Abstract
β-Cyclodextrin (CD) dimers (n = 11) were synthesized and tested against eight enzymes, seven of which were dimeric or tetrameric, for inhibitor activity. Initial screening showed that only l-lactate dehydrogenase and citrate synthase were inhibited but only by two specific CD dimers in which two β-CDs were linked on the secondary face by a pyridine-2,6-dicarboxylic group. Further investigation suggested that these CD dimers inhibit the activity of l-lactate dehydrogenase and citrate synthase at least in part by disruption of protein–protein aggregation.
The aggregation of proteins, often involving the interaction of hydrophobic patches, is central to many important biological phenomena (1). For example, many enzymes are active only in their dimeric form, and the multimerization of proteins in the formation of fibers also frequently involves the interaction of hydrophobic patches (2–4). Thus, it would be of great interest to learn how to inhibit such aggregation selectively.
We have reported that certain linked dimers of cyclodextrins (CDs) can selectively bind the hydrophobic side chains in polypeptides (5, 6). Thus, we examined the possibility that such CD dimers can bind to the hydrophobic patches of proteins so as to inhibit aggregation. We have now found that this inhibition indeed occurs and does so selectively with respect to both the CD dimer and the protein involved. Such selective inhibition of protein aggregation may have important uses in biology and medicine.
To examine this possibility, we have studied a group of enzymes, which are listed in Table 1. Several of them are active only as dimers [glucose-6-phosphate dehydrogenase (7), d-galactose dehydrogenase (8), phosphohexose isomerase (9), and citrate synthase (CS; ref. 10)]; some are active as tetramers [l-lactate dehydrogenase (LDH; ref. 11), fumarase (12), and sorbitol dehydrogenase (13)], and adenosine deaminase is a monomeric enzyme (14). Our preliminary screen simply looked at whether CD dimers inhibited the enzyme's activity.
Table 1.
The enzymes studied and their active forms
| Enzyme | Active form |
|---|---|
| Adenosine deaminase | Monomer |
| Glucose-6-phosphate dehydrogenase | Dimer |
| d-Galactose dehydrogenase | Dimer |
| Phosphohexose isomerase | Dimer |
| CS | Dimer |
| LDH | Tetramer |
| Fumarase | Tetramer |
| Sorbitol dehydrogenase | Tetramer |
Materials and Methods
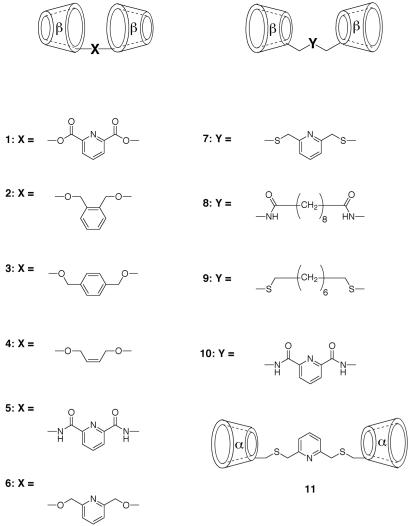
The CD dimers examined, compounds 1-11, are shown in Scheme S1. We have previously reported (5) the syntheses of 2 and 4; 3 was prepared in an analogous way by alkylating β-CD with p-xylylene dibromide. Dimer 6 was prepared by direct alkylation with 2,6-bis(bromomethyl)pyridine of the most acidic C-2 hydroxyl group of β-CD after forming its oxide anion with NaH. Dimer 1, from acylation of the β-CD anion with pyridine-2,6-dicarboxylic acid chloride, is expected to equilibrate between the C-2 and C-3 positions by acyl migration; thus, it must be a positional mixture. Dimers 8 and 10 were prepared by acylating 6-amino-6-deoxycycloheptaamylose (15) with the bis-pentafluorophenyl ester of the linker diacid, and dimer 5 was prepared by acylating 3-amino-3-deoxycycloheptaamylose (16) with pyridine-2,6-dicarboxylic acid chloride. Dimers 7 and 9 were prepared by reacting 6-deoxy-6-iodocycloheptaamylose (17) with 2,6-bis(mercaptomethyl)-pyridine and 1,8-octanedithiol, respectively, in dimethylformamide with K2CO3, and dimer 11 was obtained by the same procedure used for dimer 7 but with 6-deoxy-6-iodocyclohexaamylose. All of the compounds had the expected NMR and mass spectra for the structures assigned.
Scheme 1.
CD dimers studied.
The enzymes were assayed by using standard protocols (18) with a 15-min preassay incubation. The final assay concentrations of substrates and cofactors are as follows: for LDH, 0.5 mM NADH/1.9 mM pyruvate; for d-galactose dehydrogenase, 2.5 mM NAD/17 mM d-galactose; for glucose-6-phosphate dehydrogenase, 7.1 mM NAD/9.6 mM glucose-6-phosphate; for phosphohexose isomerase (coupled with glucose-6-phosphate dehydrogenase), 0.4 mM NADP/3 mM fructose-6-phosphate; for sorbitol dehydrogenase, 0.2 mM NADH/150 mM d-fructose; for adenosine deaminase, 0.05 mM adenosine; for fumarase, 50 mM l-malate; and for CS, 0.2 mM acetyl CoA/0.2 mM oxaloacetate.
For nondenaturing size exclusion chromatography, all runs were performed at 4°C with a 108-cm × 1-cm diameter Sepharcyl 200-h column. Flow rate was maintained at 8.4 ml/h.
Results and Discussion
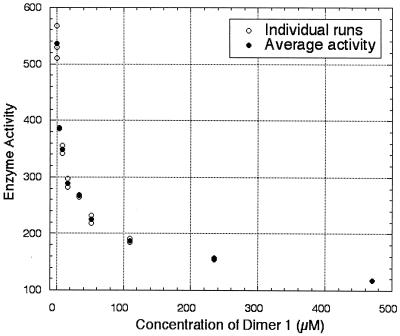
Of the enzymes studied, only CS and LDH were inhibited by any of the CD dimers examined, even at concentrations as high as 480 mM, and inhibition occurred only with two specific CD dimers, 1 and 5. Dimer 1 inhibited the catalytic activity of CS with an IC50 of 140 μM and inhibited LDH with an IC50 of 30 μM. As Fig. 1 shows, a 28% inhibition of LDH was observed with only 4 μM of dimer 1. Because the concentrations of NADH and of pyruvate are over 100 times the concentration of the CD dimer used, the inhibition must reflect interaction of the CD dimer with the enzyme and not simply scavenging of the substrate or the cofactor. Furthermore, dimer 1 did not inhibit sorbitol dehydrogenase, which also uses NADH as a cofactor. Dimer 1 did not bind to acetyl CoA, as evidenced by titration microcalorimetry.
Figure 1.
Plot of initial rate of LDH versus concentration of CD dimer 1. Initial rate is expressed in units/liter; 1 unit of LDH is defined as the amount of LDH required to reduce 1.0 μmol pyruvate to l-lactate per min at pH 7.5 and at 37°C.
The finding that the diester 1 and diamide 5 were active inhibitors of CS and LDH but that the related diether 6 was inactive is interesting. We do not yet understand the reasons for this selectivity. However, of course, ether 6 is doubly linked to the oxygen at C-2; diamide 5 is doubly linked to a nitrogen at C-3; and diester 1 should be a mixture with 2,2′ links, 3,3′ links, and 2,3′ links. Diamide 10, linked on the CD primary side, was inactive. Pyridine, β-CD, and pyridine-2,6-dicarboxylic acid showed no inhibition of CS or of LDH at concentrations up to 1 mM, either individually or in combinations, nor did a CD dimer linked with isophthalic acid instead of pyridine-2,6-dicarboxylic acid. The dimethyl ester of pyridine-2,6-dicarboxylic acid did show some inhibition of the activity of LDH (≈30% of that of CD dimer 1) but did not show any inhibition of CS.
Apparently, the diester 1 does not acylate a protein side chain, because matrix-assisted laser desorption ionization/time of flight mass spectroscopy indicates no gain in mass for the monomer of LDH (mass = 36,644 Da; after incubation with 1, mass = 36,642 Da) or of CS (mass = 49,804 Da; after incubation with 1, mass = 49,794 Da). Thus, it seems likely that the inhibition of CS and of LDH by 1 reflects dissociation of the enzyme to inactive monomer induced by binding of 1 to the dimerization (CS) or tetramerization (LDH) sites. Evidence for such dissociation was seen with nondenaturing size exclusion chromatography for each enzyme. A distinct species with a longer retention time (lower molecular weight) than that of the multimeric forms of CS and of LDH was observed when 1 was present but not otherwise, and when isolated, this species had the molecular weight of the protein monomer on SDS/PAGE (denaturing PAGE), which was identical to the monomeric peak seen with the original protein when it is disaggregated on SDS/PAGE.
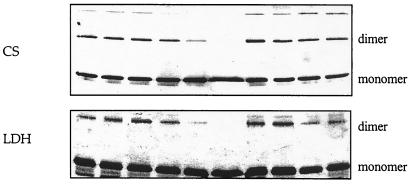
We also performed chemical cross-linking experiments. A decrease in cross-linking efficiency by reagents such as bis(sulfosuccinimidyl)suberate (BS) has been used to demonstrate the dissociation of protein dimers (19, 20). We saw (Fig. 2) a decrease in cross-linked species on SDS/PAGE (denaturing PAGE) when LDH or CS were incubated for 2 h with 1 before exposure to BS. The cross-linked species seen in lane 1 decreases almost to invisibility when the enzymes had been incubated with 1 (lanes 5 and 6), supporting the idea that 1 dissociates the normally multimeric CS and LDH. The dimethyl ester of pyridine-2,6-dicarboxylic acid had no such effect, suggesting that it inhibits LDH to a minor extent by decreasing the catalytic rate of the enzyme without disaggregating it.
Figure 2.
CD dimer 1 decreases BS cross-linking efficiency, whereas its components (β-CD, pyridine 2,6-dicarboxylic acid, and dimethyl-2,6-pyridine dicarboxylate) do not, as evidenced by SDS/PAGE of cross-linking experiments with 0.1 μM enzymes (LDH and CS): lane 1, enzyme + 100 μM BS; lane 2, enzyme + 100 μM BS; lane 3, enzyme + 100 μM BS + 77 μM CD dimer; lane 4, enzyme + 100 μM BS + 385 μM CD dimer; lane 5, enzyme + 100 μM BS + 1.5 mM CD dimer; lane 6, enzyme + 100 μM BS + 7.7 mM CD dimer; lane 7, enzyme + 100 μM BS + 15 mM β-CD; lane 8, enzyme + 100 μM BS + 7 mM pyridine 2,6-dicarboxylic acid; lane 9, enzyme + 100 μM BS + 7 mM dimethyl-2,6-pyridine dicarboxylate; lane 10, same as lane 1.
The effect of 1 on cross-linking cannot be caused by binding of the cross-linking agent by 1, because the cross-linker can be present in a 10-fold or more excess over 1 (compare Fig. 3) without affecting the results shown in Fig. 2. Of course, dimer 1 could in principle be binding to the protein near the surface lysines to block cross-linking without disaggregating the protein. Indeed, our proposal is that 1 does block cross-linking by binding to the protein but as a result of inducing protein disaggregation. Thus, the cross-linking experiment is not definitive by itself, but taken with the nondenaturing size exclusion chromatography result, the experiment supports the idea that our dimer 1 is indeed inducing protein disaggregation.
Figure 3.
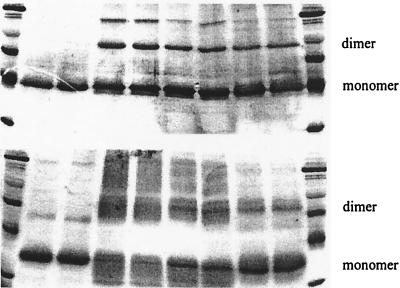
Both CD dimer 1 and dimer 5 decrease cross-linking efficiency, as evidenced by SDS/PAGE of cross-linking experiments with CS (Upper) and LDH (Lower). Lanes 1 and 10, molecular weight markers; lanes 2 and 3, enzyme alone; lanes 4 and 5, 1.0 μM enzyme + 7.0 mM BS; lanes 6 and 7, 1.0 μM enzyme + 0.7 mM dimer 5 + 7.0 mM BS; lanes 8 and 9, 1.0 μM enzyme + 0.7 mM dimer 1 + 7.0 mM BS.
Amide 5 is also apparently a disaggregating agent. As the SDS/PAGE gel in Fig. 3 shows, the inclusion of 5 greatly decreases the amount of dimeric protein captured in cross-linking experiments. However, dimer 5 was approximately three times less effective than was 1 as an inhibitor of LDH but was equally effective as 1 in inhibiting CS. We do not yet understand this difference. Nor do we understand the fact that Lineweaver–Burk plots indicate that the inhibition of LDH by 1 is apparently noncompetitive with substrate, whereas that with 5 is competitive. Of course, substrate binding to the protein monomer can induce a conformational change that favors aggregation, and this change would make the dissociation induced by 5 competitive with substrate binding.
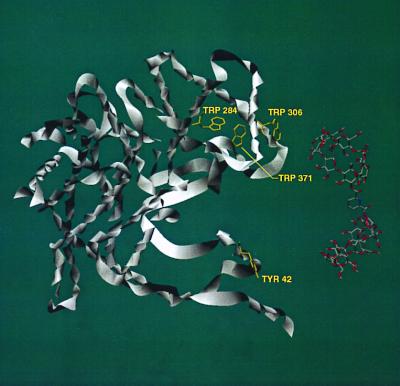
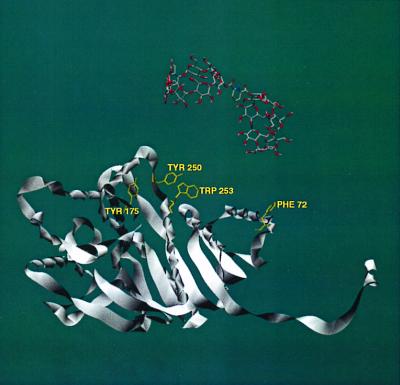
By examining the molecular structures of multimeric CS and LDH (from the Protein Data Bank, www.rcsb.org), we have identified several possible regions where binding of our dimers to protein hydrophobic residues could cause disaggregation of the proteins. In CS (Fig. 4), our CD dimers 1 and 5 have the correct geometry to bridge between Tyr-42 and Trp-284, Trp-306, or Trp-371 within the same monomer unit. In all these cases, the CD dimer would interfere with close packing of a CS dimer. In LDH (Fig. 5), our CD dimers 1 and 5 can bridge between Phe-72 and Tyr-175, Tyr-250, or Trp-253 within the same monomer unit, again intruding into the protein aggregation region. Further work is needed to make definite assignments.
Figure 4.
Crystal structure of CS. Only one monomeric unit is shown for clarity, but the geometry is that seen in the dimer of CS. A computer model of CD dimer 1 is shown in close proximity to the protein–protein interfaces. Several residues of the proteins are highlighted for their potential to bind with CD dimers 1 and 5, which would disrupt protein aggregation.
Figure 5.
Crystal structure of LDH. Only one monomeric unit is shown for clarity, but the geometry is that seen in the tetramer of LDH. A computer model of CD dimer 1 is shown in close proximity to the protein–protein interfaces. Several residues of the proteins are highlighted for their potential to bind with CD dimers 1 and 5, which would disrupt protein aggregation.
Conclusions
Other approaches to blocking protein–protein interactions have been reported recently (1, 2, 19–29), many with peptides or peptide mimics. Our work suggests that the specific binding of hydrophobic side chains by CD dimers that we had reported earlier can be extended to proteins and that it is another way to block protein aggregation. We have also made CD trimers and tetramers (30–32) with varying geometries controlled by the linkers. Thus, it will be interesting to see whether the effects reported herein can be made even stronger and more selective and whether they will have useful applications in biology and medicine.
Acknowledgments
We thank Prof. Esther Breslow (Cornell University, Ithaca, NY), Prof. Jean Chmielewski (Purdue University, West Lafayette, IN), and Prof. Sam Gellman (University of Wisconsin, Madison, WI) for helpful discussions and suggestions. We also thank Dr. Yasuhiro Itagaki (Columbia University) for assistance with the mass spectroscopy experiments. Support of this work by the National Institutes of Health and National Science Foundation is gratefully acknowledged. D.K.L. is an M.D./Ph.D. student and is grateful for the support from the National Institutes of Health Medical Scientist Training Program.
Abbreviations
- CD
cyclodextrin
- LDH
l-lactate dehydrogenase
- CS
citrate synthase
- BS
bis(sulfosuccinimidyl)suberate
References
- 1.Zutshi R, Brickner M, Chmielewski J. Curr Opin Chem Biol. 1998;2:62–66. doi: 10.1016/s1367-5931(98)80036-7. [DOI] [PubMed] [Google Scholar]
- 2.Daugherty D L, Gellman S H. J Am Chem Soc. 1999;121:4325–4333. [Google Scholar]
- 3.Bunn H F. N Engl J Med. 1997;337:762–769. doi: 10.1056/NEJM199709113371107. [DOI] [PubMed] [Google Scholar]
- 4.Pallitto M M, Ghanta J, Heinzelman P, Kiessling L L, Murphy R M. Biochemistry. 1999;38:3570–3578. doi: 10.1021/bi982119e. [DOI] [PubMed] [Google Scholar]
- 5.Breslow R, Yang Z, Ching R, Trojandt G, Odobel F. J Am Chem Soc. 1998;120:3536–3537. [Google Scholar]
- 6.Maletic M, Wennemers H, McDonald D Q, Breslow R, Still W C. Angew Chem Int Ed Engl. 1996;35:1490–1492. [Google Scholar]
- 7.Maurer P, Lessmann D, Kurz G. In: Methods in Enzymology. Wood W A, editor. Vol. 89. New York: Academic; 1982. p. 267. [DOI] [PubMed] [Google Scholar]
- 8.Blachnitzky E O, Wengenmayer F, Kurz G. Eur J Biochem. 1974;47:235–250. doi: 10.1111/j.1432-1033.1974.tb03687.x. [DOI] [PubMed] [Google Scholar]
- 9.Noltmann E A. In: The Enzymes. 3rd Ed. Boyer P D, editor. VI. New York: Academic; 1972. p. 279. [Google Scholar]
- 10.Spector L B. In: The Enzymes. 3rd Ed. Boyer P D, editor. VII. New York: Academic; 1972. p. 358. [Google Scholar]
- 11.Holbrook J J, Liljas A, Steindel S J, Rossmann M G. In: The Enzymes. Boyer P D, editor. XI. New York: Academic; 1975. pp. 199–292. [Google Scholar]
- 12.Beeckmans S, Van Driessche E. J Biol Chem. 1998;273:31661–31669. doi: 10.1074/jbc.273.48.31661. [DOI] [PubMed] [Google Scholar]
- 13.Lindstad R I, McKinley-McKee J S. FEBS Lett. 1997;408:57–61. doi: 10.1016/s0014-5793(97)00372-4. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Quiocho F A. Biochemistry. 1998;37:8314–8324. doi: 10.1021/bi980324o. [DOI] [PubMed] [Google Scholar]
- 15.Hanessian S, Benalil A, Laferriere C. J Org Chem. 1995;60:4786–4797. [Google Scholar]
- 16.Chen W H, Hayashi S, Tahara T, Nogami Y, Koga T, Fujita K. Chem Pharm Bull. 1999;47:588–589. doi: 10.1248/cpb.47.588. [DOI] [PubMed] [Google Scholar]
- 17.Ueno A, Moriwaki F, Osa T, Hamada F, Murai K. Tetrahedron. 1987;43:1571–1578. [Google Scholar]
- 18.Bergmeyer H U. Methods of Enzymatic Analysis. 3rd Ed. II and III. Deerfield Beach, FL: Verlag Chemie; 1983. [Google Scholar]
- 19.Zutshi R, Franciskovich J, Shultz M, Schweitzer B, Bishop P, Wilson M, Chmielewshi J. J Am Chem Soc. 1997;119:4841–4845. [Google Scholar]
- 20.Ghosh I, Chmielewski J. Chem Biol. 1998;5:439–445. doi: 10.1016/s1074-5521(98)90160-0. [DOI] [PubMed] [Google Scholar]
- 21.Hamuro Y, Calama M C, Park H, Hamilton A D. Angew Chem Int Ed Engl. 1997;36:2680–2683. [Google Scholar]
- 22.Alberg D G, Schreiber S L. Science. 1993;262:248–250. doi: 10.1126/science.8211144. [DOI] [PubMed] [Google Scholar]
- 23.Tauton J, Collins J L, Schreiber S L. J Am Chem Soc. 1996;118:10412–10422. [Google Scholar]
- 24.Park H, Lin Q, Hamilton A D. J Am Chem Soc. 1999;121:8–13. [Google Scholar]
- 25.Prasanna V, Bhattacharjya S, Balaram P. Biochemistry. 1998;37:6883–6893. doi: 10.1021/bi9720989. [DOI] [PubMed] [Google Scholar]
- 26.Tiley J W, Chen L, Fry D C, Emerson S D, Powers G D, Biondi D, Varnell T, Trilles R, Guthrie R, Mennona F, et al. J Am Chem Soc. 1997;119:7589–7590. [Google Scholar]
- 27.Judice J K, Tom J Y K, Huang W, Wrin T, Vennari J, Petropoulos C J, McDowell R S. Proc Natl Acad Sci USA. 1997;94:13426–13430. doi: 10.1073/pnas.94.25.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan X, Flentke G R, Rich D H. J Am Chem Soc. 1998;120:8893–8894. [Google Scholar]
- 29.Gamboni S, Chaperon C, Friedrich K, Baehler P J, Reymond C D. Biochemistry. 1998;37:12189–12194. doi: 10.1021/bi980028b. [DOI] [PubMed] [Google Scholar]
- 30.Breslow R, Zhang X, Xu R, Maleic M, Merger R. J Am Chem Soc. 1996;118:11678–11681. [Google Scholar]
- 31.Breslow R, Huang Y, Zhang X, Yang J. Proc Natl Acad Sci USA. 1997;94:11156–11158. doi: 10.1073/pnas.94.21.11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breslow R, Gabriele B, Yang J. Tetrahedron Lett. 1998. , 2887–2890. [Google Scholar]