Scholar]
- Arendt D, Tessmar K, de Campos-Baptista MI, Dorresteijn A, Wittbrodt J. Development of pigment-cup eyes in the polychaete Platynereis dumerilii and evolutionary conservation of larval eyes in Bilateria. Development. 2002;129:1143–1154. doi: 10.1242/dev.129.5.1143. [DOI] [PubMed] [Google Scholar]
- Arendt D, Wittbrodt J. Reconstructing the eyes of Urbilateria. Philos Trans R Soc Lond B Biol Sci. 2001;356:1545–1563. doi: 10.1098/rstb.2001.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Nelson WS, Sugita H. A special history of “living fossils”: molecular evolutionary patterns in horseshoe crabs. Evolution. 1994;48:1986–2001. doi: 10.1111/j.1558-5646.1994.tb02228.x. [DOI] [PubMed] [Google Scholar]
- Barlow R. The American Horseshoe Crab. Cambridge, MA: Harvard University Press; 2003. [Google Scholar]
- Barlow RB, Hitt JM, Dodge FA. Limulus vision in the marine environment. Biol Bull. 2001;200:169–176. doi: 10.2307/1543311. [DOI] [PubMed] [Google Scholar]
- Barlow RB., Jr Inhibitory fields in the Limulus lateral eye. J Gen Physiol. 1969;54:383–396. doi: 10.1085/jgp.54.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batelle B-A. The eyes of Limulus polyphemus (Xiphosura, Chelicerata) and their afferent and efferent projections. Arthropod Structure & Development. 2006;35:261–274. doi: 10.1016/j.asd.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Battelle BA, Andrews AW, Calman BG, Sellers JR, Greenberg RM, Smith WC. A myosin III from Limulus eyes is a clock-regulated phosphoprotein. J Neurosci. 1998;18:4548–4559. doi: 10.1523/JNEUROSCI.18-12-04548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battelle BA, Dabdoub A, Malone MA, Andrews AW, Cacciatore C, Calman BG, Smith WC, Payne R. Immunocytochemical localization of opsin, visual arrestin, myosin III, and calmodulin in Limulus lateral eye retinular cells and ventral photoreceptors. J Comp Neurol. 2001;435:211–225. doi: 10.1002/cne.1203. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Halder G, Gehring WJ. PAX-6 in development and evolution. Annu Rev Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- Carriere C, Plaza S, Martin P, Quatannens B, Bailly M, Stehelin D, Saule S. Characterization of quail Pax-6 (Pax-QNR) proteins expressed in the neuroretina. Mol Cell Biol. 1993;13:7257–7266. doi: 10.1128/mcb.13.12.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, Dick M, Buss LW. HOM/Hox type homeoboxes in the chelicerate Limulus polyphemus. Mol Phylogenet Evol. 1993;2:185–192. doi: 10.1006/mpev.1993.1019. [DOI] [PubMed] [Google Scholar]
- Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- Cook CE, Smith ML, Telford MJ, Bastianello A, Akam M. Hox genes and the phylogeny of the arthropods. Curr Biol. 2001;11:759–763. doi: 10.1016/s0960-9822(01)00222-6. [DOI] [PubMed] [Google Scholar]
- Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- D’Aniello S, D’Aniello E, Locascio A, Memoli A, Corrado M, Russo MT, Aniello F, Fucci L, Brown ER, Branno M. The ascidian homolog of the vertebrate homeobox gene Rx is essential for ocellus development and function. Differentiation. 2006;74:222–234. doi: 10.1111/j.1432-0436.2006.00071.x. [DOI] [PubMed] [Google Scholar]
- Dalal JS, Jinks RN, Cacciatore C, Greenberg RM, Battelle BA. Limulus opsins: diurnal regulation of expression. Vis Neurosci. 2003;20:523–534. doi: 10.1017/s095252380320506x. [DOI] [PubMed] [Google Scholar]
- Davis GK, D’Alessio JA, Patel NH. Pax3/7 genes reveal conservation and divergence in the arthropod segmentation hierarchy. Dev Biol. 2005;285:169–184. doi: 10.1016/j.ydbio.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Davis GK, Jaramillo CA, Patel NH. Pax group III genes and the evolution of insect pair-rule patterning. Development. 2001;128:3445–3458. doi: 10.1242/dev.128.18.3445. [DOI] [PubMed] [Google Scholar]
- Dearden P, Grbic M, Donly C. Vasa expression and germ-cell specification in the spider mite Tetranychus urticae. Dev Genes Evol. 2003;212:599–603. doi: 10.1007/s00427-002-0280-x. [DOI] [PubMed] [Google Scholar]
- Dearden PK, Donly C, Grbic M. Expression of pair-rule gene homologues in a chelicerate: early patterning of the two-spotted spider mite Tetranychus urticae. Development. 2002;129:5461–5472. doi: 10.1242/dev.00099. [DOI] [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Fisher DC. The Xiphosurida: archetypes of bradytely? In: Eldredge N, Stanley SM, editors. Living Fossils. New York: Springer; 1984. pp. 196–213. [Google Scholar]
- French KA. The development of photoreception in Limulus polyphemus: morphology, electrophysiology and behavior. Boston University Graduate School; Boston, MA: 1980. pp. 1–168. [Google Scholar]
- Friedrich M. Ancient mechanisms of visual sense organ development based on comparison of the gene networks controlling larval eye, ocellus and compound eye specification in Drosophila. Arthropod Structure & Development. 2006;35:357–378. doi: 10.1016/j.asd.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Kozak CA, Cepko CL. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci U S A. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ. The master control gene for morphogenesis and evolution of the eye. Genes Cells. 1996;1:11–15. doi: 10.1046/j.1365-2443.1996.11011.x. [DOI] [PubMed] [Google Scholar]
- Glardon S, Holland LZ, Gehring WJ, Holland ND. Isolation and developmental expression of the amphioxus Pax-6 gene (AmphiPax-6): insights into eye and photoreceptor evolution. Development. 1998;125:2701–2710. doi: 10.1242/dev.125.14.2701. [DOI] [PubMed] [Google Scholar]
- Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human Pax6 gene. Nature Genetics. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- Hartline HK, Floyd Ratcliff HGW. Inhibition in the eye of Limulus. Journal of General Physiology. 1956;39:651–673. doi: 10.1085/jgp.39.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995a;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. New perspectives on eye evolution. Curr Opin Genet Dev. 1995b;5:602–609. doi: 10.1016/0959-437x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- Hanson IM. Mammalian homologues of the Drosophila eye specification genes. Semin Cell Dev Biol. 2001;12:475–484. doi: 10.1006/scdb.2001.0271. [DOI] [PubMed] [Google Scholar]
- Hargrave M, Koopman P. In situ hybridization of whole-mount embryos. Methods Mol Biol. 2000;123:279–289. doi: 10.1385/1-59259-677-0:279. [DOI] [PubMed] [Google Scholar]
- Harzsch S, Vilpoux K, Blackburn DC, Platchetzki D, Brown NL, Melzer R, Kempler KE, Battelle BA. Evolution of arthropod visual systems: development of the eyes and central visual pathways in the horseshoe crab Limulus polyphemus Linnaeus, 1758 (Chelicerata, Xiphosura) Dev Dyn. 2006;235:2641–2655. doi: 10.1002/dvdy.20866. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology. 1993;42:182–192. [Google Scholar]
- Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc Natl Acad Sci U S A. 1998;95:13720–13725. doi: 10.1073/pnas.95.23.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- Lakowski J, Majumder A, Lauderdale JD. Mechanisms controlling Pax6 isoform expression in the retina have been conserved between teleosts and mammals. Dev Biol. 2007;307:498–520. doi: 10.1016/j.ydbio.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Maxmen A, Browne WE, Martindale MQ, Giribet G. Neuroanatomy of sea spiders implies an appendicular origin of the protocerebral segment. Nature. 2005;437:1144–1148. doi: 10.1038/nature03984. [DOI] [PubMed] [Google Scholar]
- Mishra R, Gorlov IP, Chao LY, Singh S, Saunders GF. PAX6, paired domain influences sequence recognition by the homeodomain. J Biol Chem. 2002;277:49488–49494. doi: 10.1074/jbc.M206478200. [DOI] [PubMed] [Google Scholar]
- Mittmann B. Early neurogenesis in the horseshoe crab Limulus polyphemus and its implication for arthropod relationships. Biol Bull. 2002;203:221–222. doi: 10.2307/1543407. [DOI] [PubMed] [Google Scholar]
- Mittmann B, Scholtz G. Distal-less expression in embryos of Limulus polyphemus (Chelicerata, Xiphosura) and Lepisma saccharina (Insecta, Zygentoma) suggests a role in the development of mechanoreceptors, chemoreceptors, and the CNS. Dev Genes Evol. 2001;211:232–243. doi: 10.1007/s004270100150. [DOI] [PubMed] [Google Scholar]
- Mittmann B, Scholtz G. Development of the nervous system in the “head” of Limulus polyphemus (Chelicerata: Xiphosura): morphological evidence for a correspondence between the segments of the chelicerae and of the (first) antennae of Mandibulata. Dev Genes Evol. 2003;213:9–17. doi: 10.1007/s00427-002-0285-5. [DOI] [PubMed] [Google Scholar]
- Mu X, Klein WH. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Semin Cell Dev Biol. 2004;15:115–123. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Nolte J, Brown JE. The spectral sensitivities of single cells in the median ocellus of Limulus. J Gen Physiol. 1969;54:636–649. doi: 10.1085/jgp.54.5.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J, Kozmik Z. Cubozoan jellyfish: an Evo/Devo model for eyes and other sensory systems. Int J Dev Biol. 2004;48:719–729. doi: 10.1387/ijdb.041851jp. [DOI] [PubMed] [Google Scholar]
- Pichaud F, Treisman J, Desplan C. Reinventing a common strategy for patterning the eye. Cell. 2001;105:9–12. doi: 10.1016/s0092-8674(01)00292-6. [DOI] [PubMed] [Google Scholar]
- Pioro HL, Stollewerk A. The expression pattern of genes involved in early neurogenesis suggests distinct and conserved functions in the diplopod Glomeris marginata. Dev Genes Evol. 2006;216:417–430. doi: 10.1007/s00427-006-0078-3. [DOI] [PubMed] [Google Scholar]
- Prpic NM. Duplicated Pax6 genes in Glomeris marginata (Myriapoda: Diplopoda), an arthropod with simple lateral eyes. Zoology (Jena) 2005;108:47–53. doi: 10.1016/j.zool.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Rudkin DM, Young GA, Nowlan GS. The oldest horseshoe crab: a new Xiphorsurid from late Ordovician Konservat-Lagerstatten deposits, Manitoba, Canada. Palaeontology. 2008;51:1–9. [Google Scholar]
- Sekiguchi K, Yamamichi Y, Costlow JD. Horseshoe crab developmental studies I. Normal embryonic development of Limulus polyphemus compared with Tachypleus tridentatus. Prog Clin Biol Res. 1982;81:53–73. [PubMed] [Google Scholar]
- Smith WC, Price DA, Greenberg RM, Battelle BA. Opsins from the lateral eyes and ocelli of the horseshoe crab, Limulus polyphemus. Proc Natl Acad Sci U S A. 1993;90:6150–6154. doi: 10.1073/pnas.90.13.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodderly DM, Jr, Barlow RB., Jr Projection of the lateral eye of Limulus to the brain. Nature. 1970;227:284–286. doi: 10.1038/227284a0. [DOI] [PubMed] [Google Scholar]
- Stollewerk A, Weller M, Tautz D. Neurogenesis in the spider Cupiennius salei. Development. 2001;128:2673–2688. doi: 10.1242/dev.128.14.2673. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Thomas RH. Expression of homeobox genes shows chelicerate arthropods retain their deutocerebral segment. Proc Natl Acad Sci U S A. 1998;95:10671–10675. doi: 10.1073/pnas.95.18.10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomarev SI, Callaerts P, Kos L, Zinovieva R, Halder G, Gehring W, Piatigorsky J. Squid Pax-6 and eye development. Proc Natl Acad Sci U S A. 1997;94:2421–2426. doi: 10.1073/pnas.94.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton CCT, et al. Positional cloning and characterization of paired box- and homeobox-containing gene from the Aniridia region. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- van Heyningen V, Williamson KA. PAX6 in sensory development. Hum Mol Genet. 2002;11:1161–1167. doi: 10.1093/hmg/11.10.1161. [DOI] [PubMed] [Google Scholar]
- Vetter ML, Brown NL. The role of basic helix-loop-helix genes in vertebrate retinogenesis. Semin Cell Dev Biol. 2001;12:491–498. doi: 10.1006/scdb.2001.0273. [DOI] [PubMed] [Google Scholar]
- Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- Wilcox TP, Zwicki DJ, Heath TA, Hillis DM. Phylogenetic relationships of the dwarf boas and a comparison of Bayesian and boostrap measures of phylogenetic support. Molecular Phylogenetics and Evolution. 2002;25:361–371. doi: 10.1016/s1055-7903(02)00244-0. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zhang X, Heaney S, Yoon A, Michelson AM, Maas RL. Regulation of Pax6 expression is conserved between mice and flies. Development. 1999;126:383–395. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- zur Lage PI, Prentice DR, Holohan EE, Jarman AP. The Drosophila proneural gene amos promotes olfactory sensillum formation and suppresses bristle formation. Development. 2003;130:4683–4693. doi: 10.1242/dev.00680. [DOI] [PubMed] [Google Scholar]