Summary
Impaired mitochondrial function has been implicated in the pathogenesis of type 2 diabetes, heart failure and neurodegeneration as well as during aging. Studies with the PGC-1 transcriptional coactivators have demonstrated that these factors are central components of the regulatory network that controls mitochondrial function in mammalian cells. Here we describe a genome-wide coactivation assay to globally identify transcription factors and cofactors in this pathway. These analyses revealed a molecular signature of the PGC-1α transcriptional network and identified BAF60a (SMARCD1) as a molecular link between the SWI/SNF chromatin-remodeling complexes and hepatic lipid metabolism. Adenoviral-mediated expression of BAF60a stimulates fatty acid β-oxidation in cultured hepatocytes and ameliorates hepatic steatosis in vivo. PGC-1α mediates the recruitment of BAF60a to PPARα binding sites, leading to transcriptional activation of peroxisomal and mitochondrial fat oxidation genes. These results define a role for the SWI/SNF complexes in the regulation of lipid homeostasis.
Introduction
Metabolic syndrome is emerging as a global epidemic and is characterized by clustering of several disorders, including obesity, type 2 diabetes, fatty liver, and dyslipidemia (Flier, 2004; Zimmet et al., 2001). Ectopic accumulation of triglycerides (TG) in the liver is an early pathogenic event in the development of nonalcoholic steatohepatitis (NASH), which is characterized by chronic inflammation and liver damage. Hepatic steatosis also contributes to the pathogenesis of insulin resistance and influences systemic glucose metabolism. The liver regulates several major aspects of lipid metabolism, including lipogenesis, fatty acid β-oxidation (FAO) as well as lipoprotein uptake and secretion. These pathways are under the control of nutritional and hormonal signals to maintain hepatic and systemic fuel homeostasis. In particular, hepatic fat oxidation is enhanced in the fasted state as a result of increased availability of fatty acids and the transcriptional activation of genes involved in peroxisomal and mitochondrial FAO (Eaton, 2002; Reddy and Hashimoto, 2001).
The transcriptional regulatory networks comprise transcription factors (TFs) and cofactors that are stably or transiently assembled into multiprotein complexes. These physical interactions form the basis for robust yet highly dynamic networks that control gene expression. Recent studies have demonstrated that transcriptional coactivators play an important role in the regulation of diverse biological programs, including mitochondrial oxidative phosphorylation (OXPHOS), smooth muscle differentiation, and B-cell development (Christian et al., 2006; Feige and Auwerx, 2007; Spiegelman and Heinrich, 2004). PPARγ Coactivator-1α (PGC-1α) is an inducible coactivator for nuclear hormone receptors and other TFs and belongs to a small family of coactivators that includes PGC-1β and PGC-1 related coactivator (Finck and Kelly, 2006; Lin et al., 2005a). PGC-1α regulates several major metabolic pathways, including mitochondrial OXPHOS (Lehman et al., 2000; St-Pierre et al., 2003; Wu et al., 1999), hepatic gluconeogenesis (Herzig et al., 2001; Yoon et al., 2001), and reactive oxygen species metabolism (St-Pierre et al., 2006; Valle et al., 2005). This factor also regulates circadian clock function, slow-twitch muscle fiber formation and heme homeostasis (Handschin et al., 2005; Lin et al., 2002; Liu et al., 2007). Mice deficient in PGC-1α have impaired mitochondrial function in several tissues and develop spongiform neurodegeneration (Leone et al., 2005; Lin et al., 2004). These observations suggest that PGC-1α is a key component of the transcriptional regulatory network that maintains energy homeostasis in mammals.
PGC-1α exerts its effects on gene expression through recruiting chromatin-remodeling complexes to its transcriptional partners, leading to a switch in chromatin structure to an active state. Although several TFs and cofactors have been found to mediate the effects of PGC-1α on energy metabolism, a comprehensive list of transcriptional partners for PGC-1α remains not available. We therefore reasoned that global identification of transcriptional partners for PGC-1α would reveal the architecture of an important metabolic regulatory network. The present studies were undertaken to develop a genome-wide high-throughput coactivation screen and to examine the biological role of PGC-1α partners in metabolic regulation. Our analyses revealed a set of TFs and cofactors that physically interact with PGC-1α and identified BAF60a as a key regulator of hepatic lipid homeostasis.
Results
Identification of Transcriptional Partners for PGC-1α Through Genome-wide Coactivation Screen
PGC-1α stimulates the transcriptional activity of its cognate TF partners when they are fused to a heterologous DNA-binding domain, such as the Gal4 DNA-binding domain (GalDBD) (Knutti et al., 2000; Vega et al., 2000), suggesting that a coactivation screen based on this reporter system could uncover biologically relevant TF partners for PGC-1α. To develop a platform for a genome-wide coactivation assay, we first compiled a non-redundant list of human TF and cofactor genes based on three recent studies on global TF identification (Gray et al., 2004; Kanamori et al., 2004; Kummerfeld and Teichmann, 2006) (Table S1). This list includes 2,385 genes encoding DNA-binding proteins and transcriptional cofactors. Among them, 1,146 are present in the Human ORFeome v3.1, a collection of over 12,212 sequence-validated ORF clones in Gateway vector (Lamesch et al., 2007; Walhout et al., 2000). We arrayed the ORF clones encoding these transcriptional regulators in 96-well plates to generate a Transcription Factor ORF Collection (TFORC), which covers 48% of all predicted transcription factors and cofactors in the human genome. TFORC contains all major classes of transcriptional regulators that closely mirror their representation in the genome (Table S2).
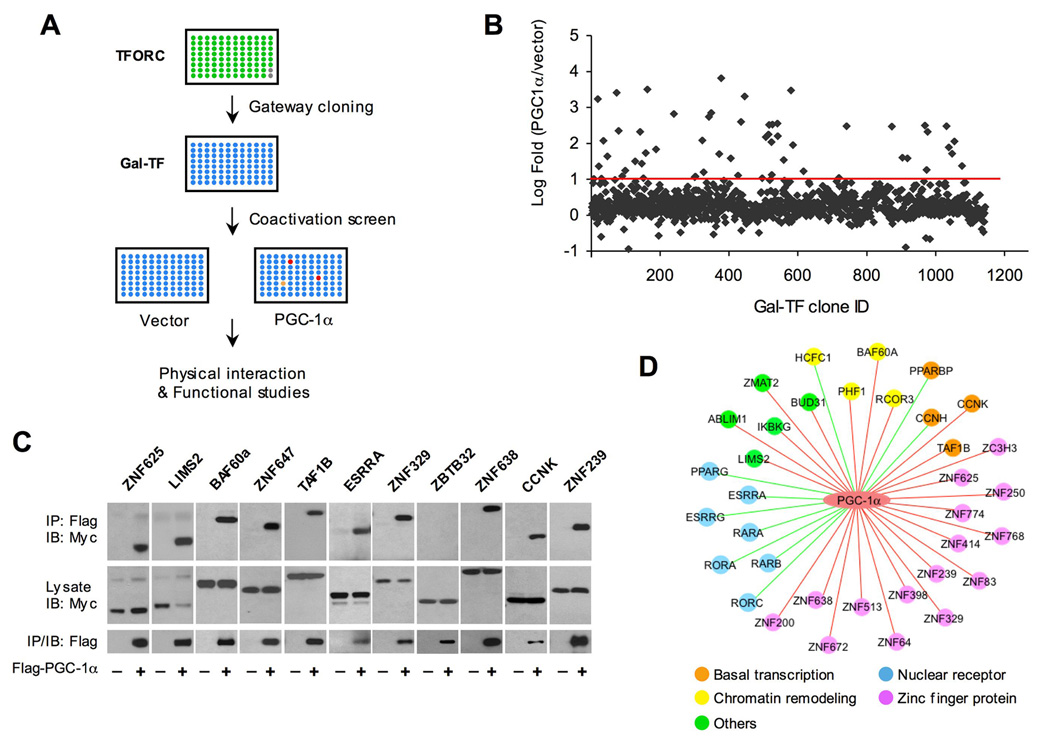
We next fused each of the 1,146 TF ORFs to the GalDBD vector using the Gateway cloning system to generate Gal-TF plasmids for coactivation studies. BOSC cells seeded in 96-well plates were transiently transfected with UAS-luciferase and individual Gal-TF plasmids in the presence or absence of PGC-1α (Figure 1A). This cell line has been used for assaying coactivation of TFs by PGC-1α in previous studies and is suitable for high-throughput reporter assays because of high transfection efficiency (Handschin et al., 2007; Yoon et al., 2001). Independent transfection experiments using Gal-TF from the same plate indicate that although absolute luciferase values vary among experiments, fold-coactivation by PGC-1α for individual factors is remarkably consistent (data not shown). We identified 40 factors whose transcriptional activity could be augmented by PGC-1α more than 10-fold in these coactivation reporter assays (Figure 1B). PGC-1α exerts its effects on transcription factors and cofactors through physical interactions. As such, we performed coimmunoprecipitation (CoIP) studies as a secondary screen to identify bona fide partners for PGC-1α (Figure 1C). A total of 35 TF and cofactors scored positive in both coactivation and CoIP assays, including 10 factors previously reported to function in concert with PGC-1α (Figure 1D). The lack of physical interaction for several candidate factors is likely due to low levels of fusion protein expression and/or the transient nature of interactions that may be lost during immunoprecipitation. The identification of known PGC-1α targets (Figure 1D, green edges) validates our approach and suggests that many of the newly identified factors are likely biologically relevant partners for PGC-1α.
Figure 1. Identification of Transcriptional Partners for PGC-1α Through Genome-wide Coactivation Screen.
(A) Schematic diagram of high-throughput coactivation screen of PGC-1α.
(B) Log-transformed fold-coactivation by PGC-1α for individual Gal-TF clones. Red line denotes 10-fold increase in luciferase activity by PGC-1α.
(C) Representative graphs of CoIP between PGC-1α and 11 putative TF partners. Immunoblotting was performed on total lysates and immunoprecipitated complexes from BOSC cells transiently transfected with Myc-TF with pcDNA3 vector or Flag-PGC-1α.
(D) Schematic diagram of the PGC-1α transcriptional regulatory network. Previously reported (green line) and new factors identified in this study (red line) are indicated.
Among the factors coactivated by PGC-1α are seven nuclear hormone receptors, including estrogen-receptor related receptor α (ERRα), ERRγ, RAR-related orphan receptor α (RORα) and RORγ. ERRα and ERRγ have been demonstrated to regulate the expression of nuclear-encoded mitochondrial genes (Huss et al., 2002; Mootha et al., 2004; Schreiber et al., 2004), whereas RORα mediates the effects of PGC-1α on metabolic and clock gene expression (Lau et al., 2004; Liu et al., 2007). In addition to nuclear receptors, PGC-1α interacts with 15 zinc-finger proteins, whose biological function remains largely unknown. PGC-1α is known to recruit other chromatin-remodeling proteins, including histone acetyltransferase (CBP/p300 and GCN5) and deacetylase complexes (Lerin et al., 2006; Puigserver et al., 1999; Rodgers et al., 2005). Several factors implicated in chromatin remodeling were uncovered in our screen, including host cell factor C1 (HCFC1) and BRG1-associated factor 60a (BAF60a). HCFC1 has been demonstrated to associate with histone methyltransferase (Wysocka et al., 2003), while BAF60a is a subunit of the SWI/SNF nucleosome-remodeling complexes (Wang et al., 1996). Together, these results suggest that PGC-1α may exert diverse effects on chromatin structure through nucleosome remodeling and histone modifications.
BAF60a Induces Peroxisomal and Mitochondrial FAO Genes
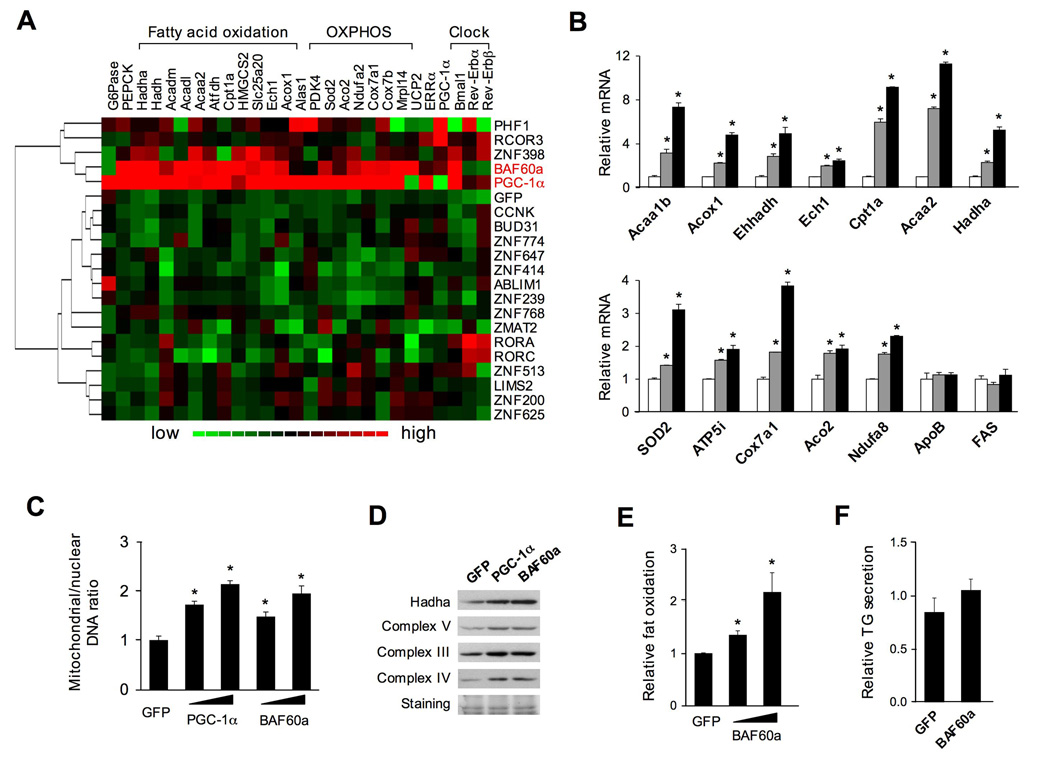
To explore the biological activities of these PGC-1α partners in regulating glucose and lipid metabolism, we generated 19 recombinant adenoviruses each expressing a different TF. We transduced cultured primary hepatocytes with adenoviruses expressing GFP, PGC-1α or individual factors and perform quantitative realtime PCR (qPCR) analyses to examine their effects on known PGC-1α target genes (see the list in Table S3). As expected, PGC-1α strongly induces mRNA expression of genes involved in fatty acid β-oxidation, mitochondrial OXPHOS, gluconeogenesis, and clock function (Figure S1). Clustering analysis of the expression data indicates that BAF60a and PGC-1α have nearly identical activities in the induction of this set of genes (Figure 2A), whereas other factors examined have modest effects. BAF60a is a subunit of the SWI/SNF complexes, which comprise a core ATPase (BRG1 or BRM) and several BRG1-associated factors (BAF), including BAF170, BAF155, BAF60, BAF57, and BAF53a (Martens and Winston, 2003; Wang et al., 1996). The SWI/SNF complexes regulate the transcription of target genes through binding to specific TFs and altering local chromatin structure. Recent studies have implicated the BAF60 family members, including BAF60a, BAF60b and BAF60c, in mediating the interaction between the SWI/SNF complexes and target TFs. For example, BAF60a interacts with glucocorticoid receptor (GR) and c-Jun (Hsiao et al., 2003; Ito et al., 2001), whereas BAF60c binds to several nuclear receptors, including PPARγ, estrogen receptor α (ERα), and RORα (Debril et al., 2004). BAF60a mRNA is present in several tissues where PGC-1α is also expressed, including brain, skeletal muscle, and the liver (data not shown). While BAF60c plays a role in the regulation of heart development and muscle differentiation (Lickert et al., 2004; Simone et al., 2004), the biological function of BAF60a remains largely unknown.
Figure 2. Regulation of Fatty Acid β-oxidation by BAF60a in Primary Hepatocytes.
(A) Clustering analysis of gene expression in primary hepatocytes transduced with adenoviruses expressing GFP, PGC-1α, or individual TFs. Relative mRNA expression of genes involved in hepatic gluconeogenesis, fatty acid β-oxidation, mitochondrial OXPHOS, and circadian clock were analyzed using qPCR.
(B) qPCR analysis of gene expression in hepatocytes transduced with GFP (open) or BAF60a adenoviruses at multiplicity of infection of 1 (grey) or 3 (filled).
(C) qPCR analysis of relative mitochondrial/nuclear DNA content.
(D) Immunoblotting analysis of transduced primary hepatocytes.
(E–F) Relative rate of fatty acid oxidation (E) and secretion of TG-containing lipoproteins (F) in transduced hepatocytes. All data represent mean ± stdev. *p<0.05.
Further analyses indicate that BAF60a induces the expression of genes involved in peroxisomal fatty acid β-oxidation in a dose-dependent manner, including acetyl-Coenzyme A acyltransferase 1B (Acaa1b), palmitoyl acyl-Coenzyme A oxidase 1 (Acox1), L-bifunctional enzyme (Ehhadh), and enoyl Coenzyme A hydratase 1(Ech1) (Figure 2B). BAF60a also induces the expression of mitochondrial FAO genes, such as carnitine palmitoyl-CoA transferase 1 (Cpt1a), acetyl-CoA acyltransferase 2 (Acaa2), and hydroxyacyl-CoA dehydrogenase (Hadha). Compared to control, the expression of several nuclear-encoded mitochondrial genes, such as superoxide dismutase 2 (SOD2) and subunits of the electron transport chain, is also increased by BAF60a. In contrast, the mRNA levels of fatty acid synthase (FAS) and apolipoprotein B (ApoB) remain similar in transduced hepatocytes, suggesting that BAF60a exerts specific effects on downstream target genes. Similar to PGC-1α, BAF60a increases mitochondrial DNA content and the expression levels of several proteins of the respiratory complexes (Figure 2C–D). To determine whether BAF60a enhances fatty acid β-oxidation, we incubated transduced hepatocytes in the presence of 3H-labeled palmitate and measured 3H2O release. Consistent with gene expression data, adenoviral-mediated expression of BAF60a dose-dependently increases fat oxidation rate (Figure 2E). In contrast, BAF60a does not alter the secretion of TG-containing lipoproteins in transduced primary hepatocytes (Figure 2F).
BAF60a Reduces Liver Triglycerides in High-fat Fed Mice
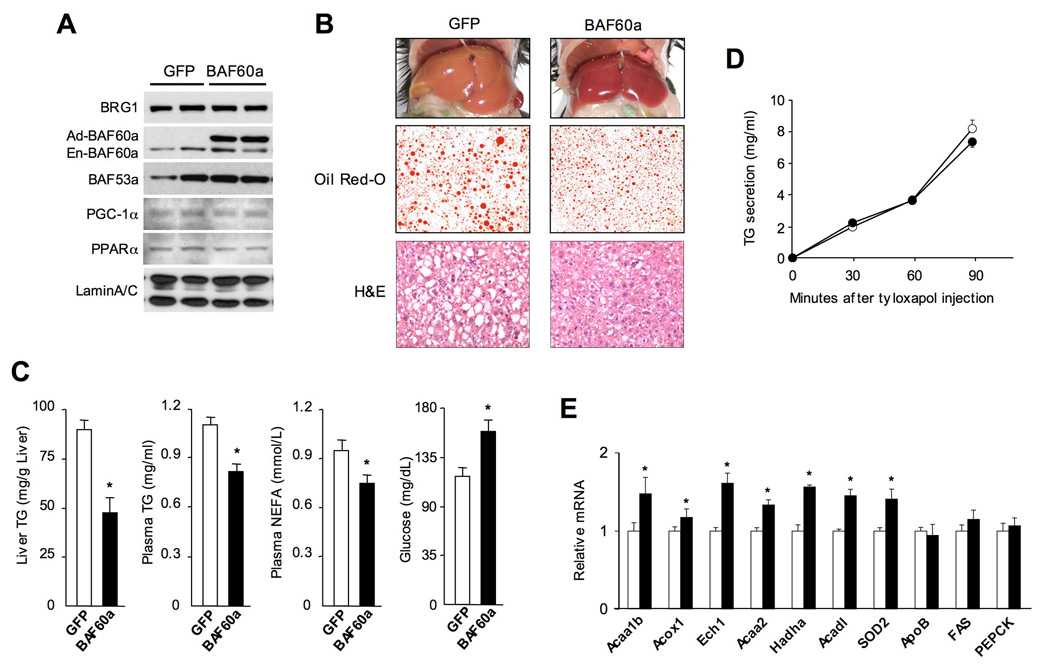
Fatty acid β-oxidation is an important component of hepatic lipid homeostasis. Hepatic steatosis results from imbalance of fat availability and clearance in the liver and contributes to the pathogenesis of insulin resistance and NASH in obesity. Because BAF60a stimulates FAO gene expression and fat oxidation in hepatocytes, it is possible that this factor may regulate hepatic TG content through affecting lipid catabolism. To determine the function of BAF60a in vivo, we transduced high-fat fed obese mice with GFP or BAF60a adenoviruses through tail vein injection. Immunoblotting analysis of nuclear extracts from transduced livers indicates that BAF60a protein levels were elevated by an approximately three to five-fold over the endogenous levels (Figure 3A). The protein levels of BRG1, BAF53a, PGC-1α, and PPARα remain similar in both groups. Livers from BAF60a-transduced mice appear red and are smaller than those from control mice (Figure 3B). In fact, liver weight to body weight ratio is reduced by approximately 18% in BAF60a-transduced mice. Measurement of total hepatic TG content indicates that BAF60a expression leads to 45% decrease in TG (Figure 3C). Consistent with these results, lipid droplets are significantly smaller in the livers expressing BAF60a, as revealed by Oil Red O and H&E staining. Similar effects on hepatic TG content were also observed in ob/ob mice transduced with BAF60a adenovirus (data not shown).
Figure 3. Adenoviral-mediated Expression of BAF60a Lowers Hepatic TG Content.
(A) Immunoblots of liver nuclear extracts from transduced mice using indicated antibodies. The expression of endogenous (En-BAF60a) and adenoviral (Ad-BAF60a) BAF60a proteins is indicated.
(B) Liver morphology and histological staining of liver sections from mice six days after tail vein injection.
(C) Liver TG, plasma lipid and glucose concentrations in mice transduced with GFP or BAF60a adenoviruses.
(D) Increase in plasma TG following intravenous injection of tyloxapol (500 mg/kg) in mice transduced with GFP (open circle) or BAF60a (filled circle) adenoviruses.
(E) qPCR analysis of hepatic gene expression in mice transduced with GFP (open) or BAF60a (filled) adenoviruses. Data in C–E represent mean ± SEM, n=5. *p<0.05.
To rule out the possibility that BAF60a may lower liver TG through stimulation of lipoprotein secretion, we measured the concentrations of plasma TG and non-esterified fatty acids in transduced mice. Compared to GFP control mice, BAF60a lowers plasma TG and fatty acid concentrations by 29% and 20% in high-fat fed mice, respectively (Figure 3C). Blood glucose levels are slightly increased in response to BAF60a. To directly measure the rate of VLDL secretion, we injected tyloxapol, an inhibitor of plasma lipases, into transduced mice and measured the increase in plasma TG concentrations. No significant difference in VLDL secretion was observed (Figure 3D). Consistently, BAF60a has modest effect on the expression of genes involved in lipoprotein metabolism, including apolipoprotein (apo) AV, apoB and apoCIII. In contrast, BAF60a significantly increases mRNA expression of genes involved in peroxisomal and mitochondrial fatty acid oxidation (Figure 3E). The mRNA levels of PEPCK remain unaffected by BAF60a. Taken together, our results strongly suggest that BAF60a ameliorates hepatic steatosis through activation of fatty acid oxidation.
Role of PGC-1α in the Induction of FAO Genes by BAF60a
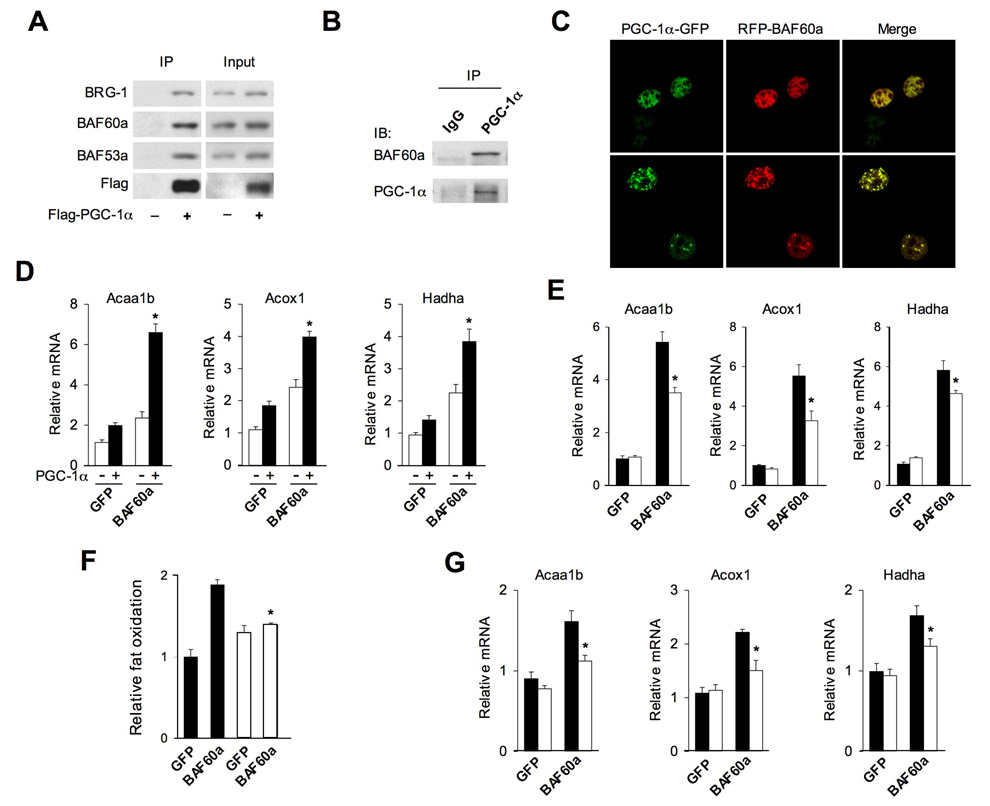
BAF60a is a core subunit of the SWI/SNF chromatin-remodeling complexes that activate or repress the transcription of diverse target genes. Our studies indicate that BAF60a and PGC-1α physically interact in the cell (Figure 1C). The surprisingly specific transcriptional effects of BAF60a raise the possibility that it may regulate FAO gene expression through directing the SWI/SNF complexes to PGC-1α. To further determine whether PGC-1α associates with endogenous BAF60a and other SWI/SNF subunits, we performed coimmunoprecipitation (CoIP) assays in BOSC cells transiently transfected with Flag-tagged PGC-1α We found that, in addition to BAF60a, PGC-1α is also associated with BRG1 and BAF53a (Figure 4A). The presence of BAF53a in the PGC-1α protein complex was also observed in previous affinity purification studies (Lerin et al., 2006). CoIP experiments with liver nuclear extracts indicate that BAF60a associates with PGC-1α in vivo (Figure 4B). Additionally, confocal microscopy studies revealed that PGC-1α and BAF60a colocalize to nuclear speckles as well as to the nucleoplasm in the cell (Figure 4C).
Figure 4. Physical and Functional Interaction Between BAF60a and PGC-1α.
(A) Immunoblots of total lysates or immunoprecipitated proteins from BOSC cells transiently transfected with pcDNA3 vector or Flag-PGC-1α plasmids. Note that PGC-1α associates with endogenous BAF60a, BRG-1, and BAF53a.
(B) Immunoblots of precipitated liver nuclear complexes using IgG and PGC-1α antibodies.
(C) Confocal microscopy images of H2.35 hepatoma cells transiently transfected with GFP-PGC-1α and RFP-BAF60a plasmids.
(D) qPCR analysis of gene expression in primary hepatocytes transduced with adenoviruses expressing GFP, BAF60a, PGC-1α, or the combination of BAF60a and PGC-1α. Data represent mean ± stdev. *p<0.01.
(E) qPCR analysis of gene expression in wild type (filled) and PGC-1α null (open) hepatocytes transduced with GFP or BAF60a adenoviruses, as indicated.
(F) FAO rate in transduced hepatocytes isolated from wild type (filled) and PGC-1α null (open) mice.
(G) qPCR analysis of gene expression in wild type (filled) and PGC-1α null (open) mouse livers transduced with GFP or BAF60a adenoviruses. Data in (E–G) represent mean ± stdev. *p<0.05 WT vs. PGC-1α null.
We next examined whether BAF60a and PGC-1α functionally interact in the induction of FAO genes in hepatocytes. While both BAF60a and PGC-1α increase mRNA levels of several FAO genes, including Acaa1b, Acox1 and Hadha, simultaneous expression of these two factors leads to significantly higher induction of target genes (Figure 4D). To determine whether PGC-1α is required for the induction of FAO genes by BAF60a, we isolated primary hepatocytes from wild type and PGC-1α null mice and transduced these hepatocytes with GFP or BAF60a adenoviruses. qPCR analysis indicates that BAF60a increases FAO gene expression in both genotypes. However, the stimulatory effect of BAF60a on these genes is significantly blunted in hepatocytes lacking PGC-1α (Figure 4E). Further, the increase in fat oxidation in response to BAF60a is significantly reduced in PGC-1α null hepatocytes (Figure 4F). Consistent with these findings, BAF60a induction of FAO genes is also diminished in PGC-1α null mouse livers (Figure 4G). We conclude from these studies that PGC-1α is necessary for the normal function of BAF60a in the regulation of FAO gene expression and fat oxidation.
Crosstalk Between BAF60a and PPARα
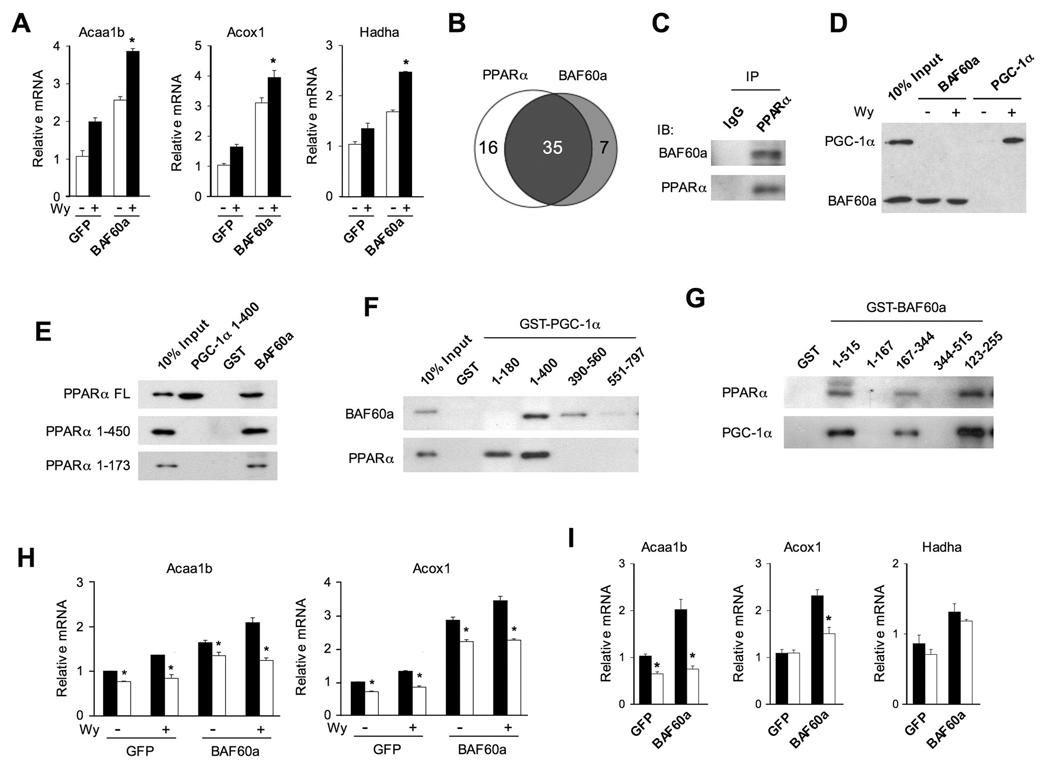
Transcriptional activation of peroxisomal and mitochondrial fat oxidation genes by BAF60a is reminiscent of PPARα activation. Previous studies have demonstrated that PPARα is a key transcriptional regulator of FAO genes and is essential for hepatic fat oxidation during starvation (Hashimoto et al., 2000; Kersten et al., 1999). In addition, PGC-1α physically interacts with PPARα and augments its transcriptional activity (Vega et al., 2000). To determine whether BAF60a and PPARα cooperatively regulate FAO genes, we treated transduced hepatocytes with vehicle (DMSO) or Wy14,643, a synthetic agonist for PPARα. Gene expression analysis indicates that the induction of Acaa1b, Acox1, and Hadha by BAF60a is significantly augmented in the presence of Wy14,643 (Figure 5A), suggesting that BAF60a and PPARα functionally crosstalk in the regulation of FAO gene transcription. Previous transcriptional profiling studies have demonstrated that Wy14,643 induces the expression of many peroxisomal and mitochondrial fat oxidation genes in cultured hepatocytes (Gene Expression Omnibus dataset GSE8302). To determine the extent to which BAF60a regulates the expression of PPARα target genes, we performed microarray analysis on primary hepatocytes transduced with GFP or BAF60a adenoviruses. Analysis of these datasets reveals that PPARα and BAF60a induce the expression of 51 and 42 FAO genes by more than 1.6-fold, respectively (Figure 5B). Among these, 35 FAO genes are induced by both factors (see the list in Table S4), suggesting that they likely function in a common transcriptional pathway.
Figure 5. Crosstalk Between PPARα and BAF60a in the Regulation of Hepatic Fat Oxidation.
(A) qPCR analysis of gene expression in hepatocytes transduced with GFP or BAF60a adenoviruses followed by vehicle (−) or 10 µM Wy14,643 (+) treatments. Data represent mean ± stdev. *p<0.05 Wy14,643 vs. vehicle.
(B) Venn diagram representation of FAO genes that are induced more than 1.6-fold by BAF60a or Wy14,643 treatment in hepatocytes.
(C) Immunoblots of precipitated liver nuclear complexes using IgG and PPARα antibodies.
(D) Immunoblots of Flag-tagged BAF60a and PGC-1α. GST-PPARα fusion protein was incubated with in vitro transcribed and translated Flag-BAF60a or Flag-PGC-1α in the absence or presence of 20 µM Wy14,643.
(E) Immunoblots of Flag-tagged full length (FL) or truncated PPARα proteins. Recombinant GST, GST-PGC-1α (1–400) or GST-BAF60a proteins were incubated with in vitro transcribed and translated PPARα followed by immunoblotting analysis.
(F) Immunoblots of Flag-tagged BAF60a and PPARα. GST fusion proteins containing various domains of PGC-1α were incubated with in vitro transcribed and translated Flag-BAF60a or Flag-PPARα followed by immunoblotting analysis.
(G) Immunoblots of Flag-tagged PPARα and PGC-1α. GST fusion proteins containing various domains of BAF60a were incubated with in vitro transcribed and translated Flag-PPARα or Flag-PGC-1α followed by immunoblotting analysis.
(H) qPCR analysis of gene expression in wild type (filled) or PPARα null (open) hepatocytes transduced with GFP or BAF60a adenoviruses, as indicated. Data represent mean ± stdev. *p<0.05 WT vs. PPARα null.
(I) qPCR analysis of gene expression in wild type (filled) or PPARα null (open) mouse livers transduced with GFP or BAF60a adenoviruses. Data represent mean ± stdev. *p<0.05 WT vs. PPARα null.
BAF60a has been reported to bind to nuclear receptors and other transcription factors; however, whether BAF60a interacts with PPARα remains unknown. To test this possibility, we examined physical association between BAF60a and PPARα in the liver. CoIP studies indicate that these two factors are present in common protein complexes (Figure 5C). By incubating in vitro transcribed and translated Flag-BAF60a with glutathione-S-transferase (GST)-PPARα fusion protein, we found that PPARα binds to BAF60a in the absence of Wy14,643, in contrast to its ligand-dependent interaction with PGC-1α (Figure 5D) (Vega et al., 2000). Consistent with this observation, while PGC-1α fails to interact with PPARα lacking AF2 domain (a.a. 1–450), BAF60a associates with PPARα even when the ligand-binding and AF2 domains are deleted (Figure 5E), suggesting that PGC-1α and BAF60a interact with PPARα through distinct domains. As expected, the N-terminus of PGC-1α (a.a. 1–180), which contains conserved LXXLL motifs for docking nuclear receptors, is sufficient for binding PPARα. However, this domain fails to bind BAF60a (Figure 5F). On the contrary, a region between amino acids 180–560 of PGC-1α appears to interact with BAF60a. While BAF60a contains two canonical LXXLL motifs (a.a 301–305 and 361–365), they appear to be dispensable for its interaction with PPARα and PGC-1α (Figure 5G). Instead, in vitro protein interaction assays demonstrate that PGC-1α and PPARα binding involves a region located within a.a. 123–255 of BAF60a. The exact protein interaction surface for PGC-1α and PPARα within this region is currently unknown.
To determine whether PPARα is required for the induction of FAO genes by BAF60a, we transduced wild type and PPARα null primary hepatocytes with GFP or BAF60a adenoviruses. Gene expression analysis indicates that BAF60a robustly increases the expression of Acaa1b and Acox1 in wild type hepatocytes (Figure 5H). In contrast, the induction of these genes is significantly blunted, but not abolished, in PPARα deficient cells. Similar findings were observed when transduced hepatocytes were treated with Wy14,643. We next examined the effect of BAF60a on FAO gene expression in wild type and PPARα null mouse livers following adenoviral transduction. Compared with wild type control, the induction of Acaa1b and Acox1, but not Hadha, in response to BAF60a is significantly impaired in PPARα null mouse livers (Figure 5I). These results indicate that PPARα is a key transcription factor that mediates the induction of some, but not all, FAO genes by BAF60a.
PGC-1α Augments the Recruitment of BAF60a to PPARα Binding Sites
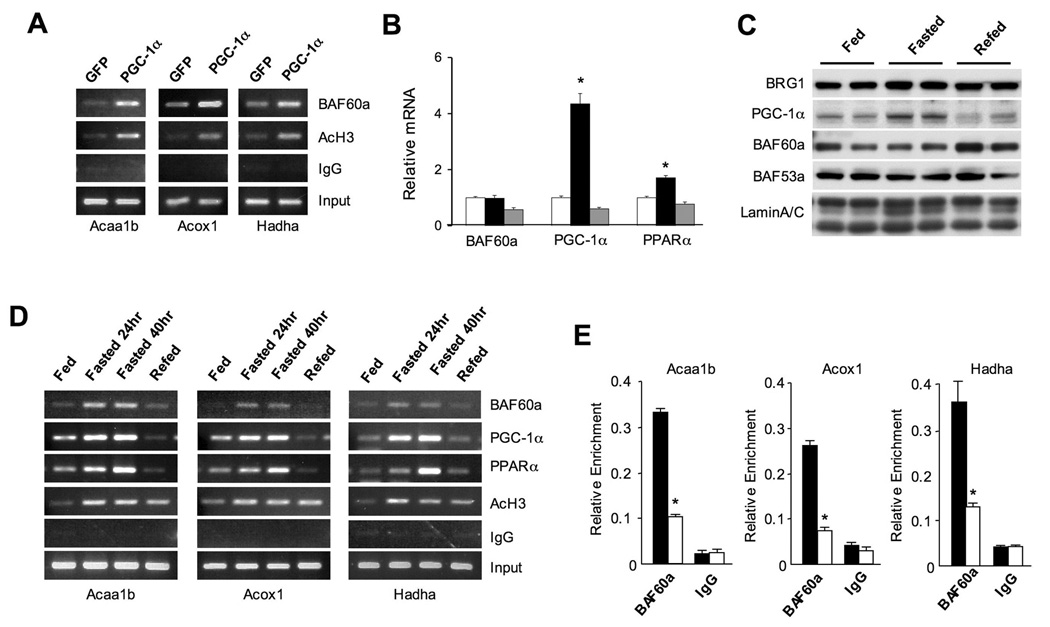
PGC-1α has been previously demonstrated to interact with and coactivate PPARα in FAO gene regulation (Finck et al., 2006; Vega et al., 2000). As described above, PGC-1α interacts with endogenous BAF60a and other subunits of the SWI/SNF complexes. To determine whether PGC-1α enhances the recruitment of BAF60a to the PPARα binding sites (PPREs), we performed chromatin immunoprecipitation (ChIP) assays in H2.35 mouse hepatoma cells transduced with GFP or PGC-1α adenoviruses. As expected, PGC-1α increases acetyl histone H3 levels in the proximity of PPREs present on Acaa1b, Acox1 and Hadha gene promoters in H2.35 hepatoma cells (Figure 6A). Compared to GFP control, the association of endogenous BAF60a with these regulatory regions is significantly enhanced by adenoviral-mediated expression of PGC-1α. These results are consistent with protein-protein interaction data and suggest that PGC-1α may promote the formation of activating transcriptional complexes that include PPARα, PGC-1α and BAF60a on the FAO promoters.
Figure 6. Recruitment of BAF60a to FAO Genes is Induced by Fasting.
(A) ChIP assay using BAF60a or AcH3 antibodies in H2.35 cells transduced with GFP or PGC-1α adenoviruses. PCR primers flanking PPRE present in the promoters of Acaa1b, Acox1, and Hadha were used.
(B) qPCR analysis of BAF60a, PGC-1α, and PPARα expression in fed (open), fasted (24 hrs, filled), and fasted/refed (24/20 hrs, grey) mouse livers. Data represent mean ± SEM, n=4. *p<0.05.
(C) Immunoblots of liver nuclear extracts using indicated antibodies.
(D) ChIP assays with chromatin extracts prepared from fed, fasted and refed mouse livers using indicated antibodies. PCR primers are the same as in (A).
(E) ChIP-qPCR assay with chromatin lysates prepared from 24-hr fasted wild type (filled) or PGC-1α null (open) mouse livers using BAF60a antibody. Data represent mean ± stdev, *p<0.01 wild type vs. PGC-1α null.
The induction of hepatic fat oxidation is an important aspect of metabolic adaptation in the liver in response to starvation. The increase in fat oxidation in the fasting state is accompanied by transcriptional activation of peroxisomal and mitochondrial FAO genes. To determine whether BAF60a expression is regulated by nutritional status, we examined its mRNA and protein levels in the livers from fed, fasted or fasted/refed mice. As expected, mRNA levels of PPARα and PGC-1α are increased in the liver during fasting (Figure 6B). To our surprise, BAF60a mRNA expression remains largely unchanged under these feeding conditions. Immunoblotting analyses of liver nuclear extracts indicate that PGC-1α protein level is increased in the fasted liver, while endogenous BAF60a protein levels are not altered by starvation (Figure 6C). In addition, protein levels of other subunits of the SWI/SNF chromatin-remodeling complex, including BRG1 and BAF53a, remain similar in all feeding conditions.
Because PGC-1α enhances the recruitment of BAF60a to the PPARα binding sites on the promoters of FAO genes, we next examined whether the recruitment of BAF60a to PPREs is regulated by feeding status. We performed ChIP assays using chromatin extracts from fed, fasted, or fasted/refed mouse livers. Compared to the fed mice, the association of BAF60a with Acaa1b, Acox1 and Hadha promoters is significantly increased following 24- or 40-hr fasting (Figure 6D). Upon refeeding, BAF60a dissociates from each of these promoters. Fasting-induced recruitment of BAF60a occurs concomitantly with enhanced association of PPARα and PGC-1α with these chromatin loci. In addition, the levels of acetylated histone are also elevated in the fasted livers, coinciding with the induction of these genes. To determine whether PGC-1α is required for the recruitment of BAF60a during starvation, we performed ChIP assays in wild type and PGC-1α null mouse livers following 24-hr fasting. ChIP-qPCR analysis of promoter occupancy by BAF60a indicates that the recruitment of BAF60a to FAO gene promoters is significantly impaired in PGC-1α null livers (Figure 6E). These data are consistent with a role for PGC-1α in mediating the recruitment of BAF60a to its target chromatin loci in the liver.
BAF60a is Required for Hepatic Fat Oxidation During Starvation
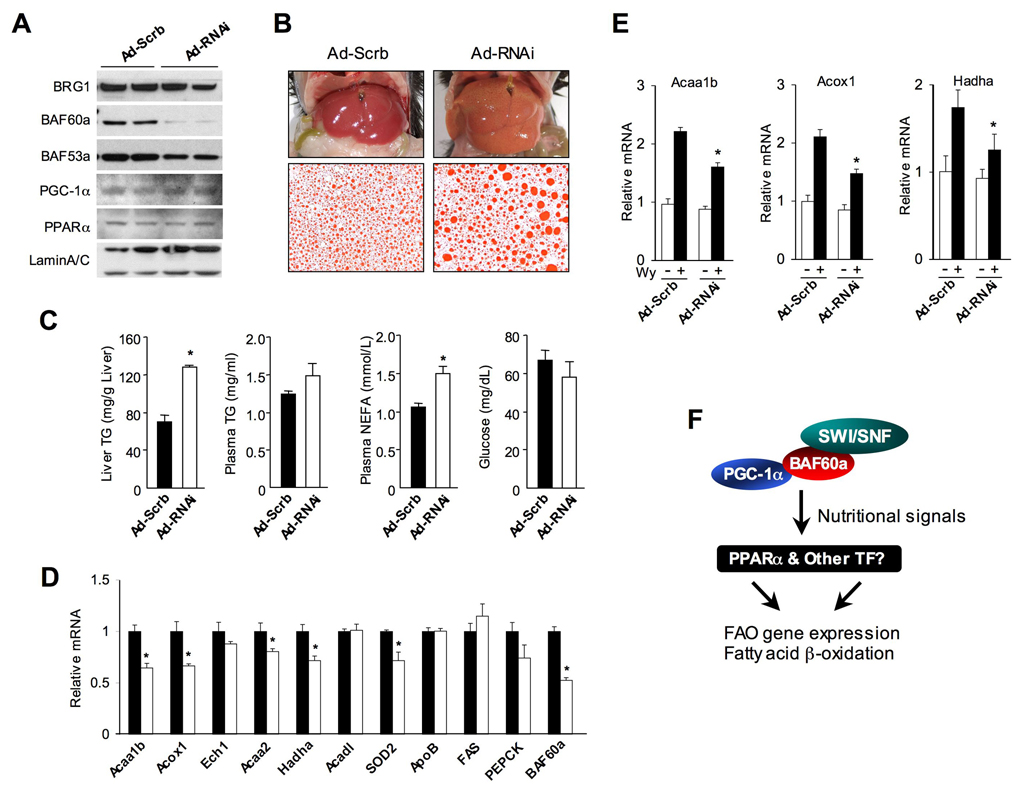
Fasting-induced association of BAF60a with FAO gene promoters suggests that it may participate in the formation of transcriptional complexes that induce the expression of FAO genes during starvation. To determine whether BAF60a is required for FAO gene expression and fat oxidation in vivo, we performed RNAi knockdown in the liver using a recombinant adenovirus expressing shRNA directed toward BAF60a. Mice were fasted for 40 hrs two days after tail vein transduction. Immunoblotting analyses of liver nuclear extracts indicate that BAF60a protein levels are significantly decreased following the injection of RNAi adenovirus (Figure 7A). The expression of BRG1, BAF53a, PGC-1α, and PPARα is only modestly affected. The animals appear normal, however, the livers from mice receiving RNAi adenovirus are significantly enlarged (0.97±0.02 g control versus 1.14±0.05 g RNAi, Mean ± SEM, p<0.01) and appear light yellow, characteristic of hepatic steatosis (Figure 7B). Measurements of hepatic lipids indicate that reduced expression of BAF60a results in approximately 2-fold increase of TG content compared to control (Figure 7C). In addition, Oil Red O staining of liver sections revealed significantly larger fat droplets in the BAF60a deficient mice. Although plasma glucose and TG levels remain similar in both groups, the concentrations of free fatty acids are elevated in the knockdown mice. These observations are consistent with impaired hepatic fatty acid oxidation when BAF60a is knocked down. Gene expression analysis revealed that the mRNA levels of key enzymes in the FAO pathway, including Acaa1b, Acox1, Acaa2 and Hadha, are significantly lower in the livers of knockdown mice (Figure 7D). Interestingly, the expression of Ech1 and Acadl remains similar in both groups, suggesting that BAF60a is differentially required for the expression of FAO genes during fasting. The impairment of hepatic fat oxidation in response to RNAi knockdown of BAF60a suggests that this factor may be required for PPARα function. To assess this, we transduced mice with control or BAF60a RNAi adenoviruses followed by oral gavage of Wy14,643. As expected, PPARα activation increases mRNA expression of FAO genes in the control livers (Figure 7E), whereas this stimulatory effect is significantly blunted in BAF60a knockdown livers. Together, these results indicate that BAF60a is required for physiological regulation of hepatic fat oxidation during metabolic stresses such as starvation.
Figure 7. BAF60a is Required for Hepatic Fat Oxidation During Starvation.
(A) Immunoblots of liver nuclear extracts from mice transduced with control (Ad-Scrb) or BAF60a RNAi (Ad-RNAi) adenoviruses.
(B) Liver morphology and histological staining of liver sections in transduced mice. Mice were subjected to 40-hr fasting before metabolic measurements and gene expression analysis.
(C) Hepatic TG, plasma lipid and glucose concentrations in mice transduced with control (filled) or BAF60a RNAi (open) adenoviruses.
(D) qPCR analysis of hepatic gene expression in mice transduced with control (filled) or BAF60a RNAi (open) adenoviruses. Data in (C–D) represent mean ± SEM (n=4). *p<0.05.
(E) qPCR analysis of hepatic gene expression in mice transduced with control or BAF60a RNAi adenoviruses followed by oral gavage with vehicle (open) or 300 mg Wy14,643 (filled). Data represent mean ± stdev, *p<0.05.
(F) Schematic model of the regulation of hepatic FAO genes by BAF60a and its associated SWI/SNF complexes. Note the nutritional regulation of BAF60a recruitment to FAO genes through PPARα and other transcription factors.
Discussion
The SWI/SNF complexes have been implicated in the regulation of diverse biological processes, including embryogenesis, cell cycle, differentiation, and tumorigenesis (Kwon and Wagner, 2007; Roberts and Orkin, 2004; Sudarsanam and Winston, 2000). While it has been recognized that the SWI/SNF complexes can stimulate or inhibit the transcription of their target genes, how they are targeted to specific chromatin loci remains poorly understood. Several subunits of the SWI/SNF complexes physically interact with transcription factors (Simone, 2006), which are postulated to link chromatin remodeling to specific biological functions. Using a genome-wide coactivation screen, here we identified BAF60a as a factor that links the SWI/SNF complexes to transcriptional coactivator PGC-1α and revealed a role for BAF60a in hepatic lipid metabolism. Adenoviral-mediated expression of BAF60a stimulates the entire program of peroxisomal and mitochondrial fat oxidation and lowers liver triglyceride content in mouse models of hepatic steatosis. Further, BAF60a is required for the activation of hepatic fat oxidation during fasting.
The PGC-1 coactivators have emerged as versatile regulators of energy metabolism and diverse biological processes. Using high-throughput transfection and reporter gene assays, we were able to interrogate this coactivator network with high sensitivity and coverage. Because of the stringent criteria (10-fold coactivation and CoIP assays) that we used in our screen, several factors known to interact with PGC-1α (MEF2c and GR, for example) scored negative in this study. In the case of GR, the absence of ligand in our initial screen may contribute to its weak coactivation by PGC-1α. In addition to known transcriptional partners for PGC-1α, our analyses revealed a signature of the PGC-1α transcriptional network that includes nuclear receptors, zinc-finger proteins as well as factors involved in different aspects of chromatin biology. This quantitative assay of individual TF activity can be applied to globally identify the nuclear targets of metabolic signaling pathways.
Several lines of evidence link BAF60a to the PPARα pathway. BAF60a and PPARα share a large number of target genes involved in peroxisomal and mitochondrial fat oxidation pathways. Pharmacological activation of PPARα by Wy14,643 augments the induction of FAO genes by BAF60a. In addition, the transcriptional function of BAF60a is significantly impaired in PPARα null hepatocytes. Despite this, BAF60a is still capable of activating the expression of FAO genes in the absence of PPARα, suggesting that both PPARα-dependent and PPARα-independent pathways mediate the metabolic effects of BAF60a on hepatic fat oxidation (Figure 7F). At the molecular level, BAF60a is recruited to PPARα binding sites on the FAO gene promoters in a PGC-1α dependent manner. Given that PGC-1α interacts with PPARα and BAF60a through different domains, these data strongly suggest that PGC-1α promotes the formation of a transcriptional complex involving all three factors in the proximity of FAO gene promoters.
While the induction of FAO genes by BAF60a is significantly blunted in PGC-1α null hepatocytes, the transcriptional effect of BAF60a is not completely abolished in the absence of PGC-1α. PGC-1β interacts with BAF60a in CoIP studies (data not shown) and may compensate for the loss of PGC-1α. Alternatively, BAF60a may be recruited to the FAO gene promoters through its direct interaction with PPARα. In fact, BAF60a binds to the N-terminus of PPARα in a ligand-independent manner, suggesting that PGC-1α and BAF60a may interact with PPARα through different binding surface. Perhaps the most interesting observation from these studies is that although the levels of BAF60a mRNA and protein remain largely unchanged in response to fasting, its recruitment to the promoters of FAO genes is significantly increased during starvation. While increased PGC-1α expression contributes to BAF60a association with chromatin, BAF60a itself may also be directly modulated by nutritional signals at the posttranslational levels. In addition, because PGC-1α interacts with other chromatin-remodeling proteins, we cannot rule out the possibility that PGC-1α may generate permissive environment that facilitates the association of BAF60a with chromatin. Lipin 1, a gene mutated in the fatty liver dystrophy mice, has been shown to participate in the formation of PPARα/PGC-1α transcriptional complex (Finck et al., 2006; Peterfy et al., 2001). Whether Lipin 1 modulates the transcriptional activity of BAF60a, particularly in the context of FAO gene expression, remains unknown.
Hepatic steatosis results from an imbalance of fat availability and the ability of the liver to catabolize and/or secrete lipoproteins. Adenoviral-mediated expression of BAF60a ameliorates hepatic steatosis in diet-induced obese and ob/ob mouse livers. The decrease in hepatic TG content is accompanied by the induction of FAO genes, suggesting that activation of the BAF60a pathway is sufficient to alter the balance of hepatic lipid metabolism through promoting fat oxidation. Interestingly, we did not observe an increase of circulating ketone bodies in BAF60a transduced mice. This may be due to rapid clearance of ketone bodies and/or complete oxidation of lipids in the liver. RNAi knockdown of BAF60a impairs fat oxidation and leads to hepatic steatosis following starvation, implicating BAF60a as an essential factor for the activation of FAO genes during fasting. Together, BAF60a defines a critical link between the SWI/SNF chromatin-remodeling complexes and hepatic lipid metabolism.
Experimental Procedures
TFORC and coactivation screen
TFORC was obtained by arraying individual TF entry clones from the Human ORFeome v3.1 onto 96-well plates. These entry clones were subsequently transferred to a GalDBD destination vector using LR recombinase of to generate Gal-TF fusion plasmids (Invitrogen). For coactivation assay, BOSC cells seeded in 96-well plates were transiently transfected with UAS-luciferase reporter (10 ng) and GAL-TF plasmids (10–20 ng) in the presence of vector or pcDNA3 Flag-PGC-1α (100 ng) using Lipofectamine and Plus reagent. Luciferase activity was measured 30 hrs after transfection.
Coimmunoprecipitation assay
Individual TF was cloned into a Gateway destination vector to generate N-terminal Myc fusion proteins. These plasmids were co-transfected into BOSC cells with either pcDNA3 vector or pcDNA3 Flag-PGC-1α. Immunoprecipitated protein complexes and lysates were analyzed by immunoblotting using Myc and Flag antibodies (Sigma). For interaction between PGC-1α and endogenous SWI/SNF subunits, BOSC cells were transiently transfected with pcDNA3 vector or Flag-PGC-1α for 40 hrs. Total lysates or immunoprecipitated proteins were analyzed by immunoblotting using antibodies against BRG-1 (Santa Cruz Biotech), BAF60a (BD Biosciences), or BAF53a (Proteintech Group). For in vitro protein interaction assay, similar amounts of GST fusion proteins were used in the binding assay in the presence of in vitro transcribed and translated Flag-tagged proteins generated using TNT coupled system (Promega). The precipitated proteins were analyzed by immunoblotting.
Primary hepatocyte studies
The maintenance and adenoviral transduction of primary hepatocytes was performed as previously described (Liu et al., 2007). Total RNA was isolated using Trizol reagents 48 hrs following transduction, reverse transcribed, and analyzed by qPCR using primers listed in Table S5. Primers for ribosomal protein 36B4 were included for normalization. For the measurements of fatty acid oxidation, transduced hepatocytes were incubated with 800 µl (9,10(n)-3H) palmitic acid (Perkin-Elmer) bound to fatty acid-free BSA (final concentration, 125 µM; palmitate to BSA 1:1) in the presence of 1.0 mM carnitine for 2 hrs. The release of 3H2O was measured using a scintillation counter following trichloroacetic acid precipitation and ion-exchange chromatography, as previously described (Finck et al., 2006). Total radioactivity was normalized to protein content for the calculation of FAO rate. For the measurements of TG secretion, hepatocytes were incubated with [1,2,3-3H] glycerol (5.0 µCi/ml) for 3 hr. TG in the medium was extracted for scintillation counting followed by normalization to cellular protein content.
ChIP assay
Chromatin immunoprecipitation was performed essentially as described by the Upstate Biotechnology. Chromatin lysates were prepared from H2.35 hepatoma cells after crosslinking with 1% formaldehyde followed by brief sonication. For chromatin lysates from mouse livers, liver nuclei were isolated as previously described (Calfee-Mason et al., 2002) and then cross-linked in 1% formaldehyde for 10 minutes followed by sonication. After pre-cleared with Protein-G agarose beads, chromatin lysates were immunoprecipitated using antibodies against BAF60a, acetylated histone H3 (Upstate Biotechnology), PGC-1α (Santa Cruz Biotech), PPARα (Chemicon) or normal mouse IgG in the presence of BSA and salmon sperm DNA. Beads were extensively washed before reverse cross-linking. DNA was purified using a PCR purification kit (Qiagen) and subsequently analyzed by PCR using primers surrounding PPRE on Acox1, Acaa1b, and Hadha promoters (Lemay and Hwang, 2006; Nicolas-Frances et al., 2000; Tugwood et al., 1992) (Table S5).
In vivo adenoviral transduction and metabolic analyses
C57/Bl6J male mice were fed a high-fat diet (D12492, Research Diets) for 10 weeks to induce obesity. GFP or BAF60a adenoviruses were administered into mice through tail vein injection (0.15 OD per mouse), as previously described (Lin et al., 2005b). Liver triglycerides and plasma lipid concentrations were measured 5–7 days following tail vein injection using commercial assay kits (Sigma, Wako Diagnostics). Frozen or paraffin-embedded sections were prepared for Oil Red O and H&E staining, respectively.
For VLDL secretion assay, transduced mice were injected with a single dose of tyloxapol (500 mg/kg) through tail vein. Tail blood was sampled 30, 60, and 90 minutes after injection and assayed for TG concentrations.
Microarray data has been deposited to the NCBI Gene Expression Omnibus database (GSE9523).
Supplementary Material
Acknowledgements
We thank S. Gu and A. Baker for technical assistance, L. Chang, X. Chen and S. He for assistance with microscopy, and the members of the Lin lab for discussions. We also thank Dr. Puigserver for providing GST-PGC-1α plasmids, and Drs. O. MacDougald, A. Saltiel, and J. Schwartz for comments on the manuscript. This work was supported by NIH (DK077086 and DK065584, J.D.L.), the Juvenile Diabetes Research Foundation (J.D.L.), the Michigan Diabetes Research and Training Center, and the Ellison Foundation (M.V.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Calfee-Mason KG, Spear BT, Glauert HP. Vitamin E inhibits hepatic NF-kappaB activation in rats administered the hepatic tumor promoter, phenobarbital. J Nutr. 2002;132:3178–3185. doi: 10.1093/jn/131.10.3178. [DOI] [PubMed] [Google Scholar]
- Christian M, White R, Parker MG. Metabolic regulation by the nuclear receptor corepressor RIP140. Trends Endocrinol Metab. 2006;17:243–250. doi: 10.1016/j.tem.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279:16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- Eaton S. Control of mitochondrial beta-oxidation flux. Prog Lipid Res. 2002;41:197–239. doi: 10.1016/s0163-7827(01)00024-8. [DOI] [PubMed] [Google Scholar]
- Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Jr, Kelly DP. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004;116:337–350. doi: 10.1016/s0092-8674(03)01081-x. [DOI] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Handschin C, Kobayashi YM, Chin S, Seale P, Campbell KP, Spiegelman BM. PGC-1alpha regulates the neuromuscular junction program and ameliorates Duchenne muscular dystrophy. Genes Dev. 2007;21:770–783. doi: 10.1101/gad.1525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Lin J, Rhee J, Peyer AK, Chin S, Wu PH, Meyer UA, Spiegelman BM. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1alpha. Cell. 2005;122:505–515. doi: 10.1016/j.cell.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Cook WS, Qi C, Yeldandi AV, Reddy JK, Rao MS. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J Biol Chem. 2000;275:28918–28928. doi: 10.1074/jbc.M910350199. [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210–2220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucinerich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, Murakami M, Iba H. Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J Biol Chem. 2001;276:2852–2857. doi: 10.1074/jbc.M009633200. [DOI] [PubMed] [Google Scholar]
- Kanamori M, Konno H, Osato N, Kawai J, Hayashizaki Y, Suzuki H. A genome-wide and nonredundant mouse transcription factor database. Biochem Biophys Res Commun. 2004;322:787–793. doi: 10.1016/j.bbrc.2004.07.179. [DOI] [PubMed] [Google Scholar]
- Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutti D, Kaul A, Kralli A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol Cell Biol. 2000;20:2411–2422. doi: 10.1128/mcb.20.7.2411-2422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummerfeld SK, Teichmann SA. DBD: a transcription factor prediction database. Nucleic Acids Res. 2006;34:D74–D81. doi: 10.1093/nar/gkj131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CS, Wagner D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 2007;23:403–412. doi: 10.1016/j.tig.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Nixon SJ, Parton RG, Muscat GE. RORalpha regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: caveolin-3 and CPT-1 are direct targets of ROR. J Biol Chem. 2004;279:36828–36840. doi: 10.1074/jbc.M404927200. [DOI] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay DG, Hwang DH. Genome-wide identification of peroxisome proliferator response elements using integrated computational genomics. J Lipid Res. 2006;47:1583–1587. doi: 10.1194/jlr.M500504-JLR200. [DOI] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005a;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lin J, Yang R, Tarr PT, Wu PH, Handschin C, Li S, Yang W, Pei L, Uldry M, Tontonoz P, et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell. 2005b;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, Roskams T, Trautwein C, Pasparakis M. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119–132. doi: 10.1016/j.ccr.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nicolas-Frances V, Dasari VK, Abruzzi E, Osumi T, Latruffe N. The peroxisome proliferator response element (PPRE) present at positions −681/−669 in the rat liver 3-ketoacyl-CoA thiolase B gene functionally interacts differently with PPARalpha and HNF-4. Biochem Biophys Res Commun. 2000;269:347–351. doi: 10.1006/bbrc.2000.2249. [DOI] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Reddy JK, Hashimoto T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu Rev Nutr. 2001;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone C. SWI/SNF: the crossroads where extracellular signaling pathways meet chromatin. J Cell Physiol. 2006;207:309–314. doi: 10.1002/jcp.20514. [DOI] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Winston F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- Tugwood JD, Issemann I, Anderson RG, Bundell KR, McPheat WL, Green S. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. Embo J. 1992;11:433–439. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle I, Alvarez-Barrientos A, Arza E, Lamas S, Monsalve M. PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc Res. 2005;66:562–573. doi: 10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.